Here, Hu et al. investigated the mechanisms by which IKAROS promotes B-cell differentiation and prevents leukemogenesis. By using chromatin, transcription, and gene inactivation studies in primary wild-type and IKAROS-deficient B-cell precursors, they show that a dual mechanism of IKAROS regulation promotes differentiation while safeguarding against a hybrid stem–epithelial–B-cell phenotype that underlies high-risk B-ALL.
Keywords: TEAD, YAP1, LHX2, PRC2, self-renewal
Abstract
IKAROS is required for the differentiation of highly proliferative pre-B-cell precursors, and loss of IKAROS function indicates poor prognosis in precursor B-cell acute lymphoblastic leukemia (B-ALL). Here we show that IKAROS regulates this developmental stage by positive and negative regulation of superenhancers with distinct lineage affiliations. IKAROS defines superenhancers at pre-B-cell differentiation genes together with B-cell master regulators such as PAX5, EBF1, and IRF4 but is required for a highly permissive chromatin environment, a function that cannot be compensated for by the other transcription factors. IKAROS is also highly enriched at inactive enhancers of genes normally expressed in stem–epithelial cells. Upon IKAROS loss, expression of pre-B-cell differentiation genes is attenuated, while a group of extralineage transcription factors that are directly repressed by IKAROS and depend on EBF1 relocalization at their enhancers for expression is induced. LHX2, LMO2, and TEAD–YAP1, normally kept separate from native B-cell transcription regulators by IKAROS, now cooperate directly with them in a de novo superenhancer network with its own feed-forward transcriptional reinforcement. Induction of de novo superenhancers antagonizes Polycomb repression and superimposes aberrant stem–epithelial cell properties in a B-cell precursor. This dual mechanism of IKAROS regulation promotes differentiation while safeguarding against a hybrid stem–epithelial–B-cell phenotype that underlies high-risk B-ALL.
Cell lineages are ultimately defined by subroutines of signaling pathways and transcription factors that coordinately regulate growth and differentiation. B-cell differentiation is marked by a progressive loss in commitment to alternate lymphoid cell fates and a stepwise gain in immunoglobulin gene rearrangements that is coupled to activation of discrete downstream signaling pathways. These signaling events first support and subsequently restrain expansion of pre-B-cell receptor-bearing precursors (pre-B cells) by promoting their differentiation to an immune-competent B cell (Herzog et al. 2009). A network of transcription factors (e.g., E2A, EBF1, PAX5, IRF4, IKAROS, and its family member, AIOLOS) coordinates the gene expression events required for transition through these early stages of B-cell differentiation, and its deregulation is associated with leukemic transformation (Nutt and Kee 2007; Bryder and Sigvardsson 2010; Mandel and Grosschedl 2010). Human pre-B-cell precursor leukemias (B-cell acute lymphoblastic leukemia [B-ALL]) harbor multiple mutations that, on one hand, constitutively activate pre-BCR and IL7R signaling while, on the other, inactivate transcription factors supporting B-cell differentiation (Mullighan et al. 2007; Mullighan 2012; Roberts et al. 2012; Inaba et al. 2013). Several of these signaling and transcriptional regulators are also engaged at later stages of the pathway that take place in peripheral lymphatics, such as antibody affinity maturation at germinal centers and differentiation of high-affinity B cells into long-lived antibody-producing plasma cells (Johnson and Calame 2003; Rickert 2013). Thus, regulatory mechanisms operating at both the early and late stages of B-cell differentiation may be responsible for the pathogenesis of different types of B-lymphoid neoplasms from precursor (B-ALL) to mature B cell (chronic lymphocytic leukemia) to plasma cell (multiple myeloma).
Among the key transcriptional regulators of B-cell differentiation, inactivating mutations in the IKZF1 gene that encodes IKAROS are uniquely associated with a high frequency of leukemia relapse, drug resistance, and poor prognosis (Martinelli et al. 2009; Mullighan et al. 2009; Kuiper et al. 2010). The most frequent IKAROS mutations generate dominant-negative protein isoforms that interfere with both IKAROS and AIOLOS activity in early B-cell precursors. However, both long-lived antibody-producing plasma cells and their malignant counterparts in multiple myeloma are dependent on the activity of the Ikzf1 gene family for growth and survival (Cortes and Georgopoulos 2004; Kronke et al. 2014; Lu et al. 2014). IKAROS is one of the earliest-acting lymphoid lineage transcription factors required for priming of lymphoid lineage gene expression and providing lymphoid lineage differentiation potential to multipotent hematopoietic progenitors (Ng et al. 2009; Yoshida et al. 2010). Following commitment into the lymphoid lineage, IKAROS and its family member, AIOLOS, are required for transition from the highly proliferative and stromal-dependent large pre-B cell to the quiescent and stromal-independent small pre-B cell, during which immunoglobulin light chain rearrangement takes place (Heizmann et al. 2013; Joshi et al. 2014; Schwickert et al. 2014).
Engagement of wild-type large pre-B cells with bone marrow (BM) stroma supports limited self-renewal but is not necessary for proliferative expansion or survival of these cells as they differentiate to the small pre-B-cell stage (Joshi et al. 2014). In sharp contrast, large pre-B cells deficient for IKAROS activity are stromal-dependent for proliferation and survival, show a dramatic increase in self-renewal, and are unable to differentiate (Joshi et al. 2014). In line with an altered cellular phenotype, IKAROS-deficient large pre-B cells have attenuated pre-BCR signaling and dramatically increased integrin signaling and integrin-dependent adhesion to BM stroma. Notably, upon stromal detachment, IKAROS-deficient but not wild-type large pre-B cells undergo an anoikis type of cell death that is indicative of an epithelial cell-like phenotype supported by distinct mechanisms of survival (Joshi et al. 2014). Notably, these epithelial-like properties are retained after IKAROS-deficient large pre-B cells transition to a leukemic stage and may be responsible for the drug resistance and high-risk phenotype attributed to these leukemic cells (Joshi et al. 2014; Churchman et al. 2015).
Our present studies show that IKAROS is engaged in the reciprocal regulation of superenhancer networks with distinct lineage affiliations. IKAROS in the company of other B-cell master regulators defines a set of superenhancers that support expression of key signaling regulators of pre-B-cell differentiation. In the absence of IKAROS, B-cell transcription factors still recruited at these regulatory sites are unable to provide the highly permissive chromatin environment required for pre-B-cell differentiation. Inactive and poised enhancers allied with genes normally expressed in stem–epithelial cell precursors and repressed in pre-B cells are highly enriched for IKAROS in limited company of B-cell transcription factors. These genes include key hematopoietic and epithelial cell transcriptional regulators such as LMO2, LHX2, and the YAP–TEAD nuclear effectors of HIPPO signaling. Upon loss of IKAROS activity, these “extralineage” transcription factors are rapidly expressed and collaborate with native B-cell transcription factors to define a de novo landscape of superenhancers. These de novo superenhancers antagonize Polycomb repression at promoters and induce a gene expression program that provides B-cell precursors with stem–epithelial cell properties before they become neoplastic.
Results
IKAROS associates with distinct enhancer networks in pre-B-cell precursors
The transition from a highly proliferative to a quiescent pre-B-cell precursor is a tightly regulated developmental process and a primary target for leukemogenesis in humans (Fig. 1A; Cobaleda and Sanchez-Garcia 2009). Nonetheless, the underlying transcriptional and epigenetic mechanisms remain unclear. To delineate this process, we generated ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) data sets for transcriptional regulators functionally implicated in early B-cell differentiation and for histone modifications that demarcate transcriptionally active, poised, and repressed promoters and enhancers (Fig. 1A; Supplemental Fig. 1A–D; Nutt and Kee 2007; Bryder and Sigvardsson 2010; Mandel and Grosschedl 2010; Zhou et al. 2011). The cell populations used for ChIP-seq analysis included stromal-adherent large pre-B cells, a transient stage in normal B-cell differentiation, and a parallel population in which the DNA-binding domain of IKAROS was deleted, generating dominant-negative IKAROS (IKDN) isoforms analogous to mutant proteins found frequently in high-risk, treatment-resistant human B-ALL (Fig. 1A; Joshi et al. 2014). Wild-type and IKDN stromal-adherent large pre-B cells were obtained from primary cultures of BM large pre-B cells as previously described (Joshi et al. 2014). RNA sequencing (RNA-seq) was performed with wild-type and IKDN large pre-B cells both immediately ex-vivo or after limited culture on BM stroma to generate appropriate cell numbers required for the multiple ChIP assays (Joshi et al. 2014).
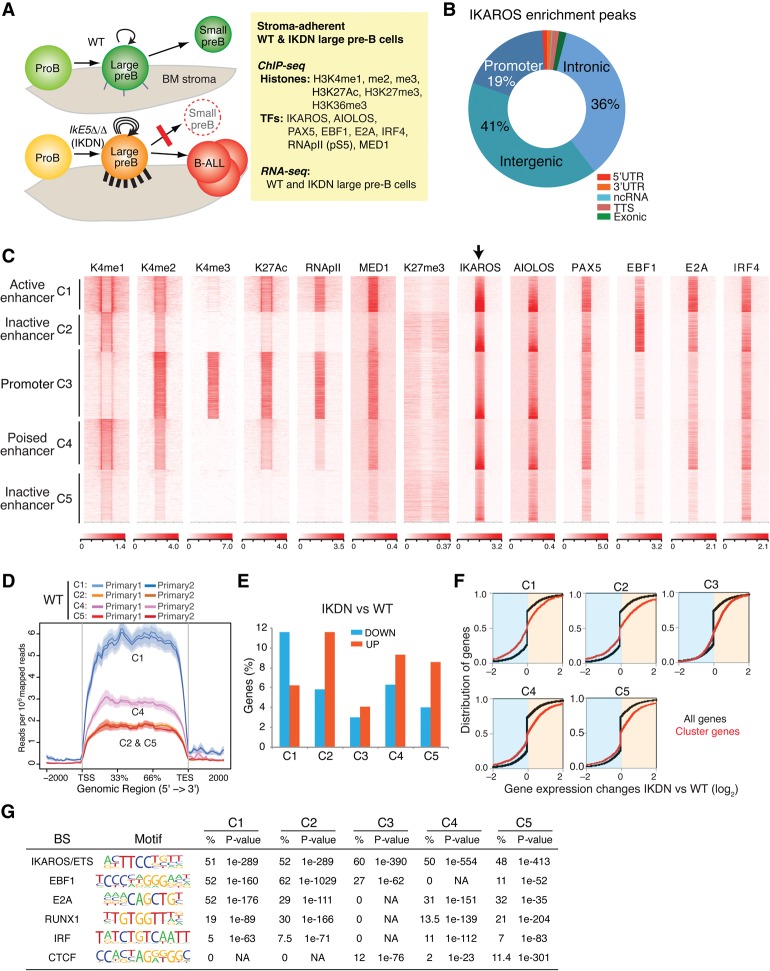
Figure 1.
Transcriptional and epigenetic landscape of wild-type adherent large pre-B cells. (A) Outline of an experimental approach to delineate the transcriptional mechanisms that guide pre-B-cell differentiation through a highly proliferative stromal-adherent phase. The effects of homozygous deletion of Ikzf1 exon 5 (IkE5Δ/Δ), encoding the DNA-binding domain and generating an IKDN isoform, on cell adhesion (cell projections on BM stroma), self-renewal (circular arrows), differentiation block (red line), and leukemogenesis (B-ALL) are depicted. (B) Relative distribution of IKAROS enrichment peaks at the indicated genomic locations in wild-type stromal-adherent large pre-B cells. (C) Heat map generated by K-means clustering of reads from ChIPs of histone modifications and transcription factors centered on IKAROS enrichment peaks (±15 kb). Read distribution within IKAROS enrichment peaks and size were normalized to the expected fragment size using the default spline fit algorithm of NGS.plot. Five clusters were generated, four of which were designated by histone modification enrichment as enhancers (C1, C2, C4, and C5) and one of which was designated as a promoter (C3). (D) Average gene expression for IKAROS gene targets in enhancer clusters C1, C2, C4, and C5, shown as exonic read distribution over the gene body (read count per million mapped reads). RNA-seq studies were performed with two independently isolated wild-type large pre-B-cell samples. (E) Percentage distribution of 1597 up-regulated and 610 down-regulated genes in IKDN pre-B cells in clusters (as in C) defined by IKAROS binding, histone modifications, and transcription factor distributions. (F) Cumulative distribution function (CDF) plots depicting differential gene expression between IKDN and wild-type pre-B cells are shown for each gene cluster (red) defined in C relative to differential gene expression of all genes (black). P-values for differences in gene expression for each cluster relative to all genes were <0.0001, calculated using a two-sided Kolmogorov Smirnov test. (G) De novo transcription factor-binding motif analysis (HOMER) of IKAROS enrichment peaks in wild-type adherent large pre-B cells is shown for each cluster defined in C. The most significantly discovered motifs, frequency (percentage), and P-values are shown.
