ATP-citrate lyase (ACLY), a key enzyme for lipid synthesis, is frequently overexpressed or activated in cancer to promote lipid synthesis and tumor progression. Zhang et al. show that Cullin3 (CUL3), a core protein of the CUL3–RING ubiquitin ligase complex, interacts with ACLY through its adaptor protein, KLHL25, to ubiquitinate and degrade ACLY in cells. Through negative regulation of ACLY, CUL3 inhibits lipid synthesis, cell proliferation, and tumor growth in lung cancer cells.
Keywords: CUL3, ACLY, KLHL25, ubiquitination, lipid synthesis, tumor progression
Abstract
Increased lipid synthesis is a key characteristic of many cancers that is critical for cancer progression. ATP-citrate lyase (ACLY), a key enzyme for lipid synthesis, is frequently overexpressed or activated in cancer to promote lipid synthesis and tumor progression. Cullin3 (CUL3), a core protein for the CUL3–RING ubiquitin ligase complex, has been reported to be a tumor suppressor and frequently down-regulated in lung cancer. Here, we found that CUL3 interacts with ACLY through its adaptor protein, KLHL25 (Kelch-like family member 25), to ubiquitinate and degrade ACLY in cells. Through negative regulation of ACLY, CUL3 inhibits lipid synthesis, cell proliferation, and xenograft tumor growth of lung cancer cells. Furthermore, ACLY inhibitor SB-204990 greatly abolishes the promoting effect of CUL3 down-regulation on lipid synthesis, cell proliferation, and tumor growth. Importantly, low CUL3 expression is associated with high ACLY expression and poor prognosis in human lung cancer. In summary, our results identify CUL3–KLHL25 ubiquitin ligase as a novel negative regulator for ACLY and lipid synthesis and demonstrate that decreased CUL3 expression is an important mechanism for increased ACLY expression and lipid synthesis in lung cancer. These results also reveal that negative regulation of ACLY and lipid synthesis is a novel and critical mechanism for CUL3 in tumor suppression.
Metabolic changes are a hallmark of cancer cells. Compelling evidence has shown that the metabolic changes of cancer cells are critical for cancer progression (Hanahan and Weinberg 2011; DeNicola and Cantley 2015; Pavlova and Thompson 2016). Increased glycolysis is the most well-studied metabolic change in cancer, which provides ATP as well as precursors and reducing equivalents for the biosynthesis of macromolecules, such as nucleotides, proteins, and lipids (DeNicola and Cantley 2015; Pavlova and Thompson 2016). In addition to increased glycolysis, increased de novo lipid synthesis is another key characteristic of many cancers (Menendez and Lupu 2007; Currie et al. 2013). Lipids are essential constituents of all biological membranes and important substrates for energy metabolism. Whereas most normal tissues preferentially use circulating lipids and display a low rate of de novo lipid synthesis, many cancers display a very high rate of de novo lipid synthesis irrespective of the levels of extracellular lipids (Menendez and Lupu 2007; Currie et al. 2013). 14C-glucose incorporation assays have demonstrated that, in tumor cells, almost all fatty acids are derived from de novo lipid synthesis despite an abundant supply of extracellular lipids (Medes et al. 1953; Ookhtens et al. 1984). Recent studies have shown that de novo lipid synthesis plays a critical role in tumor progression, which could be targeted for cancer therapy (Bauer et al. 2005; Hatzivassiliou et al. 2005; Menendez and Lupu 2007; Currie et al. 2013).
As a key enzyme in de novo lipid synthesis, ACLY (ATP-citrate lyase) connects glucose metabolism with de novo lipid synthesis (Fig. 1A; Menendez and Lupu 2007; Zaidi et al. 2012b; Currie et al. 2013). ACLY converts citrate in the cytosol to acetyl-CoA. As a precursor for de novo lipid synthesis, acetyl-CoA is used to synthesize fatty acids through the fatty acid synthesis pathway and cholesterol and isoprenoids through the mevalonate pathway in cells (Fig. 1A). ACLY is frequently overexpressed and/or activated in different types of cancer, including lung cancer, which is an important mechanism contributing to increased de novo lipid synthesis in cancer (Milgraum et al. 1997; Swinnen et al. 2004; Menendez and Lupu 2007; Migita et al. 2008; Lin et al. 2013). Targeting ACLY by RNAi or specific small molecule inhibitors, such as SB-204990, has been shown to inhibit tumor cell proliferation in vitro and in vivo, supporting a pivotal role of ACLY in tumorigenesis and the potential of ACLY as a cancer therapeutic target (Bauer et al. 2005; Hatzivassiliou et al. 2005; Zaidi et al. 2012b). Currently, the regulation of ACLY in cancer is not well understood. It has been reported that insulin and growth factors can transcriptionally induce ACLY expression through the PI3K/AKT pathway, and, furthermore, ACLY can be phosphorylated and activated by AKT (Towle et al. 1997; Berwick et al. 2002; Migita et al. 2008). More recently, it was reported that acetylation modification can stabilize ACLY protein to promote lipid synthesis and progression of lung cancer (Lin et al. 2013).
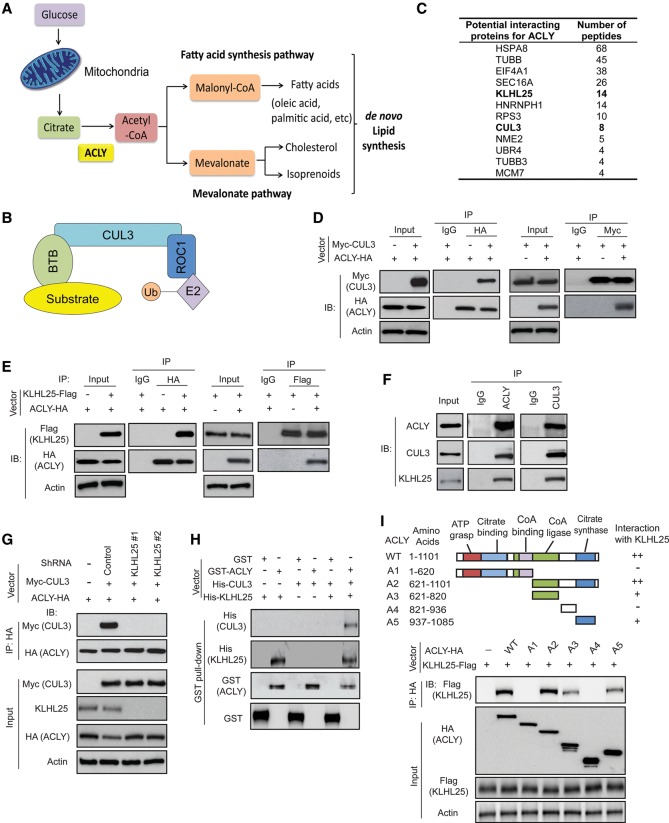
Figure 1.
ACLY forms a complex with CUL3 (Cullin3)–KLHL25 (Kelch-like family member 25). (A) Schematic diagram of carbon flux in de novo lipid synthesis. ACLY, which coverts citrate to acetyl-CoA, is a key enzyme for de novo lipid synthesis. (B) Model of the ROC1–CUL3–BTB (Broad complex/Tramtrack/Bric-a-brac) ubiquitin (Ub) ligase complex. (C) Potential ACLY-interacting proteins identified by coimmunoprecipitation (co-IP) followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. HEK293T cells expressing ACLY-HA or cells transduced with control vectors were used for co-IP with the anti-HA antibody followed by LC-MS/MS analysis. The potential ACLY-interacting proteins are listed with the number of peptides identified by LC-MS/MS analysis. (D) ACLY-HA interacted with Myc-CUL3 in cells. H1299 cells were transduced with vectors expressing Myc-CUL3 and ACLY-HA as well as control vectors for co-IP assays using the anti-HA and anti-Myc antibodies, respectively. (IP) Immunoprecipitation; (IB) immunoblot. (E) ACLY-HA interacted with KLHL25-Flag in cells. H1299 cells transduced with vectors expressing ACLY-HA and KLHL25-Flag, respectively, were used for co-IP assays using anti-HA or anti-Flag antibodies. (F) Endogenous ACLY interacted with CUL3 and KLHL25 in H1299 cells detected by co-IP assays. (G) Knockdown of endogenous KLHL25 by shRNA vectors impaired the interaction between ACLY-HA and Myc-CUL3. H1299 cells were transduced with two different shRNA vectors against KLHL25 and vectors expressing ACLY-HA and Myc-CUL3, respectively, for co-IP assays. (H) In vitro GST pull-down analysis for the direct interaction of KLHL25 with ACLY. Purified GST-ACLY immobilized on glutathione beads was incubated with purified His-CUL3 and/or His-KLHL25 proteins. Bound proteins were analyzed by Western blot assays. (I) KLHL25 bound to ACLY at its C terminus. (Top panel) Schematic representation of vectors expressing HA-tagged wild-type or serial deletion mutants of ACLY. (Bottom panel) Two different regions at the C terminus of ACLY interacted with KLHL25. H1299 cells were transduced with wild-type or deletion mutant ACLY-HA vectors together with KLHL25-Flag vectors for co-IP assays.
