Abstract
Resident macrophages support embryonic development and tissue homeostasis and repair. The mechanisms that control their differentiation remain unclear. We report here that Erythro-Myeloid Progenitors generate pre-macrophages (pMacs) that simultaneously colonize the whole embryo from embryonic day (E)9.5 in a chemokine-receptor dependent manner. The core macrophage program initiated in pMacs is rapidly diversified as expression of transcriptional regulators becomes tissue-specific in early macrophages. This process appears essential for macrophage specification and maintenance, as inactivation of Id3 impairs the development of liver macrophages and results in selective Kupffer cell deficiency in adults. We propose that macrophage differentiation is an integral part of organogenesis as colonization of organ anlagen by pMacs is followed by their specification into tissue macrophages, hereby generating the macrophage diversity observed in postnatal tissues.
Graphical abstract
Tissue-resident macrophages are a diverse family of cells found in most organs, such as brain microglia, liver Kupffer cells, lung alveolar macrophages, and epidermal Langerhans cells. They share an embryonic origin and differentiate, at least in part, from yolk sac (YS) Erythro-Myeloid Progenitors (EMPs) (1, 2), and are self-maintained in adult tissues independently of hematopoietic stem cells (HSCs) under steady state conditions (3-6). However, the mechanisms responsible for the generation of macrophage diversity observed in adult mice remain unclear. It was proposed that resident macrophage diversity reflects their exposure to specialized tissue environments (7-10), or the contribution of distinct embryonic or fetal progenitors to distinct subsets (2, 11-13). The preferential expression of transcription factors in macrophage subsets was also noted (7), and appears functionally important. Several such cases have been functionally validated by knockout mice, including Gata6 for large peritoneal macrophages (9, 10, 14), Runx3 for Langerhans cells (15), Nr1h3 for splenic marginal zone macrophages (16), SpiC for splenic red pulp macrophages (17) and Pparg for alveolar macrophages (18). To better understand how macrophage diversity is generated, we performed a molecular and spatio-temporal analysis of macrophage development in mice.
Colonization of developing tissues by EMP-derived macrophage precursors
EMPs (Csf1r+ Kit+ CD45low AA4.1+) are first detected in the YS at E8.5 (2, 19) and subsequently colonize the fetal liver (1, 2). Previous fate mapping analysis of EMP differentiation indicated that their progeny loses Kit expression and increases CD45 expression as they invade the embryo, before acquiring F4/80 expression to give rise to fetal and postnatal tissue-resident macrophages (2) (Fig. 1A). To explore the spatio-temporal and molecular determinants of macrophage differentiation and diversification, we first performed whole transcriptome sequencing of sorted CD45low Kit+ EMPs, CD45+ Kit− Lin− (Ter119, Gr1, F4/80) cells, and F4/80+ macrophages from embryonic and postnatal tissues up to 3 weeks after birth (Fig. 1A, S1A, B). We identified genes that were significantly upregulated between EMPs and CD45+ Kit− Lin− cells (adjusted p-value≤0.05, DESeq2 (20), Benjamini Hochberg (BH)-correction, Table S1). Subsequent summarization and visualization of these genes via scorecard analysis (21) (Fig. 1B), indicated that the CD45+ Kit− Lin− cells signature was also present in F4/80+ macrophages across tissues and over the entire time course analysis in the embryo and postnatal mice in the kidney, liver, and brain, albeit epidermal Langerhans cells and lung alveolar macrophage signatures were modified after birth (Fig. 1B). A second scorecard analysis of genes upregulated between EMPs and early (E10.25-E10.5) F4/80+ macrophages identified a signature that was already detectable in CD45+ Kit− Lin− cells, and conserved in later macrophages across tissues (Fig. S1C, Table S1). Unsupervised principal component analysis (PCA) showed a distinct grouping of EMPs, CD45+ Kit− Lin− cells and macrophages, irrespectively of their tissue of origin i.e. YS, liver, head or caudal region (Fig. S1D). Morphologically, CD45+ Kit− Lin− cells from the YS, fetal liver, head and caudal embryo displayed a similar morphology, as did macrophages from the same tissues (Fig. 1C). CD45+ Kit− Lin− cells resembled EMPs, albeit with the presence of occasional phagocytic vacuoles, while phagocytic features become prominent in F4/80+ cells (Fig. 1C).
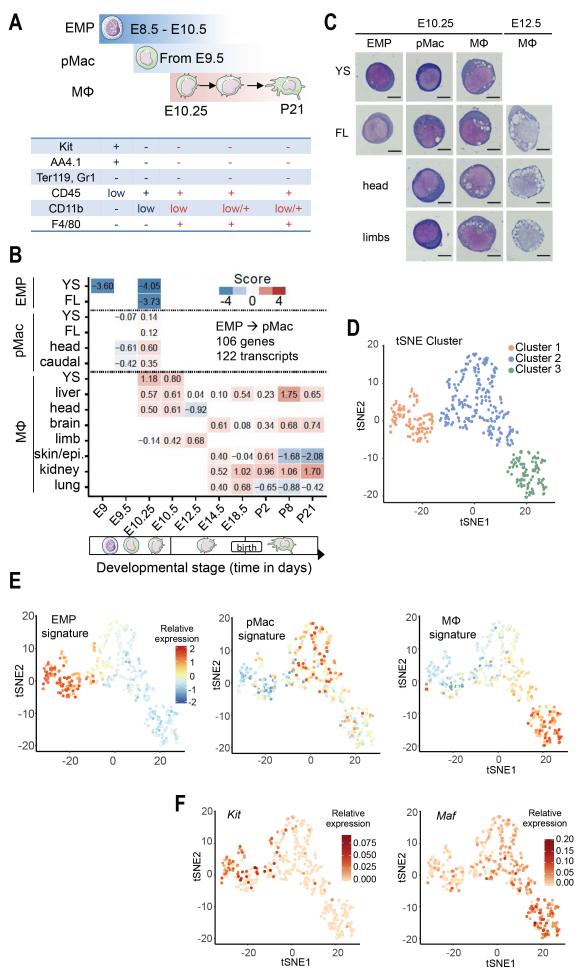
Fig. 1. A core macrophage program is initiated simultaneously in pMacs in all tissues.
(A) Summary of surface phenotype used for EMPs, pMacs, and macrophages. (B) Scorecard visualization of differentially upregulated genes (DESeq2 Wald test, adjusted p-value < 0.05, BH-correction) in pMacs (E9.5 and E10.25) in comparison to EMPs. The table shows the relative enrichment of differentially upregulated genes in pMacs across cell types and tissues (y-axis) and developmental time points (x-axis, from E9 to P21). See Table S1, Fig. S1, and Methods for details of the scorecard. (C) May-Gruenwald-Giemsa stained cytospin preparations of sorted EMPs, pMacs and early macrophages from yolk sac (YS), head, limbs and fetal liver (FL) at E10.25 and E12.5. n=3 independent experiments. (D) tSNE plot of scRNA-seq data showing distribution of CD45low/+ cells from E10.25 embryos into three clusters (see also Fig. S2). Cluster distribution based on DBScan is overlaid onto the graph. (E) Superimposition of EMP-, pMac-, or macrophage-specific signatures defined by the bulk RNA sequencing on the tSNE plot shown in D. (F) tSNE plot as in (D) overlaid with the relative expression values for Kit and Maf.
These data suggested that a macrophage differentiation program was initiated simultaneously in the whole embryo in CD45+ Kit− Lin− cells, which will be referred to below as pre-Macrophage (pMac). To further test this hypothesis, we performed independent unbiased whole transcriptome single-cell RNA sequencing (scRNA-seq) of CD45low/+ cells purified from the whole embryo at E10.25 (30-34 somite pairs). Nonlinear dimensionality reduction in combination with unsupervised clustering of cells indicated that these cells are best described by three major clusters (Fig. 1D, Fig. S2). Overlay of EMP, pMac, and macrophage signatures from differentially expressed genes in bulk RNA-seq analysis (Table S2) indicated superimposition on cluster 1, 2, and 3 respectively (Fig. 1E) albeit intermediate differentiation states in EMPs, pMacs, and macrophages were clearly apparent, which suggested a gradual differentiation path from EMP to macrophages via pMacs (Fig. 1E), consistent with the scorecard analysis.
Core macrophage transcriptional program
Analysis of genes differentially expressed in EMPs, pMacs, and early macrophages by scRNA-seq (Fig. 1F, 2A, Table S2) and bulk RNA-seq analysis (Fig. 2B, Table S1) confirmed that a core macrophage transcriptional program was initiated in pMacs. As Kit, Gata1, and Gata2 expression was lost, pMacs upregulated expression of Csf1r, and the transcription factors Maf, Batf3, Pparg, Irf8, and Zeb2, the chemokine receptor Cx3cr1, cytokine receptors, complement and complement receptors, pattern-recognition receptors, phagocytic receptors, Fc gamma receptors, the inhibitory receptor Sirpa, MerTK, cathepsins, Aif1 (Iba1), Emr1 (F4/80) and Grn (Granulin). Expression of selected cytokine receptors for Il-4, Il-13, Interferon gamma and Tumor necrosis factor, phagocytic and activating receptors Mrc1 (CD206), Trem2, Dectin-1 (Clec7a), Fc-gamma receptors (Fcgr1, Fcgr2/3, Fcgr4), Iba1, and Grn was confirmed at the protein level by flow cytometry and by immunofluorescence in situ on EMP-derived pMacs and early macrophages from the YS as well as from the head, caudal, limbs, and liver of the embryo proper, fetal macrophages and adult tissue macrophages in Csf1rMeriCreMer; Rosa26LSL-YFP mice pulsed with OH-TAM at E8.5 (Fig. 3A, B, Fig. 5A, Fig. S3, S4). Expression of these proteins was first detected in pMacs and increased as pMacs differentiated into F4/80+ macrophages (Fig. 3A,B, Fig. S3). Of note, pMacs and macrophages represented 70-90% of EMP-derived cells in the head and caudal embryo, while 80-90% of YFP+ cells in the E10.25 fetal liver represented progenitors (Fig. 3B, Fig. S4).
Fig. 2. Differentially expressed genes during differentiation from EMP to macrophage.
(A) (upper panel) Developmental pseudotime diagram (q-value<0.05) showing down regulation of EMP-specific genes (differentially expressed compared to the macrophage and pMac cluster, p-value<0.05, FC>1.4) over the differentiation path from EMP to pMacs and macrophages. (lower panel) Similar plot depicting macrophage-specific genes significantly regulated over pseudotime (q-value<0.05) and differentially expressed compared to the EMP and pMac cluster (p-value<0.05, FC>1.4)). See Fig. S2G. (B) Heatmap representation of selected genes differentially regulated between EMPs vs. pMacs and EMPs vs. early macrophages in bulk RNA-seq analysis. Black boxes were drawn around those samples used for differential expression analysis. See also Table S1, S2.
Fig. 3. EMP-derived pMacs colonize the embryo to generate macrophages.
(A) Flow cytometry analysis of E10.25 Csf1rMeriCreMer; Rosa26LSL-YFP (OH-TAM at E8.5) tissues showing expression of Il4ra, Il13ra1, CD16.2, CD64, Ifngr, Tnfr2, Tim4, and CD206 on YFP+ Kit+ progenitors, pMacs, and macrophages. MFI: mean fluorescent intensity. Data are representative of n=4 independent experiments with 4-6 embryos per marker. See also Fig. S3A. (B) Quantification of immunostainings on cryosection from E10.25 Csf1rMeriCreMer; Rosa26LSL-YFP embryos, pulse-labeled with OH-TAM at E8.5 with antibodies against YFP, Iba1 and CD16/32, Dectin-1, Trem2, F4/80, CD206 or Granulin. n=2-4 embryos and 2 sections per embryo per marker. See Fig. S4. (C) tSNE plot as in (1D) overlaid with the relative expression values for Tnfrsf11a and Cx3cr1. (D) YFP labeling efficiency of Tnfrsf11aCre+; Rosa26LSL-YFP in pMacs and F4/80+ macrophages in YS and whole embryo at E10.25, fetal liver HSCs (long term (LT, Lin−Kit+Sca1+CD150+CD48−), short term (ST, Lin−Kit+Sca1+CD150−CD48−) and multipotent progenitor (MPP, Lin−Kit+Sca1+CD150−CD48+)) and tissue macrophages at E14.5 and 6 weeks, and blood leukocytes (B-cells (CD19+), T-cells (CD19−Ly6G−CD115−CD3+), NK cells (CD19−Ly6G−CD115−CD3−NKp46+), neutrophils (CD19−Ly6G+) and Ly6Chi monocytes (CD19−CD115+Ly6G−Ly6Chi), and tissue CD11bhigh myeloid cells from 6 week-old mice. Circles represent individual mice. n=4 independent experiments. See Fig. S5.
Fig. 5. Early specification of tissue-resident macrophages.