Analysis of IKAROS chromatin distribution in wild-type stromal-adherent large pre-B cells revealed that the majority of IKAROS enrichment peaks (77% of 13,843) were located at nonpromoter regions and displayed a very similar distribution to its family member, AIOLOS (Fig. 1B,C). Co-clustering of histone modifications at IKAROS peaks indicated that a majority was associated with active or poised enhancers (clusters C1, C2, C4, and C5), and a minority was associated with active promoters (cluster C3) (Fig. 1C). IKAROS peaks in cluster C1 were highly enriched for H3K4me2, H3K27ac, RNApII, and the Mediator complex subunit 1 (MED1) that together define transcriptionally active enhancers (Fig. 1C, C1 [K4me1hi K4me2hi K27achi RNApIIhi MED1hi]; Heintzman et al. 2009; Creyghton et al. 2010; De Santa et al. 2010; Malik and Roeder 2010; Whyte et al. 2013). Strong enrichment for B-cell lineage master transcriptional regulators PAX5, E2A, and IRF4 and moderate enrichment for EBF1 were also observed in cluster C1. Clusters C4, C2, and C5 displayed a progressive reduction in H3K4me1, H3K4me2, H3K27ac, RNApII, MED1, and pre-B-cell transcription factors, consistent with a poised to inactive enhancer status (i.e., Fig. 1C, C4 [K4me1int K4me2int K27acint], C2 [K4me1int-low K4me2int-low K27acint-low], C5 [K4me1low K4me2low K27aclow]). Notably, EBF1 was strongly enriched in cluster C2 but not in any of the other clusters associated with repressed genes in wild-type pre-B cells (e.g., C4 or C5).
IKAROS peaks in cluster C3 identify active promoters strongly enriched for H3K4me3, H3K4me2, H3K27ac, RNApII, MED1, PAX5, and IRF4 (Fig. 1C, C3). Lower enrichment of EBF1 and E2A at the C3 promoter cluster was consistent with the reported enhancer-specific targeting of these factors (Lin et al. 2010; Li et al. 2012). In contrast to other histone modifications associated with active and poised regulatory enhancers and promoters, H3K27me3, a mark of PRC2 complex activity, was not detected in the immediate vicinity of IKAROS peaks associated with either promoters or poised enhancers (Fig. 1C, H3K27me3). However, strong PRC2 enrichment was consistently seen at the promoters of genes that were dependent on IKAROS for repression in wild-type stromal-adherent large pre-B cells, as described in later sections.
Genes associated with the active enhancer cluster C1 were, on average, more highly expressed compared with genes associated with the poised/inactive enhancer clusters C4, C2, and C5 in wild-type stromal-adherent large pre-B cells (Fig. 1D). A higher proportion of genes dependent on IKAROS for expression in wild-type pre-B cells was present in C1 compared with other clusters (Fig. 1E). Conversely, a higher proportion of genes dependent on IKAROS for repression was associated with C2, C4, and C5. The differential gene expression changes between IKDN and wild-type pre-B cells were tested for each individual cluster relative to all expressed genes (Fig. 1F). A significant shift in the cumulative distribution function curve to the left was seen for C1, consistent with more down-regulated genes, and to the right for C2, C4, and C5, consistent with more up-regulated genes in response to IKAROS inactivation.
De novo motif analysis of IKAROS enrichment peaks (within 200 base pairs [bp] from the defined IKAROS chromatin-binding site) in each cluster confirmed that IKAROS targets chromatin through its own DNA-binding activity and in close proximity to B-cell transcription regulators (Fig. 1G). In addition, differences in motif composition in the vicinity of IKAROS-binding sites were seen in each cluster. For example, the active enhancer cluster C1 showed a high frequency for EBF- and E2A-binding motifs (51%–52%), consistent with their strong chromatin enrichment at these sites (Fig. 1C). C2 had the highest occurrence of EBF1 motifs (62%), again consistent with the strongest EBF1 enrichment in this cluster. In contrast, C5 and C4 were progressively depleted of EBF1-binding motifs and chromatin enrichment. Similar levels of E2A-binding motifs were detected in C2, C4, and C5 (29%–32%), albeit at lower levels compared with C1 (52%). Finally, motifs for the chromatin organizer CTCF were detected only in C5 and the promoter cluster C3.
Thus, at the highly proliferative, stromal-adherent phase of B-cell differentiation, IKAROS appears to be engaged in two mechanisms of transcriptional regulation. The presence of IKAROS at active enhancers in the context of E2A, EBF1, PAX5, and IRF4 may contribute to the positive regulation of gene expression required for pre-B-cell precursor differentiation. At poised/inactive enhancers, IKAROS may contribute to the repression of genes that might interfere with further differentiation. Here, IKAROS-specific interactions with transcription factors such as EBF1 may promote repression of these genes.
Pre-B-cell differentiation is controlled by IKAROS superenhancers
The co-occupancy of IKAROS with other B-cell master regulators and MED1 at active enhancer sites (e.g., C1) in wild-type large pre-B cells suggested their possible participation at superenhancer regulatory domains. Superenhancers are defined as clusters of binding sites for master transcription regulators that are highly enriched for active histone modifications and the Mediator complex (Hnisz et al. 2013, 2015; Whyte et al. 2013). It has been proposed that superenhancers are responsible for the high level of gene expression required of developmentally regulated genes to specify cell identity (Whyte et al. 2013). We therefore examined the participation of B-cell transcription factors in putative superenhancers that are associated with genes that support the large-to-small pre-B-cell transition and are down-regulated in IKDN stromal-adherent large pre-B cells (Joshi et al. 2014; Schwickert et al. 2014). Many of the 610 down-regulated genes are functional components of pathways supporting various aspects of lymphocyte differentiation (e.g., VDJ recombination and pre-BCR signaling) (Fig. 2A) and, in their majority, were associated with enrichment peaks for IKAROS, EBF1, PAX5, and IRF4 in wild-type stromal-adherent large pre-B cells (Fig. 2B).
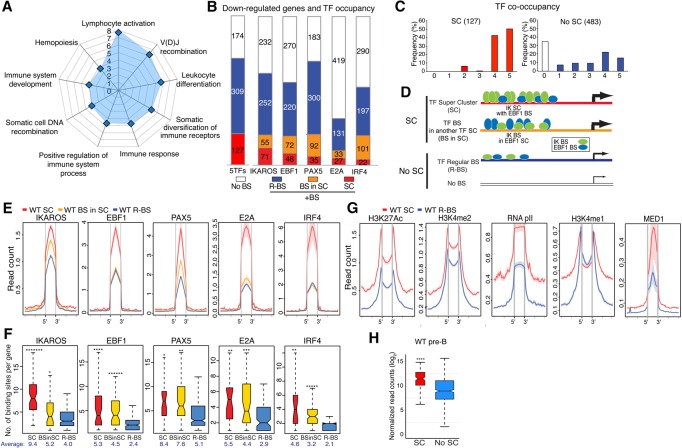
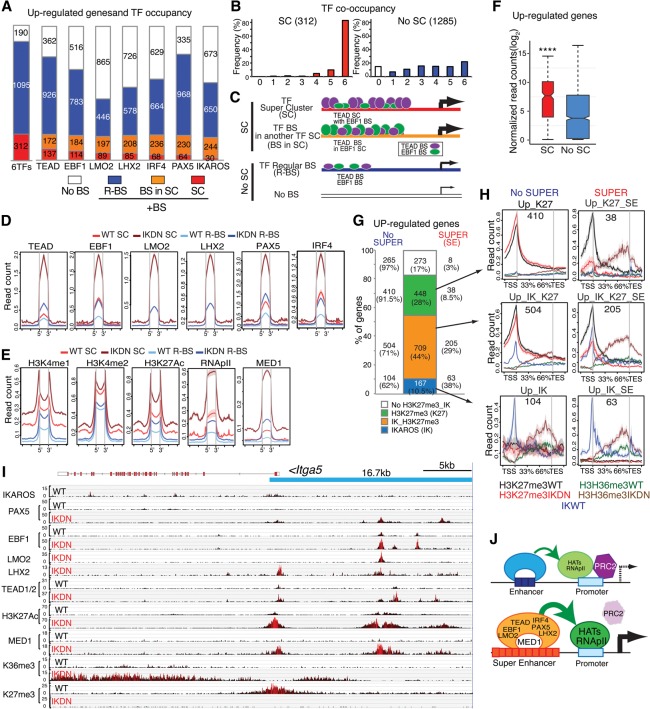
Figure 2.
IKAROS-based enhancer superclusters (SCs) support the large-to-small pre-B-cell transition. (A) Gene ontology (GO) of functional pathways for genes with superenhancers down-regulated in IkE5Δ/Δ (IKDN) stromal-adherent large pre-B cells and associated with IKAROS-binding sites in wild-type pre-B cells is shown. P-values (−log10 transformed) for pathway discovery are indicated. (B) Histogram depicting the frequency (in wild-type large pre-B cells) of occupancy by the indicated B-cell transcription factors of 610 genes down-regulated upon loss of IKAROS activity. Genes bound by a specific transcription factor are further subdivided by whether they are associated with regular binding sites (R-BS), binding sites in a SC configuration, or another factor's SC (BS in SC). The histogram at the left (5TFs) indicates the overall number of genes associated with any transcription factor SC (127), associated with a regular enhancer (R-BS; 309), or not associated with any transcription factor-binding sites (No BS; 174). (C) The frequency of occupancy of down-regulated genes with (SC) or without (no SC) SCs by the five transcription factors (from 0 to 5) is shown. (D) Schematic representation of idealized examples of IKAROS-binding sites (green ovals) in a SC configuration (SC), within a SC of another transcription factor (shown here as EBF1 in blue ovals; BS in SC), in a regular enhancer (R-BS), or in a gene with no binding sites (No BS) for these factors. The frequency of transcription factor occupancy is represented by the size of the ovals, whereas the relative density of binding sites is depicted by the number of ovals. The relative transcription level at these gene subsets is depicted by the thickness of an arrow at the transcription start site (TSS). (E) Comparative enrichment of transcription factors at binding sites associated with SCs ([red] SC; [yellow] BS in SC) or regular enhancers (R-BS) at down-regulated genes. Read densities (read count per million mapped reads) for IKAROS, EBF1, PAX5, E2A, and IRF4 are provided for wild-type pre-B cells. The start (5′) and end (3′) of transcription factor-binding sites are indicated. (F) Box plots depicting the distribution of the number of binding sites for the transcription factors in B at SCs, binding sites associated with SCs, and regular enhancer configurations. Box plot whiskers extend to 1.5 times the interquartile range. The average number of binding sites per gene subset is shown below each plot. P-values for differential distributions in the SC or BS in SC group relative to the R-BS group were obtained using an unpaired two-tailed t-test with variance determined by an F-test. Statistical significance compared with the R-BS subset are shown as P-values. (*******) P < 10−9; (******) P < 10−6; (*****) P < 10−5; (****) P < 10−4; (***) P < 10−3; (**) P < 10−2; (*) P < 0.05. (G) Comparative enrichment of permissive histone modifications (H3K27ac, H3K4me2, H3K4me1, RNApII, and MED1) at constituent SC-binding sites (SC) or regular binding sites (R-BS) as in D. (H) Expression in wild-type pre-B cells of down-regulated genes with or without SCs (SC or no SC) is shown in a box plot of log2 transformed normalized exonic read counts. Box plot whiskers extend to 1.5 times the interquartile range. A highly significant difference in expression between the two subsets in wild-type large pre-B cells is supported by a P-value of <10−4 (****) obtained by two-tailed unpaired t-test.
We used a rank order superenhancer algorithm that stiches together transcription factor enrichment peaks into 12.5-kb regions and sorts them according to enrichment value (Whyte et al. 2013; Hah et al. 2015) to identify clusters of binding sites for each B-cell transcription factor in wild-type stromal-adherent large pre-B cells (Supplemental Fig. 2A). We defined these as transcription factor superclusters (SCs). SCs for each B-cell transcription factor were assigned to genes by proximity, and association with the IKAROS-dependent down-regulated genes was deduced (Fig. 2B, SC). One-hundred-twenty-seven unique down-regulated genes were obtained, each associated with a transcription factor SC defined by at least one of these B-cell factors (Supplemental Fig. 2B; Supplemental Table S1). Down-regulated genes with transcription factor SCs were identified primarily by IKAROS (56%; 71 of 127) and, to a progressively lesser extent, EBF1 (38%; 48 of 127), PAX5 (28%; 35 of 127), E2A (21%; 27 of 127), and IRF4 (17%; 22 of 127) (Fig. 2B).