Ubiquitination is an important post-translational modification of cellular proteins. Cullin–RING ubiquitin (Ub) ligases are the largest known class of Ub ligases. Cullin3 (CUL3) is a protein of Cullin family. The CUL3–RING Ub ligase complex is composed of CUL3, which acts as a core scaffolding protein; a RING domain containing E3 Ub ligase protein ROC1; and an adaptor protein containing the BTB (Broad complex/Tramtrack/Bric-a-brac) domain, which serves as both the substrate adaptor and the substrate recognition protein (Fig. 1B; Lee and Zhou 2010; Genschik et al. 2013). Through interaction with different BTB domain-containing proteins, CUL3 forms different ROC1–CUL3–BTB Ub ligase complexes to regulate the levels of specific substrate proteins, and thus, are involved in regulation of different biological processes in cells. For example, KEAP1 is the most well-known adaptor protein for CUL3. CUL3–KEAP1 targets transcriptional factor Nrf2 for ubiquitination and degradation to regulate oxidative stress in cells (Itoh et al. 1999; Cullinan et al. 2004). Recently, KLHL25 (Kelch-like family member 25) was reported to form a complex with CUL3 as an adaptor protein to regulate ubiquitination and degradation of hypophosphorylated 4E-BP1 and thereby maintain translation homeostasis in cells (Yanagiya et al. 2012). CUL3 expression is frequently down-regulated in different types of cancer, including lung, breast, and liver cancer (Kossatz et al. 2010; Lee and Zhou 2010; Thu et al. 2011; Haagenson et al. 2012; Dorr et al. 2015). A recent study using a transposon mutagenesis screen in mice indicates that CUL3 is a tumor suppressor in lung cancer (Dorr et al. 2015). Currently, the role and mechanism of CUL3 in cancer metabolism remain unclear.
In this study, we identify CUL3 as a novel negative regulator of ACLY and lipid synthesis. CUL3 interacts with ACLY through its adaptor protein, KLHL25, to ubiquitinate and degrade ACLY. Through negative regulation of ACLY, CUL3 reduces acetyl-CoA levels and inhibits lipid synthesis. Negative regulation of ACLY by CUL3 contributes greatly to the tumor-suppressive role of CUL3 in lung cancer. Decreased CUL3 expression in lung cancer cells promotes lipid synthesis, cell proliferation, and tumor growth, which can be greatly abolished by targeting ACLY using RNAi and ACLY inhibitor SB-204990. Importantly, low CUL3 expression is associated with high ACLY expression and poor prognosis in human lung cancer. These results reveal a critical role of CUL3–KLHL25-mediated ACLY degradation in lipid metabolism and tumor suppression.
Results
ACLY interacts with CUL3 and KLHL25 to form a complex
ACLY is frequently overexpressed and activated in different types of cancer, including lung cancer, as a critical mechanism contributing to increased lipid synthesis in cancer. However, the mechanism underlying ACLY regulation in cancer is not well understood. To investigate the mechanism underlying ACLY regulation in cancer cells, we screened for proteins interacting with ACLY using coimmunoprecipitation (co-IP) followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) assays in human kidney HEK293T cells transduced with the retroviral pLPCX-ACLY-HA vector to express ACLY-HA and control cells transduced with the empty vector. Through this approach, CUL3 was identified as a potential binding protein for ACLY (Fig. 1C). The interaction between CUL3 and ACLY was confirmed by co-IP followed by Western blot assays in human lung cancer H1299 cells cotransduced with pLPCX-ACLY-HA and pLPCX-Myc-CUL3 retroviral vectors to express ACLY-HA and Myc-CUL3, respectively (Fig. 1D).
BTB domain-containing proteins function as substrate adaptors for the ROC1–CUL3–BTB Ub ligase complex to bring substrate proteins for ubiquitination and degradation (Fig. 1B). Recently, BTB domain-containing protein KLHL25 was reported to form a complex with CUL3 to regulate ubiquitination and degradation of 4E-BP1 (Yanagiya et al. 2012). Interestingly, KLHL25 was identified as a potential binding protein of ACLY by our LC-MS/MS assays (Fig. 1C), suggesting that KLHL25 may function as a substrate adaptor to bridge ACLY to CUL3. Therefore, we investigated whether KLHL25 can mediate CUL3–ACLY interaction. First, pLPCX retroviral expression vectors encoding ACLY-HA and KLHL25-Flag, respectively, were transduced into H1299 cells for co-IP assays to investigate whether KLHL25 binds to ACLY. Indeed, we found that KLHL25-Flag bound to ACLY-HA (Fig. 1E). The interaction between endogenous ACLY and CUL3 as well as KLHL25 was observed in H1299 cells by co-IP followed by Western blot assays (Fig. 1F). Notably, knockdown of endogenous KLHL25 by two different shRNA vectors largely abolished the interaction of Myc-CUL3 with ACLY-HA (Fig. 1G), suggesting that KLHL25 is required for the interaction between CUL3 and ACLY. This was confirmed by in vitro GST pull-down assays using recombinant proteins purified from bacteria, which showed that ACLY, KLHL25, and CUL3 formed a complex in vitro (Fig. 1H). Results from GST pull-down assays further showed that ACLY directly interacted with KLHL25 but not CUL3, and KLHL25 is required for the interaction between ACLY and CUL3 (Fig. 1H).
To identify the domain of ACLY that interacts with KLHL25, different HA-tagged deletion mutants of ACLY were constructed (Fig. 1I). Co-IP assays in H1299 cells showed that KLHL25 bound to ACLY at its C terminus; fragment A2 (amino acids 621–1101) bound to KLHL25, whereas fragment A1 (amino acids 1–620) did not bind to KLHL25 (Fig. 1I). Furthermore, KLHL25 bound to ACLY at two different domains of its C terminus, including fragments A3 (amino acids 621–820) and A5 (amino acids 937–1085) (Fig. 1I). These two domains have been reported to be crucial for ACLY activity (Sun et al. 2010). These results together demonstrate that ACLY directly interacts with KLHL25 to form a complex with CUL3 in cells.
CUL3 down-regulates ACLY protein levels, and CUL3 expression is associated with ACLY expression in human lung cancer
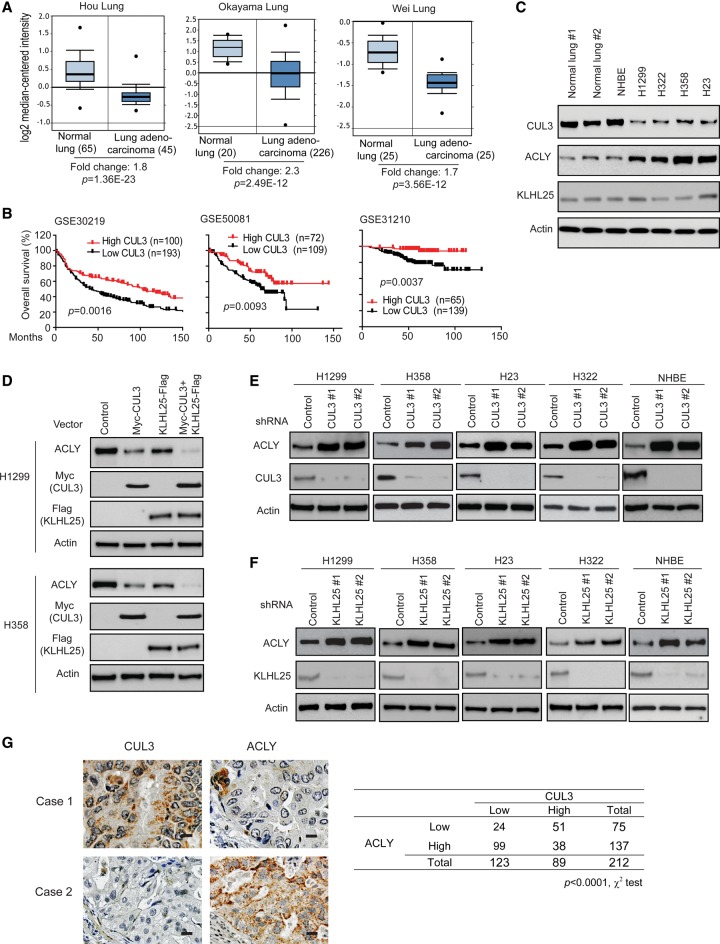
CUL3 has been suggested as a tumor suppressor in lung cancer, and its expression was reported to be frequently decreased in lung cancer (Thu et al. 2011; Dorr et al. 2015). Consistent with previous reports (Thu et al. 2011; Dorr et al. 2015), compared with normal lung tissues, significantly lower levels of CUL3 mRNA were observed in different cohorts of lung cancer from Oncomine (http://www.oncomine.org), a publicly available cancer database (Fig. 2A). Furthermore, low CUL3 expression is associated with poor prognosis of cancer patients in different cohorts of human lung cancer from the publicly available Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) (Fig. 2B). Compared with normal lung tissues, KLHL25 expression levels in lung cancer tissues did not show significant differences in the above-mentioned cohorts from the Oncomine database (Supplemental Fig. S1).
Figure 2.
CUL3 negatively regulates ACLY protein levels in cells, and low CUL3 expression is associated with high ACLY expression in human lung cancer. (A) CUL3 mRNA levels are decreased in different cohorts of human lung cancer obtained from the Oncomine database. The expression levels of CUL3 are expressed in terms of a log2 median-centered intensity, which was calculated by normalizing the intensity of the CUL3 probe to the median of the probe intensities across the entire array. (B) Low CUL3 expression is associated with poor prognosis in human lung cancer. The survival information and mRNA levels of CUL3 were obtained from the publicly available GEO databases. (C) The protein levels of CUL3, ACLY, and KLHL25 in normal lung tissues, primary NHBE cells, and different human lung cancer cells. (D) Ectopic expression of Myc-CUL3 and KLHL25-Flag negatively regulated the levels of endogenous ACLY in human lung H1299 and H358 cells. (E,F) Knockdown of endogenous CUL3 (E) or KLHL25 (F) by shRNA vectors increased protein levels of endogenous ACLY in human lung cancer cells and NHBE cells. (G) A significant inverse correlation between the CUL3 and ACLY expression levels in human lung cancer specimens detected by immunohistochemistry (IHC). (Left panels) Representative IHC staining images in two lung cancer specimens. (Right panel) Summary of IHC staining results in human lung cancer tissue microarrays (TMAs). n = 212, U.S. Biomax. Bar, 20 µM.