(A) Flow cytometry analysis of Csf1rMeriCreMer; Rosa26LSL-YFP (OH-TAM at E8.5) liver, brain, lung and skin F4/80+ cells from postnatal mice (4 weeks old) showing expression of Il4ra, Il13ra1, Tnfr2, Ifngr, Dectin-1, CD64, Tim4, and CD206 (black dotted on whole population and green on YFP+ cells). Gray histograms show the fluorescence intensity of the FMO controls. (B, C) Scorecard analysis of all differentially upregulated genes in postnatal macrophages. The scorecards show the relative enrichment of each set of upregulated genes across each cell type (y-axis) and developmental time point (x-axis). See Methods for details of the score card. Numbers for each population indicate differentially up-regulated transcripts in postnatal (P2-P21) brain, liver, kidney, epidermis or lung macrophages when comparing one population vs. the others. See also Table S3. (D) Heatmap representing all differentially upregulated transcriptional regulators (2-fold change, adj. p-value<0.05, BH-correction) between postnatal macrophages from brain, liver, kidney, skin and lung macrophages, and their relative expression in tissue macrophages from E10.25 to P21.
Detection of the early expression of the cytokine receptor Tnfrsf11a in pMacs by RNA-seq and scRNA-seq analyses (Fig. 2B, 3C) predicted that pMacs or their progeny may express YFP in Tnfrsf11aCre (22); Rosa26LSL-YFP mice. Indeed, we observed YFP labeling by flow cytometry in ~80% pMacs and early macrophages from the YS and embryo of Tnfrsf11aCre; Rosa26LSL-YFP mice (Fig. 3D, S5). Fetal macrophages in all tissues also expressed YFP at comparable levels at E10.25 and E14.5 (Fig. 3D). In addition, ~80% of brain, lung, epidermis, kidney, and liver macrophages from 6-week old mice expressed YFP (Fig. 3D). Therefore, the whole resident macrophage lineage is labeled in Tnfrsf11aCre; Rosa26LSL-YFP mice, although Tnfrsf11a expression itself is lost in postnatal Langerhans cells and alveolar macrophages (Fig. 2B). Moreover, we noted that YFP expression was observed only in ~15% of fetal HSCs, adult HSC, and HSC-derived cells in the blood and tissues of adult mice (Fig. 3D, S5). Tnfrsf11aCre; Rosa26LSL-YFP mice thus represent an efficient and relatively specific model for genetic labeling of tissue-resident macrophages in fetuses and adult mice.
Early Cx3cr1 expression by pMacs (Fig. 2B, Fig. 3C) is in line with previous reports showing Cx3cr1 expression in macrophage precursors (23), and suggested that this chemokine receptor may be involved in colonization of embryo tissues by pMacs. Time course analysis of GFP expression in Cx3cr1gfp/+ mice from E8.5 (19-21 somite pairs - sp) to E10.5 (38-39sp) indicated that GFP expression is not detected in Kit+ progenitors, but is upregulated in Kitlow Dectin-1+ pMacs and is highest in F4/80+ macrophages that appear at E10.25 (Fig. 4A). Next, we found that colonization of the head and caudal tissues is delayed in Cx3cr1-deficient embryos as pMacs and macrophages numbers are decreased in the head and caudal/limbs tissues of E9.5 and E10.5 embryos in comparison with Cx3cr1+/− littermates, while they accumulate in the YS and fetal liver (Fig. 4B). Nevertheless, tissue macrophage numbers even out in the consecutive days of embryonic development in most tissues, with the exception of the kidney where a 50% lower number in resident macrophages is still observed in adult mice in line with previous research (24) (Fig. S6).
Fig. 4. Tissue colonization by pMacs is Cx3cr1-dependent.
(A) Expression of GFP and Dectin-1 in Cx3cr1gfp/+ mice during development (E8.5-10.5) in Kit+ cells (CD45low, Kit+), pMacs and macrophages. sp: somite pairs. Data are representative of n=9 independent experiments. Biological replicates have been aggregated per cell type, time point and tissue. (B) Flow cytometry analysis in Cx3cr1+/− and Cx3cr1−/− of pMacs and macrophages from yolk sac (YS), head, and caudal at E9.5 (upper panel) and liver, YS, head, and limbs at E10.5 (lower panel). Circles represent individual mice. Data are representative of n=6 independent experiments. P-values were calculated using Student’s t test. sp: somite pairs.
A progressive enrichment of pMac and macrophage specific gene expression signatures was observed in gene set enrichment analysis (GSEA) of bulk RNA-seq data (Fig. S7A,B). Moreover, the core macrophage signature identified in adult mice by the Immgen consortium (25) was already enriched in the genes upregulated in pMacs and early macrophages compared to EMPs (Fig. S7C,D). In silico investigation of transcription factor binding sites identified using ChIP-seq in the proximity of upregulated genes (transcription start site (TSS) +/− 20kb) in pMacs and macrophages further supports the proposition that pMacs undergo a coordinated macrophage differentiation program: a LOLA analysis (26) yielded a statistically significant association (adjusted p-value≤0.001, Benjamini Yekutieli-correction) with binding sites for Spi1, Egr1, Irf1, Irf8, Maf, Jun, Stat1, Stat3, Stat5b, Stat6, Rela, and Relb (Fig. S8A). These factors are expressed in our dataset (Fig. S8B, Table S1, S5) and their binding sites align at the same loci in enhancers and super-enhancers associated with differentially regulated genes, such as Thrombospondin1 (Thbs1) (27), Cx3cr1 (8), F4/80 (Emr1), and Mrc1, but not with control genes such as Gata1 and MyoD (Fig. S8C). Comparable results, with a higher statistical significance, were found for genes differentially upregulated in early macrophages when compared to EMPs (Fig. S8A).
Altogether, these results characterize at the cellular and molecular level the EMP-derived macrophage precursors (pMacs) that acquire a core macrophage transcriptional program as they colonize the head and caudal embryo from E9.5 in a Cx3cr1-dependent manner to give rise to tissue-resident macrophages. These data are consistent with our previous demonstration that the vast majority of resident macrophages in these tissues originate from a progenitor that does not express the stem cell and endothelial marker Tie2 after E10.5 (2), but do not exclude the possibility that later precursors may also contribute to the resident macrophage pool.
Early specification of tissue-resident macrophages
Following the acquisition of this core program, F4/80+ macrophages soon display heterogeneity between different tissues (Fig. 2B, Fig. 5A). For example, expression of Timd4 is lost at the transcriptional and protein level in microglia, alveolar macrophages, and Langerhans cells, but maintained in Kupffer cells throughout development and postnatally. Expression of the mannose receptor (CD206) is lost in microglia and Langerhans cells. Dectin-1 and CD64 expression is maintained in all subsets except Langerhans cells (Fig. 2B, Fig. 5A, Fig. S3). To systematically investigate the kinetics and molecular determinants of macrophage diversification, we first characterized tissue-specific signatures of genes differentially upregulated in postnatal microglia, kidney macrophages, Langerhans cells, alveolar macrophages and Kupffer cells in the bulk RNA-seq dataset. Unsupervised clustering analysis suggested that macrophages in different tissues undergo characteristic differentiation trajectories (Fig. S9A). Supervised analyses identified lists of genes differentially upregulated in each cell population (Table S3, Fig. S9B, C). The signatures of adult tissue-resident macrophages of the brain, lung, and liver previously defined by independent research (7, 25, 28) were progressively enriched in our developing resident macrophage populations (Fig. S10A-D). Finally, scorecard (21) and GSEA analysis of differentially upregulated genes for each postnatal macrophage population in all tissues and over time from E9 to P21 (Fig. 5B,C, S10C) revealed that the tissue-specific signature of postnatal microglia, Kupffer cells, and kidney macrophages could be traced back to fetal macrophages, as early as E12.5 (Fig. 5B). The signatures of Langerhans cells and alveolar macrophages reflected important postnatal changes in gene expression (Fig. 5C), noted in previous studies (29), which may reflect their anatomical location at epithelial barriers.
Tissue-specific sets of transcriptional regulators define macrophage diversity
A heat-map visualization of all transcriptional regulators present in the postnatal tissue-specific signatures (2-fold change, adj. p-value<0.05, BH-correction, Fig. 5D) confirmed the tissue-specific expression of the transcription factors Sall1 and Sall3 in microglia (7), Nr1h3 (Lxra) in Kupffer cells (30), Pparg in lung alveolar macrophages (18, 31), and Runx3 and Ahr in Langerhans cells (15, 32) (Fig. 5D), and identified additional tissue-specific transcriptional regulators such as Id1 and Id3 in Kupffer cells (Fig. 5D). In addition, this analysis showed that many of these transcriptional regulators start to be differentially expressed in early tissue macrophages, as early as E10.25, for example in Sall1 and Sall3 in head macrophages, Nr1h3, Id1 and Id3 in the liver, and Ahr in limb macrophages (Fig. 5D). Some genes, like Id1 and Sall3 are expressed by progenitors and pMacs before their expression becomes restricted to macrophages in the liver and the head, respectively (Fig. S10E), while expression of other genes such as Id3 and Sall1 is low in progenitors and upregulated in pMacs (Fig. S10E). In situ immunofluorescence confirmed expression of Id3 by E10.25 macrophages in the liver and head and of Id1 in liver macrophages (Fig. 6A). These data altogether suggested that the transcriptional programs of tissue-specific resident macrophages start to be established early on, as soon as macrophages or pMacs are present in tissues and identified a number of novel ‘candidate’ tissue-specific transcriptional regulators.
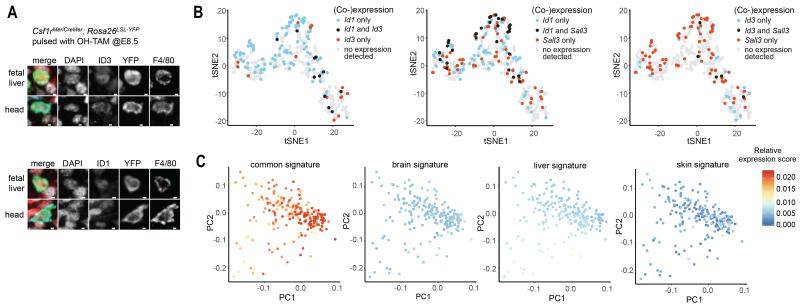
Fig. 6. Tissue-specific macrophage signatures are not detected in pMacs.
(A) Immunostaining with antibodies against Id1 or Id3 (red), F4/80 (cyan) and YFP (green) on cryosections from E10.25 Csf1rMeriCreMer; Rosa26LSL-YFP embryos (OH-TAM at E8.5) (upper panel). Nuclei are counterstained with DAPI (white). Scale bar represents 2 μm. (B) tSNE plots of scRNA-seq data from CD45low/+ cells from E10.25 embryo showing co-expression of Id1, Id3, and Sall3. See Fig. S11. (C) PCA plot of scRNA-seq data of cells from cluster 2 (pMacs) with superimposed fetal tissue macrophage-specific signatures. See Fig. S11, Table S2 and methods.
We thus investigated whether specification of tissue macrophages takes place in F4/80+ macrophages or at the level of their pMac or EMP precursors. We plotted transcriptional co-expression of Id1, Id3, and Sall3 onto the tSNE representation of single CD45low/+ cells from our scRNA-seq dataset (see Fig. 1D, Fig. 6B, Fig. S11). Co-expression of Id1 and Id3 was found in pMacs and macrophages. However, Id1 and Sall3 were co-expressed in EMPs and pMacs and Id3 and Sall3 were co-expressed in pMacs, suggesting that cells at the pMac state are not completely committed to exclusive expression of tissue-specific transcription factors. These data confirmed that Id1 and Sall3 are expressed by progenitors and pMacs (Fig. S10E), while their expression is ultimately lost by tissue macrophages outside the liver and the brain, respectively (Fig. 5D, Fig. 6B and Fig. S10E). When common and tissue-specific E14.5-E18.5 macrophage signatures (Table S2) were superimposed on the pMac population, we found that the common macrophage signature was expressed in pMacs with a gradual enrichment within this population (Fig. 6C). However, we did not observe pMac subsets expressing tissue-specific signatures (Fig. 6C). The lack of tissue specificity within pMacs was confirmed using an alternative bioinformatic approach based on analysis of multimodal expression followed by hierarchical clustering of genes with subsequent analysis of enrichment of tissue signatures within these clusters in pMacs (Fig. S11E).
Taken together, these data suggest that expression of Id1/Id3 or Sall3 does not specify pMac subsets pre-committed to give rise to either Kupffer cells or microglia, and more generally that diversification of tissue macrophages takes place after pMacs have colonized tissues and differentiated into F4/80+ macrophages.