Genes with a SC defined by one factor were also associated with binding sites for other factors that were not in a SC configuration (Fig. 2B–D, model). However, in general, both the density of binding sites and, in some cases, their occupancy for these other factors were greater than the corresponding density and occupancy at genes with regular enhancers (Fig. 2E,F). For example, both the frequency of occupancy by IKAROS protein and the density of IKAROS-binding sites were significantly higher in the 44% (55 of 127) of genes associated with SCs that were not defined by IKAROS as opposed to genes with regular enhancers (Fig. 2B,E,F [respectively, cf. BS in SC vs. R-BS], modeled in D). Furthermore, the density of binding sites for EBF1, PAX5, IRF4, and E2A was significantly enriched at genes with SCs defined by IKAROS and other transcription factors relative to genes with regular enhancers (Fig. 2F [BS in SC vs. R-BS], modeled in D). The detected occupancy of PAX5 on its binding sites at genes with SCs defined by other factors was also significantly higher than at genes with regular PAX5 enhancers (Fig. 2E, PAX5 BS in SC vs. R-BS). Notably, 92% of the down-regulated genes associated with SCs were bound by at least four and frequently five transcription factors (Fig. 2C). Thus, there is a strong cooperation of all five B-cell transcription factors at all of the SCs identified in this analysis. It is possible that the stringency of the algorithm has created an artificial distinction within this group between the clusters (for example) with higher IKAROS density and occupancy and those just below the threshold. Nonetheless, our approach of defining SCs by individual transcription factors has identified a distinct class of regulatory domains with strong collective occupancy for at least five B-cell master regulators that is not seen at regular enhancers.
Analysis of chromatin configuration at individual transcription factor-binding sites that define SCs (referred to here as constituent SC-binding sites) revealed that they were highly enriched for active enhancer histone modifications (e.g., H3K27ac, H3K4me2, and RNApII) and MED1 compared with regular enhancers associated with down-regulated genes (Fig. 2G). Only H3K4me1, a histone modification that is prevalent at inactive or poised enhancers, showed a small decrease at SCs compared with regular enhancer-binding sites (Fig. 2G). Down-regulated genes affiliated with SCs were normally highly expressed in wild-type pre-B cells relative to down-regulated genes with no SCs, providing functional evidence for increased activity at the associated clusters of enhancers (Fig. 2H).
Taken together, our data identify a class of regulatory elements with properties of superenhancers. These include high enrichment with the Mediator complex, B-cell master regulators of transcription, a highly permissive chromatin environment, and association with highly expressed developmentally regulated genes. Superenhancers identified here are associated with genes dependent on IKAROS for strong expression in pre-B cells and are defined predominantly by dense clusters of IKAROS-binding sites in conjunction with at least four other B-cell master regulators. The cooperative activity of these factors across this entire set of superenhancers defines a critical for pre-B-cell differentiation gene expression program.
The epigenetic landscape of pre-B-cell superenhancers is dependent on IKAROS
We next examined the effects of IKAROS loss on the chromatin and transcription factor landscape of superenhancers associated with pre-B-cell differentiation genes. Notably, among the down-regulated genes associated with IKAROS superenhancers were key regulators of pre-B-cell signaling and differentiation, such as Sykb, CD79b, Blnk, Foxo1, and Ccnd3. Loss-of-function mutations in these genes have been reported to arrest pre-B-cell differentiation (Supplemental Fig. 3A; Cheng et al. 1995; Gong and Nussenzweig 1996; Minegishi et al. 1999; Pappu et al. 1999; Cooper et al. 2006; Dengler et al. 2008). As shown for the genes encoding SYK and BLNK, loss of IKAROS correlated with a reduction in transcriptionally permissive histone modifications such as H3K27ac, H3K4me3, and the basal transcription complex (RNApII) in the vicinity of IKAROS-binding sites (Fig. 3A). Surprisingly, occupancy by pre-B-cell transcription factors such as PAX5, EBF1, and IRF4 and the transcriptional coactivator MED1 was not reduced (Fig. 3A).
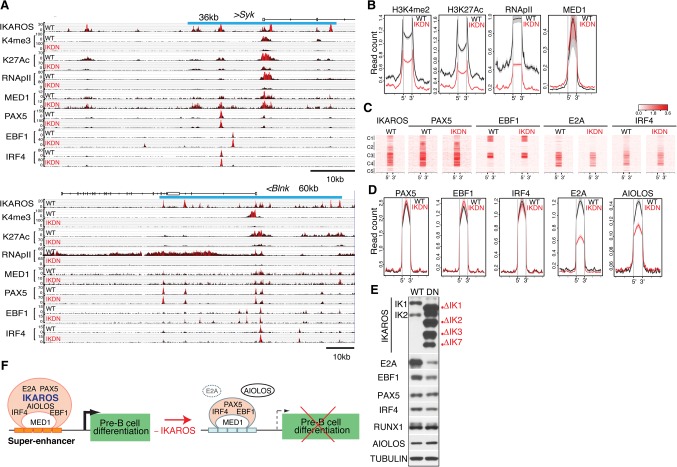
Figure 3.
The permissive chromatin state of pre-B-cell superenhancers is dependent on IKAROS. (A) Genome browser tracks are shown for ChIP-seq of IKAROS, H3K4me3, H3K27ac, RNApIIS5, MED1, PAX5, EBF1, and IRF4 at the Sykb (top panel) and Blnk (bottom panel) genes in wild-type and IKDN pre-B cells. The associated IKAROS superenhancers are demarcated by blue lines (Sykb, 36 kb; Blnk, 60 kb). Black histograms depict sequencing read distribution, with red indicating a higher read depth. (B) Comparative analysis of read densities for permissive histone modifications (H3K27me2, H3K27ac, RNApII, and MED1) at transcription factor-binding sites that constitute superenhancers affiliated with down-regulated genes is shown in wild-type (black) and IKDN (red) pre-B cells. (C,D) Comparative analysis of B-cell transcription factor enrichment at superenhancer-binding sites as described in B. (C) Heat maps were generated by K-means clustering of reads from ChIPs for transcription factors centered at the constituent binding sites of superenhancers associated with 127 down-regulated genes described in Figure 2B. (D) Read densities of transcription factors at superenhancer-binding sites as in B and C (±15 kb). (E) Immunoblot analysis of B-cell transcription factors in wild-type and IKDN stromal-adherent large pre-B cells. The two major IKAROS isoforms (IK1 and IK2) expressed in wild-type pre-B cells are identified at the left. Mutant isoforms are identified at the right. (F) A model of regulation of pre-B-cell differentiation genes by IKAROS superenhancers. Transcriptionally permissive IKAROS-based superenhancers (orange bars) are associated with highly expressed genes promoting pre-B-cell differentiation (black arrow). Loss of IKAROS causes restriction in the chromatin configuration at superenhancers (light-blue bars), transcription is attenuated (broken arrow), and pre-B-cell differentiation is blocked. However, with the exception of AIOLOS and E2A, binding of PAX5, EBF1, IRF4, and MED1 at these regulatory sites was not affected.
The chromatin and transcription factor landscape was evaluated at all of the superenhancers associated with down-regulated genes in IKDN pre-B cells (Fig. 3B–D). The high level of permissive histone modifications (e.g., H3K4me2 and H3K27ac) seen at the constituent enhancer sites of these superenhancers in wild-type pre-B cells was greatly reduced in IKDN pre-B cells (Fig. 3B). Notably, although RNApII occupancy was greatly reduced, MED1 occupancy was unchanged. Enrichment of the B-cell transcription factors PAX5, EBF1, and IRF4 was also not altered (Fig. 3C,D). In contrast, E2A and AIOLOS enrichment was greatly reduced (Fig. 3C,D). Although a decrease in E2A protein was seen in IKDN pre-B cells, AIOLOS protein levels were slightly increased (Fig. 3E). A further examination of all AIOLOS enrichment peaks identified in wild-type pre-B cells revealed extensive co-occupancy with IKAROS and an overall reduction in IKDN pre-B cells (Supplemental Fig. 4A–C). A reduction in transcriptionally permissive histone modifications and binding of E2A and AIOLOS was also seen at regular enhancers, but PAX5, EBF1, IRF4, or MED1 binding was unchanged (Supplemental Fig. 3B–D).
In summary, our data indicate that IKAROS is a key regulator of superenhancers that control expression of genes required for pre-B-cell differentiation (Fig. 3F). IKAROS supports their highly permissive chromatin environment and recruitment of RNApII. Nonetheless, targeting of B-cell master regulators such as PAX5, EBF1, and IRF4 and the Mediator complex at these superenhancers is not dependent on IKAROS.
IKAROS represses a transcriptional network of nonlymphoid and lymphoid regulators
Aberrant induction of genes normally expressed in neuro-epithelial cells and involved in molecular and cellular processes, such as adhesion-mediated receptor signaling, actin cytoskeleton, axon guidance, and cell morphogenesis, is observed in IKDN pre-B cells (Joshi et al. 2014). Expression of these genes is likely responsible for the prominent stromal adhesion and high clonogenic potential of IKDN pre-B cells and their leukemic counterparts that were described previously (Joshi et al. 2014). We therefore evaluated the role of IKAROS in the repression of this gene signature in wild-type pre-B cells and tested which transcription regulators supported its induction in IKDN pre-B cells.
A set of transcription factor genes normally expressed in stem cells and precursors of epithelial origin was induced in IKDN pre-B cells (Fig. 4A). These included Lhx2, Tbx19, Lmo2, Hoxb5–8, Tead1, Tead2, and the TEAD coactivator YAP1 (Yap1). Expression of these genes was also examined in leukemic cells generated by transformation of primary IKDN pre-B cells by the oncogenic tyrosine kinase BCR-ABL1 (Roumiantsev et al. 2001). These leukemic cells retain the “neuro-epithelial” gene signature and epithelial cell-like phenotypes of primary IKDN pre-B cells (Churchman et al. 2015; N Jena, I Joshi, T Yoshida, Z Zhang, Z Liu, P Tata, A Raufi, IM Shapiro, D Weever, JA Pachter, et al., unpubl.). Expression of Tead1/2, Hoxb5, Hoxb6, Hoxb7, and Hoxb8 was increased further in IKDN BCR-ABL1+ leukemic pre-B cells, while the level of native B-cell transcription factors such as Ebf1 was not significantly changed (Fig. 4A). These transcription factors, which are not normally expressed in pre-B cells (referred to here as “extralineage” factors), were readily detected at the protein level, supporting a functional contribution to transcription (Fig. 4B). All of these transcription factor genes were directly repressed by IKAROS (Fig. 4C, Tead1; Supplemental Fig. 5A,B, Lmo2 and Lhx2; data not shown). They were also repressed by PRC2, as indicated by the presence of H3K27me3 at promoters and through the gene body in wild-type pre-B cells (Fig. 4C; Supplemental Fig. 5A,B).
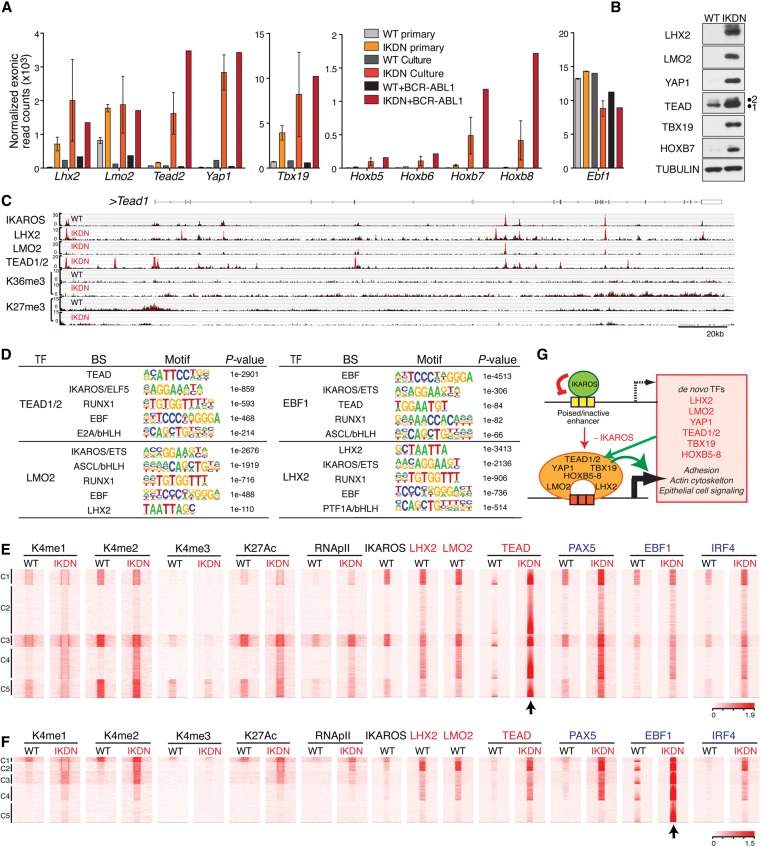
Figure 4.