The CUL3–KLHL25 Ub ligase complex was reported recently to target 4E-BP1 for ubiquitination and degradation (Yanagiya et al. 2012). To investigate whether CUL3–KLHL25 regulates ACLY protein levels, different human lung cancer cell lines, including H1299, H358, H322, and H23, were used. These cells showed lower levels of CUL3 protein and higher levels of ACLY protein compared with normal human lung tissues and primary NHBE (normal human bronchial epithelial) cells (Fig. 2C). We first examined whether ectopic expression of CUL3 and KLHL25 down-regulates ACLY protein in cells by transducing H1299 and H358 cells with pLPCX retroviral vectors expressing Myc-CUL3 and KLHL25-Flag, respectively. As shown in Figure 2D, ectopic expression of Myc-CUL3 or KLHL25-Flag down-regulated the endogenous ACLY protein levels in cells. Furthermore, coexpression of Myc-CUL3 and KLHL25-Flag displayed a more pronounced effect on ACLY protein levels. To investigate whether endogenous CUL3 and KLHL25 negatively regulate ACLY in cells, the endogenous CUL3 and KLHL25 were knocked down by two different shRNA vectors, respectively, in different human lung cancer cell lines, including H1299, H358, H23, and H322 cells. Knockdown of CUL3 or KLHL25 greatly increased the ACLY protein levels in these cells (Fig. 2E,F). Consistent results were observed in NHBE cells (Fig. 2E,F). Furthermore, ectopic expression of CUL3 or KLHL25 or knockdown of endogenous CUL3 or KLHL25 did not clearly affect the mRNA levels of ACLY (Supplemental Fig. S2A–C), indicating that the CUL3–KLHL25 ligase complex negatively regulates ACLY at the protein but the not mRNA level. These results demonstrate that the CUL3–KLHL25 ligase complex negatively regulates ACLY protein in cells.
The correlation between the CUL3 expression and ACLY levels was further investigated in human lung cancer specimens in tissue microarrays (TMAs; n = 212, U.S. Biomax). The levels of CUL3 and ACLY proteins in cancer samples were determined by immunohistochemistry (IHC) staining. As shown in Figure 2G, there is a significant correlation between low CUL3 expression and high ACLY expression in human lung cancer (P < 0.0001).
CUL3–KLHL25 targets ACLY for ubiquitination and degradation
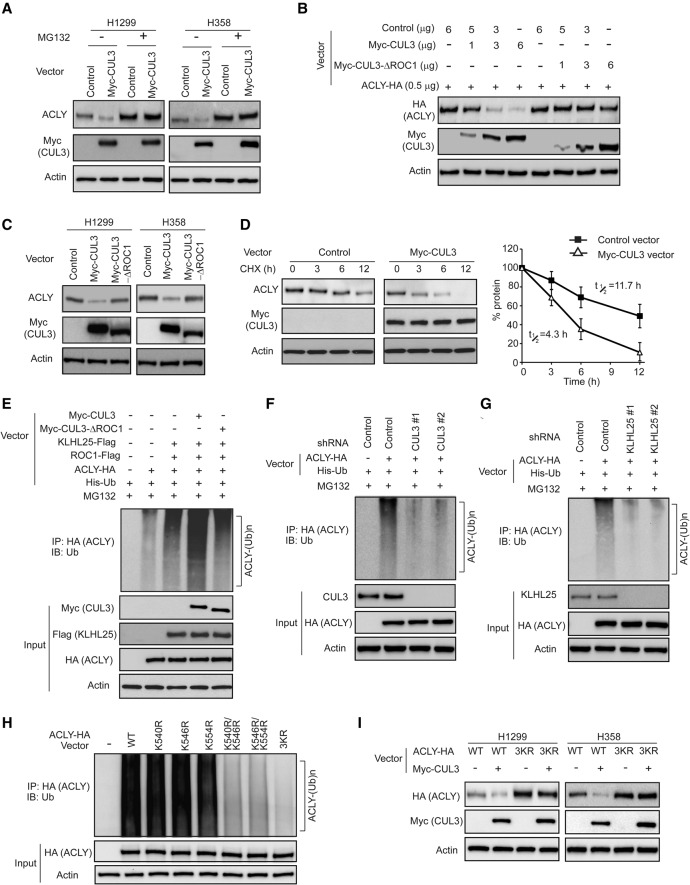
We further investigated whether the CUL3–KLHL25 complex negatively regulates ACLY through the Ub–proteasome degradation. First, H1299 and H358 cells transduced with pLPCX-Myc-CUL3 expression vectors were treated with proteasome inhibitor MG132 to examine whether blocking Ub–proteasome degradation can inhibit the down-regulation of ACLY protein levels by Myc-CUL3 in cells. MG132 treatment largely abolished the inhibitory effect of Myc-CUL3 on endogenous ACLY levels in cells (Fig. 3A). Deletion of the ROC1-binding site of CUL3 (amino acids 597–615; CUL3–ΔROC1) disrupts the interaction of CUL3 with E3 Ub ligase protein ROC1 (Furukawa et al. 2003; Lee et al. 2010). Therefore, the pLPCX-Myc-CUL3–ΔROC1 vector was used to examine whether disruption of CUL3–ROC1 interaction can abolish the effect of CUL3 on ACLY protein levels. Whereas expression of wild-type Myc-CUL3 clearly reduced the protein levels of both exogenous ACLY-HA and endogenous ACLY in cells, expression of the Myc-CUL3–ΔROC1 mutant did not clearly affect the levels of exogenous ACLY-HA or endogenous ACLY in cells (Fig. 3B,C). We further determined whether CUL3 decreases the half-life of ACLY protein by treating H1299 cells transduced with control or Myc-CUL3 vectors with protein synthesis inhibitor cyclohexamide (CHX) for different time periods. Compared with control cells, ectopic expression of Myc-CUL3 greatly reduced the half-life of ACLY protein (Fig. 3D).
Figure 3.
CUL3–KLHL25 ubiquitinates and degrades ACLY. (A) The down-regulation of endogenous ACLY by ectopic Myc-CUL3 was blocked by proteasome inhibitor MG132 in H1299 and H358 cells. (B) The ectopic Myc-CUL3–ΔROC1 mutant failed to down-regulate exogenous ACLY-HA in H1299 cells. H1299 cells were transduced with vectors expressing ACLY-HA together with different amounts of wild-type Myc-CUL3 or Myc-CUL3–ΔROC1 vectors. (C) The ectopic Myc-CUL3–ΔROC1 mutant failed to down-regulate endogenous ACLY in H1299 and H358 cells. (D) Ectopic Myc-CUL3 expression decreased the half-life of endogenous ACLY protein. H1299 cells transduced with the Myc-CUL3 vector or control vector were treated with 50 µg/mL CHX for the indicated time periods. Data are presented as mean ± SD. n = 3. (E) Ectopic expression of Myc-CUL3 but not Myc-CUL3–ΔROC1 promoted the ubiquitination of ACLY in cells. H1299 cells were transduced with the indicated vectors. Cell lysates were immunoprecipitated with an anti-HA antibody and then immunoblotted with an anti-Ub antibody. (F) Knockdown of endogenous CUL3 decreased ubiquitination of ACLY in H1299 cells. (G) Knockdown of endogenous KLHL25 decreased ubiquitination of ACLY in H1299 cells. (H) Double or triple mutations (3KR) at K540, K546, and/or K554 greatly reduced the ubiquitination of ACLY by CUL3–KLHL25 in H1299 cells. Cells were transfected with vectors expressing Myc-CUL3, KLHL25-Flag, ROC1-Flag, and His-Ub together with wild-type or mutant ACLY-HA and then treated with MG132 for in vivo ubiquitination assays. (I) The 3KR mutation largely abolished the negative regulation of ACLY protein by ectopic Myc-CUL3 in H1299 and H358 cells. Cells were transduced with vectors expressing wild-type or 3KR ACLY-HA together with Myc-CUL3 vectors.
We next investigated whether CUL3–KLHL25 promotes ACLY degradation through ubiquitination by in vivo ubiquitination assays. Compared with cells transfected with vectors to express KLHL25-Flag, ROC1-Flag, ACLY-HA, and His-Ub, respectively, the expression of wild-type Myc-CUL3 together with these above-mentioned vectors greatly promoted ubiquitination of ACLY-HA in H1299 cells, whereas expression of the Myc-CUL3–ΔROC1 mutant did not clearly promote the ubiquitination of ACLY-HA (Fig. 3E). Consistently, knockdown of CUL3 or KLHL25 by shRNA vectors greatly reduced ubiquitination of ACLY-HA in H1299 cells (Fig. 3F,G). These results demonstrate that ACLY is a direct substrate of the ROC1–CUL3–KLHL25 complex.
Recently, ACLY was reported to be ubiquitinated by UBR4 at K540, K546, and K554 (Lin et al. 2013). Interestingly, we found that CUL3–KLHL25 also ubiquitinated ACLY at K540, K546, and K554. Results from ubiquitination assays in H1299 cells showed that whereas single mutations at K540, K546, and K554 (K540R, K546R, or K554R) did not clearly reduce ubiquitination of ACLY by CUL3–KLHL25 compared with wild-type ACLY, double mutations (K540R/K546R or K546R/K554R) clearly reduced ubiquitination of ACLY by CUL3–KLHL25, and triple mutations (K540R/K546R/K554R [3KR]) dramatically reduced ubiquitination of ACLY by CUL3–KLHL25 (Fig. 3H). This result suggests that K540, K546, and K554 of ACLY are the ubiquitination target residues of CUL3–KLHL25. Consistently, 3KR mutations largely abolished the effect of Myc-CUL3 on ACLY degradation in cells (Fig. 3I). Compared with knockdown of CUL3 or UBR4 individually, knockdown of CUL3 and UBR4 simultaneously displayed a more pronounced effect on ACLY protein levels in different cells (Supplemental Fig. S3A,B), suggesting that CUL3 and UBR4 coordinate to regulate ACLY in cells.