The transcriptional regulator Id3 is essential for Kupffer cell development
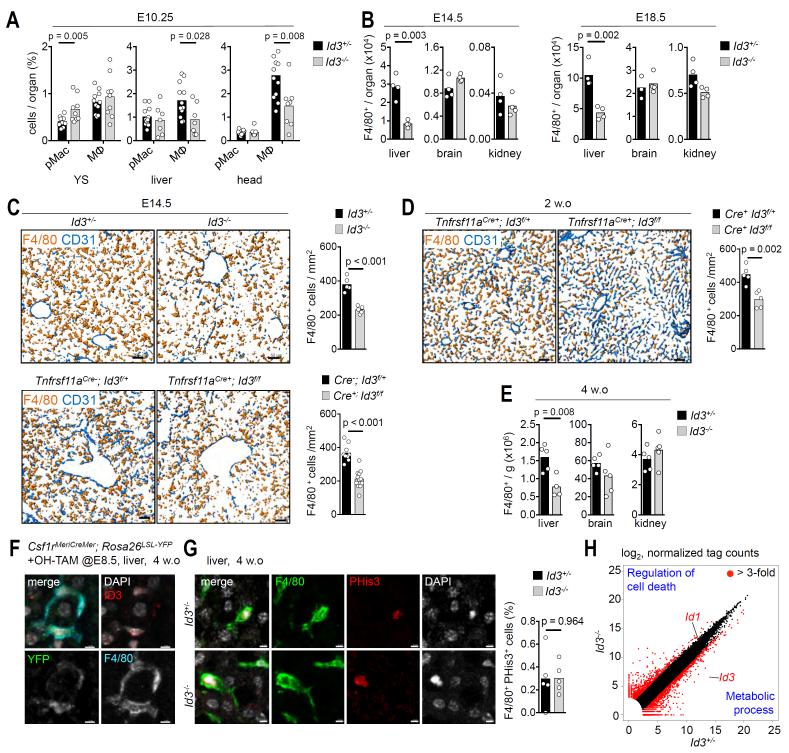
To functionally validate the role of early tissue-specific transcriptional regulators on the development and specification of tissue macrophages, and thereby the significance of our findings, we studied the role of Id3 expression in Kupffer cell differentiation. E10.25 Id3-deficient embryos had normal or increased numbers of pMacs/early macrophages in the YS, but macrophages were reduced in numbers in the embryo proper (liver and head) in comparison with littermate controls (Fig. 7A). The further development of liver macrophages was severely impaired in E14.5 and E18.5 Id3-deficient embryos as determined by flow cytometry and histology, and 4-week old Id3-deficient mice still presented with a marked Kupffer cell deficiency, while development of microglia and kidney macrophages appeared normal (Fig. 7B-D, S12C). The role of Id3 in Kupffer cells appears cell autonomous as targeted deletion of an Id3 floxed allele in pMacs (Tnfrsf11aCre+; Id3f/f) recapitulated the phenotype of the Id3-deficient mice in embryo and postnatal mice (Fig. 7C, E). Expression of Id3 in postnatal Kupffer cells was confirmed by qRT-PCR (Fig. S12A), and by immunofluorescence in Kupffer cells from Csf1rMeriCreMer; Rosa26LSL-YFP mice pulse-labeled with OH-TAM at E8.5 (Fig. 7F, Fig. S12B). In contrast, fate-mapped microglia do not express Id3 (Fig. S12B), in line with our RNA-seq data (Fig. 5D). Of note, the partial Kupffer cell deficiency observed in Id3-deficient and Tnfrsf11aCre+; Id3f/f embryo and adult was not associated with an abnormal liver lobular architecture or vasculature (Fig. 7C,E, Fig. S12D). Kupffer cell proliferation in the steady state was not affected by Id3 deficiency (Fig. 7G), but RNA-seq analysis of Id3−/− and Id3+/− adult Kupffer cells indicated the upregulation (>3-fold) of Id1 expression, and GO-term analysis evidenced that Id3−/− cells overexpressed genes involved in the control of cell death and cytokine responses and down-regulated genes involved in metabolic processes (Fig. 7H, Fig. S12E,F, Table S6). These data suggest that Id3 is important for the development and maintenance of Kupffer cells in the liver, but dispensable for other macrophage subsets. As Id1 and Id3 are co-expressed, it will be interesting to investigate whether up-regulation of Id1 partially compensates for Id3-deficiency (33, 34).
Fig. 7. Id3 is important for Kupffer cells development.
(A) Flow cytometry analysis of pMacs and macrophages in yolk sac (YS) and liver from E10.25 Id3−/− and Id3+/− embryos. Circles represent individual mice. n=3 independent experiments. (B) Flow cytometry analysis of F4/80+ macrophages in liver, brain and kidney from E14.5 and E18.5 Id3−/− and Id3+/− mice. Circles represent individual mice. n=4 independent experiments. (C) Immunostaining with antibodies against CD31 and F4/80 on liver cryosections from E14.5 Id3−/− and Id3+/−, and Tnfrsf11aCre−;Id3f/+ and Tnfrsf11aCre+; Id3f/f mice. The figure displays isovolume-rendered images. Bar graphs represent F4/80+ cells/mm2. Circles represent individual images. n=3 independent experiments. (D) Flow cytometry analysis of F4/80+ macrophages in liver, brain and kidney from 4 week-old Id3−/− and Id3+/− mice. Circles represent individual mice. n=2 independent experiments. (E) Immunostaining with antibodies against CD31 and F4/80 on liver cryosections from 2 week-old Tnfrsf11aCre+;Id3f/+ and Tnfrsf11aCre+; Id3f/f mice. The figure displays isovolume-rendered images. Bar graphs represent F4/80+ cells/mm2. Circles represent individual images. (F) Immunostaining with antibodies against Id3 (red), F4/80 (cyan) and YFP (green) on cryosections from on livers from 4 week-old Csf1rMeriCreMer; Rosa26LSL-YFP (OH-TAM at E8.5) mice. Nuclei are counterstained with DAPI (white). Scale bar represents 5 um. (G) Immunostaining with antibodies against F4/80 (green) phospho-histone 3 (PHis3, red) and DAPI (gray) on liver cryosections from 4 week-old Id3−/− and Id3+/− mice. Scale bar represents 10 um. Data are representative of 5 mice per genotype. Each dot represents the mean of PHis3+ cells (in %) of total F4/80+ found in 5 sampling areas (830 um2) for each individual liver. (H) Scatterplot comparison of gene expression of 3 week-old Id3−/− and Id3+/− Kupffer cells. Both axes (in log2 scale) represent normalized gene expression values (average value from three Id3+/− and two Id3−/− replicates). Red circles mark the 3-fold cut-off in both directions in gene expression level. Top GO terms for genes enriched in either Id3+/− or Id3−/− are indicated. P-values were calculated using Student’s t test..See also Fig. S12.
Conclusions
We show here that EMPs rapidly differentiate into a population of cells that we call pMacs, because they simultaneously colonize the whole embryo from E9.5 in a Cx3cr1-dependent manner while differentiating into macrophages. pMacs do not yet have macrophage morphology but are in the process of establishing a full core macrophage differentiation program that includes cytokine receptors, phagocytic and pattern recognition receptors, and complement. Starting from E10.5, almost immediately after colonization of embryonic tissues, tissue-specific expression of transcriptional regulators in F4/80+ macrophages initiates their specification into adult type resident macrophages (Fig. 8). Our data therefore suggest that a broad or core macrophage program is progressively restricted or refined to a tissue-specific one in response to the absence and presence of tissue-specific cues, rather than the alternative possibility that ‘committed’ subsets of early macrophages, pMacs or even EMPs choose their future tissue of residence. The present results are consistent with an EMP origin of tissue-resident macrophages (2) albeit they do not exclude the additional contribution of ‘primitive’ precursors (23), fetal HSCs or of a second wave of EMPs (11, 12), which would adopt a similar differentiation program when entering tissues. However, we note that the novel transcriptional dataset we provide here may lead to re-interpretation of fate-mapping models used to characterize the contribution of such later waves. For example, the progressive YFP expression by fetal macrophages in S100a4Cre; Rosa26LSL-YFP mice (12) is compatible with our present findings, and do not require the existence of a ‘second wave’ of precursors, because expression of S100a4 and several other S100a family members was found to be part of the pMac transcriptional profile (Fig. S13).
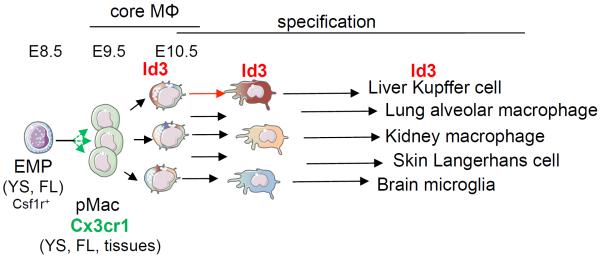
Fig. 8. Graphic summary of the establishment of the core macrophage program and subsequent specification.
Cx3cr1 is expressed in pMacs and is important for colonization of the embryo. Id3 is a liver macrophage-specific gene, which is essential for Kupffer cell development.
In summary, we propose that establishment of a core macrophage differentiation program in EMP-derived pMacs, as they colonize the embryo, is followed by the initiation of macrophage specification via the acquisition of tissue-specific transcriptional regulators, such as Id3 in Kupffer cells. This process initiated simultaneously in the whole embryo during the first two days of organogenesis, from E8.5 to E10.5. Differentiation of resident macrophages is thus a developmental process and an integral part of organogenesis, independent of postnatal changes in the environment in kidney, liver, and brain, but not in skin and lung. These results also identify a developmental window where the molecular mechanisms of macrophage specification can be best studied, tools to selectively label resident macrophages, and sets of tissue specific transcriptional regulators expressed by developing and adult macrophages that may control their differentiation, maintenance and function, as well as a molecular road map that will support efforts to differentiate specialized macrophages: microglia, Kupffer cells, kidney macrophages, alveolar macrophages and Langerhans cells in vitro from multipotent progenitors. Finally, our work provides a framework to analyze and understand the consequence(s) of genetic variation for macrophage contribution to disease pathogenesis in different tissues.
Material and Methods
Mice
Csf1rMeriCreMer, Csf1riCre, Rosa26YFP-LSL, Cx3cr1gfp/+ and Tnfrsf11aCre mice (2, 5, 22, 35) and Id3−/−; Id1fl/fl and Id3f/f mice -kindly provided by R. Benezra (34, 36, 37)- were maintained under SPF conditions. Animal procedures were performed in adherence to our project licence issued by the United Kingdom Home Office under the Animals (Scientific Procedures) Act 1986 and by the Institutional Review Board (IACUC 15-04-006) from MSKCC. Genotyping was performed according to protocols described previously for Csf1riCre (38) Csf1rMeriCreMer (5), Id3−/−; Id1fl/fl (36), and Tnfrsf11aCre (22) mice. Genotyping of Cx3cr1gfp/+ was performed as recommended by Jackson Laboratory, Bar Harbor, Maine with the following primers: Cx3cr1 F 5′-CCC AGA CAC TCG TTG TCC TT-3′, Cx3cr1 R 5′-GTC TTC ACG TTC GGT CTG GT and Cx3cr1 R mut 5’CTC CCC CTG AAC CTG AAA C-3′. Cre recombination in Csf1rMeriCreMer; Rosa26LSL-YFP embryos was induced by single injection at E8.5 of 75 mg per g (body weight) of 4-hydroxytamoxifen (OH-TAM, Sigma) into pregnant females as described (2). The OH-TAM was supplemented with 37.5 mg per g (body weight) progesterone (Sigma) to counteract the mixed oestrogen agonist effects of tamoxifen, which can result in fetal abortions. Embryonic development was estimated as previously considering the day of vaginal plug formation as 0.5 days post-coitum (dpc), and staged by developmental criteria. E9: 20-25sp, E9.5: 26-29sp, E10.25: 30-35sp, E10.5: 36-44sp.
Preparation of cell suspensions and cell sorting
Pregnant females were killed by cervical dislocation or by exposure to CO2. Embryos ranging from E9 to E18.5 were removed from the uterus, washed in 4°C phosphate-buffered saline (PBS, Invitrogen) and dissected under a Leica M80 microscope. Yolk sacs (YS) were harvested from embryos between E9 and E10.5. To obtain single-cell suspensions for FACS sorting, tissues were included in cold PBS and mechanically disrupted under a 100um filter. Postnatal tissues were collected following the same procedure. For collection of Langerhans cells, epidermal sheets (from E18.5 to P21) were separated from the dermis after incubation for 45 min at RT in 4.8mg/ml of dispase (Invitrogen), 3% fetal calf serum (FCS, Invitrogen) and 1uM of flavopiridol (Sigma). The epidermis was further digested for 30min at RT in PBS containing 2 mg/ml of collagenase D (Roche), 200U/ml DNase I (Sigma), 4.8 mg/ml of dispase (Invitrogen), 3% FCS (Invitrogen) and 1uM of flavopiridol (Sigma) followed by mechanical disruption under a 100um filter. For blood phenotyping, mice were anaesthetized and blood was collected by cardiac puncture. For flow cytometry experiments, organs were incubated in PBS containing 1mg/ml collagenase D (Roche), 100U/ml DNase I (Sigma), 2.4mg/ml of dispase (Invitrogen) and 3% FCS (Invitrogen) at 37°C for 30 min prior to mechanical disruption. Epidermal sheets were obtained as previously described (2). For embryonic tissue incubation time at 37°C was reduced to 20 min. Cell suspensions were centrifuged at 320g for 7 min, resuspended in FACS buffer (PBS, 0.5% BSA and 2 mm EDTA) containing purified anti-CD16/32 (FccRIII/II) (1:100 dilution) and incubated for 15min at 4°C. Samples were immunostained with antibodies mixes for 30 min at 4°C. The full list of antibodies used can be found in Table S4.