Direct repression of an extralineage transcription factor network by IKAROS. (A) Induction of mRNA expression for Lhx2, Lmo2, Tead2, Yap1, Tbx19, and Hoxb5–8 in IKDN preleukemic and leukemic (+BCR-ABL1) pre-B cells. Normalized exonic raw reads are shown for the average of two wild-type and two IKDN ex vivo isolated large pre-B-cell populations (primary), one wild-type and two IKDN cultured stromal-adherent large pre-B-cell populations (Culture), one wild-type + BCR–ABL1 and one IKDN + BCR–ABL1 cultured large pre-B leukemia cell population. Ebf1 expression is shown as a control. (B) Immunoblot analysis of protein expression for the transcription factors from A. (C) Genome browser tracks are shown at the Tead1 locus for ChIP-seq of IKAROS, LHX2, LMO2, TEAD 1/2, H3K36me3 (K36me3), and H3K27me3 (K27me3) in wild-type and IKDN pre-B cells. (D) De novo motif discovery (HOMER) for peaks of TEAD (1/2), EBF1, LHX2, and LMO2 specifically enriched in IKDN stromal-adherent large pre-B cells. The most significant discovered motifs and log10 P-values are shown. (E,F) Heat maps were generated by K-means clustering of reads from ChIPs for histone modifications (H3K4me1, H3K4me2, H3K4me3, and H3K27ac) and transcription factors (RNApII, IKAROS, LHX2, LMO2, TEAD, PAX5, EBF1, and IRF4) centered (arrow) at de novo TEAD (E) or EBF1 (F) peaks (C1–C5). Extralineage and B-cell transcription factors are marked in red and blue, respectively. (G) Model of IKAROS repression at inactive/poised enhancers associated with master transcription regulators of stem and epithelial cell origin. Induction of these transcription factors triggered by loss of IKAROS initiates a feed-forward cross-regulatory loop that augments their own expression and that of other genes involved in epithelial cell functions.
Derepression of these extralineage transcription factors is likely responsible for reorganizing the transcriptional landscape in IKAROS-deficient pre-B cells. To further evaluate this possibility, we generated ChIP-seq data sets for TEAD (1/2), LHX2, and LMO2 in IKDN pre-B cells and deduced high-confidence enrichment peaks (Supplemental Fig. 1B,D). Analysis of transcription factor-binding motifs in the vicinity (200 bp) of extralineage transcription factor peaks confirmed chromatin targeting through their own DNA-binding activities and indicated proximity to each other and native B-cell transcription factors, including IKAROS-binding sites (Fig. 4D). As shown for TEAD (1/2), association with de novo enhancers was indicated by an increase in one or more enhancer modifications (e.g., H3K4me1, H3K4me2, and H3K27ac) or RNApII detected at the majority of its de novo enrichment peaks in IKDN pre-B cells (Fig. 4E, C1–C5). Analysis of LMO2 and LHX2 de novo enrichment peaks also showed an increase in active enhancer modifications, albeit in a more restricted manner (Supplemental Fig. 5C,D, clusters C3 and C5). Several of the LMO2 and LHX2 de novo sites correlated with poised enhancers in wild-type pre-B cells that did not change in activity in IKDN pre-B cells (Supplemental Fig. 5C, C2; Supplemental Fig. 5D, C4). Both of these clusters were highly enriched for IKAROS.
Notably, many of the de novo TEAD enhancers in IKAROS-deficient pre-B cells showed strong occupancy by not only other extralineage factors (e.g., LMO2 and LHX2) but also native B-cell master regulators such as PAX5, EBF1, and IRF4 (Fig. 5A, C1–C5), suggesting that these extralineage factors do not simply overlay their developmental imprint on an independently controlled B-cell program. Three of the TEAD enhancer clusters—highly enriched by both extralineage and B-cell transcription factors in IKDN pre-B cells—were also strongly enriched for IKAROS in wild-type pre-B cells (Fig. 5A, C1, C3, and C5). In contrast, EBF1 de novo peaks in IKDN pre-B cells showed a widespread overlap with TEAD, LHX2, and LMO2 but limited overlap with IKAROS in wild-type pre-B cells (Fig. 5F).
Figure 5.
Induction of de novo superenhancers and Polycomb eviction. (A) Histogram depicting the frequency (in IKDN large pre-B cells) of occupancy of 1597 up-regulated genes by the indicated extralineage and B-cell transcription factors. Genes bound by a specific transcription factor are further subdivided by whether they are associated with regular binding sites (R-BS) or binding sites in SCs (SC or BS in SC). (B) The frequency of transcription factor co-occupancy (from 0 to 6) in up-regulated genes with SCs or regular enhancer binding sites (R-BS) is shown. (C) Schematic representation of idealized examples of binding of TEAD (purple ovals) and EBF1 (green ovals) at up-regulated genes in a SC configuration (SC), within a SC of another transcription factor (BS in SC), or in a regular enhancer (R-BS) or at a gene with no binding sites (No BS). The frequency of transcription factor occupancy inferred from D is represented by the size of the ovals, while the relative density of binding sites obtained from Supplemental Figure 6D is depicted by the number of ovals. (D,E) Occupancy as determined by read density (read count per million mapped reads) for transcription factors (EBF1, PAX5, LMO2, LHX2, PAX5, and IRF4) (D) and histone modifications (H3K4me1, H3K4me2, H3K27ac) as well as RNApII and MED1 at transcription factor-binding sites in SCs or regular enhancers (R-BS) associated with up-regulated genes (E) is shown for wild-type ([red] SC; [light blue] R-BS) and IKDN ([brown] SC; [dark blue] R-BS) pre-B cells. (F) Expression in IKDN large pre-B cells of up-regulated genes with or without SCs (SC or No SC) is shown in a box plot of log2 transformed normalized exonic read counts. A highly significant difference in expression between the two subsets in IKDN large pre-B cells is supported by a P-value of <0.0001 (****) obtained by a two-tailed unpaired t-test. (G) Bar graph distribution of up-regulated genes (IKDN pre-B) subdivided into four groups according to their association with PRC2 activity (H3K27me3/K27) or IKAROS (IK) binding. Further subdivision of these subsets according to superenhancer association is provided by numbers and frequencies at the left (no SUPER) or right (SUPER/SE) side of the bar graph. (H) PRC2 (H3K27me3), transcriptional activity (H3K36me3), and IKAROS association are shown over the body of up-regulated gene subsets with (right column) or without (left column) superenhancers in wild-type and IKDN pre-B cells. (I) A 16.7-kb superenhancer contributed by EBF1, TEAD, and LMO2 at the 5′ prime end of the Itga5 gene is shown on the genome browser. ChIP-seq enrichment tracks are shown for IKAROS, PAX5, EBF1, LMO2, LHX2, TEAD (1/2), H3K27ac, MED1, H3K36me3, and H3K27me3. (J) Model of gene derepression by de novo enhancers and superenhancers in IKDN pre-B cells. Recruitment of permissive chromatin modifiers by the extralineage and B-cell transcription factors at enhancers and subsequent promoter interactions may support eviction of the PRC2-repressive complex. Cooperative interactions among transcription and chromatin regulators at superenhancers may augment this process, leading to rapid PRC2 eviction and gene activation.
Taken together, these results demonstrate induction of a de novo network of enhancers that are occupied by both extralineage and B-cell transcription factors in IKDN pre-B cells. IKAROS acts to prevent induction of this aberrant transcription network by both repressing expression of extralineage transcription factors and binding to and repressing a subset of their enhancer-binding site targets in wild-type pre-B cells.
De novo superenhancers comprised of extralineage and B-cell transcription factors
We next examined the role that this network of extralineage transcription factors plays in the regulation of the ∼1600 genes induced in IKAROS-deficient pre-B cells. The majority of up-regulated genes was bound by both extralineage (TEAD1/2, LMO2, and LHX2) and B-cell (EBF1, IRF4, and PAX5) transcription factors (Fig. 5A). Using an approach similar to that used with down-regulated genes, we identified 312 unique up-regulated genes associated with a SC defined by at least one of the extralineage or B-cell transcription factors (Fig. 5B,C; Supplemental Fig. 6A–C; Supplemental Table S2). Strikingly, 82% of up-regulated genes with SCs were occupied by all six extralineage and B-cell transcription factors in this analysis (Fig. 5B). This cooperation between transcription factor groups was also observed in the up-regulated genes not associated with SCs but was less striking, with only 26% of the up-regulated genes in this group showing binding of all six factors (Fig. 5B). Again, in genes with a SC defined by one factor, although the frequency for occupancy and density of binding sites for the remaining transcription factors did not always reach the threshold for SC designation, they were nonetheless found to be associated at significantly higher levels compared with genes with regular enhancers (modeled in Fig. 5C; Supplemental Fig. 6D).
Chromatin configuration analysis of up-regulated genes determined that transcription factor-binding sites defining SCs had a higher occupancy for poised and active enhancer marks (e.g., H3K4me1, H3K27ac, H3K4me2, RNApII, and MED1) compared with regular enhancers (Fig. 5E). Occupancy by both extralineage and B-cell transcription factors and frequency of permissive histone modifications at enhancers and superenhancer sites were greatly induced in IKDN compared with wild-type pre-B cells (Fig. 5D,E, IKDN SC vs. wild-type SC, and IKDN R-BS vs. wild-type R-BS). Consistent with the higher occupancy by transcription factors and their permissive chromatin state, up-regulated genes with SCs were expressed at higher levels in IKDN pre-B cells relative to up-regulated genes without SCs (Fig. 5F). Thus, the transcriptional and epigenetic makeup of de novo SCs for extralineage and B-cell transcription factors in IKDN pre-B cells strongly supports their role as superenhancers, and they will be referred to as such henceforth.
The up-regulated genes in IKDN pre-B cells were subdivided into four subsets defined by the presence or absence of PRC2 activity (i.e., H3K27me3) and/or IKAROS in wild-type pre-B cells (Fig. 5G). Each of these subsets was further subdivided by whether it was associated with superenhancers in IKDN pre-B cells (Fig. 5G). The distribution of H3K27me3 and H3K36me3 (transcription elongation) at the gene body of these gene subsets was tested in wild-type and IKDN pre-B cells (Fig. 5H). Genes defined through association with PRC2 activity had strong H3K27me3 at their promoters in wild-type pre-B cells (Fig. 5H, top and middle panels). A strong reduction in H3K27me3 and increase in H3K36me3 were observed at genes associated with superenhancers in IKDN pre-B cells whether or not they were bound by IKAROS (Fig. 5H, right top and middle panels). Nonetheless, up-regulated genes without superenhancers that were associated with H3K27me3 and bound by IKAROS in wild-type pre-B cells also displayed a reduction in H3K27me3 and increase in H3K36me3 in IKDN pre-B cells, albeit to a much smaller extent compared with their counterparts with superenhancers (Fig. 5H, middle left vs. right panel, respectively). In sharp contrast, up-regulated genes without superenhancers and without IKAROS binding in wild-type pre-B cells displayed no change in H3K27me3, although a small increase in H3K36me3 was detected (Fig. 5H, left top panel). IKAROS association with up-regulated genes was predominantly through enhancers, although a smaller fraction of genes with IKAROS at both promoters and enhancers was observed (Supplemental Fig. 6E). Examples of up-regulated genes (e.g., Itga5, Amotl2, Rgs10, and Plxnb1) with superenhancers for TEAD, EBF1, and LMO2 that show PRC2 eviction in IKDN pre-B cells with or without IKAROS binding are provided in Figure 5I (Itga5) and Supplemental Figure 7 (Amotl2, Rgs10, and Plxnb1).
Taken together, our data argue that loss of IKAROS leads to the induction of a de novo network of enhancers that is supported by both extralineage and B-cell transcription factors (Fig. 5J). Cooperation between these factors sets up a network of de novo superenhancers that effectively evicts PRC2, allowing expression of genes that define a hybrid B-cell–epithelial cell identity.