CUL3 inhibits de novo lipid synthesis through negative regulation of ACLY
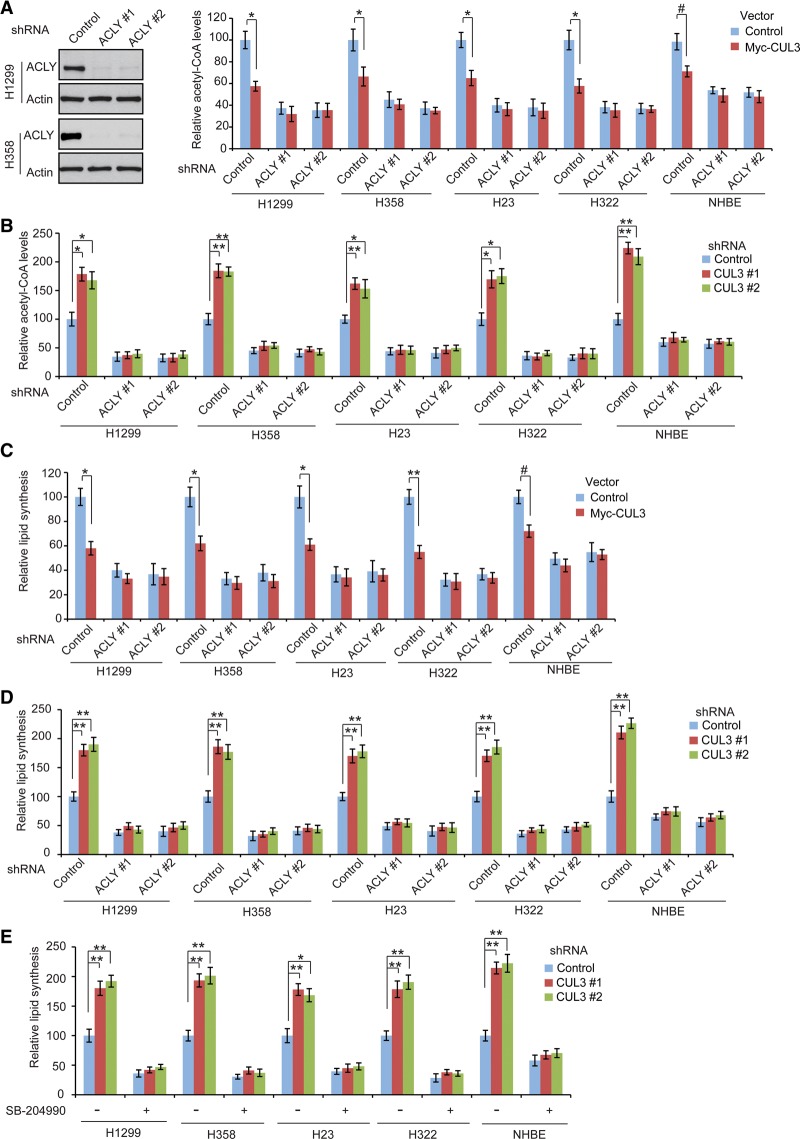
ACLY connects glucose metabolism with de novo lipid synthesis by converting citrate in the cytosol to acetyl-CoA, which is the building block for lipid synthesis (Fig. 1A; Menendez and Lupu 2007; Zaidi et al. 2012b). Our finding that the CUL3–KLHL25 ligase complex ubiquitinates and degrades ACLY suggests that CUL3–KLHL25 may reduce acetyl-CoA levels and therefore inhibits lipid synthesis in cells. We first investigated the impact of CUL3 expression on acetyl-CoA levels in cells. Ectopic expression of Myc-CUL3 significantly decreased the acetyl-CoA levels in the above-mentioned human lung cancer cells and NHBE cells (Fig. 4A). Consistently, CUL3 knockdown by shRNA vectors significantly enhanced acetyl-CoA levels in cells (Fig. 4B). These results indicate that CUL3 negatively regulates acetyl-CoA levels in cells. To further investigate whether CUL3 reduces acetyl-CoA levels through its negative regulation of ACLY, the endogenous ACLY was knocked down by shRNA vectors, and the effect of Myc-CUL3 expression on acetyl-CoA levels in cells was examined. Consistent with previous reports (Hatzivassiliou et al. 2005; Wang et al. 2009), knockdown of ACLY greatly reduced acetyl-CoA levels in cells (Fig. 4A; Supplemental Fig. S4A). Notably, the effect of CUL3 on acetyl-CoA levels was much less pronounced in cells with ACLY knockdown compared with control cells (Fig. 4A,B). These results demonstrate that CUL3 reduces acetyl-CoA levels in cells through negative regulation of ACLY.
Figure 4.
CUL3 reduces acetyl-CoA levels and inhibits lipid synthesis through negative regulation of ACLY. (A) Ectopic Myc-CUL3 expression reduced acetyl-CoA levels in human lung cells, and this effect was much less pronounced in cells with ACLY knockdown. (Left panels) Representative immunoblots of ACLY knockdown by two different shRNA vectors in H1299 and H358 cells. The knockdown of ACLY in other cells is shown in Supplemental Figure S4A. (B) CUL3 knockdown enhanced acetyl-CoA levels in cells, which was largely abolished by ACLY knockdown. The efficiency of CUL3 knockdown is shown in Figure 2E. (C) Ectopic Myc-CUL3 expression inhibited lipid synthesis in human lung cells, and this effect was much less pronounced in cells with ACLY knockdown. (D) CUL3 knockdown enhanced lipid synthesis in cells, which was largely abolished by ACLY knockdown. (E) ACLY inhibition by specific ACLY inhibitor SB-204990 largely abolished the promoting effect of CUL3 knockdown on lipid synthesis in lung cells. Cells with CUL3 knockdown were treated with 30 µM SB-204990 or vehicle for 12 h before assays. In A–E, data are presented as mean ± SD. n = 3. (#) P < 0.05; (*) P < 0.01; (**) P < 0.001, Student's t-tests.
We next investigated the impact of CUL3 on de novo lipid synthesis by measuring the incorporation of 14C-glucose into total lipids in cells, which is a widely used method to measure the rate of de novo lipid synthesis in cells (Hatzivassiliou et al. 2005; Daemen et al. 2015; Das et al. 2015). Ectopic expression of Myc-CUL3 greatly inhibited de novo lipid synthesis in different cells (Fig. 4C). Furthermore, CUL3 knockdown greatly increased de novo lipid synthesis in these cells (Fig. 4D). These results demonstrate that CUL3 inhibits lipid synthesis in human lung cells. Notably, the effect of CUL3 on lipid synthesis was much less pronounced in cells with ACLY knockdown compared with control cells (Fig. 4C,D), suggesting that CUL3 inhibits lipid synthesis through negative regulation of ACLY. To confirm these results, we treated cells with SB-204990, a small molecule inhibitor for ACLY (Hatzivassiliou et al. 2005; Riviere et al. 2009). Consistent with previous reports (Hatzivassiliou et al. 2005; Riviere et al. 2009), SB-204990 greatly inhibited lipid synthesis in these cells (Fig. 4E). Notably, SB-204990 largely abolished the promoting effect of CUL3 knockdown on lipid synthesis (Fig. 4E). We also investigated the impact of KLHL25 upon acetyl-CoA levels and de novo lipid synthesis. Consistent with the effect of CUL3 knockdown, KLHL25 knockdown by shRNA vectors greatly increased acetyl-CoA levels and de novo lipid synthesis in cells. Furthermore, these effects were largely abolished by ACLY knockdown (Supplemental Fig. S4B,C). Collectively, these results indicate that the CUL3–KLHL25 complex reduces acetyl-CoA levels and inhibits lipid synthesis through negative regulation of ACLY.