Cell sorting was performed using an Aria II BD cell sorter. Single live cells were gated on the basis of dead cell exclusion (DAPI), side (SSC-A) and forward scatter (FSC-A) gating, and doublet exclusion using forward scatter width (FSC-W) against FSC-A. For bulk sequencing, EMPs were identified after gating on Kit+CD45lo cells based on AA4.1 expression. pMacs were identified after gating on Kit−CD45+ based on CD11b expression and no expression of F4/80. Additionally, Gr1+ or Ter119+ cells were excluded from the F4/80−CD11b+ gate. Macrophage populations were identified after gating on CD45 based on expression of F4/80 and CD11b. 100 cells for each sample were directly sorted into a 96 well plate (Biorad) in 4ul of H2O containing 0.2% of triton TXT (Sigma) and 0.8U/ul of RNAse inhibitor (Clontech), and processed as indicated below (Generation of ‘bulk’ transcriptomes from candidate EMP, pMac, and macrophage populations from E9 to P21). For single cell sequencing, all CD45low/+ single cells from a E10.25 (30-34sp) embryo proper were sorted into 384-well plates filled with 2 μl lysis buffer (Triton-X 0.2% (Sigma) in molecular biology grade H2O (Sigma) supplemented with 0.4 U/μl protein-based RNase inhibitor (Takara) and barcoded poly-T primers, and processed as described below (Generation of single cell transcriptomes).
Cytology
Cells were collected into FCS and centrifuged (800 rpm, 10 min, low acceleration) onto Superfrost slides (Thermo Scientific) using a Cytospin 3 (Thermo Shandon). Slides were air-dried for at least 30 min, and fixed for 5 min in methanol, stained in 50 % May-Grunwald solution for 5 min, 14 % Giemsa for 15 min, washed with Sorensons buffered distilled water (pH 6.8) for 5 min and rinsed with Sorensons buffered distilled water (pH 6.8). Slides were air-dried and mounted with Entellan New (Merck) and representative pictures were taken using an Axio Lab.A1 microscope (Zeiss) under a N-Achroplan 100x/01.25 objective.
Immunofluorescence, imaging, analysis and illustrations
E10.25 Embryos were fixed for 4 hours in 4% formaldehyde (Sigma) under agitation and >E10.25 embryos were fixed overnight. After fixation, embryos were incubated overnight in 30% sucrose and embedded in OCT compound (Sakura Finetek). Cryoblocks were cut at a thickness of 10 -12μm and then blocked with PBS containing 5% normal goat serum (Invitrogen); 1 % BSA (w/v); 0.3% Triton X-100 for 1 hour at room temperature. Samples were incubated overnight at 4°C with rat anti-mouse F4/80 (1:200, Biorad), rabbit anti-mouse Iba1 (1:200; Wako), chicken anti-GFP for YFP detection (1:500, Invitrogen), rat anti-mouse Dectin-1 (1:200, Biorad), anti-Granulin (1:200, abcam), rat anti-mouse CD206 (1:200, Biorad), armenian hamster anti-mouse PECAM-1 (1:300, Thermo Scientific), rabbit anti-mouse/human ID3 (1:500, Biocheck), rabbit anti-mouse ID1 (1:500, Biocheck), rat anti-mouse CD16/CD32 (BD Biosciences), goat anti-mouse Trem2 (1:200, Abcam), armenian hamster anti-mouse CD119 (1:200, Clone 2E2, eBioscience), armenian hamster anti-mouse CD120b (1:200, clone 55R-286, Biolegend). Secondary antibodies used were anti-rabbit Cy3 (1:500; Invitrogen), anti-chicken Alexa Fluor 488 (1:500; Invitrogen), anti-rat Alexa Fluor 555 (1:500; Invitrogen), anti-rat Alexa Fluor 488 (1:500; Invitrogen), anti-rat Alexa Fluor 647 (1:500; Invitrogen), anti-goat Alexa Fluor 568 and anti-armenian hamster Dylight 649 (Jackson ImmunoResearch Laboratories). Samples were then mounted with Fluoromount mounting medium with DAPI (eBiosciences) and visualized using a LSM880 Zeiss microscope with 20x/0.5 (dry), performing a tile scan and Z-stack on whole embryos or tissues. Image analysis and cell quantification was performed using Imaris (Bitplane) software. To determine the CD31+ area in Id3+/− and Id3−/− liver sections, maximum intensity Z-projections pictures were converted into binary images and the CD31+ area was measured using Image J (NIH, Bethesda, MD, USA) (39). Results were normalized per mm2 of tissue. Illustrations were created by adapting templates from Servier Medical Arts (http://www.servier.com/Powerpoint-image-bank, licensed under a Creative Commons Attribution 3.0 Unported License).
Generation and analysis of Kupffer cells in Id3−/− and littermate controls
For qRT-PCR experiments, cells were sorted as described above. Hepatocytes were enriched by centrifugation of the whole liver cell suspension at 50 g for 3 min (Sorvall Legend XTR centrifuge). Supernatant was taken for further staining of macrophages (CD45+, CD11blo, F4/80+). Hepatocytes were sorted using the FSC-A and SSC-A gate with subsequent exclusion of doublets and CD45+ cells. Cells were sorted into RNA lysis buffer and RNA extraction was performed as per manufacturers protocol (Macherey-Nagel). cDNA was synthesized using the QuantiTect Reverse Transcription Kit (Qiagen) as per manufacturers protocol. qRT-PCR was performed on a QuantStudio 6 Flex using TaqMan Fast Advanced Master Mix (applied biosystems) and TaqMan probes for Id3 (Mm00492575_m1), Nr1h3 (Mm00443451_m1), and GAPDH (Mm99999915_g1) (Life Technology). For RNA-seq of Id3+/− and Id3−/−, Kupffer cells were sorted into Trizol as described above. RNA from sorted cells was extracted using RNeasy mini kit (Qiagen) according to instructions provided by the manufacturer. After ribogreen quantification and quality control of Agilent BioAnalyzer, 400pg of total RNA underwent amplification (12 cycles) using the SMART-seq V4 (Clonetech) ultra low input RNA kit for sequencing. 10 ng of amplified cDNA was used to prepare Illumina hiseq libraries with the Kapa DNA library preparation chemistry (Kapa Biosystems) using 8 cycles of PCR. Samples were barcoded and run on a Hiseq 2500 1T in a 50bp/50bp Paired end run, using the TruSeq SBS Kit v3 (Illumina). An average of 54 million paired reads were generated per sample and the percent of mRNA bases was closed to 77% on average.The output data (FASTQ files) were mapped to the target genome using the rnaStar aligner (40) that maps reads genomically and resolves reads across splice junctions. 2 pass mapping method was used, outlined in Engstrom et al.(41) in which the reads are mapped twice. The first mapping pass uses a list of known annotated junctions from Ensemble. Novel junctions found in the first pass are then added to the known junctions and a second mapping pass is done (on the second pass the RemoveNoncanoncial flag is used). After mapping we post process the output SAM files using the PICARD tools to: add read groups, AddOrReplaceReadGroups which in additional sorts the file and coverts it to the compressed BAM format. We then compute the expression count matrix from the mapped reads using HTSeq and one of several possible gene model databases. The raw count matrix generated by HTSeq are then processed using the R/Bioconductor package DESeq, which was used to both normalize the full dataset and analyze differential expression between sample groups. Gene Ontology analysis was performed using the GO analysis function in GeneSpring GX 13.0 (Agilent), with the p-value calculated using a hypergeometric test with Benjamini–Yekutieli correction. For that, genes with a fold change difference of ± 2 between Id3+/− and Id3−/− Kupffer cells were selected. Significantly regulated genes (t-test p< 0.05; FDR <0.05) from this selection were grouped into gene ontology (GO) terms.
Generation and analysis of ‘bulk’ transcriptomes from candidate EMP, pMac, and macrophage populations from E9 to P21
RNA Isolation and Library Construction
cDNA synthesis and enrichment was performed following the Smart-seq2 protocol as described (42, 43). ERCC spike-in RNA (Ambion) was added to the lysis buffer in a final dilution of 1:1,000,000. After the cDNA was synthesized and amplified from single cells using 18 cycles, quantitative PCR was performed with GoTaq-PCR master mix (Promega) on a C1000 Touch Thermal Cycler qPCR instrument (Bio-Rad) to test for house keeping gene expression. Library preparation was conducted on 1ng of cDNA using the Nextera XT library preparation kit (Illumina) as described (43). Sequencing was performed by the Biomedical Sequencing Facility at CeMM using a 50bp single-read setup on the Illumina HiSeq 2500 platform.
RNA-seq Analysis
We first trimmed off sequencing adapter from the reads generated, and then aligned the reads using Bowtie v 1.1.1, (44) parameters: -q -p 6 -a -m 100 --minins 0 --maxins 5000 --fr --sam --chunkmbs 200) to the cDNA reference transcriptome (mm10 cDNA sequences from Ensembl). For genome browser track visualization, we generated a second alignment with Tophat2 (45) (v 2.0.13, parameters: --b2-L 15 --library-type fr-unstranded --mate-inner-dist 150 --max-multihits 100 --no-coverage-search --GTF) against the reference genome (mm10). Next, we removed duplicate reads before quantifying transcript levels with BitSeq (46) (v 1.12.0). The raw transcript counts were loaded into R and processed further. At this stage, we removed samples with a substandard quality, that is, all samples that had less than 2 million reads, less than 33% of reads aligned, or less than 5,000 transcripts detected (with >= 25 reads). 20 out of 178 datasets failed these criteria (11.2%). Of the remaining datasets, we merged technical replicates creating 93 unique biological samples. We took forward only transcripts that were detected reliably (>=50 reads) in at least 4 samples (n = 37,521). For statistical analysis, we used raw read counts as input for DESeq2 (20), factoring in the flowcell identifier as a covariate to reduce the effect of technical variation. To identify genes of particular interest to the development of tissue-resident macrophages over time and in different tissues, we performed two types of comparisons: (a) Cell type-specific: Pairwise comparisons (Wald test) between EMPs, pre-macrophages, and macrophages (up to E10.5) independent of their tissue of origin (treating time and tissue as covariates). (b) Tissue-specific: Pairwise comparisons (Wald test) between macrophages from all tissues stratified and stage (post-natal). We considered genes with an FDR-corrected p-value <= 0.05 as differentially expressed. For visualization and illustration purposes (PCA, supplementary tables, heatmaps), we used values adjusted by variance stabilizing transformation from DESeq2 in which batch-effects had been corrected for with ComBat (47).
Evaluation of lists of differentially expressed genes from the RNAseq data
We bioinformatically investigated the differentially expressed transcripts (Table S5) from our statistical comparisons in several ways: To identify transcription factors specifically regulating each set of transcripts, we use LOLA(26) (v 0.99.4) together with its core database of ChIP-seq binding peaks from CODEX (48) to identify enrichment of experimentally-derived transcription factor binding locations in a window around the promoter regions (TSS +/− 20kb) of differentially expressed transcripts. We corrected for multiple testing using the Benjamini & Yekutieli method. To visualize and summarize the expression patterns of many genes (lists of differentially expressed genes) in many different conditions (different tissues at different time points) and across replicates, we sought to use an adaptation of lineage scorecards(21). Briefly, we considered each list of differentially upregulated genes as a set of marker genes and determined the relative enrichment of these marker sets in each individual condition (tissue by cell type by time) in comparison to all other datasets using a modified version of parametric gene set enrichment analysis in R (package: PGSEA). We also used gene set enrichment analysis (GSEA) to test for the relative overrepresentation of gene signatures in sorted gene lists. To this end, we extracted lists of genes sorted by logarithmic fold change between the mean expression levels in any one tissue Mac, pMac, or EMP sample stratified by stage (early = E9 - E10.5, fetal = E12.5 - E18.5, postnatal = P2 - P21) compared to all other samples at the same stage. Additionally, we extracted lists of all genes we found differentially upregulated in any tissue-specific signature, cell type-specific signature, or in lists of genes extracted from the publications of Gorgani et al. (2008), Gautier et al. (2012), and Lavin et al. (2014) (7, 28, 49). External gene lists were translated to Ensembl Transcript IDs using g:profiler (50). Both sets of data were loaded into and analyzed using the GSEA tool (51) and the results read and summarized using the metaGSEA R library. We also incorporated ChIP-seq binding profiles (bigWig) for factors identified in the LOLA analysis from the CODEX database. Heatmaps were generated using GeneSpring GX 13.0 (Agilent). All other analyses and plotting was performed in R.