Functional specialization of transcription factors at de novo superenhancers
The functional contribution of the de novo network of transcription factors to the transcriptional regulation and biology of IKDN pre-B cells was tested (Fig. 6A). Knockdown studies for EBF1, YAP1, LMO2, and LHX2 were performed with two independent factor-specific shRNAs or scrambled shRNA controls (Fig. 6B). Four days to 6 d after lentiviral infection, IKDN pre-B cells were harvested, and the effect of knocking down specific factors on protein and mRNA expression was examined. To determine potential effects on the previously described loss-of-IKAROS-mediated cellular phenotypes, cells were also tested for cell adhesion, growth, cell cycle, and self-renewing potential (Joshi et al. 2014).
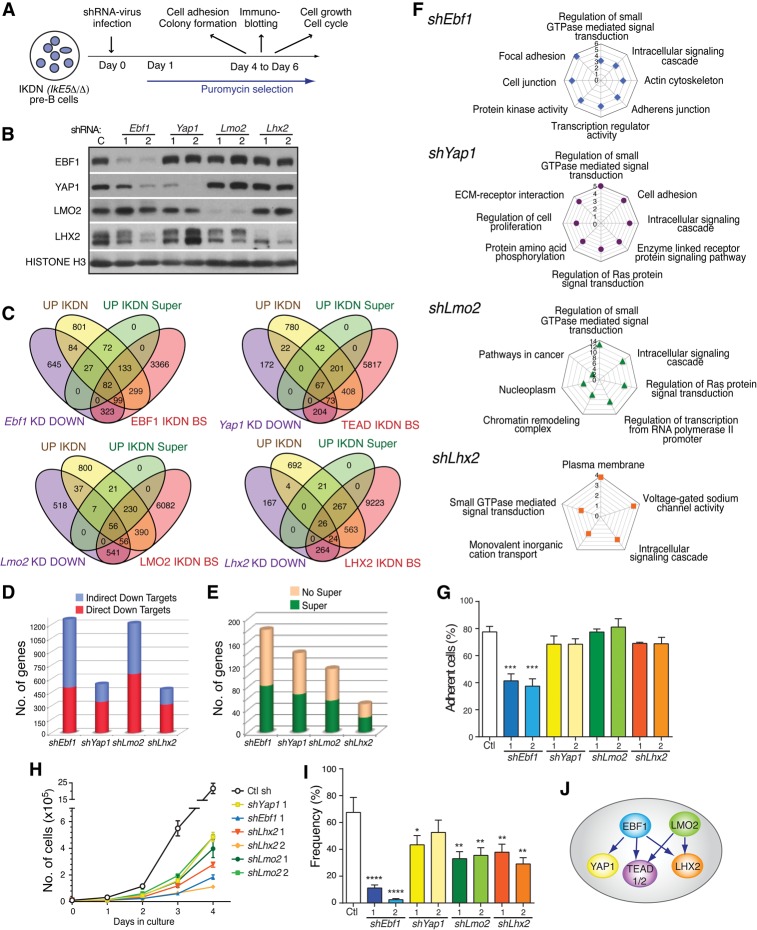
Figure 6.
Differential regulation of cell adhesion, growth, and self-renewal by a de novo network of transcription factors. (A) Schematic depiction of studies on transcription factor knockdown in IKDN pre-B cells. IKDN pre-B cells were transduced with lentivirus expressing two independent shRNAs each for Ebf1, Yap1, Lhx2, and Lmo2 and were placed under puromycin selection 1 d later. Following 4–6 d of selection, IKDN pre-B cells were tested for protein and mRNA expression changes as well as changes in adhesion, cell growth, cell cycle, and colony formation in a limiting dilution assay. (B) Immunoblot analysis of IKDN pre-B cells treated with factor-specific shRNA vectors or controls. (C) Venn diagrams depicting the overlap between genes down-regulated (DOWN) by factor-specific knockdowns (KD), genes associated with de novo sites for these factors in IKDN pre-B cells (IKDN BS), all genes up-regulated upon loss of IKAROS in pre-B cells (Up IKDN), and the subset associated with superenhancers (UP IKDN Super). (D) Bar graphs representing the subdivision of down-regulated direct target genes by factor-specific knockdowns into indirect and direct targets deduced from C. (E) Bar graph depicting the subdivision of genes directly dependent on EBF1, YAP1, LHX2, and LMO2 for expression and IKAROS for repression by whether they associate with superenhancers (Super) in IKDN pre-B cells or not (No Super) as deduced from C. (F) GO of functional pathways enriched for down-regulated genes by factor-specific knockdowns. The P-values (−log10 transformation) for pathway discovery are indicated. (G) Effect of transcription factor inactivation on integrin-mediated cell adhesion of IKDN pre-B cells. The standard error of mean for cell adhesion is shown for each factor and the significant P-value for Ebf1 sh2. (***) P = 0.0007. (H) The effect of transcription factor inactivation on growth of IKDN pre-B cells. Growth was measured from 1 to 4 d after replating of puromycin-selected cells on BM stroma. Standard deviations and P-values are provided for Ebf1 sh2 (P = 0.0042), Lhx2 sh1 (P = 0.0053) and sh2 (P = 0.004), Lmo2 sh1 (P = 0.0009) and sh2 (P = 0.0083), and Yap1 sh1 (P = 0.0054). (I) The effect of inactivation of transcription factors on the self-renewing properties of IKDN pre-B cells was measured by a limiting dilution colony-forming assay. P-values are provided for Ebf1 sh1 (P = 6.6 × 10−15), sh2 ([****] P = 9.85 × 10−35), Lhx2 sh1 ([**] P = 0.00753) and sh2 ([**] P = 0.0001), Lmo2 sh1 ([**] P = 0.001) and sh2 ([**] P = 0.0033), and Yap1 sh1 ([*] P = 0.042) and sh2 (P = 0.273). (J) A model of Yap1, TEAD1/2, and Lhx2 regulation by EBF1 and LMO2 supported by protein and gene expression studies is depicted.
Direct functional gene targets for each of these transcription factors were deduced by comparing changes in gene expression conferred by their knockdown with the genes associated with their de novo binding sites in IKDN pre-B cells (Fig. 6C,D; Supplemental Fig. 8A). A further comparison was performed between up-regulated and down-regulated genes caused by factor knockdown in IKDN pre-B cells and genes activated or repressed by IKAROS (Fig. 6C; Supplemental Fig. 8A; Supplemental Table S3). A substantial overlap was seen between genes that were dependent on these factors for expression (down-regulated by knockdown in IKDN) and genes that were dependent on IKAROS for repression (up-regulated in IKDN) (Fig. 6C). A major fraction of these direct functional gene targets was controlled by superenhancers (Fig. 6C,E). In contrast, with the exception of EBF1, there was little overlap between genes that were dependent on these factors for repression (up-regulated by factor knockdown in IKDN) and genes that were dependent on IKAROS for expression (down-regulated in IKDN) (Supplemental Fig. 8A).
Gene ontology (GO) analysis of direct functional targets dependent on EBF1, YAP1, LMO2, and LHX2 for expression in IKDN pre-B cells confirmed their participation in functional pathways similar to the genes induced by loss of IKAROS (Fig. 6F; Joshi et al. 2014). Pathways supporting cell adhesion, actin cytoskeleton, inorganic cation transport, Ras signal transduction, small GTPase, and transcription regulation were highly enriched by gene targets of the extralineage–B-cell transcription factor network. Notably, EBF1 contributed the most to multiple pathways of cell adhesion. In addition, knockdown of EBF1 reduced expression of most members of the de novo transcription network, including YAP, LHX2 TEAD1/2, and TBX19, with the effect seen at both the protein and mRNA level (Fig. 6B; data not shown).
Adhesion of IKAROS-deficient pre-B cells to integrin ligands was dependent on EBF1, while the other factors were individually dispensable for this property (Fig. 6G). Similarly, knockdown of LHX2 had a pronounced effect on S–G2M accumulation of the IKDN pre-B cells, suggesting a block in G1, whereas knockdown of other factors individually had minimal effect on this cell cycle transition (Supplemental Fig. 8B). In contrast, other properties required the cooperative action of all of the factors. Both the general accumulation of IKDN pre-B cells in culture and the aberrant self-renewal properties of IKAROS-deficient pre-B cells (Joshi et al. 2014) measured in a limiting dilution colony-forming assay were dependent on each of the individual factors, suggesting that each contributed to a general effect on cell cycle or cell survival (Fig. 6H,I). Consistent with the knockdown of these transcription factors causing a growth reduction in IKDN pre-B cells, overgrowth by a population of IKDN pre-B cells in which these factors were not efficiently depleted was observed at later time points of cell culture (data not shown).
Thus, the “altered cell” identity of IKAROS-deficient pre-B cells, defined by biological properties such as cell adhesion, rapid growth, and increased self-renewal, is under the control of an extralineage B-cell network of transcription factors. EBF1 is at the center of this hybrid transcriptional network, directly affecting expression of most of the de novo transcriptional factors and their downstream targets (Fig. 6J).
IKAROS directly represses an ‘epithelial-like’ cell fate in large pre-B cells
In the pre-B-cell populations analyzed above, IKAROS was conditionally inactivated starting at the lympho–myeloid (LMPP) progenitor stages (Joshi et al. 2014). Thus, the properties of IKAROS mutant large pre-B cells may reflect epigenetic memory of defects arising from IKAROS requirement in an earlier multipotent progenitor. Alternatively, IKAROS may actively repress the functional pathways that support niche interactions, growth, and self-renewal in large pre-B cells.
To determine whether IKAROS was actively involved in the repression of stem and neuro-epithelial cell properties in large pre-B cells, a mouse strain (IkE5fl/fl;Rosa26-ERT2-Cre) carrying a ubiquitously expressed tamoxifen-activated Cre recombinase was generated that permitted IKAROS deletion after wild-type large pre-B cells were established in short-term stromal cultures (Fig. 7A). This approach allowed us to characterize the time course of aberrant gene expression following loss of IKAROS. Pre-B cells were harvested after 1–16 d of 4-hydroxy tamoxifen (4OHT) treatment to induce ERT2-Cre activity and were tested for IkE5 deletion, protein and RNA expression, and adhesion to integrin ligands. After 2 d of ERT2-Cre induction, only IKAROS mutant proteins that lacked the DNA-binding domain were detected by immunoblotting (Fig. 7B).
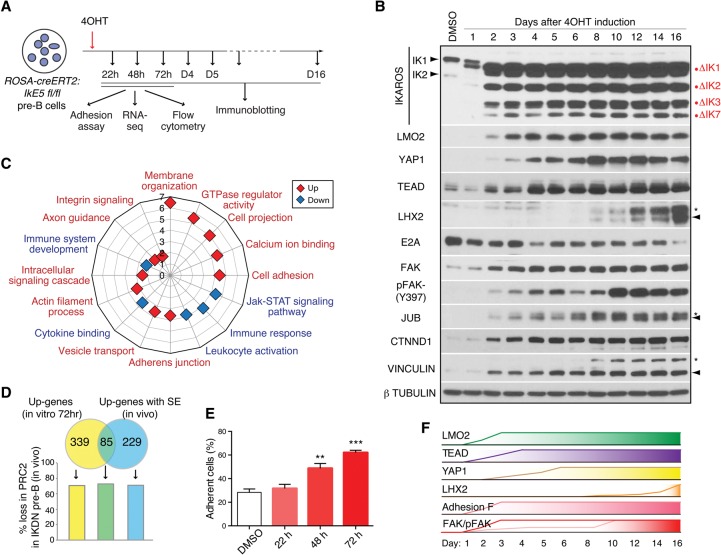
Figure 7.
Active repression of an “epithelial-like” cell fate in wild-type pre-B cells. (A) Schematic representation of in vitro IKAROS inactivation in IkE5fl/fl;Rosa26-ERT2-Cre stromal-adherent large pre-B cells by 4-OHT treatment. The effects on cell adhesion, RNA expression, and immune phenotype were measured following 22–72 h of treatment. Protein expression was determined from 1–16 d of treatment. (B) Immunoblot analysis is shown for IKAROS, FAK, FAKY397, LHX2, TEAD1/2, YAP1, LMO2, AJUBA, CTNND1, VINCULLIN, E2A, and TUBULIN (control) either in untreated IkE5fl/fl;Rosa26-ERT2-Cre pre-B cells or after 4-OHT treatment for 1–16 d. (C) GO analysis of functional pathways enriched by up-regulated (red) and down-regulated (blue) genes in IkE5fl/fl;Rosa26-ERT2-Cre pre-B cells after 72 h of 4-OHT treatment. The P-values (−log10 transformation) for pathway discovery are indicated. (D) Venn diagram depicting the overlap between significantly up-regulated genes after in vitro IKAROS deletion in wild-type pre-B cells (as in C) and up-regulated genes with transcription factor superenhancers (SE) in IKDN pre-B cells generated in vivo. The frequency of loss in PRC2 activity in IKDN pre-B (generated in vivo) is correlated with the three up-regulated gene subsets (as defined by Venn). (E) The effect of in vitro IKAROS inactivation on pre-B-cell integrin-mediated cell adhesion. Integrin ligand adhesion was measured in untreated cells (DMSO) and cells treated for 20–70 h with 4-OHT. The standard error of mean for cell adhesion and P-values for change in cell adhesion at 48 h ([**] P = 0.0034) and 70 h ([***] P = 0.0002) relative to DMSO control are shown. (F) A schematic diagram of the rapid changes in key transcription factors, adhesion, and signaling molecules that occur shortly after in vitro IKAROS deletion in large pre-B cells.