CUL3 inhibits the proliferation and anchorage-independent growth of lung cancer cells through negative regulation of ACLY
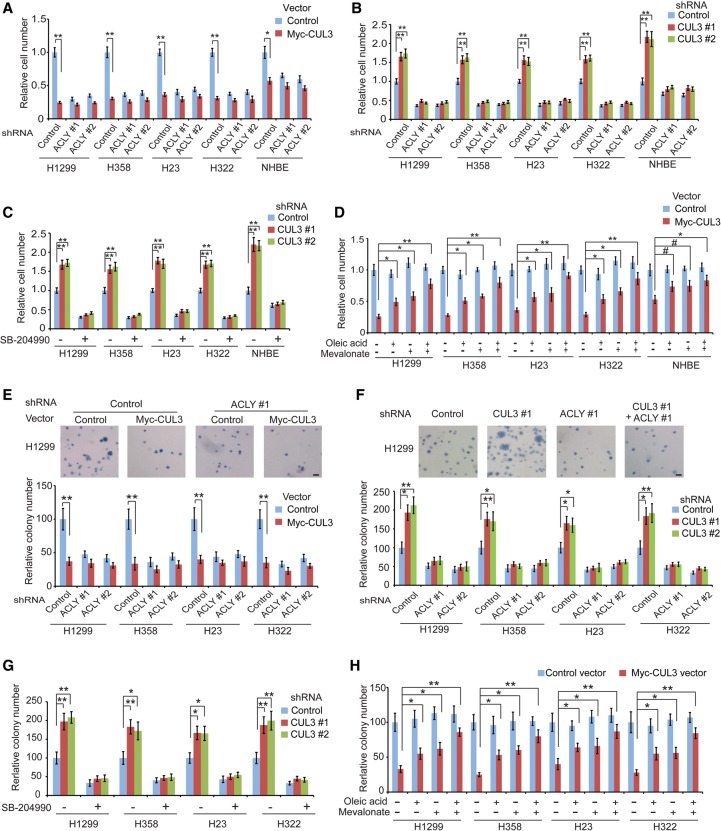
ACLY plays a critical role in promoting cancer cell proliferation, and targeting ACLY by RNAi or specific small molecule inhibitors such as SB-204990 inhibits proliferation of cancer cells in vitro and in vivo (Bauer et al. 2005; Hatzivassiliou et al. 2005; Migita et al. 2008; Zaidi et al. 2012a). Here, we investigated whether CUL3 inhibits proliferation of lung cancer cells through negative regulation of ACLY in different lung cancer cells as well as normal lung NHBE cells in vitro by MTT assays. Ectopic expression of Myc-CUL3 significantly inhibited cell proliferation (Fig. 5A), whereas CUL3 knockdown significantly promoted cell proliferation (Fig. 5B). Furthermore, negative regulation of ACLY by CUL3 contributes greatly to the inhibitory effect of CUL3 on cell proliferation. ACLY knockdown inhibited cell proliferation (Fig. 5A). Furthermore, the inhibitory effect of Myc-CUL3 on cell proliferation was much less pronounced in cells with ACLY knockdown compared with control cells (Fig. 5A). Inhibiting ACLY by shRNA or SB-204990 treatment greatly abolished the promoting effect of CUL3 knockdown on cell proliferation (Fig. 5B,C). KLHL25 displayed an effect on cell proliferation similar to that of CUL3; KLHL25 knockdown significantly promoted cell proliferation, and this effect was largely abolished in cells with ACLY knockdown (Supplemental Fig. S5A).
Figure 5.
CUL3 inhibits cell proliferation and anchorage-independent cell growth through negative regulation of ACLY. (A) Ectopic Myc-CUL3 expression inhibited the proliferation of human lung cells, and this effect was much less pronounced in cells with ACLY knockdown. (B) CUL3 knockdown promoted the proliferation of human lung cells, which was greatly abolished by simultaneous knockdown of ACLY. (C) ACLY inhibition by SB-204990 greatly abolished the promoting effect of CUL3 knockdown on cell proliferation. (D) Treating cells with oleic acid and/or mevalonate reduced the inhibitory effect of ectopic Myc-CUL3 expression on the proliferation of human lung cells. Cells were treated with 20 µM oleic acid and/or 20 µM mevalonate. In A–D, cell proliferation was analyzed by MTT assays. The relative cell number at day 4 after seeding, which was quantified by comparing the cell number of each group with the control group, is shown. Data are presented as mean ± SD. n = 4. (#) P < 0.05; (*) P < 0.01; (**) P < 0.001, Student's t-tests. (E) Ectopic Myc-CUL3 expression inhibited the anchorage-independent growth of human lung cancer cells in soft agar, and this effect was much less pronounced in cells with ACLY knockdown. (F) CUL3 knockdown promoted the anchorage-independent growth of human lung cancer cells, which was greatly abolished by ACLY knockdown. (G) ACLY inhibition by SB-204990 greatly abolished the promoting effect of CUL3 knockdown on anchorage-independent growth in human lung cancer cells. (H) Treating cells with oleic acid and/or mevalonate reduced the inhibitory effect of ectopic Myc-CUL3 expression on anchorage-independent growth in human lung cancer cells. The top panels in E and F show representative images of the anchorage-independent growth of H1299 cells in soft agar. Bar, 100 µm. Data are presented as mean ± SD. n = 4. (*) P < 0.01; (**) P < 0.001, Student's t-tests.
To further confirm that CUL3–KLHL25 inhibits cell proliferation through down-regulating ACLY, H1299, and H358, cells were transduced with shRNA vectors to knock down endogenous ACLY and then transduced with vectors expressing wild-type or 3KR ACLY that are resistant to shRNA against ACLY (rACLY) (Supplemental Fig. S5B). Whereas Myc-CUL3 significantly inhibited proliferation of cells expressing ectopic wild-type rACLY, Myc-CUL3 did not clearly affect proliferation of cells expressing 3KR rACLY in these cells (Supplemental Fig. S5C). Collectively, these results indicate that the negative regulation of ACLY contributes greatly to the inhibitory effect of CUL3–KLHL25 on the proliferation of human lung cells.
Promoting lipid synthesis is critical for the role of ACLY in tumor progression (Bauer et al. 2005; Hatzivassiliou et al. 2005; Migita et al. 2008; Zaidi et al. 2012a). It has been reported that supplementation of cells with metabolites from the fatty acid synthesis pathway (e.g., oleic acid and palmitic acid) and the mevalonate pathway (e.g., mevalonate and cholesterol) rescued the growth inhibition of cancer cells caused by inhibition of lipid synthesis, including ACLY knockdown (Nakakuki et al. 2007; Zaidi et al. 2012a; Young et al. 2013). To investigate whether CUL3 inhibits cell proliferation through down-regulation of lipid synthesis, we evaluated the effect of supplementation of oleic acid and mevalonate on the proliferation of lung cells with ectopic Myc-CUL3 expression. Whereas treating cells with oleic acid and/or mevalonate did not clearly affect the proliferation of control cells, treating cells with oleic acid and/or mevalonate significantly reduced the inhibitory effect of ACLY knockdown on cell proliferation (Supplemental Fig. S5D), suggesting that promoting lipid synthesis is critical for ACLY's function in promoting cell proliferation. Notably, supplementation of oleic acid or mevalonate significantly reduced the inhibitory effect of Myc-CUL3 expression on cell proliferation (Fig. 5D). This effect was more pronounced when cells were treated with oleic acid and mevalonate together (Fig. 5D).
It is known that ACLY is not the only enzyme that produces acetyl-CoA in cells. Acetyl-CoA synthetase 2 (ACSS2) can use acetate as a substrate to produce acetyl-CoA in cells (Supplemental Fig. S5E; Menendez and Lupu 2007; Currie et al. 2013). It has been reported that acetate supplementation can rescue cells from growth inhibition and other defects resulting from ACLY knockdown (Wellen et al. 2009; Zaidi et al. 2012a). Therefore, we investigated whether acetate supplementation can rescue the cell proliferation arrest caused by the down-regulation of ACLY and acetyl-CoA levels in cells by CUL3 expression. Consistent with the effects of oleic acid and/or mevalonate supplementation, acetate supplementation greatly reduced the inhibitory effect of ectopic Myc-CUL3 expression on the proliferation of human lung cancer cells (Supplemental Fig. S5F). Taken together, these results strongly suggest that inhibition of lipid synthesis contributes greatly to the inhibitory effect of CUL3 on the proliferation of human lung cells.
We further investigated whether CUL3 inhibits anchorage-independent cell growth through negative regulation of ACLY. Myc-CUL3 expression significantly inhibited the anchorage-independent growth of different human lung cancer cell lines in soft agar, whereas knockdown of CUL3 significantly promoted the anchorage-independent growth of these cells (Fig. 5E,F). Furthermore, the effect of CUL3 on the anchorage-independent growth of cells was much less pronounced in cells with ACLY knockdown compared with control cells (Fig. 5E,F). Consistently, ACLY inhibition by SB-204990 greatly abolished the promoting effect of CUL3 knockdown on the anchorage-independent growth of cells (Fig. 5G). KLHL25 displayed an effect on colony formation similar to that of CUL3; KLHL25 knockdown significantly promoted colony formation, and this effect was largely abolished in cells with ACLY knockdown (Supplemental Fig. S6A). Furthermore, Myc-CUL3 expression significantly reduced the anchorage-independent growth of lung cancer cells expressing ectopic wild-type rACLY but not 3KR rACLY (Supplemental Fig. S6B). Supplementation of oleic acid and/or mevalonate significantly reduced the inhibitory effects of ACLY knockdown (Supplemental Fig. S6C) and Myc-CUL3 expression (Fig. 5H) on the anchorage-independent growth of lung cancer cells. Similarly, acetate supplementation greatly reduced the inhibitory effect of ectopic Myc-CUL3 expression on the anchorage-independent growth of lung cancer cells (Supplemental Fig. S6D).
KEAP1 is a well-known adaptor protein of CUL3. CUL3–KEAP1 negatively regulates Nrf2, contributing to the tumor-suppressive function of CUL3–KEAP1 (Itoh et al. 1999; Cullinan et al. 2004). Whereas H1299, H358, H23, and NHBE cells lines do not contain a KEAP1 mutation, H322 cells contain a KEAP1 R460S mutation in the Kelch domain (Dai et al. 2013). Many mutations in the KEAP1 Kelch domain have been reported to disrupt KEAP1 function (Dai et al. 2013). Ectopic expression of Myc-CUL3 in A549, a human lung cancer cell line with a KEAP1 inactivation mutation (Dai et al. 2013), significantly inhibited cell proliferation and anchorage-independent cell growth, whereas CUL3 knockdown significantly promoted cell proliferation and anchorage-independent cell growth in A549 cells (Supplemental Fig. S7A–D). These effects were much less pronounced when endogenous ACLY was knocked down in A549 cells (Supplemental Fig. S7A–D). Consistently, while knockdown of KEAP1 slightly promoted cell proliferation in H1299 and H358 cells that express wild-type KEAP1 protein, knockdown of KEAP1 did not clearly affect the effects of Myc-CUL3 expression or CUL3 knockdown on cell proliferation in cells (Supplemental Fig. S7E–G). These results suggest a role of CUL3 in the inhibition of proliferation and anchorage-independent growth of lung cancer cells in the absence of KEAP1. Taken together, these results strongly suggest that the negative regulation of ACLY-mediated lipid synthesis by CUL3–KLHL25 greatly contributes to the inhibitory effect of CUL3 on the proliferation and anchorage-independent growth of lung cancer cells.