Generation and analysis of single cell transcriptomes
For single-cell RNA sequencing the MARS-Seq approach described by Jaitin et al. (52) was applied using the Biomek FXP system (Beckman Coulter). In brief, single cells were sorted into each well of 384-well plates filled with 2 μl lysis buffer (Triton-X 0.2% (Sigma) in molecular biology grade H2O (Sigma) supplemented with 0.4 U/μl protein-based RNase inhibitor (Takara)) and barcoded poly-T primers (400 nM (IDT); for barcode details see Jaitin et al., 2014). Samples were pre-incubated (3 min at 80°), reverse transcriptase mix (10 mM DTT (Invitrogen), 4 mM dNTPs (NEB), 2.5 U/μl Superscript III RT enzyme (Invitrogen) in 50 mM Tris-HCl (pH 8.3; Sigma), 75 mM KCl (Sigma), 3 mM MgCl2 (Sigma), ERCC RNA Spike-In mix (Life Technologies) at 1:80×107 dilution per cell) was added to each well and mRNA was reverse transcribed to cDNA (2 min at 42°C, 50 min at 50°C, 5 min at 85°C). Excess primers were digested (Exo I (NEB); 37°C for 30 min then 10 min at 80°C) followed by a 1.2 × SPRI bead (Beckman Coulter) cleanup, samples were pooled and second strands synthesized (second strand synthesis kit (NEB); 2.5 h at 16°C) followed by a 1.4 × SPRI bead (Beckman Coulter) cleanup. Samples were linearly amplified by T7-promoter guided in vitro transcription (T7 High Yield RNA polymerase IVT kit (NEB); 37°C for 12h). DNA templates were digested (Turbo DNase I (Ambion); 15 min at 37°C) followed by a 1.2 × SPRI bead (Beckman Coulter) cleanup and the RNA was fragmented (Zn2+ RNA fragmentation solution (Ambion); 1.5 min at 70°C) followed by a 2 × SPRI bead (Beckman Coulter) cleanup. Barcoded ssDNA adapters (IDT; for barcode details see Jaitin et al., 2014, (52)) were ligated to the fragmented RNA (9.5% DMSO (Sigma), 1 mM ATP, 20% PEG8000 and 1 U/μl T4 RNA ligase I (NEB) in 50 mM Tris HCl pH7.5 (Sigma), 10 mM MgCl2 and 1mM DTT; 22°C for 2 h) and a second RT reaction (Affinity Script RT buffer, 10 mM DTT, 4 mM dNTP, 2.5 U/μl Affinity Script RT enzyme (Agilent); 2 min at 42°C, 45 min at 50°C, 5 min at 85°C) was performed followed by a 1.5 × SPRI bead (Beckman Coulter) cleanup. Final libraries were generated by subsequent nested PCR reaction (0.5 μM of each Illumina primer (IDT; for primers details see Jaitin et al., 2014) and KAPA HiFi HotStart ready mix (Kapa Biosystems) for 15 cycles according to manufacturer’s protocol followed by a 0.7 × SPRI bead (Beckman Coulter) cleanup. Library quantity and quality were assessed using the Agilent 2200 Tapestation system and libraries were subjected to next generation sequencing using an Illumina HiSeq1500 instrument (PE with no index; read 1: 61 bases (3 bases random nucleotides, 4 bases pool barcode, 53 bases specific sequence), read 2: 13 bases (6 bases cell barcode, 6 bases unique molecular identifier)).
Pre-processing, quality assessment and control of single cell transcriptome data
From sequenced data, pool barcodes, cell specific tags and Unique molecular identifiers (UMI) were extracted (576 cells sequenced). Subsequently, sequencing reads with ambiguous plate/cell-specific tags or UMI sequence with low quality (Phred < 27) and reads which map to E. coli were eliminated using Bowtie with parameters “-M 1 -t --best --chunkmbs 64 –strata”. Next, fastq files were demultiplexed using the fastx_barcode_splitter from the fastx_toolkit and R1 reads (after trimming of pool barcode sequences) were mapped to the mouse mm10 & ERCC pseudo genome assembly using Bowtie “-m 1 -t --best --chunkmbs 64 –strata”.
Valid reads were then counted using unique molecular identifiers if they mapped to the exon based gene model derived from Ensembl’s biomart, mm10. Following this, a gene expression matrix was generated containing the number of unique UMIs associated with valid reads for every cell and every gene. Additionally, UMI sequencing errors were corrected for and filtered as described in (52).
Filtering single cell transcriptome data
In order to avoid biases introduced by low quality data we performed the following filtering of single cell data. Removal of cells with a ratio of mitochondrial versus endogenous genes exceeding 0.15 and cells with less than 320 molecule counts or less than 150 unique genes were removed from the analysis. Prior to analysis expression tables were filtered for mitochondrial, ribosomal and predicted genes to reduce noise.
Analysis of MARS-seq single cell transcriptome data
Analysis of the normalized and filtered single cell gene expression data (8657 genes across 408 single cell transcriptomes used in the final expression table) was done using several functions of the SEURAT single cell analysis package (53) and Monocle 2 (54). First, highly variable genes were determined as genes exceeding the dispersion threshold of 0.75. To infer the structure of the gene expression data a PCA was performed on the basis of highly variable genes. Following t-distributed stochastic neighbor embedding (tSNE) DBScan clustering was performed to identify clusters of cells. The optimal number of clusters was identified by calculating several cluster indices by the NbClust R package(55). Relative expression for a cell was calculated as gene expression of a gene/gene set in relation to the total molecule counts in this cell. To identify clusters within the MARS-seq data the relative gene expression profiles of cell type specific gene signatures were overlaid to the tSNE plots. Pseudotime analysis was performed by the Monocle 2 algorithm by genes exceeding the average expression cutoff of 1 while having an empirical dispersion higher than 1. To analyze expression of single genes, relative expression was overlaid onto the tSNA plots or visualized as dot plots.
Generation of cell signatures for analysis of single-cell data
In order to unambiguously identify cell state specific genes for EMPs, pMacs and early macrophages we generated exclusive gene signatures. Here, differentially expressed (DE) genes were identified by a 1-way ANOVA model (∣FC∣ > 1.4, FDR-adjusted p-value < 0.05 (56)) between EMPs (E10.25 yolk sac and fetal liver), pMacs (E10.25 yolk sac, fetal liver, head and limbs) and early macrophages (E10.25 and E10.5 yolk sac, fetal liver, head and limbs). The non-overlapping DE genes between these three contrasts were chosen as exclusive gene signatures for further analysis.
Generation of tissue signatures for analysis of single-cell data
To identify genes that are upregulated in tissue macrophages in relation to early macrophages a 1-way ANOVA model (∣FC∣ > 1.5, FDR-adjusted p-value < 0.05 (31)) was calculated. Upregulated non-overlapping DE genes between early macrophages (E10.25 and E10.5, fetal liver, head and limbs/skin) vs brain macrophages (E14.5 and E18.5 brain), liver macrophages (E14.5 and E18.5 liver) or limb/skin macrophages (E14.5 and E18.5 limb/skin) were used as tissue macrophage specific gene signatures. Furthermore, a common early macrophage signature was defined as being upregulated in early macrophages (E10.25 and E10.5 fetal liver, head and limbs) vs. all other late tissue macrophages (E14.5 and E18.5 liver, head and limbs/skin). We assessed enrichment of these signatures in scRNA-seq data from pMacs by calculating an relative enrichment score for each signature in pMacs (molecule count of signature genes/(total molecule count * number of signature genes).
Enrichment of tissue signatures in scRNA-seq data
To assess enrichment of tissue macrophage-specific signatures or differentiation signatures in scRNA-seq data from pMacs, we identified genes with multimodal expression using Hartigan’s Dip test statistic for unimodality. Next, multimodal genes were grouped by hierarchical clustering and enrichment of signatures within the clusters tested using hypergeometric testing with FDR correction (Benjamini-Hochberg).
Supplementary Material
Fig. S1: Quality control and analysis of bulk RNA-seq. (A) Number of reliably detected transcripts (covered with a least 25 reads) in each library. The colored circles underneath indicate the batch (sequencing flow cell), time, cell type, and tissue of each sample with the color code as given below panel B. (B) Hierarchical clustering with Euclidean distance and complete linkage of all samples based on their RNA-seq gene expression profiles. The colored circles underneath the cluster dendrogram indicate the batch (sequencing flow cell), time, cell type, and tissue of each sample with the color code as given below. (C) Scorecard analysis of differentially up-regulated genes (DESeq2 Wald test, adjusted p-value<0.05, BH-correction) in early macrophages (E10.25, E10.5) in comparison to EMPs. The table shows the relative enrichment of differentially upregulated genes in macrophages across cell types and tissues (y-axis) and developmental time points (x-axis, from E9 to P21). See Methods for details of the scorecard. (D) Principal component analysis (PCA) plot of EMPs (red, E9-E10.25), pMacs (yellow, E9.5-E10.25) and macrophages (purple, E10.25-E10.5) from the head, caudal, fetal liver (FL) and yolk sac (YS). The shape of each dot indicates the tissue the sample was taken from. The first and second principal component explain 18.9% and 11.1% of the entire variation in the data, respectively.
Fig. S2: Quality control and analysis of single-cell RNA-seq. (A) Workflow of the MARS-seq single cell data analysis. (B) Mean-variability plot shows average expression and dispersion for each gene. This analysis was used to determine highly variable genes (labeled by gene symbol). These 138 highly variable genes were used to perform a dimensionality reduction of the single-cell data by a principal component analysis. (C) The highest gene loadings in the first and second principal component from the PCA of 408 high quality cells, colored by batch association, showed even distribution of cells among the PCA plot based on the 138 most highly variable genes. (D) Heatmap of 138 highly variable genes among single-cell clusters as defined by DBScan clustering. (E) Optimal cluster number was identified by calculation of diverse indices for determining the best clustering scheme using the NbClust R package. (F) PCA plot of 408 single cells colored by cluster association. Clusters were defined by PCA + DBScan clustering. (G) Kinetic diagram shows the pseudotemporal ordering of single cells as determined by Monocle 2. Dots indicate individual cells and are colored according to the cluster association as in (F). Black line indicates the progression of single cells over developmental pseudotime.
Fig. S3 Expression of surface markers on EMP-derived cells during development. (A) Flow cytometry analysis of E10.25 Csf1rMeriCreMer; Rosa26LSL-YFP (OH-TAM at E8.5) tissues showing expression of Il4ra, Il13ra1, Tnfr2, Ifngr, CD16.2, CD64, Tim4, and CD206 on YFP+ Kit+ progenitors (gray), pMacs (blue) and macrophages (orange). Histograms represent the fluorescence intensity for each antibody in each cell subset. Data are representative of n=4 independent experiments with 4-6 embryos per marker. (B,C) Flow cytometry analysis of Csf1rMeriCreMer; Rosa26LSL-YFP (OH-TAM at E8.5) liver, brain, lung, and skin F4/80+ cells from E14.5 embryos showing expression of Il4ra, Il13ra1, Tnfr2, Ifngr, Dectin-1, CD64, Tim4, and CD206 (black dotted on whole population and green on YFP+ cells). Gray histograms show the fluorescence intensity of the FMO controls.
Fig. S4 Expression of the core macrophage program on EMP-derived cells. (A) Immunostaining on cryosections from E10.25 Csf1rMeriCreMer; Rosa26LSL-YFP embryos, pulse-labeled with OH-TAM at E8.5 with antibodies against YFP (green), Iba1 (red/cyan), and CD206 (red), Ifngr (red), Tnfr2 (red), Dectin-1 (red), Trem2 (red), CD16/32 (red), Granulin (Grn, red), or F4/80 (cyan). Scale bars represent 10 μm. Data are representative of n=3 embryos for each marker.
(B) Whole mount immunostaining of E9.5 Csf1riCre; Rosa26LSL-YFP embryo labeled with antibodies against YFP (green), Iba1 (red), F4/80 (cyan) and DAPI (white). Scale bars represent 10 μm. Data are representative of n=3 embryos. (C) Immunostaining on cryosections from E10.25 Csf1rMeriCreMer; Rosa26LSL-YFP embryo liver, pulse-labeled with OH-TAM at E8.5 with antibodies against YFP (green), Dectin-1 (red) and Iba1 (cyan) (upper panel) or YFP (green), Kit (red) and F4/80 (cyan) (lower panel) Scale bars represent 15 μm. (D, E) Immunostaining on cryosection from E14.5 (D) and E18.5 (E) Csf1rMeriCreMer; Rosa26LSL-YFP mouse embryos stained with antibodies against YFP (green), Iba1 (red), and F4/80 (cyan). (F) Immunostaining on cryosection from E14.5 Csf1rMeriCreMer; Rosa26LSL-YFP mouse embryo stained with antibodies against YFP (green), Granulin (Grn, red) and F4/80 (cyan). Scale bars represent 10 μm.