Analysis of gene expression at day 3 revealed a reciprocal effect on genes affiliated with pre-B-cell versus epithelial cell functional pathways (Fig. 7C; Supplemental Table S4). These early changes in gene expression were qualitatively similar to the steady-state changes seen in IKDN pre-B cells obtained in vivo (Figs. 2A, 5A; Joshi et al. 2014). Notably, the majority (302 of 424; 71%) of these early-induced genes had evidence of PRC2 activity in wild-type pre-B cells that was lost in IKDN pre-B cells generated in vivo (Fig. 7D). Furthermore, approximately one-third (85 of 314; 27%) of the up-regulated genes associated with superenhancers identified from IKDN pre-B cells generated in vivo were rapidly induced upon in vitro deletion of IKAROS in established wild-type pre-B-cell cultures (Fig. 7D). Of these superenhancer-associated genes, the majority (62 of 85; 73%) showed evidence of PRC2 repression in wild-type pre-B cells and loss of PRC2 activity in IKDN pre-B cells (Fig. 7D).
Concomitant with these early changes in gene expression was the induction of the extralineage transcription factors LMO2, YAP1, and TEAD at both the RNA and protein levels (Fig. 7B; data not shown). LMO2, YAP1, and TEAD proteins were first detected at day 2 or 3 and reached a maximum at days 3–8 after 4-OHT treatment. In contrast, induction of LHX2 was a later event, first detected at day 8 only after the other extralineage factors had accumulated, consistent with the dependence of LHX2 expression on these factors seen in the knockdown studies (Fig. 7B). A progressive decrease in E2A expression was also seen reaching a minimum at day 16 (Fig. 7B).
A significant increase in pre-B-cell adhesion to fibronectin and dependence on BM stroma for survival was first detected at day 2 (Fig. 7E; data not shown). In line with the increase in cell adhesion, an early increase (day 2) in focal adhesion kinase (FAK) protein and pY397 phosphorylation, indicative of FAK activation, was seen that was further augmented by days 10–12 (Fig. 7B). Expression of Ajuba (JUB), Catenin δ-1 (CTNDD1), and Vinculin (VCL)—proteins involved in the regulation of actin cytoskeleton and implicated in cell adhesion and migration—was also induced as early as day 2 of IKAROS deletion (Fig. 7B).
Thus, IKAROS deletion at either the LMPP stage or the large pre-B-cell stage results in a qualitatively similar phenotype. In vitro depletion of IKAROS activity in wild-type large pre-B cells caused a rapid induction of extralineage transcription factors as well as key membrane and signaling factors normally repressed by IKAROS and PRC2 and induced by superenhancers. Working together, this de novo transcription and signaling network appears to trigger a self-propagating feed-forward loop that alters pre-B-cell identity toward an aberrant “epithelial cell-like” phenotype.
Discussion
The central requirement for IKAROS to provide pluripotent hematopoietic stem cells with lymphoid differentiation potential defines this factor as a master regulator of lymphocyte differentiation (Yoshida et al. 2006; Ng et al. 2007). After B-cell lineage specification, IKAROS is again required for the orderly transition through the highly proliferative pre-B-cell stage. Here the importance of IKAROS is underscored by the consequence of reduction in IKAROS activity, which arrests pre-B cells in a premalignant state that can progress to B-ALL that is refractory to anti-leukemic therapy (Joshi et al. 2014; Churchman et al. 2015). Our current investigation into the molecular mechanisms by which IKAROS controls this critical phase in B-cell precursor differentiation sets a new paradigm for regulation of lineage progression and fidelity that is based on simultaneous positive and negative regulation of superenhancer networks. Our studies reveal that the high-risk properties frequently encountered in pre-B-cell leukemias are rapidly established upon IKAROS loss in a preleukemic pre-B cell because IKAROS not only is required for the expression of B-cell differentiation genes but also actively represses cryptic superenhancers that direct alternative lineage differentiation. An unexpected key contributor in the derepression of IKAROS gene targets is EBF1, whose de novo activity is unleashed upon IKAROS inactivation.
Previous genome-wide studies on IKAROS regulation in B-cell precursors used an extended Bio-ChIP tag at the IKAROS C terminus to identify IKAROS-binding sites that were located mostly at promoters (Schwickert et al. 2014). Motif analysis of these sites failed to reveal association with other master B-cell transcription regulators that preferentially associate with enhancers. Co-occupancy of such factors with a fraction of the promoter-distal IKAROS-binding sites was coerced by combined analysis with previously published ChIP-seq data sets on these factors. An increase of permissive promoter and enhancer (i.e., H3K4me3 and H3K9Ac) chromatin modifications was reported at genes whose expression was dependent on IKAROS, albeit in a manner that was mostly independent of direct association with IKAROS. No further insight into the mechanisms of regulation of these genes was provided. The difference in gene occupancy by IKAROS in the past and current studies may be a technical issue arising from differences in ChIP of the Bio-Chip-tagged versus native protein and high-quality validated antibodies. While we cannot fully evaluate the validity of past data at this developmental point, our own data on predominant association of IKAROS with a range of active, poised, and inactive enhancers were validated by chromatin, gene expression, and functional studies of a network of IKAROS B-cell and extralineage transcription factors that act in concert at this key stage of B-cell differentiation.
Our approach first focused on the IKAROS-binding sites in the genome that were associated with gene expression loss when IKAROS was deleted. This defined a network of superenhancers at genes such as Syk, Blnk, CD79b, Ccnd3, and Foxo1 that are required for pre-B-cell differentiation. These superenhancers were co-occupied by a group of B-cell transcription factors that have been identified as critical for normal differentiation. By definition, gene expression was suppressed when IKAROS was removed, and this was associated with a loss of the highly permissive chromatin environment at these loci. Notably, the high occupancy by B-cell transcription factors such as PAX5, EBF1, and IRF4 and the Mediator complex was maintained upon IKAROS loss, but that was not sufficient to maintain gene expression. Thus, IKAROS supports the activity of superenhancers that are essential for pre-B-cell differentiation working either downstream from or independently of these transcription factors to support recruitment of permissive chromatin modifiers and the RNApII complex. IKAROS function downstream from Mediator complex recruitment at these sites provides new insight into a critical step in the regulatory process that supports superenhancer activation. IKAROS loss does deplete AIOLOS and E2A from superenhancers, suggesting that an IKAROS–AIOLOS higher-order complex (Kim et al. 1999) may act with E2A to enable recruitment of permissive chromatin modifiers and the basal transcription complex. Nevertheless, IKAROS complex activity is directly required at superenhancers of key pre-B-cell differentiation genes whose expression is rapidly attenuated upon IKAROS loss prior to E2A reduction.
Recently, BRG1 and IKAROS were implicated in the regulation of superenhancers associated with MYC and its downstream gene targets involved in ribosome biogenesis in pre-pro-B cells (Bossen et al. 2015). In contrast to the activity reported here, IKAROS is not required for the activity of the MYC and other related superenhancers in pre-pro-B cells, as loss of IKAROS does not arrest differentiation at this stage or interfere with proliferation, MYC expression, or ribosome biogenesis—all phenotypes detected upon loss of BRG1. We showed previously that IKAROS associates with BRG1 and the SWI/SNF chromatin remodeling complex (Kim et al. 1999), but, in this context, BRG1 activity is not strictly dependent on IKAROS, although it may collaborate with IKAROS to promote activity of superenhancers during pre-B-cell differentiation. Superenhancers were previously identified in mouse pro-B-cell precursors by strong enrichment of Pu.1 and Mediator and were associated with several key early B-cell regulators (Whyte et al. 2013). With the exception of a few genes (i.e., Ccnd3, Rag1, etc.) the majority of superenhancers identified by our study was associated with a distinct set of genes that were critical for the large-to-small pre-B-cell transition and were dependent on IKAROS.
Our work also identifies a group of cryptic superenhancers that are actively repressed by IKAROS in pre-B cells. We reported previously that loss of IKAROS is associated with gene activation in T cells (Zhang et al. 2011). However, in that case, the mechanism of gene regulation was quite different, and the “repression mechanism” was indirect. Activated gene loci were not occupied by IKAROS in the wild-type cells. Rather, upon loss of IKAROS, components of the chromatin remodeling complex associated with IKAROS in T cells were redistributed to loci bearing the PRC2 mark that were released from their repressed state. The work here identifies a novel mechanism by which IKAROS occupies and actively represses superenhancer-associated sites whose activation would interfere with B-cell differentiation.
IKAROS is highly enriched at poised and inactive enhancers associated with an array of genes involved in cell adhesion, axon guidance, secretion, and G-protein-coupled receptor signaling—functional pathways that are normally active in neuro-epithelial cells. Among the IKAROS-repressed gene targets are transcription factors that are normally expressed in stem–epithelial cells and are rapidly induced upon IKAROS loss in pre-B cells. These extralineage transcription factors, acting on their own and in concert with native B-cell transcription factors, generate a novel landscape of enhancers and superenhancers and induce a gene expression program that creates the pathophysiological milieu, supporting expansion and survival of preleukemic pre-B cells. Importantly, the B-lineage transcription factor EBF1 plays a central role in this de novo landscape of enhancers and superenhancers. Loss in EBF1 causes loss in expression of de novo enhancer- and superenhancer-affiliated genes responsible for the aberrant epithelial-like properties of IKDN pre-B cells.
In their majority, the genes repressed by IKAROS exhibit IKAROS binding at poised enhancers and PRC2 activity at promoters, which is reduced upon IKAROS loss. IKAROS is involved in maintaining PRC2 activity at these gene loci in either a direct or indirect manner. A direct IKAROS association with PRC2 and targeting of this complex were reported in T-cell precursors (Oravecz et al. 2015). However, we failed to detect a direct association between IKAROS and the PRC2 complex by immunoprecipitation in B-cell precursors (data not shown). Our studies also dispute any in situ chromatin interaction between IKAROS and PRC2, as their enrichment sites (IKAROS and H3K27me3) are mutually exclusive. Nonetheless, an IKAROS downstream mechanism that involves antagonism between de novo enhancers and PRC2 repression has emerged from our studies. The increase in permissive chromatin modifications and RNApII recruitment detected at de novo enhancers is accompanied by similar increases at affiliated promoters together with reduction in PRC2 activity. Notably, PRC2 inhibition was best affiliated with local induction of superenhancers. Our findings are consistent with previous reports of transcriptionally permissive chromatin environments leading to reversal of PRC2 repression (Schmitges et al. 2011; Vernimmen et al. 2011; Kondo et al. 2014).
Extralineage transcription factors directly repressed by IKAROS and occupying the de novo enhancer network included LMO2, a pioneer factor in iHSC that is associated with hematopoietic stem cell self-renewal, early lineage decisions, and B-cell precursor leukemias (Riddell et al. 2014; Chambers and Rabbitts 2015); YAP1 and TEAD (1/2), the nuclear effectors of the Hippo pathway implicated in growth and self-renewal in a variety of nonlymphoid cell types; and LHX2, a pioneer factor in neuronal and skin epithelial stem cells (Rhee et al. 2006; Salvatierra et al. 2014; Adam et al. 2015; Yu et al. 2015). Notably, with the exception of LMO2, these extralineage transcription factors are direct targets of EBF1 in IKDN pre-B cells. The rapid induction of extralineage transcription factors upon IKAROS loss is likely initiated by rapid relocalization of EBF1 at de novo enhancer sites and amplified into a full-fledged aberrant gene expression program, as these factors further reinforce their own expression and induce downstream mediators of an aberrant transcriptional cascade. The presence of such a feed-forward loop is supported by ChIP-seq data that reveal a direct cross-targeting between several of these factors, occupancy by EBF1, and down-regulation in gene expression in factor-specific knockdown studies in IKDN pre-B cells. As an example of downstream amplification of an aberrant response, the Lhx2 enhancer is a target of IKAROS in wild-type pre-B cells and EBF1 in IKDN pre-B cells, but Lhx2 induction upon IKAROS deletion is observed only after a first wave of extralineage transcription regulators is expressed. Expression of Lhx2 is dependent on these factors, as knockdown of at least Lmo2 in IKAROS-deficient pre-B cells resulted in loss of Lhx2 expression. The same feed-forward loops that encourage “safe” commitment to a cell fate choice in an appropriate context are co-opted to promote progression toward leukemia in this chimeric regulatory environment in the absence of IKAROS.