Negative regulation of ACLY contributes to the tumor-suppressive function of CUL3 in vivo
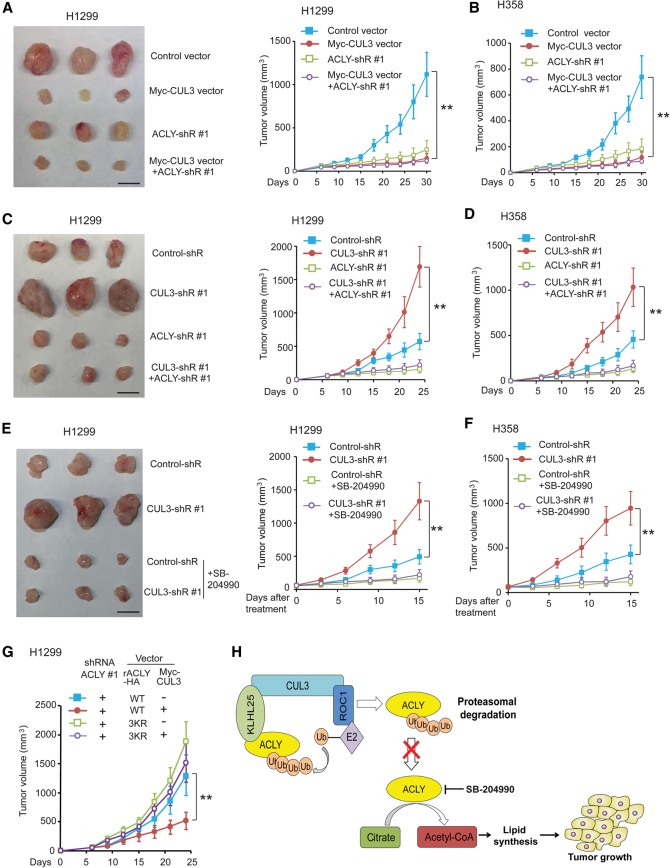
To investigate whether negative regulation of ACLY contributes to the tumor-suppressive function of CUL3 in lung cancer, subcutaneous xenograft tumorigenesis mouse models were used. Ectopic expression of Myc-CUL3 significantly reduced the growth of xenograft tumors formed by H1299 or H358 cells compared with tumors formed by control cells transduced with empty vectors (Fig. 6A,B). Knockdown of ACLY by different shRNA vectors in H1299 or H358 cells greatly inhibited the growth of xenograft tumors. Notably, the effect of Myc-CUL3 on the growth of xenograft tumors was much less pronounced in cells with ACLY knockdown (Fig. 6A,B). Furthermore, knockdown of CUL3 greatly promoted the growth of xenograft tumors formed by H1299 or H358 cells (Fig. 6C,D). The promoting effect of CUL3 knockdown on tumor growth was greatly abolished by further knockdown of endogenous ACLY in cells (Fig. 6C,D). We further investigated whether ACLY inhibitor SB-204990 inhibits the promoting effect of CUL3 knockdown on the growth of xenograft tumors. When the volume of xenograft tumors formed by H1299 and H358 cells with or without CUL3 knockdown reached ∼60 mm3, mice were injected with SB-204990 for 14 d. SB-204990 greatly abolished the promoting effect of CUL3 knockdown on the growth of xenograft tumors formed by H1299 cells and H358 cells (Fig. 6E,F). Furthermore, ectopic expression of 3KR mutant rACLY greatly abolished the inhibitory effect of Myc-CUL3 on the growth of xenograft tumors formed by H1299 cells (Fig. 6G). Ectopic expression of Myc-CUL3 in human lung A549 cells containing a KEAP1 inactivation mutation significantly inhibited the growth of xenograft tumors formed by A549 cells, and this effect was much less pronounced in A549 cells with ACLY knockdown (Supplemental Fig. S8). These results strongly suggest that the negative regulation of ACLY by CUL3 contributes greatly to the tumor-suppressive function of CUL3 in lung cancer (Fig. 6H).
Figure 6.
CUL3 inhibits the growth of xenograft lung tumors in mice through negative regulation of ACLY. (A,B) Ectopic expression of Myc-CUL3 in H1299 (A) and H358 (B) cells inhibited the growth of xenograft tumors, and this effect was much less pronounced in cells with ACLY knockdown. The left panel in A shows representative images of xenograft tumors formed by H1299 cells at day 30 after inoculation of cells. The right panel in A and B show the growth curves of xenograft tumors. (C,D) CUL3 knockdown promoted the growth of xenograft tumors formed by H1299 (C) and H358 (D) cells, which was greatly abolished by ACLY knockdown. The left panel in C shows representative images of xenograft tumors formed by H1299 cells at day 24 after inoculation of cells. (E,F) ACLY inhibition by SB-204990 greatly abolished the promoting effect of CUL3 knockdown on the growth of xenograft tumors formed by H1299 (E) and H358 (F) cells. When the tumor volume reached ∼60 mm3, mice were injected (intraperitoneally [i.p.]) with SB-204990 daily for 14 d. The left panel in E shows representative images of xenograft tumors formed by H1299 cells at the end of treatment. (G) Ectopic expression of 3KR mutant ACLY greatly abolished the inhibitory effect of Myc-CUL3 on the growth of xenograft tumors formed by H1299 cells. Cells were transduced with shRNA to knock down endogenous ACLY and were then transduced with vectors to express wild-type or 3KR rACLY together with Myc-CUL3 vectors. In A–F, two shRNA vectors against CUL3 and ACLY, respectively, were used, and similar results were observed. For the sake of clarity, only results from one shRNA vector are shown. Data are presented as mean ± SD. n = 8. (**) P < 0.001, ANOVA followed by Student's t-tests. (H) Model of inhibition of de novo lipid synthesis and tumor growth by CUL3–KLHL25-mediated ACLY ubiquitination and degradation.
Discussion
Overexpression and activation of ACLY are frequently observed in human cancers, which are critical mechanisms contributing to the increased lipid synthesis in human cancer (Milgraum et al. 1997; Kuhajda 2000; Swinnen et al. 2004; Yahagi et al. 2005). However, the mechanism of ACLY regulation in cancer is poorly understood. In this study, we found that KLHL25 functions as a substrate adaptor to bridge ACLY to CUL3. Thus, the CUL3–KLHL25 Ub ligase complex targets ACLY for ubiquitination and degradation, which is a new mechanism for ACLY regulation in cells. CUL3 is frequently down-regulated in different types of cancer, including lung cancer (Kossatz et al. 2010; Thu et al. 2011; Haagenson et al. 2012; Dorr et al. 2015). Considering the important role of CUL3 in the negative regulation of ACLY, our results strongly suggest that the down-regulation of CUL3 in human lung cancer is an important mechanism contributing to the frequent ACLY overexpression in human lung cancer. Indeed, the results from this study show that decreased expression of CUL3 is associated with high expression of ACLY and poor prognosis in human lung cancer.
CUL3 has been suggested as a tumor suppressor in human lung cancer (Dorr et al. 2015). However, the mechanism by which CUL3 exerts its tumor-suppressive function in cancer is not well understood. As a key metabolic change in cancer cells, increased de novo lipid synthesis plays a critical role in tumor progression (Menendez and Lupu 2007; Santos and Schulze 2012; Currie et al. 2013). In this study, we found that CUL3 plays an unidentified and critical role in the negative regulation of lipid synthesis through ubiquitination and degradation of ACLY. Ectopic expression of CUL3 reduced the levels of ACLY protein in cells, which in turn reduced acetyl-CoA levels in cells and inhibited lipid synthesis and proliferation of cancer cells both in vitro and in vivo. In contrast, knockdown of CUL3 increased the levels of ACLY protein in human lung cancer cells, which in turn increased acetyl-CoA levels in cells and promoted lipid synthesis and proliferation of cancer cells. Furthermore, the promoting effects of CUL3 knockdown on lipid synthesis and the proliferation of cancer cells both in vitro and in vivo can be largely abolished by inhibiting ACLY using RNAi and SB-204990, a specific ACLY inhibitor. These results strongly suggest that the negative regulation of ACLY protein levels by the CUL3–KLHL25 Ub ligase complex to inhibit lipid synthesis is an unidentified and important mechanism contributing to the tumor-suppressive function of CUL3 in human lung cancer. Our results also suggest that targeting ACLY by specific small molecule ACLY inhibitors could be a potential therapeutic strategy for human lung cancer with decreased CUL3 expression.
It is worth noting that our results show that ACLY inhibition by RNAi or ACLY inhibitor SB-204990 greatly but not completely abolished the promoting effects of CUL3 knockdown on the proliferation, anchorage-independent growth, and xenograft tumorigenesis of human lung cancer cells. This suggests that, in addition to the negative regulation of ACLY and lipid synthesis, additional mechanisms also contribute to the tumor-suppressive function of CUL3 in human lung cancer. It is known that CUL3 interacts with different adaptor proteins to regulate ubiquitination and degradation of different substrate proteins. For instance, CUL3 was reported to ubiquitinate and degrade Cyclin E to regulate cell cycle (Singer et al. 1999; Kossatz et al. 2010). CUL3–KEAP1 ubiquitinates Nrf2 to regulate oxidative stress and ubiquitinates NF-κB activator IKKβ to inhibit NF-κB activation (Itoh et al. 1999; Cullinan et al. 2004; Lee et al. 2009). CUL3–SPOP was recently reported to target oncoprotein ERG to suppress prostate cancer progression (Gan et al. 2015). These functions of CUL3 could contribute to the tumor-suppressive function of CUL3 in different tissues in a context-dependent manner. It remains unclear how these different functions of CUL3 are coordinated to suppress tumor progression in different tissues, which should be addressed in the future.