Fig. S5 Analysis of Tnfrsf11aCre; Rosa26LSL-YFP mice. (A) Gating strategy for Tnfrsf11aCre; Rosa26LSL-YFP embryos in E10.25 YS pMacs (Kit− CD45+ F4/80− CD11blow Gr1− Ter119−; green) and macrophages (CD45+F4/80+CD11blo; blue) (upper panel), and in E14.5 fetal liver LT-HSCs (Lin−Kit+Sca1+CD150+CD48−; orange), ST-HSCs (Lin−Kit+Sca1+CD150−CD48−; blue) and MPPs (Lin−Kit+Sca1+CD150−CD48+; purple) (lower panel). Histograms represent YFP expression in Tnfrsf11aCre−; Rosa26LSL-YFP (grey) and Tnfrsf11aCre+; Rosa26LSL-YFP (color for cell type indicated in gating strategy). (B) Immunostaining on cryosection from E14.5 Tnfrsf11aCre; Rosa26LSL-YFP embryo, with antibodies against YFP (green), Iba1 (red) and F4/80 (cyan). Scale bars represent 10 μm.
Fig. S6 Analysis of Cx3cr1-deficient mice. (A) Immunostaining on cryosection from E10.25 Cx3cr1gfp/+ embryo with antibodies against GFP (green), Iba1 (red) and F4/80 (cyan). (B, C, D) Flow cytometry analysis in Cx3cr1+/− and Cx3cr1−/− of F4/80+ macrophages from liver, brain, and limbs at E12.5 (B), liver, brain, limbs, and lung from at E14.5 (C) and liver, brain, limbs, lung, and epidermis from 12 week-old mice (D). Circles represent individual mice. Data are representative of n=7 independent experiments.
Fig. S7 Gene set enrichment analysis (GSEA) of cell type-specific expression patterns. (A) GSEA plots illustrating the relative enrichment of genes that we found statistically significantly upregulated (see Methods) in the comparison of macrophage vs. pMac (top row), macrophage vs. EMP (middle row), or pMac vs. EMP (bottom row). The black bars in the middle of each plot indicate the transcripts which are in each respective lists. The order in which transcripts were input into the analysis was defined by the relative expression change (logarithmic fold change) in macrophage vs. pMac (left), macrophage vs. EMP (center), or pMac vs. EMP (right), respectively. (B) The GSEA results from panel A are summarized here by the normalized enrichment score (NES) of each analysis. The colored squares indicate the two cell types compared with the color code as defined at the bottom right of this figure. Asterisks adjacent to bars indicate significance (FDR-corrected p-value); *: q≤0.05, **: q≤0.01, ***: q≤0.001. (C) Comparison of transcripts upregulated in macrophage vs. pMac, macrophage vs. EMP, and pMac vs. EMP, and the core macrophage signature extracted from Gautier et al. (2012). After translating gene denominators to Ensembl transcript IDs, Gautier's list matches 179 transcripts, only 27 of which occur in one of the lists of upregulated genes in this study. (D) GSEA results of Gautier's core macrophage signature genes in the comparisons of macrophage vs. pMac, macrophage vs. EMP, or pMac vs. EMP from this study (same gene rankings used in panels A and B). In all three comparisons, the core macrophage signature is enriched significantly (q≤0.01).
Fig. S8 LOLA enrichment analysis and enhancer regions. (A) Results of a LOLA enrichment analysis of promoter-adjacent regions (TSS +/− 20kb) of upregulated genes during transition from EMPs to pMacs (upper panel) or EMPs to early macrophages (E10.25-E10.5; lower panel). Each dot represents one single ChIP-seq experiment with the size relative to the quantity of enrichment (log odds ratio) and colors indicating the cell type used in the respective experiment. The key below the plots denominates the color coding. The numbers (“x/y”) given behind the cell type specify the number of enriched (x) out of all available datasets (y) from the respective cell type. Shown are all transcription factors of the genomic binding locations, which are significantly enriched (adjusted p-value <0.001, Benjamini Yekutieli correction) in at least one dataset in either comparison. All transcription factors shown are expressed in at least one sample examined, with the exception of Gcgr and Maff (greyed out and marked with an asterisk). (B) Heat map representation of the expression of the transcription factors shown in (A) in EMPs, pMacs and early macrophages (E10.25-E10.5) (C) Genome browser tracks showing ChIP-seq signals for the indicated transcription factors at the Emr1, Cx3cr1, Mrc1, Thbs1, Gata1 and Myod1 loci. The tracks display ChIP-seq data from macrophage populations, except for Maf (T cells), and Irf8 (dendritic cells).
Fig. S9 Coordinated changes of gene expression identified during the specification of tissue-resident macrophages. (A) Top panel shows an unsupervised, low-dimensional projection via principal component analysis of the RNA-seq data in this study. The color of each dot indicates the tissue the sample was taken from. Bottom panels show only the macrophage datasets from one tissue at a time (gray dots). The mean of the coordinates at each time point was calculated and indicated as a colored dot (with the same color code as above). Consecutive time points were then connected with arrows to visualize the differentiation trajectory described by the transcriptional profiles of these samples. (B) Line plots illustrating average expression of tissue-specific gene signatures in macrophages over time. Each panel corresponds to one list of differentially upregulated tissue-specific genes (Table S3) and each line to the average of all macrophage samples of one tissue at the given time. Shaded areas indicate +/− standard deviation. (C) Heatmap showing the expression levels of all RNA-seq datasets across all transcript differentially upregulated in any tissue in either stage (early embryonic, fetal, postnatal). Expression values have been scaled as z-scores per row, resulting in a color scheme in which red values represent highly expressed and blue values represent lowly expressed transcripts. Hierarchical Ward clustering with Euclidean distance was used to arrange the samples and genes and the resulting dendrogram was cut at equal height into seven (column-wise) or ten (row-wise) clusters to highlight the strongest clusters of genes and samples more clearly. The color bars on top of the heatmap indicate the time, cell type, and tissue of each sample.
Fig. S10 Gene set enrichment analysis (GSEA) of tissue-specific expression patterns in adult macrophages. (A) GSEA plots illustrating the relative enrichment of genes that were found to be statistically significantly upregulated (see Methods) in the comparison to Langerhans cells, microglia, kidney macrophages, Kupffer cells, and alveolar macrophages (from left to right) in comparison to the respective other adult macrophage populations. The black bars in the middle of each plot indicate the transcripts, which are in each respective lists. The order in which transcripts were input into the analysis was defined by the relative expression change (logarithmic fold change) in macrophages in the corresponding tissue (e.g. head/brain for microglia) compared to other macrophages at the same developmental stage (from top to bottom: early, fetal, or postnatal). The colored hashes (#) beneath each plot link to the bars in panel B. (B) The GSEA results from (A) are summarized here by the normalized enrichment score (NES) of each analysis. The colored squares underneath the bar plots indicate the tissue, cell type, and developmental stage of the sample for which enrichment is shown, following the color code as used in panel C. The bars highlighted in color correspond to the plots shown in panel A. Additionally, these and other bars from samples from a tissue related to the signature at hand are indicated by shading. Asterisks adjacent to bars indicate significance (FDR-corrected p-value); *: q≤0.05, **: q≤0.01, ***: q≤0.001. (C) GSEA results against externally defined gene signatures extracted from Gautier et al. (2012), Gorgani et al. (2008), and Lavin et al. (2014). The same color code, shading, and annotations are used as in panel B. (D) Hierarchical clustering of differentially up-regulated genes (2-fold change, adj. P-value < 0.05, BH-correction) in post-natal (P2-P21) brain, liver, kidney, epidermis or lung macrophages comparing one population vs. the others (see also Table S3). Each sample represents the mean of at least two biological replicates and two technical replicates, except for E14.5 liver macrophages and P8 and P21 lung macrophages, which consist of one biological replicate and two technical replicates. (E) Heatmap representation of the expression of Id1, Id3, Sall1, and Sall3 in EMPs and pMacs from bulk RNA-seq data.
Fig. S11 scRNA-seq analysis of specification of tissue-resident macrophages. (A) Workflow for overlaying transcription factor co-expression and tissue macrophage-specific signatures onto the tSNE plots. (B) Normalized expression of Kit, Stab1, Maf, and Cx3cr1 within EMPs, pMacs, and macrophages. (C) Normalized expression of Id1, Id3, and Sall3 within EMPs, pMacs, and macrophages. (D) tSNE plots showing expression of Id1, Id3, and Sall3. (E) Heatmap depicting enrichment of tissue macrophage-specific signatures as in Fig. 3F or differentiation signatures as in Fig. 1E in pMacs on gene clusters defined by multimodal gene expression with subsequent hierarchical clustering of scRNA-seq data.
Fig. S12 Role of Id3 in Kupffer cell development (A) Relative expression of Id3 and Nr3h1 transcript by qRT-PCR (normalized to GAPDH) in sorted Kupffer cells and hepatocytes of C57BL/6 mice n=3. (B) Immunostaining with antibodies against YFP (green), ID3 (red) and F4/80 (cyan) on cryosection from adult (4 week-old) liver (upper panel) or brain (lower panel) from a pulse labeled Csf1rMeriCreMer; Rosa26LSL-YFP mouse (OH-TAM at E8.5). Nuclei were counterstained with DAPI (white). Scale bar represents 10 μm. (C) Immunostaining with antibodies against CD31 and F4/80 on liver cryosections from 4 week-old Id3−/− and Id3+/− mice. The figure displays isovolume-rendered images. Scale bars represent 150 μm for the overview of the adult tissue and 50 μm for insets. Data are representative of 5 adult mice. (D) CD31+ area quantification on liver sections from Id3+/− (n=3) and Id3−/− (n=4) 4 week-old mice (left panels) or from Tnfrsf11aCre+; Id3+/f (n=6) and Tnfrsf11aCre+; Id3f/f (n=6) 2 week-old mice (right panels). (E) Unsupervised hierarchical clustering on whole transcriptome from Id3+/− and Id3−/− Kupffer cells (Distance metric: Euclidian, linkage rule: Ward’s, number of genes: 18882) (F) Significantly enriched Gene Ontology (GO) terms identified for 2-fold up-regulated (left table) or down-regulated (right table) genes in Id3−/− vs. Id3+/− Kupffer cells enriched by a t-test (P<0.05; FDR<0.05). GO analysis was performed using GeneSpring. GO terms are depicted and raked by corrected P-value (FDR false discovery rate corrected for multiple testing).
Fig. S13 Heat map representation of the normalized counts (log2) reads of S100a mRNA found in our dataset.
Specification of tissue-resident macrophages.
Erythro-myeloid progenitors (EMP) from the yolk sac colonize the fetal liver and give rise to macrophage precursors (pMacs) that acquire a core macrophage transcriptional program and colonize the embryo from E9.5 in a Cx3cr1-dependent manner (green arrows). Specification of F4/80+ resident macrophages (brown arrows), starting from E10.25, is initiated by the expression of tissue-specific transcriptional regulators. Id3 (red) is important for Kupffer cell development. Transcription factors noted in blue have been shown to be important for the differentiation or the maintenance of the corresponding macrophage subsets.
Summary.
Introduction
Embryonic development and tissue homeostasis depend on cooperation between specialized cell types. Resident macrophages are professional phagocytes that survey their surroundings, eliminate unfit cells, microorganisms and metabolic waste, and produce a large range of bioactive molecules and growth factors. Resident macrophages also serve tissue-specific purposes; for example, microglia in the central nervous system support neuronal circuit development, Kupffer cells scavenge blood particles and dying red blood cells in the liver, alveolar macrophages uptake surfactant and remove airborne pollutants and microbes from the airways. Resident macrophage diversity in adult mice is reflected in tissue-specific gene expression profiles, which may be due to responses to specific cues from their microenvironment, different developmental processes, and the contribution of distinct progenitors cell types. Altogether, the mechanisms responsible for the generation of tissue-resident macrophage diversity remain unclear.
Rationale
Tissue-resident macrophages originate - at least in part - from mesodermal Erythro-Myeloid Progenitors (EMP) from the yolk sac, which invade the embryo proper at the onset of organogenesis. These tissue-resident macrophages are also self-maintained in postnatal tissues, independently of definitive hematopoietic stem cells (HSCs) in a steady state. We therefore hypothesized that resident macrophages represent a ‘founding’ cell type within most organ anlagen. In this model the generation of macrophage diversity, as observed in the tissues of postnatal mice, may be integral to organogenesis.