Extralineage transcription factors work together with B-cell master regulators directly on the de novo enhancer landscape of IKAROS-deficient pre-B cells. TEAD (1/2) and EBF1 are the backbone of this chimeric regulatory network and contribute to a repertoire of superenhancers associated with epithelial cell functions. Functional studies with the TEAD coactivator YAP1 as well as LMO2 and LHX2 indicate that together they support the aberrant growth and self-renewal of IKAROS-deficient pre-B cells. On the other hand, EBF1 is the leading factor in cell adhesion-associated superenhancers and was the only factor of those tested that was necessary for the increase in integrin-mediated cell adhesion observed in IKAROS-deficient pre-B cells. EBF1 is expressed in neuronal cells, and its contribution to the adhesion gene signature in IKDN pre-B cells may reflect its normal function in the neuronal lineage. Thus, IKAROS may restrict access of EBF1 and other B-cell transcriptional regulators to regulatory neighborhoods that are normally accessible in nonlymphoid cell types.
IKAROS plays two critical roles in pre-B cells. It is required for the active transcription of the gene expression program that defines B-cell differentiation. Simultaneously, it represses transcription regulators that drive expression cassettes that are critical for development in nonlymphoid lineages. In isolation, each of these normally separated collections of transcription factors mediates the orderly differentiation of its respective cell types. However, intermingling these distinct control regimens in the same cell can lead to dire consequences. Our previous studies focused on a specific target of this aberrant transcriptional cascade—the integrin signaling pathway—that allows for niche interaction-mediated survival and expansion of preleukemic and leukemic cells and identified one set of therapeutic targets. Our present work identifies the proximal transcriptional mediators that initiate this aberrant gene expression profile as a hybrid lineage melange not found in normal cells. Understanding how these two disparate transcription factor groups cooperate in IKAROS-deficient pre-B cells provides new approaches to develop therapeutic strategies directed against IKAROS mutant, high-risk human B-ALL.
Materials and methods
Animals, cell sorting, and pre-B-cell cultures
Wild-type, IkE5fl/fl; CD2-Cre, and IkE5fl/fl; Rosa26-ERT2-Cre mouse strains were used for the isolation of wild-type and IKDN large pre-B cells. All animal experiments were done according to protocols approved by the Subcommittee on Research Animal Care at Massachusetts General Hospital (Charlestown, MA) and the Institutional Animal Use and Care Committee of the University of California at Irvine and in accordance with the guidelines set forth by the National Institutes of Health.
Cell sorting
For isolation of large pre-B cells, also described as early pre-B cells that express pre-BCR and are highly proliferative (for review, see Hardy and Hayakawa 2001), BM cell preparations were depleted of cells binding to anti-Ter119, anti-Mac-1, anti-Gr-1, anti-IgM, anti-CD3, anti-CD8α, anti-TCRβ, and anti-DX5 (Supplemental Table S1) by removal with magnetic beads conjugated to BioMag goat anti-rat IgG (Qiagen, 310107). The cells remaining after depletion were labeled with fluorochrome-conjugated monoclonal antibodies to B-cell markers (anti-CD19 [eBiosciences, 25-0193], anti-CD43 [BD, 553270], anti-BP1 [Ebiosciences, 11-5995], anti-CD25 [BD, 553075], and anti-CD2 [BD, 553112]) and were used for flow cytometry. Large pre-B cells from wild-type, IkE5fl/fl CD2-Cre, or IkE5fl/fl ERT2-Cre were sorted as CD19+CD43+BP1+ using a MoFlo-Legacy (Cytomation) cell sorter.
Pre-B-cell cultures
Wild-type and IkE5Δ/Δ (IKDN) sorted large pre-B cells were co-cultured with OP9 stroma as described previously (Joshi et al. 2014). Nonadherent pre-B cells were collected by several washes with fresh medium. Stroma-adherent large pre-B cells from wild-type or IKDN cultures were detached with 0.2% trypsin (EDTA-free) for 2 min at room temperature. The harvested large pre-B cells were ∼99% of the cells in suspension.
Deletion of Ikaros in large pre-B cells in vitro
Large pre-B cells were sorted from the BM of IkE5fl/fl;Rosa26-ERT2-Cre mice and expanded for 6 d on OP9 stromal cells in the presence of 5 ng/mL IL7. They were then replated onto stromal cells with 0.2 μM 4-OHT (Sigma, H7904) or an equal volume of DMSO as control. Cells were harvested at different time points (1–16 d) of culture in 4-OHT.
Knockdown of transcription factors in IKDN pre-B cells in vitro
Small hairpins targeting Lhx2, Lmo2, and Yap1 were cloned into the pLKO.1 lentiviral vector. The Ebf1 shRNA vectors were reported previously by Griffin et al. (2013). All hairpin sequences are in Supplemental Table S5. shRNA and packaging vectors were cotransfected into HEK293T/17 cells (American Type Culture Collection, CRL-11268), and viral supernatants were collected at 48 and 72 h after transfection and concentrated by ultracentrifugation through a 20% sucrose cushion. For infection, large adherent pre-B cells were mixed with viruses, spun at 1500 rpm for 30 min onto Retronectin-coated wells of a 24-well plate, and then cultured for 5 h in the presence of 5 ng/mL IL7 and 4 μg/mL polybrene (Sigma, H9268-10G). Pre-B cells were then trypsinized and transferred onto a new plate seeded with OP9 stroma. After 24 h, the medium was removed and replaced with fresh medium containing 1 μg/mL puromycin (Sigma, P8833). Cells were selected for 3–5 d in puromycin and then used for further analysis.
Colony-forming assay
Adherent IKDN pre-B cells infected with shRNAs for extralineage and B-cell transcription factors were selected with puromycin for 3 d and then replated onto OP9 stroma-containing 96-well plates at twofold serial dilutions. Cells at concentrations of one to 16 cells per well and with 16 to 24 replicates per concentration were grown with 5 ng/mL IL-7. Wells with pre-B-cell colonies were scored between days 8 and 10. The mean frequency of colony-forming cells and P-values were calculated by the extreme limiting dilution analysis (ELDA) method (Hu and Smyth 2009).
Cell growth analysis
IKDN large adherent pre-B cells infected with shRNAs and selected with puromycin were plated in 96-well (counted on day 1), 48-well (counted on day 2), 12-well (counted on day 3), or six-well (counted on day 4) plates in the presence of OP9 stroma and 5 ng/mL IL7. From day 1 to day 4 after replating, cells were harvested, and cell number, cell cycle, and apoptosis were determined. Growth curves and statistical analysis on growth were performed with GraphPad Prism software. P-values were calculated by two-tailed unpaired t-test with Welch's correction.
Cell cycle analysis
Cells harvested at the indicated time points were fixed in 70% ethanol overnight at 4°C. Fixed cells were stained with propidium iodide (PI) staining buffer (250 μg/mL RNase A, 50 μg/mL PI) for 30 min at 37°C, and the DNA content was detected by FACSCanto. Data were analyzed with FlowJo software (Tree Star).
Cell adhesion to fibronectin
Adhesion assays were performed as described previously (Joshi et al. 2014). Briefly, non-TC-treated plates (BD) were coated with 10 μg/mL fibronectin (Invitrogen) overnight at 4°C. After blocking the plates with 2% BSA for 1 h, an equal number of cells were plated and incubated for 1 h. At the end of the assay, unbound and bound cells were harvested and enumerated. Percent adhesion was calculated by the ratio of bound cells over total cells used in the assay.
ChIP and analysis
ChIP
ChIP was performed according to previously described protocols (Zhang et al. 2011) with some modifications. Cells were fixed in 1% fresh formaldehyde for 10 min at room temperature, quenched with glycine, and washed twice with ice-cold PBS. Cells were resuspended in lysis buffer 1 (50 mM HEPES-KOH at pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100) for 10 min at 4°C, pelleted, and then resuspended in lysis buffer 2 (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM Tris at pH 8) for 5 min at 4°C. Cells were finally resuspended in RIPA buffer (50 mM HEPES at pH 7.9, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.2% SDS, 0.5 mM PMSF, 1× protease inhibitor cocktail [Roche]) to a final concentration of 5 × 107 cells per milliliter. Chromatin was sonicated to an average size of 350 bp with a Branson Sonifier 450 equipped with a microprobe. Chromatin was cleared by centrifugation at 20,000g for 10 min, incubated with 5–10 μg of antibodies prebound to Dynabeads protein G (Life Technologies), and rotated overnight at 4°C. The antibodies used for ChIP are in Supplemental Table S6. Chromatin from 5 × 106 to 10 × 106 cells was used for each histone modification, and chromatin from 2 × 107 to 10 × 107 cells was used for each transcription factor. ChIPs for each histone modifications and transcription factor were performed in parallel with wild-type and IKDN pre-B-cell chromatins. Three times more chromatin was used for TEAD ChIPs in wild-type and E2A ChIPs in IKDN pre-B cells because of their low respective expression in these cells. After extensive washing on beads, bound chromatin was eluted and decross-linked in 300 μL of elution buffer (50 mM Tris at pH 8, 10 mM EDTA, 1% SDS, 0.3 M NaCl) overnight at 65°C. Proteinases K was added to a final concentration of 200 μg/mL and incubated for 2 h at 45°C. DNA was ethanol-precipitated, resuspended in TE, and purified using the DNA Clean and Concentrator-5 kit (Zymo Research).
ChIP-seq libraries, genome mapping, and data analysis
Chromatin-immunoprecipitated DNA and input DNA were prepared for sequencing according to a modified version of the Illumina/Solexa Genomic DNA protocol. Briefly, 2.5–40 ng of DNA was end-repaired, end-adenylated, and then ligated with Illumina TruSeq indexed adaptors. The ligated DNA was purified with AMPure XP beads (Beckman Coulter) and then amplified with KAPA HiFi DNA polymerase (KAPA Biosystems) for eight to 13 cycles. After amplification, the library DNA was separated on a 2% agarose gel, and DNA fragments in the 200- to 500-bp range were purified with a gel DNA recovery kit (Zymo Research). The purified DNA was diluted to 10 nM and multiplexed for sequencing in a HiSeq2000 sequencer at the Bauer Center Systems Biology Core at Harvard University. Image analysis and base calling were performed using Illumina HiSeq 2000 software bcl2FastQ version 1.8.3. Raw sequencing data sets were uploaded to DNAnexus, a cloud-based genome informatics and data management platform.
Read alignment was performed to the mouse mm10 genome using BWA (Li and Durbin 2009) or a modified version of STAR (Dobin et al. 2013). Picard CollectMultipleMetrics tools (Broad Institute) were used for read mapping analysis. Sequencing depth was tested by analyzing high-quality biological replicates and determining whether independent data sets, when combined, yielded a significantly higher number of new peaks.
Transcription factor peaks, histone modification-enriched regions, and gene body coverage were initially identified using both MACS and findPeaks (HOMER) algorithms (Zhang et al. 2008; Heinz et al. 2010) with input chromatin as control. Comparison between the two methods for both transcription factors and histones indicated that 90% of peaks or regions identified by MACS were also identified by the HOMER method. We continued our analysis with the HOMER peak-finding and analysis tools. Peaks that were differentially enriched in either wild-type or IKDN pre-B-cell chromatin were obtained by setting one condition as test and the other as input control. Peaks were identified with a threshold of false discovery rate of 0.001 for tag clustering, a fourfold increase in tag density relative to input control in both local and average distributions (Poisson distribution P-value threshold of 0.0001), and a fourfold increase in tag density at peaks relative to a surrounding 10-kb region (local Poisson distribution P-value threshold of 0.0001).
Identification of transcription factor SCs was performed using the HOMER findPeaks “style super” method as described by Hah et al. (2015), similar to the method described by Whyte et al. (2013). Hierarchical and K-means clustering of ChIP-seq data sets was performed using NGS.plot (Shen et al. 2014) or the Homer AnnotatePeaks.pl program (Heinz et al. 2010) combined with Cluster 3.0 and Java TreeView analysis. Histograms of tag densities for histone modifications and transcription factors over regulatory sites or gene bodies were plotted using NGS.plot software (Shen et al. 2014).
The HOMER de novo motif discovery algorithm was used to analyze binding sites for frequently occurring DNA sequence (SQ) motifs. For data visualization, primary sequencing data were sorted, indexed, and uploaded onto a local IGV or a cloud-based Biodalliance genome browser (http://www.biodalliance.org). Venn diagrams were generated using the online-based Venny Tool.