In summary, our study identified CUL3–KLHL25 as a new Ub ligase complex for ACLY. Through negative regulation of ACLY, CUL3–KLHL25 reduces acetyl-CoA levels and inhibits lipid synthesis in cells, which contributes greatly to the tumor-suppressive role of CUL3 in lung cancer. Thus, our results provide a novel and critical mechanism for increased ACLY expression and lipid synthesis in cancer. These results also reveal that negative regulation of ACLY and lipid synthesis is a novel and critical mechanism for CUL3 in tumor suppression. Our results further suggest that targeting ACLY and lipid synthesis could be a potential therapeutic strategy for human lung cancer with decreased CUL3 expression.
Materials and methods
Cell culture and vectors
Human lung cancer H1299, H358, H23, H322, and A549 cells were obtained from American Type Culture Collection. Primary NHBE cells were obtained from Lonza. The pLPCX-Myc-CUL3 retroviral vector was constructed by subcloning the CUL3 DNA fragment from pcDNA3-Myc-CUL3 (Addgene) into the pLPCX vector. The pLPCX-ACLY-HA and pLPCX-KLHL25-Flag vectors were constructed by using a ACLY fragment and a KLHL25 fragment from PANT7_cGST-ACLY and pDONR221-KLHL25 vectors (DNASU Plasmid Repository), respectively. Lentiviral shRNA vectors against CUL3 (V3LHS_351781 and V3LHS_ 351782) and ACLY (V3LHS_395677 and V2LHS_94212) were purchased from Open Biosystems. pLPCX-ACLY-HA vectors with K540R, K546R, and/or K554R mutations were constructed by site-directed mutagenesis using a QuikChange II XL site-directed mutagenesis kit (Agilent Technologies). Detailed information of primers and vectors is in the Supplemental Material.
LC-MS/MS assays
To determine potential ACLY-binding proteins, co-IPs followed by LC-MS/MS assays were performed as described previously (Zhang et al. 2016). In brief, ACLY-HA protein in HEK293T cells with stable expression of ACLY-HA was pulled down by co-IP using anti-HA beads. HEK293T cells transduced with control vectors were used as a control. LC-MS/MS analysis was performed at the Biological Mass Spectrometry facility of Rutgers University.
Western blot assays
Standard Western blot assays were used to analyze protein expression. The following antibodies were used for assays: anti-Flag-M2 (Sigma, F1804), anti-β-Actin (Sigma, A5441), anti-Myc (Roche, 9E10), anti-HA (Roche, 3F10), anti-ACLY (Abcam, ab40793), anti-CUL3 (Santa Cruz Biotechnology, sc-166110), anti-KLHL25 (Santa Cruz Biotechnology, sc-131968), anti-His (Santa Cruz Biotechnology, sc-803), anti-Ub (Santa Cruz Biotechnology, sc-8017), and anti-GST (Santa Cruz Biotechnology, sc-138). The band intensity was quantified by digitalization of the X-ray film and was analyzed with ImageJ software (Scion Corporation).
GST pull-down assays
His-tagged KLHL25 and CUL3 proteins were purified from bacteria through a Ni2+-NTA (Qiagen) column. Purified His-KLHL25 proteins and/or His-CUL3 (200 ng) were incubated with the glutathione Sepharose beads (Sigma) immobilized with 200 ng of GST-ACLY or GST, respectively. The bound proteins were washed and analyzed by Western blot assays.
In vivo ubiquitination assays
Assays were performed as described previously (Liu et al. 2014). In brief, cells were transduced with different expression vectors (including Myc-CUL3, Myc-CUL3–ΔROC1, KLHL25-Flag, ACLY-HA, ROC1-Flag, and His-Ub) or shRNA against CUL3 or KLHL25 together with ACLY-HA and His-Ub expression vectors. After transduction, cells were treated with 10 µM MG132 for 8 h before assays. The levels of ACLY ubiquitination were determined by immunoprecipitation with an anti-HA antibody followed by Western blot assays with an anti-Ub antibody.
Analysis of gene expression
The expression of genes in cells was analyzed by quantitative TaqMan real-time PCR assays using TaqMan PCR mixture (Applied Biosystems) as described previously (Liu et al. 2014). All primers were obtained from Applied Biosystems. The expression of genes was normalized to actin.
IHC assays
TMAs, which included 212 lung tumor tissues, were obtained from U.S. Biomax. The IHC stainings were performed, and IHC results were scored as described previously (Reiter et al. 1998; Yang et al. 2013). In brief, signals in tumor cells were visually quantified using a scoring system from 0 to 9. The scores were obtained by multiplying the intensity of signals by the percentage of positive cells (signal: 0 = no signal, 1 = weak signal, 2 = intermediate signal, and 3 = strong signal; percentages: 0 = 0%, 1 = <25%, 2 = 25%–50%, and 3 = >50%). Low expression and high expression were defined as scores of <6 and ≥6, respectively.
Measurements of de novo lipid synthesis
The rate of de novo lipid synthesis was analyzed by 14C-labeled glucose incorporation assays to measure the incorporation of 14C-labeled glucose into total lipids in cells as described previously (Hatzivassiliou et al. 2005; Daemen et al. 2015). In brief, cells were cultured in six-well plates overnight in low-glucose and low-serum medium before 1 µCi of 14C-labeled glucose per well was added to the cells. After incubation for 8 h, cells were collected, and the total lipid fraction was extracted by chloroform and methanol (2:1 v/v). 14C-incorporation into total cellular lipids was measured in the lipid-containing phase using a scintillation counter. The obtained counts were normalized to cell number.
Measurements of acetyl-CoA levels
The intracellular levels of acetyl-CoA were measured as described previously (Hatzivassiliou et al. 2005; Zhang et al. 2011). Briefly, cells were harvested in cold PBS with 0.5 mM EDTA, and the pellet was deproteinized in 50 µL of 6% perchloric acid and neutralized with 27.2 µL of 2 M KHCO3. Acetyl-CoA in the cleared supernatants was measured by using an acetyl-CoA assay kit (Biovision) according to the manufacturer's instructions.
Cell proliferation and anchorage-independent growth assays
Cell proliferation was determined by MTT assays. In brief, cells seeded in 96-well plates were incubated with MTT solution for 4 h at 1–4 d after seeding. The absorbance at 570 nm was measured using a plate reader (PerkinElmer). Anchorage-independent growth assays in soft agar were performed as described previously (Zhang et al. 2013; Liu et al. 2014).
Xenograft tumorigenicity assays
Animal maintenance and experiments were performed in accordance with the Institutional Animal Care and Use Committee (IACUC) guidelines and with approval of the IACUC at Rutgers University. Xenograft tumorigenicity assays were performed as described previously (Zhang et al. 2013). In brief, 5 × 106 cells in 0.2 mL of PBS were injected subcutaneously (s.c.) into 8-wk-old BALB/c male athymic nude mice (Taconic; n = 8 mice per group). For SB-204990 treatment, when the tumor volume reached ∼60 mm3, mice were injected (intraperitoneally [i.p.]) with 135 mg/kg SB-204990 per day for 14 d. Control mice were injected with vehicle. Tumor volume = 1/2(length × width2).
Statistical analysis
The differences in xenograft tumor growth among groups were analyzed for statistical significance by ANOVA using GraphPad Prism software. Kaplan-Meier statistics were performed to analyze the significance of differences in the survival of patients among different groups using GraphPad Prism software. All other P-values were obtained using Student's t-test or χ2 test.
Supplementary Material
Acknowledgments
We thank Dr. Arnold Levine for helpful discussion and comments. This work was supported by grants from the National Institutes of Health (R01CA143204), New Jersey Health Foundation, and Bush Medical Research Award to Z.F.; grants from the National Institutes of Health (R01CA160558), Department of Defense (W81XWH-16-1-0358), and Ellison Medical Foundation to W.H.; grants from the National Institutes of Health (R01CA195612 and R01CA156706) to Z.S.; and a grant from the Breast Cancer Research Foundation to B.G.H. G.H. is an intern from Northern Highlands Regional High School (Allendale, NJ). Z.F. and W.H. designed experiments, analyzed the data, and wrote the manuscript. C.Z., J.L., G.H., Y.Z., X.Y., H.W., J.L., and J.Z. carried out the experiments and analyzed the data. Z.S. and B.G.H. contributed reagents.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.283283.116.