Results
To test this hypothesis and explore the molecular basis of macrophage diversity in mammals, we performed a spatio-temporal analysis of macrophage development in mice, from embryonic day E9 to three weeks after birth. Unbiased single cell RNA-seq analysis of CD45+ cells, combined with RNA-seq analyses of sorted cell populations, genetic fate-mapping, and in situ analyses, revealed that EMP give rise to a population of pre-macrophages (pMacs), which colonize the whole embryo from E9.5, as they acquire a core macrophage differentiation program that includes pattern recognition, scavengers, and cytokine receptors. The chemokine receptor Cx3cr1 is upregulated in pMacs and important for embryo colonization, which is delayed in Cx3cr1-deficient embryos. Fate-mapping of pMac using a Tnfrsf11a–Cre reporter labels homogeneously fetal and adult tissue-resident macrophages, but not HSCs and their progeny. Transcriptional regulators that identify postnatal tissue-resident macrophages in brain, liver, kidney, skin and lung were specifically upregulated immediately following colonization. These dynamic changes mark the onset of diversification into adult macrophages. We identified Id3 as a Kupffer cell-specific transcriptional regulator. Deletion of Id3 in pMacs resulted in Kupffer cell deficiency but did not affect development of microglia and kidney macrophages.
Conclusion
Our study shows that EMP-derived precursors colonize embryonic tissues and simultaneously acquire a full core macrophage program. This is followed by their diversification into tissue-specific macrophages during organogenesis, likely via the expression of distinct sets of transcriptional regulators. These results indicate that differentiation of tissue-resident macrophages is an integral part of organogenesis, and identify a spatio-temporal molecular road map for the generation of macrophage diversity in vivo. Our findings provide a conceptual framework to analyze and understand the consequence(s) of genetic variation for macrophage contribution to development, homeostasis, and disease pathogenesis in different tissues, and will support efforts to differentiate specialized macrophages in vitro.
Acknowledgement
This work was supported by the National Cancer Institute of the US National Institutes of Health (P30CA008748), and by Wellcome Trust investigator (WT101853MA), and European Research Council investigator awards (2010-StG-261299) to FG. EM is supported by an EMBO long-term Fellowship (ALTF 530-2015). FH is supported by a DFG postdoctoral fellowship (HA 7723/1-1). JK is supported by a DOC Fellowship of the Austrian Academy of Sciences. CB is supported by a New Frontiers Group award of the Austrian Academy of Sciences. MB and JLS are members of the Excellence Cluster ImmunoSensation. MB is supported by a DFG grant (BE 4427/3-1), JLS is supported by DFG grants SFB 704, SFB 645, INST 217/575-1, INST 217/576-1 and INST 217/577-1. The authors are indebted to R. Benezra, Memorial Sloan Kettering Cancer Center, New York for the Id3−/− and Id3f/f strains. We thank the Biomedical Sequencing Facility at CeMM and the MSKCC Integrated Genomics and Bioinformatics Cores for assistance with next generation sequencing. Sequencing datasets described in this work have been submitted to the Gene Expression Omnibus (GEO) repository (GEO accession number: GSE81774).
Footnotes
Additionally, a genome browser track hub, as well as additional supplementary data are available at the following URL: http://macrophage-development.computational-epigenetics.org/.
Author contribution. FG, EM designed the study and wrote the manuscript. EM, IB performed cell sorting, flow cytometry, fate-mapping, immunostaining experiments and RNA-seq analysis on Id3-deficicient and control Kupffer cells. LC and CEJG helped with analysis of Tnfrsf11aCre+; Rosa26LSL-YFP mice. EGP assisted with the design of cell sorting experiments. MF designed and prepared bulk RNA-seq libraries. FH, JK, CB performed primary and differential analysis of the bulk RNA-seq data. KH and MB generated single-cell RNA-seq libraries and performed single-cell RNA-seq, PG, MB and JLS analyzed single cell RNA-seq data. All authors contributed to the manuscript.
References and Notes
- 1.McGrath KE, et al. Distinct Sources of Hematopoietic Progenitors Emerge before HSCs and Provide Functional Blood Cells in the Mammalian Embryo. Cell reports. 2015;11:1892–1904. doi: 10.1016/j.celrep.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gomez Perdiguero E, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hashimoto D, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yona S, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schulz C, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 6.Davies LC, et al. A quantifiable proliferative burst of tissue macrophages restores homeostatic macrophage populations after acute inflammation. European journal of immunology. 2011;41:2155–2164. doi: 10.1002/eji.201141817. [DOI] [PubMed] [Google Scholar]
- 7.Lavin Y, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gosselin D, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157:832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosas M, et al. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science. 2014;344:645–648. doi: 10.1126/science.1251414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheng J, Ruedl C, Karjalainen K. Most Tissue-Resident Macrophages Except Microglia Are Derived from Fetal Hematopoietic Stem Cells. Immunity. 2015;43:382–393. doi: 10.1016/j.immuni.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 12.Hoeffel G, et al. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity. 2015;42:665–678. doi: 10.1016/j.immuni.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginhoux F, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gautier EL, et al. Gata6 regulates aspartoacylase expression in resident peritoneal macrophages and controls their survival. The Journal of experimental medicine. 2014;211:1525–1531. doi: 10.1084/jem.20140570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fainaru O, et al. Runx3 regulates mouse TGF-beta-mediated dendritic cell function and its absence results in airway inflammation. The EMBO journal. 2004;23:969–979. doi: 10.1038/sj.emboj.7600085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.N A-G, et al. The nuclear receptor LXRalpha controls the functional specialization of splenic macrophages. Nature immunology. 2013;14:831–839. doi: 10.1038/ni.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohyama M, et al. Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature. 2009;457:318–321. doi: 10.1038/nature07472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gautier EL, et al. Systemic analysis of PPARgamma in mouse macrophage populations reveals marked diversity in expression with critical roles in resolution of inflammation and airway immunity. J Immunol. 2012;189:2614–2624. doi: 10.4049/jimmunol.1200495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertrand JY, et al. Characterization of purified intraembryonic hematopoietic stem cells as a tool to define their site of origin. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:134–139. doi: 10.1073/pnas.0402270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bock C, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeda K, et al. Wnt5a-Ror2 signaling between osteoblast-lineage cells and osteoclast precursors enhances osteoclastogenesis. Nature medicine. 2012;18:405–412. doi: 10.1038/nm.2653. [DOI] [PubMed] [Google Scholar]
- 23.Bertrand JY, et al. Three pathways to mature macrophages in the early mouse yolk sac. Blood. 2005;106:3004–3011. doi: 10.1182/blood-2005-02-0461. [DOI] [PubMed] [Google Scholar]
- 24.Lionakis MS, et al. CX3CR1-dependent renal macrophage survival promotes Candida control and host survival. The Journal of clinical investigation. 2013;123:5035–5051. doi: 10.1172/JCI71307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gautier EL, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nature immunology. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheffield NC, Bock C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorgani NN, Ma Y, Clark HF. Gene signatures reflect the marked heterogeneity of tissue-resident macrophages. Immunology and cell biology. 2008;86:246–254. doi: 10.1038/sj.icb.7100131. [DOI] [PubMed] [Google Scholar]
- 29.Chorro L, et al. Langerhans cell (LC) proliferation mediates neonatal development, homeostasis, and inflammation-associated expansion of the epidermal LC network. The Journal of experimental medicine. 2009;206:3089–3100. doi: 10.1084/jem.20091586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoekstra M, Kruijt JK, Van Eck M, Van Berkel TJ. Specific gene expression of ATP-binding cassette transporters and nuclear hormone receptors in rat liver parenchymal, endothelial, and Kupffer cells. The Journal of biological chemistry. 2003;278:25448–25453. doi: 10.1074/jbc.M301189200. [DOI] [PubMed] [Google Scholar]
- 31.Schneider C, et al. Induction of the nuclear receptor PPAR-gamma by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nature immunology. 2014;15:1026–1037. doi: 10.1038/ni.3005. [DOI] [PubMed] [Google Scholar]
- 32.Jux B, Kadow S, Esser C. Langerhans cell maturation and contact hypersensitivity are impaired in aryl hydrocarbon receptor-null mice. J Immunol. 2009;182:6709–6717. doi: 10.4049/jimmunol.0713344. [DOI] [PubMed] [Google Scholar]
- 33.Jen Y, Manova K, Benezra R. Expression patterns of Id1, Id2, and Id3 are highly related but distinct from that of Id4 during mouse embryogenesis. Developmental dynamics : an official publication of the American Association of Anatomists. 1996;207:235–252. doi: 10.1002/(SICI)1097-0177(199611)207:3<235::AID-AJA1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 34.Lyden D, et al. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 1999;401:670–677. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 35.Qian BZ, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Q, et al. Developmental ablation of Id1 and Id3 genes in the vasculature leads to postnatal cardiac phenotypes. Developmental biology. 2011;349:53–64. doi: 10.1016/j.ydbio.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Z, et al. Modeling Sjogren's syndrome with Id3 conditional knockout mice. Immunol Lett. 2011;135:34–42. doi: 10.1016/j.imlet.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng L, et al. A novel mouse model of inflammatory bowel disease links mammalian target of rapamycin-dependent hyperproliferation of colonic epithelium to inflammation-associated tumorigenesis. The American journal of pathology. 2010;176:952–967. doi: 10.2353/ajpath.2010.090622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sreeramkumar V, et al. Neutrophils scan for activated platelets to initiate inflammation. Science. 2014;346:1234–1238. doi: 10.1126/science.1256478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engstrom PG, et al. Systematic evaluation of spliced alignment programs for RNA-seq data. Nature methods. 2013;10:1185–1191. doi: 10.1038/nmeth.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, et al. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep. 2015 doi: 10.15252/embr.201540946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nature protocols. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 44.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glaus P, Honkela A, Rattray M. Identifying differentially expressed transcripts from RNA-seq data with biological variation. Bioinformatics. 2012;28:1721–1728. doi: 10.1093/bioinformatics/bts260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 48.Sanchez-Castillo M, et al. CODEX: a next-generation sequencing experiment database for the haematopoietic and embryonic stem cell communities. Nucleic acids research. 2015;43:D1117–1123. doi: 10.1093/nar/gku895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gautier EL, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nature immunology. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reimand J, Arak T, Vilo J. g:Profiler--a web server for functional interpretation of gene lists (2011 update) Nucleic acids research. 2011;39:W307–315. doi: 10.1093/nar/gkr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nature biotechnology. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature biotechnology. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Charrad M, Ghazzali N, Boiteau V, Niknafs A. Nbclust: An R Package for Determining the Relevant Number of Clusters in a Data Set. J Stat Softw. 2014;61:1–36. [Google Scholar]
- 56.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1: Quality control and analysis of bulk RNA-seq. (A) Number of reliably detected transcripts (covered with a least 25 reads) in each library. The colored circles underneath indicate the batch (sequencing flow cell), time, cell type, and tissue of each sample with the color code as given below panel B. (B) Hierarchical clustering with Euclidean distance and complete linkage of all samples based on their RNA-seq gene expression profiles. The colored circles underneath the cluster dendrogram indicate the batch (sequencing flow cell), time, cell type, and tissue of each sample with the color code as given below. (C) Scorecard analysis of differentially up-regulated genes (DESeq2 Wald test, adjusted p-value<0.05, BH-correction) in early macrophages (E10.25, E10.5) in comparison to EMPs. The table shows the relative enrichment of differentially upregulated genes in macrophages across cell types and tissues (y-axis) and developmental time points (x-axis, from E9 to P21). See Methods for details of the scorecard. (D) Principal component analysis (PCA) plot of EMPs (red, E9-E10.25), pMacs (yellow, E9.5-E10.25) and macrophages (purple, E10.25-E10.5) from the head, caudal, fetal liver (FL) and yolk sac (YS). The shape of each dot indicates the tissue the sample was taken from. The first and second principal component explain 18.9% and 11.1% of the entire variation in the data, respectively.
Fig. S2: Quality control and analysis of single-cell RNA-seq. (A) Workflow of the MARS-seq single cell data analysis. (B) Mean-variability plot shows average expression and dispersion for each gene. This analysis was used to determine highly variable genes (labeled by gene symbol). These 138 highly variable genes were used to perform a dimensionality reduction of the single-cell data by a principal component analysis. (C) The highest gene loadings in the first and second principal component from the PCA of 408 high quality cells, colored by batch association, showed even distribution of cells among the PCA plot based on the 138 most highly variable genes. (D) Heatmap of 138 highly variable genes among single-cell clusters as defined by DBScan clustering. (E) Optimal cluster number was identified by calculation of diverse indices for determining the best clustering scheme using the NbClust R package. (F) PCA plot of 408 single cells colored by cluster association. Clusters were defined by PCA + DBScan clustering. (G) Kinetic diagram shows the pseudotemporal ordering of single cells as determined by Monocle 2. Dots indicate individual cells and are colored according to the cluster association as in (F). Black line indicates the progression of single cells over developmental pseudotime.