Statistical analysis
The R Bioconductor packages implemented through RStudio were used for statistical analysis and extraction and plotting of data from NGS-generated data files. The statistical significance between two array groups was determined by Student's t-test. In order to determine whether variances between two groups were equal or unequal, F-test was performed.
Gene expression analysis
Construction of RNA-seq libraries, gene expression, and pathway analysis
RNA was extracted with Trizol (Invitrogen) and purified using the PureLink RNA minikit (Ambion). The TruSeq RNA sample preparation kit was used for construction of cDNA libraries for RNA-seq (Illumina). The cDNA libraries were ligated with indexed primers and amplified by PCR for 10 cycles. The amplified libraries were multiplexed and sequenced at the Systems Biology Laboratory at Harvard University. Alignment to the mouse mm10 assembly was conducted by STAR (Dobin et al. 2013). Normalization and differential expression were performed using DeSeq2 through a HOMER implementation of the package in R. Heat maps of normalized tags for gene subsets across wild-type and IKDN pre-B-cell populations were generated with Cluster and Java TreeView software. GO analysis of deregulated genes in IKDN pre-B cells was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID).
Accession numbers
The data discussed in this study have been deposited to NCBI and are accessible through Gene Expression Omnibus series accession number GSEA86897.
Supplementary Material
Acknowledgments
We thank Dr. C. Benner for his advice and consultation on various applications of the HOMER software used throughout this project; Dr. K. White, Dr. J.M. Park, Dr. B. Morgan, Dr. T. Naito, and Dr. C. Benner for critical review of the manuscript; Dr. J. Drouin for anti-Tbx19 antibodies; and E. Wu and B. Czyzewski for mouse husbandry. This research was supported by National Institutes of Health grants R01CA162092 and R21AI124326 (K.G.), R01CA090576 (R.A.V.E.), and R01CA190964 (R.A.V.E. and K.G.). K.G. Is a Massachusetts General Hospital scholar supported by Dr. Jean de Gunzburg. High-throughput DNA sequencing was performed at the Bauer Center for Genomic Research, Harvard University, Cambridge. Y.H. performed the chromatin studies, the transcription factor knockdowns, and their effect on gene expression, cell cycle, cell adhesion, and clonogenicity. Z.Z. performed supporting protein studies. M.K., A.O.E., F.G., and K.G. performed ChIP and RNA-seq data analysis and presentation. T.Y. performed gene expression studies on primary wild-type, IKDN, BCR–ABL1+, and IKDN–BCR–ABL1+ pre-B cells and data presentation. I.J. performed adhesion assays with large pre-B cells in wild-type, IKDN, and factor knockdown pre-B cells. N.J. generated and analyzed IKDN–BCR–ABL1+ pre-B cells. R.S. and M.S. aided with the Ebf1 knockdown studies. J.F. and N.E.B. aided with the Yap1 knockdown studies. R.A.V.E. and K.G. supervised the project and wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.283762.116.
References
- Adam RC, Yang H, Rockowitz S, Larsen SB, Nikolova M, Oristian DS, Polak L, Kadaja M, Asare A, Zheng D, et al. 2015. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 521: 366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossen C, Murre CS, Chang AN, Mansson R, Rodewald HR, Murre C. 2015. The chromatin remodeler Brg1 activates enhancer repertoires to establish B cell identity and modulate cell growth. Nat Immunol 16: 775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryder D, Sigvardsson M. 2010. Shaping up a lineage—lessons from B lymphopoesis. Curr Opin Immunol 22: 148–153. [DOI] [PubMed] [Google Scholar]
- Chambers J, Rabbitts TH. 2015. LMO2 at 25 years: a paradigm of chromosomal translocation proteins. Open Biol 5: 150062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AM, Rowley B, Pao W, Hayday A, Bolen JB, Pawson T. 1995. Syk tyrosine kinase required for mouse viability and B-cell development. Nature 378: 303–306. [DOI] [PubMed] [Google Scholar]
- Churchman ML, Low J, Qu C, Paietta EM, Kasper LH, Chang Y, Payne-Turner D, Althoff MJ, Song G, Chen SC, et al. 2015. Efficacy of retinoids in IKZF1-mutated BCR-ABL1 acute lymphoblastic leukemia. Cancer Cell 28: 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobaleda C, Sanchez-Garcia I. 2009. B-cell acute lymphoblastic leukaemia: towards understanding its cellular origin. Bioessays 31: 600–609. [DOI] [PubMed] [Google Scholar]
- Cooper AB, Sawai CM, Sicinska E, Powers SE, Sicinski P, Clark MR, Aifantis I. 2006. A unique function for cyclin D3 in early B cell development. Nat Immunol 7: 489–497. [DOI] [PubMed] [Google Scholar]
- Cortes M, Georgopoulos K. 2004. Aiolos is required for the generation of high affinity bone marrow plasma cells responsible for long-term immunity. J Exp Med 199: 209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler HS, Baracho GV, Omori SA, Bruckner S, Arden KC, Castrillon DH, DePinho RA, Rickert RC. 2008. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol 9: 1388–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. 2010. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 8: e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Nussenzweig MC. 1996. Regulation of an early developmental checkpoint in the B cell pathway by Igβ. Science 272: 411–414. [DOI] [PubMed] [Google Scholar]
- Griffin MJ, Zhou Y, Kang S, Zhang X, Mikkelsen TS, Rosen ED. 2013. Early B-cell factor-1 (EBF1) is a key regulator of metabolic and inflammatory signaling pathways in mature adipocytes. J Biol Chem 288: 35925–35939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Benner C, Chong LW, Yu RT, Downes M, Evans RM. 2015. Inflammation-sensitive super enhancers form domains of coordinately regulated enhancer RNAs. Proc Natl Acad Sci 112: E297–E302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RR, Hayakawa K. 2001. B cell development pathways. Annu Rev Immunol 19: 595–621. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heizmann B, Kastner P, Chan S. 2013. Ikaros is absolutely required for pre-B cell differentiation by attenuating IL-7 signals. J Exp Med 210: 2823–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog S, Reth M, Jumaa H. 2009. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol 9: 195–205. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. 2013. Super-enhancers in the control of cell identity and disease. Cell 155: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, Bradner JE, Young RA. 2015. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol Cell 58: 362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Smyth GK. 2009. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 347: 70–78. [DOI] [PubMed] [Google Scholar]
- Inaba H, Greaves M, Mullighan CG. 2013. Acute lymphoblastic leukaemia. Lancet 381: 1943–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Calame K. 2003. Transcription factors controlling the beginning and end of B-cell differentiation. Curr Opin Genet Dev 13: 522–528. [DOI] [PubMed] [Google Scholar]
- Joshi I, Yoshida T, Jena N, Qi X, Zhang J, Van Etten RA, Georgopoulos K. 2014. Loss of Ikaros DNA-binding function confers integrin-dependent survival on pre-B cells and progression to acute lymphoblastic leukemia. Nat Immunol 15: 294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. 1999. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10: 345–355. [DOI] [PubMed] [Google Scholar]
- Kondo T, Isono K, Kondo K, Endo TA, Itohara S, Vidal M, Koseki H. 2014. Polycomb potentiates meis2 activation in midbrain by mediating interaction of the promoter with a tissue-specific enhancer. Dev Cell 28: 94–101. [DOI] [PubMed] [Google Scholar]
- Kronke J, Udeshi ND, Narla A, Grauman P, Hurst SN, McConkey M, Svinkina T, Heckl D, Comer E, Li X, et al. 2014. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343: 301–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper RP, Waanders E, van der Velden VH, van Reijmersdal SV, Venkatachalam R, Scheijen B, Sonneveld E, van Dongen JJ, Veerman AJ, van Leeuwen FN, et al. 2010. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia 24: 1258–1264. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Spolski R, Liao W, Wang L, Murphy TL, Murphy KM, Leonard WJ. 2012. BATF–JUN is critical for IRF4-mediated transcription in T cells. Nature 490: 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. 2010. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol 11: 635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Middleton RE, Sun H, Naniong M, Ott CJ, Mitsiades CS, Wong KK, Bradner JE, Kaelin WG Jr. 2014. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 343: 305–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. 2010. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 11: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel EM, Grosschedl R. 2010. Transcription control of early B cell differentiation. Curr Opin Immunol 22: 161–167. [DOI] [PubMed] [Google Scholar]
- Martinelli G, Iacobucci I, Storlazzi CT, Vignetti M, Paoloni F, Cilloni D, Soverini S, Vitale A, Chiaretti S, Cimino G, et al. 2009. IKZF1 (Ikaros) deletions in BCR–ABL1-positive acute lymphoblastic leukemia are associated with short disease-free survival and high rate of cumulative incidence of relapse: a GIMEMA AL WP report. J Clin Oncol 27: 5202–5207. [DOI] [PubMed] [Google Scholar]
- Minegishi Y, Rohrer J, Coustan-Smith E, Lederman HM, Pappu R, Campana D, Chan AC, Conley ME. 1999. An essential role for BLNK in human B cell development. Science 286: 1954–1957. [DOI] [PubMed] [Google Scholar]
- Mullighan CG. 2012. Molecular genetics of B-precursor acute lymphoblastic leukemia. J Clin Invest 122: 3407–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al. 2007. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446: 758–764. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, Ma J, Liu W, Cheng C, Schulman BA, et al. 2009. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 360: 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Yoshida T, Georgopoulos K. 2007. Ikaros and chromatin regulation in early hematopoiesis. Curr Opin Immunol 19: 116–122. [DOI] [PubMed] [Google Scholar]
- Ng SY, Yoshida T, Zhang J, Georgopoulos K. 2009. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity 30: 493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Kee BL. 2007. The transcriptional regulation of B cell lineage commitment. Immunity 26: 715–725. [DOI] [PubMed] [Google Scholar]
- Oravecz A, Apostolov A, Polak K, Jost B, Le Gras S, Chan S, Kastner P. 2015. Ikaros mediates gene silencing in T cells through Polycomb repressive complex 2. Nat Commun 6: 8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappu R, Cheng AM, Li B, Gong Q, Chiu C, Griffin N, White M, Sleckman BP, Chan AC. 1999. Requirement for B cell linker protein (BLNK) in B cell development. Science 286: 1949–1954. [DOI] [PubMed] [Google Scholar]
- Rhee H, Polak L, Fuchs E. 2006. Lhx2 maintains stem cell character in hair follicles. Science 312: 1946–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert RC. 2013. New insights into pre-BCR and BCR signalling with relevance to B cell malignancies. Nat Rev Immunol 13: 578–591. [DOI] [PubMed] [Google Scholar]
- Riddell J, Gazit R, Garrison BS, Guo G, Saadatpour A, Mandal PK, Ebina W, Volchkov P, Yuan GC, Orkin SH, et al. 2014. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell 157: 549–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KG, Morin RD, Zhang J, Hirst M, Zhao Y, Su X, Chen SC, Payne-Turner D, Churchman ML, Harvey RC, et al. 2012. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell 22: 153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumiantsev S, de Aos IE, Varticovski L, Ilaria RL, Van Etten RA. 2001. The src homology 2 domain of Bcr/Abl is required for efficient induction of chronic myeloid leukemia-like disease in mice but not for lymphoid leukemogenesis or activation of phosphatidylinositol 3-kinase. Blood 97: 4–13. [DOI] [PubMed] [Google Scholar]
- Salvatierra J, Lee DA, Zibetti C, Duran-Moreno M, Yoo S, Newman EA, Wang H, Bedont JL, de Melo J, Miranda-Angulo AL, et al. 2014. The LIM homeodomain factor Lhx2 is required for hypothalamic tanycyte specification and differentiation. J Neurosci 34: 16809–16820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. 2011. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42: 330–341. [DOI] [PubMed] [Google Scholar]
- Schwickert TA, Tagoh H, Gultekin S, Dakic A, Axelsson E, Minnich M, Ebert A, Werner B, Roth M, Cimmino L, et al. 2014. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol 15: 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, Nestler E. 2014. NGS.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernimmen D, Lynch MD, De Gobbi M, Garrick D, Sharpe JA, Sloane-Stanley JA, Smith AJ, Higgs DR. 2011. Polycomb eviction as a new distant enhancer function. Genes Dev 25: 1583–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K. 2006. Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol 7: 382–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Ng SY, Georgopoulos K. 2010. Awakening lineage potential by Ikaros-mediated transcriptional priming. Curr Opin Immunol 22: 154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Guan KL. 2015. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163: 811–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Jackson AF, Naito T, Dose M, Seavitt J, Liu F, Heller EJ, Kashiwagi M, Yoshida T, Gounari F, et al. 2011. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol 13: 86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. 2011. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12: 7–18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.