References
- Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. 2005. ATP citrate lyase is an important component of cell growth and transformation. Oncogene 24: 6314–6322. [DOI] [PubMed] [Google Scholar]
- Berwick DC, Hers I, Heesom KJ, Moule SK, Tavare JM. 2002. The identification of ATP-citrate lyase as a protein kinase B (Akt) substrate in primary adipocytes. J Biol Chem 277: 33895–33900. [DOI] [PubMed] [Google Scholar]
- Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. 2004. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3–Keap1 ligase. Mol Cell Biol 24: 8477–8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie E, Schulze A, Zechner R, Walther TC, Farese RV Jr. 2013. Cellular fatty acid metabolism and cancer. Cell Metab 18: 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daemen A, Peterson D, Sahu N, McCord R, Du X, Liu B, Kowanetz K, Hong R, Moffat J, Gao M, et al. 2015. Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. Proc Natl Acad Sci 112: E4410–E4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai B, Yoo SY, Bartholomeusz G, Graham RA, Majidi M, Yan S, Meng J, Ji L, Coombes K, Minna JD, et al. 2013. KEAP1-dependent synthetic lethality induced by AKT and TXNRD1 inhibitors in lung cancer. Cancer Res 73: 5532–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Morvan F, Jourde B, Meier V, Kahle P, Brebbia P, Toussaint G, Glass DJ, Fornaro M. 2015. ATP citrate lyase improves mitochondrial function in skeletal muscle. Cell Metab 21: 868–876. [DOI] [PubMed] [Google Scholar]
- DeNicola GM, Cantley LC. 2015. Cancer's fuel choice: new flavors for a picky eater. Mol Cell 60: 514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr C, Janik C, Weg M, Been RA, Bader J, Kang R, Ng B, Foran L, Landman SR, O'Sullivan MG, et al. 2015. Transposon mutagenesis screen identifies potential lung cancer drivers and CUL3 as a tumor suppressor. Mol Cancer Res 13: 1238–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa M, He YJ, Borchers C, Xiong Y. 2003. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol 5: 1001–1007. [DOI] [PubMed] [Google Scholar]
- Gan W, Dai X, Lunardi A, Li Z, Inuzuka H, Liu P, Varmeh S, Zhang J, Cheng L, Sun Y, et al. 2015. SPOP promotes ubiquitination and degradation of the ERG oncoprotein to suppress prostate cancer progression. Mol Cell 59: 917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P, Sumara I, Lechner E. 2013. The emerging family of CULLIN3–RING ubiquitin ligases (CRL3s): cellular functions and disease implications. EMBO J 32: 2307–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagenson KK, Tait L, Wang J, Shekhar MP, Polin L, Chen W, Wu GS. 2012. Cullin-3 protein expression levels correlate with breast cancer progression. Cancer Biol Ther 13: 1042–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, Hingorani SR, Tuveson DA, Thompson CB. 2005. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer cell 8: 311–321. [DOI] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 13: 76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossatz U, Breuhahn K, Wolf B, Hardtke-Wolenski M, Wilkens L, Steinemann D, Singer S, Brass F, Kubicka S, Schlegelberger B, et al. 2010. The cyclin E regulator cullin 3 prevents mouse hepatic progenitor cells from becoming tumor-initiating cells. J Clin Invest 120: 3820–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhajda FP. 2000. Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition 16: 202–208. [DOI] [PubMed] [Google Scholar]
- Lee J, Zhou P. 2010. Cullins and cancer. Genes Cancer 1: 690–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DF, Kuo HP, Liu M, Chou CK, Xia W, Du Y, Shen J, Chen CT, Huo L, Hsu MC, et al. 2009. KEAP1 E3 ligase-mediated downregulation of NF-κB signaling by targeting IKKβ. Mol Cell 36: 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YR, Yuan WC, Ho HC, Chen CH, Shih HM, Chen RH. 2010. The Cullin 3 substrate adaptor KLHL20 mediates DAPK ubiquitination to control interferon responses. EMBO J 29: 1748–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Tao R, Gao X, Li T, Zhou X, Guan KL, Xiong Y, Lei QY. 2013. Acetylation stabilizes ATP-citrate lyase to promote lipid biosynthesis and tumor growth. Mol Cell 51: 506–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhang C, Wang XL, Ly P, Belyi V, Xu-Monette ZY, Young KH, Hu W, Feng Z. 2014. E3 ubiquitin ligase TRIM32 negatively regulates tumor suppressor p53 to promote tumorigenesis. Cell Death Differ 21: 1792–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medes G, Thomas A, Weinhouse S. 1953. Metabolism of neoplastic tissue. IV. A study of lipid synthesis in neoplastic tissue slices in vitro. Cancer Res 13: 27–29. [PubMed] [Google Scholar]
- Menendez JA, Lupu R. 2007. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer 7: 763–777. [DOI] [PubMed] [Google Scholar]
- Migita T, Narita T, Nomura K, Miyagi E, Inazuka F, Matsuura M, Ushijima M, Mashima T, Seimiya H, Satoh Y, et al. 2008. ATP citrate lyase: activation and therapeutic implications in non-small cell lung cancer. Cancer Res 68: 8547–8554. [DOI] [PubMed] [Google Scholar]
- Milgraum LZ, Witters LA, Pasternack GR, Kuhajda FP. 1997. Enzymes of the fatty acid synthesis pathway are highly expressed in in situ breast carcinoma. Clin Cancer Res 3: 2115–2120. [PubMed] [Google Scholar]
- Nakakuki M, Shimano H, Inoue N, Tamura M, Matsuzaka T, Nakagawa Y, Yahagi N, Toyoshima H, Sato R, Yamada N. 2007. A transcription factor of lipid synthesis, sterol regulatory element-binding protein (SREBP)-1a causes G(1) cell-cycle arrest after accumulation of cyclin-dependent kinase (cdk) inhibitors. FEBS J 274: 4440–4452. [DOI] [PubMed] [Google Scholar]
- Ookhtens M, Kannan R, Lyon I, Baker N. 1984. Liver and adipose tissue contributions to newly formed fatty acids in an ascites tumor. Am J Physiol 247: R146–R153. [DOI] [PubMed] [Google Scholar]
- Pavlova NN, Thompson CB. 2016. The emerging hallmarks of cancer metabolism. Cell Metab 23: 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter RE, Gu Z, Watabe T, Thomas G, Szigeti K, Davis E, Wahl M, Nisitani S, Yamashiro J, Le Beau MM, et al. 1998. Prostate stem cell antigen: a cell surface marker overexpressed in prostate cancer. Proc Natl Acad Sci 95: 1735–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere L, Moreau P, Allmann S, Hahn M, Biran M, Plazolles N, Franconi JM, Boshart M, Bringaud F. 2009. Acetate produced in the mitochondrion is the essential precursor for lipid biosynthesis in procyclic trypanosomes. Proc Natl Acad Sci 106: 12694–12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos CR, Schulze A. 2012. Lipid metabolism in cancer. FEBS J 279: 2610–2623. [DOI] [PubMed] [Google Scholar]
- Singer JD, Gurian-West M, Clurman B, Roberts JM. 1999. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev 13: 2375–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Hayakawa K, Bateman KS, Fraser ME. 2010. Identification of the citrate-binding site of human ATP-citrate lyase using X-ray crystallography. J Biol Chem 285: 27418–27428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinnen JV, Heemers H, van de Sande T, de Schrijver E, Brusselmans K, Heyns W, Verhoeven G. 2004. Androgens, lipogenesis and prostate cancer. J Steroid Biochem Mol Biol 92: 273–279. [DOI] [PubMed] [Google Scholar]
- Thu KL, Pikor LA, Chari R, Wilson IM, Macaulay CE, English JC, Tsao MS, Gazdar AF, Lam S, Lam WL, et al. 2011. Genetic disruption of KEAP1/CUL3 E3 ubiquitin ligase complex components is a key mechanism of NF-κB pathway activation in lung cancer. J Thorac Oncol 6: 1521–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towle HC, Kaytor EN, Shih HM. 1997. Regulation of the expression of lipogenic enzyme genes by carbohydrate. Annu Rev Nutr 17: 405–433. [DOI] [PubMed] [Google Scholar]
- Wang Q, Jiang L, Wang J, Li S, Yu Y, You J, Zeng R, Gao X, Rui L, Li W, et al. 2009. Abrogation of hepatic ATP-citrate lyase protects against fatty liver and ameliorates hyperglycemia in leptin receptor-deficient mice. Hepatology 49: 1166–1175. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. 2009. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324: 1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahagi N, Shimano H, Hasegawa K, Ohashi K, Matsuzaka T, Najima Y, Sekiya M, Tomita S, Okazaki H, Tamura Y, et al. 2005. Co-ordinate activation of lipogenic enzymes in hepatocellular carcinoma. Eur J Cancer 41: 1316–1322. [DOI] [PubMed] [Google Scholar]
- Yanagiya A, Suyama E, Adachi H, Svitkin YV, Aza-Blanc P, Imataka H, Mikami S, Martineau Y, Ronai ZA, Sonenberg N. 2012. Translational homeostasis via the mRNA cap-binding protein, eIF4E. Mol Cell 46: 847–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Sun Y, Hu L, Zheng H, Ji P, Pecot CV, Zhao Y, Reynolds S, Cheng H, Rupaimoole R, et al. 2013. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer Cell 23: 186–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RM, Ackerman D, Quinn ZL, Mancuso A, Gruber M, Liu L, Giannoukos DN, Bobrovnikova-Marjon E, Diehl JA, Keith B, et al. 2013. Dysregulated mTORC1 renders cells critically dependent on desaturated lipids for survival under tumor-like stress. Genes Dev 27: 1115–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi N, Royaux I, Swinnen JV, Smans K. 2012a. ATP citrate lyase knockdown induces growth arrest and apoptosis through different cell- and environment-dependent mechanisms. Mol Cancer Ther 11: 1925–1935. [DOI] [PubMed] [Google Scholar]
- Zaidi N, Swinnen JV, Smans K. 2012b. ATP-citrate lyase: a key player in cancer metabolism. Cancer Res 72: 3709–3714. [DOI] [PubMed] [Google Scholar]
- Zhang C, Lin M, Wu R, Wang X, Yang B, Levine AJ, Hu W, Feng Z. 2011. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci 108: 16259–16264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Liu J, Liang Y, Wu R, Zhao Y, Hong X, Lin M, Yu H, Liu L, Levine AJ, et al. 2013. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun 4: 2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Liu J, Zhao Y, Yue X, Zhu Y, Wang X, Wu H, Blanco F, Li S, Bhanot G, et al. 2016. Glutaminase 2 is a novel negative regulator of small GTPase Rac1 and mediates p53 function in suppressing metastasis. Elife 5: e10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.