Fig. S3 Expression of surface markers on EMP-derived cells during development. (A) Flow cytometry analysis of E10.25 Csf1rMeriCreMer; Rosa26LSL-YFP (OH-TAM at E8.5) tissues showing expression of Il4ra, Il13ra1, Tnfr2, Ifngr, CD16.2, CD64, Tim4, and CD206 on YFP+ Kit+ progenitors (gray), pMacs (blue) and macrophages (orange). Histograms represent the fluorescence intensity for each antibody in each cell subset. Data are representative of n=4 independent experiments with 4-6 embryos per marker. (B,C) Flow cytometry analysis of Csf1rMeriCreMer; Rosa26LSL-YFP (OH-TAM at E8.5) liver, brain, lung, and skin F4/80+ cells from E14.5 embryos showing expression of Il4ra, Il13ra1, Tnfr2, Ifngr, Dectin-1, CD64, Tim4, and CD206 (black dotted on whole population and green on YFP+ cells). Gray histograms show the fluorescence intensity of the FMO controls.
Fig. S4 Expression of the core macrophage program on EMP-derived cells. (A) Immunostaining on cryosections from E10.25 Csf1rMeriCreMer; Rosa26LSL-YFP embryos, pulse-labeled with OH-TAM at E8.5 with antibodies against YFP (green), Iba1 (red/cyan), and CD206 (red), Ifngr (red), Tnfr2 (red), Dectin-1 (red), Trem2 (red), CD16/32 (red), Granulin (Grn, red), or F4/80 (cyan). Scale bars represent 10 μm. Data are representative of n=3 embryos for each marker.
(B) Whole mount immunostaining of E9.5 Csf1riCre; Rosa26LSL-YFP embryo labeled with antibodies against YFP (green), Iba1 (red), F4/80 (cyan) and DAPI (white). Scale bars represent 10 μm. Data are representative of n=3 embryos. (C) Immunostaining on cryosections from E10.25 Csf1rMeriCreMer; Rosa26LSL-YFP embryo liver, pulse-labeled with OH-TAM at E8.5 with antibodies against YFP (green), Dectin-1 (red) and Iba1 (cyan) (upper panel) or YFP (green), Kit (red) and F4/80 (cyan) (lower panel) Scale bars represent 15 μm. (D, E) Immunostaining on cryosection from E14.5 (D) and E18.5 (E) Csf1rMeriCreMer; Rosa26LSL-YFP mouse embryos stained with antibodies against YFP (green), Iba1 (red), and F4/80 (cyan). (F) Immunostaining on cryosection from E14.5 Csf1rMeriCreMer; Rosa26LSL-YFP mouse embryo stained with antibodies against YFP (green), Granulin (Grn, red) and F4/80 (cyan). Scale bars represent 10 μm.
Fig. S5 Analysis of Tnfrsf11aCre; Rosa26LSL-YFP mice. (A) Gating strategy for Tnfrsf11aCre; Rosa26LSL-YFP embryos in E10.25 YS pMacs (Kit− CD45+ F4/80− CD11blow Gr1− Ter119−; green) and macrophages (CD45+F4/80+CD11blo; blue) (upper panel), and in E14.5 fetal liver LT-HSCs (Lin−Kit+Sca1+CD150+CD48−; orange), ST-HSCs (Lin−Kit+Sca1+CD150−CD48−; blue) and MPPs (Lin−Kit+Sca1+CD150−CD48+; purple) (lower panel). Histograms represent YFP expression in Tnfrsf11aCre−; Rosa26LSL-YFP (grey) and Tnfrsf11aCre+; Rosa26LSL-YFP (color for cell type indicated in gating strategy). (B) Immunostaining on cryosection from E14.5 Tnfrsf11aCre; Rosa26LSL-YFP embryo, with antibodies against YFP (green), Iba1 (red) and F4/80 (cyan). Scale bars represent 10 μm.
Fig. S6 Analysis of Cx3cr1-deficient mice. (A) Immunostaining on cryosection from E10.25 Cx3cr1gfp/+ embryo with antibodies against GFP (green), Iba1 (red) and F4/80 (cyan). (B, C, D) Flow cytometry analysis in Cx3cr1+/− and Cx3cr1−/− of F4/80+ macrophages from liver, brain, and limbs at E12.5 (B), liver, brain, limbs, and lung from at E14.5 (C) and liver, brain, limbs, lung, and epidermis from 12 week-old mice (D). Circles represent individual mice. Data are representative of n=7 independent experiments.
Fig. S7 Gene set enrichment analysis (GSEA) of cell type-specific expression patterns. (A) GSEA plots illustrating the relative enrichment of genes that we found statistically significantly upregulated (see Methods) in the comparison of macrophage vs. pMac (top row), macrophage vs. EMP (middle row), or pMac vs. EMP (bottom row). The black bars in the middle of each plot indicate the transcripts which are in each respective lists. The order in which transcripts were input into the analysis was defined by the relative expression change (logarithmic fold change) in macrophage vs. pMac (left), macrophage vs. EMP (center), or pMac vs. EMP (right), respectively. (B) The GSEA results from panel A are summarized here by the normalized enrichment score (NES) of each analysis. The colored squares indicate the two cell types compared with the color code as defined at the bottom right of this figure. Asterisks adjacent to bars indicate significance (FDR-corrected p-value); *: q≤0.05, **: q≤0.01, ***: q≤0.001. (C) Comparison of transcripts upregulated in macrophage vs. pMac, macrophage vs. EMP, and pMac vs. EMP, and the core macrophage signature extracted from Gautier et al. (2012). After translating gene denominators to Ensembl transcript IDs, Gautier's list matches 179 transcripts, only 27 of which occur in one of the lists of upregulated genes in this study. (D) GSEA results of Gautier's core macrophage signature genes in the comparisons of macrophage vs. pMac, macrophage vs. EMP, or pMac vs. EMP from this study (same gene rankings used in panels A and B). In all three comparisons, the core macrophage signature is enriched significantly (q≤0.01).
Fig. S8 LOLA enrichment analysis and enhancer regions. (A) Results of a LOLA enrichment analysis of promoter-adjacent regions (TSS +/− 20kb) of upregulated genes during transition from EMPs to pMacs (upper panel) or EMPs to early macrophages (E10.25-E10.5; lower panel). Each dot represents one single ChIP-seq experiment with the size relative to the quantity of enrichment (log odds ratio) and colors indicating the cell type used in the respective experiment. The key below the plots denominates the color coding. The numbers (“x/y”) given behind the cell type specify the number of enriched (x) out of all available datasets (y) from the respective cell type. Shown are all transcription factors of the genomic binding locations, which are significantly enriched (adjusted p-value <0.001, Benjamini Yekutieli correction) in at least one dataset in either comparison. All transcription factors shown are expressed in at least one sample examined, with the exception of Gcgr and Maff (greyed out and marked with an asterisk). (B) Heat map representation of the expression of the transcription factors shown in (A) in EMPs, pMacs and early macrophages (E10.25-E10.5) (C) Genome browser tracks showing ChIP-seq signals for the indicated transcription factors at the Emr1, Cx3cr1, Mrc1, Thbs1, Gata1 and Myod1 loci. The tracks display ChIP-seq data from macrophage populations, except for Maf (T cells), and Irf8 (dendritic cells).
Fig. S9 Coordinated changes of gene expression identified during the specification of tissue-resident macrophages. (A) Top panel shows an unsupervised, low-dimensional projection via principal component analysis of the RNA-seq data in this study. The color of each dot indicates the tissue the sample was taken from. Bottom panels show only the macrophage datasets from one tissue at a time (gray dots). The mean of the coordinates at each time point was calculated and indicated as a colored dot (with the same color code as above). Consecutive time points were then connected with arrows to visualize the differentiation trajectory described by the transcriptional profiles of these samples. (B) Line plots illustrating average expression of tissue-specific gene signatures in macrophages over time. Each panel corresponds to one list of differentially upregulated tissue-specific genes (Table S3) and each line to the average of all macrophage samples of one tissue at the given time. Shaded areas indicate +/− standard deviation. (C) Heatmap showing the expression levels of all RNA-seq datasets across all transcript differentially upregulated in any tissue in either stage (early embryonic, fetal, postnatal). Expression values have been scaled as z-scores per row, resulting in a color scheme in which red values represent highly expressed and blue values represent lowly expressed transcripts. Hierarchical Ward clustering with Euclidean distance was used to arrange the samples and genes and the resulting dendrogram was cut at equal height into seven (column-wise) or ten (row-wise) clusters to highlight the strongest clusters of genes and samples more clearly. The color bars on top of the heatmap indicate the time, cell type, and tissue of each sample.
Fig. S10 Gene set enrichment analysis (GSEA) of tissue-specific expression patterns in adult macrophages. (A) GSEA plots illustrating the relative enrichment of genes that were found to be statistically significantly upregulated (see Methods) in the comparison to Langerhans cells, microglia, kidney macrophages, Kupffer cells, and alveolar macrophages (from left to right) in comparison to the respective other adult macrophage populations. The black bars in the middle of each plot indicate the transcripts, which are in each respective lists. The order in which transcripts were input into the analysis was defined by the relative expression change (logarithmic fold change) in macrophages in the corresponding tissue (e.g. head/brain for microglia) compared to other macrophages at the same developmental stage (from top to bottom: early, fetal, or postnatal). The colored hashes (#) beneath each plot link to the bars in panel B. (B) The GSEA results from (A) are summarized here by the normalized enrichment score (NES) of each analysis. The colored squares underneath the bar plots indicate the tissue, cell type, and developmental stage of the sample for which enrichment is shown, following the color code as used in panel C. The bars highlighted in color correspond to the plots shown in panel A. Additionally, these and other bars from samples from a tissue related to the signature at hand are indicated by shading. Asterisks adjacent to bars indicate significance (FDR-corrected p-value); *: q≤0.05, **: q≤0.01, ***: q≤0.001. (C) GSEA results against externally defined gene signatures extracted from Gautier et al. (2012), Gorgani et al. (2008), and Lavin et al. (2014). The same color code, shading, and annotations are used as in panel B. (D) Hierarchical clustering of differentially up-regulated genes (2-fold change, adj. P-value < 0.05, BH-correction) in post-natal (P2-P21) brain, liver, kidney, epidermis or lung macrophages comparing one population vs. the others (see also Table S3). Each sample represents the mean of at least two biological replicates and two technical replicates, except for E14.5 liver macrophages and P8 and P21 lung macrophages, which consist of one biological replicate and two technical replicates. (E) Heatmap representation of the expression of Id1, Id3, Sall1, and Sall3 in EMPs and pMacs from bulk RNA-seq data.
Fig. S11 scRNA-seq analysis of specification of tissue-resident macrophages. (A) Workflow for overlaying transcription factor co-expression and tissue macrophage-specific signatures onto the tSNE plots. (B) Normalized expression of Kit, Stab1, Maf, and Cx3cr1 within EMPs, pMacs, and macrophages. (C) Normalized expression of Id1, Id3, and Sall3 within EMPs, pMacs, and macrophages. (D) tSNE plots showing expression of Id1, Id3, and Sall3. (E) Heatmap depicting enrichment of tissue macrophage-specific signatures as in Fig. 3F or differentiation signatures as in Fig. 1E in pMacs on gene clusters defined by multimodal gene expression with subsequent hierarchical clustering of scRNA-seq data.
Fig. S12 Role of Id3 in Kupffer cell development (A) Relative expression of Id3 and Nr3h1 transcript by qRT-PCR (normalized to GAPDH) in sorted Kupffer cells and hepatocytes of C57BL/6 mice n=3. (B) Immunostaining with antibodies against YFP (green), ID3 (red) and F4/80 (cyan) on cryosection from adult (4 week-old) liver (upper panel) or brain (lower panel) from a pulse labeled Csf1rMeriCreMer; Rosa26LSL-YFP mouse (OH-TAM at E8.5). Nuclei were counterstained with DAPI (white). Scale bar represents 10 μm. (C) Immunostaining with antibodies against CD31 and F4/80 on liver cryosections from 4 week-old Id3−/− and Id3+/− mice. The figure displays isovolume-rendered images. Scale bars represent 150 μm for the overview of the adult tissue and 50 μm for insets. Data are representative of 5 adult mice. (D) CD31+ area quantification on liver sections from Id3+/− (n=3) and Id3−/− (n=4) 4 week-old mice (left panels) or from Tnfrsf11aCre+; Id3+/f (n=6) and Tnfrsf11aCre+; Id3f/f (n=6) 2 week-old mice (right panels). (E) Unsupervised hierarchical clustering on whole transcriptome from Id3+/− and Id3−/− Kupffer cells (Distance metric: Euclidian, linkage rule: Ward’s, number of genes: 18882) (F) Significantly enriched Gene Ontology (GO) terms identified for 2-fold up-regulated (left table) or down-regulated (right table) genes in Id3−/− vs. Id3+/− Kupffer cells enriched by a t-test (P<0.05; FDR<0.05). GO analysis was performed using GeneSpring. GO terms are depicted and raked by corrected P-value (FDR false discovery rate corrected for multiple testing).
Fig. S13 Heat map representation of the normalized counts (log2) reads of S100a mRNA found in our dataset.