ABSTRACT
Storm water runoff is a major source of pollution, and understanding the components of storm water discharge is essential to remediation efforts and proper assessment of risks to human and ecosystem health. In this study, culturable Escherichia coli and ampicillin-resistant E. coli levels were quantified and microbial source tracking (MST) markers (including markers for general Bacteroidales spp., human, ruminant/cow, gull, and dog) were detected in storm water outfalls and sites along the Humber River in Toronto, Ontario, Canada, and enumerated via endpoint PCR and quantitative PCR (qPCR). Additionally, chemical source tracking (CST) markers specific for human wastewater (caffeine, carbamazepine, codeine, cotinine, acetaminophen, and acesulfame) were quantified. Human and gull fecal sources were detected at all sites, although concentrations of the human fecal marker were higher, particularly in outfalls (mean outfall concentrations of 4.22 log10 copies, expressed as copy numbers [CN]/100 milliliters for human and 0.46 log10 CN/100 milliliters for gull). Higher concentrations of caffeine, acetaminophen, acesulfame, E. coli, and the human fecal marker were indicative of greater raw sewage contamination at several sites (maximum concentrations of 34,800 ng/liter, 5,120 ng/liter, 9,720 ng/liter, 5.26 log10 CFU/100 ml, and 7.65 log10 CN/100 ml, respectively). These results indicate pervasive sewage contamination at storm water outfalls and throughout the Humber River, with multiple lines of evidence identifying Black Creek and two storm water outfalls with prominent sewage cross-connection problems requiring remediation. Limited data are available on specific sources of pollution in storm water, though our results indicate the value of using both MST and CST methodologies to more reliably assess sewage contamination in impacted watersheds.
IMPORTANCE Storm water runoff is one of the most prominent non-point sources of biological and chemical contaminants which can potentially degrade water quality and pose risks to human and ecosystem health. Therefore, identifying fecal contamination in storm water runoff and outfalls is essential for remediation efforts to reduce risks to public health. This study employed multiple methods of identifying levels and sources of fecal contamination in both river and storm water outfall sites, evaluating the efficacy of using culture-based enumeration of E. coli, molecular methods of determining the source(s) of contamination, and CST markers as indicators of fecal contamination. The results identified pervasive human sewage contamination in storm water outfalls and throughout an urban watershed and highlight the utility of using both MST and CST to identify raw sewage contamination.
INTRODUCTION
Storm water runoff has been identified as one of the most prominent non-point sources of both biological and chemical contaminants, degrading water quality and posing risks to public health in impacted recreational waters (1–3). The high level of fecal contamination present in storm water runoff has been noted as one the leading causes for beach closures and advisories in the United States (4) and has been directly linked to disease outbreaks (5, 6). The major sources of human fecal contamination in storm water runoff are failing sewage infrastructures and cross-connections between sewage and storm water networks (7, 8). However, in addition to sewage contamination, storm water runoff can also carry other forms of animal waste, as well as a variety of pesticides and other chemical contaminants (2, 9, 10). Understanding the composition of the resultant runoff is therefore essential to tracking storm water pollution and assessing overall risk to public health.
While surface waters are monitored using concentrations of fecal indicator bacteria (FIB), such as Escherichia coli and enterococci (11, 12), the FIB paradigm is imperfect. Among other problems, the presence of FIB does not always correlate with the occurrence of pathogens, particularly viral or protozoan pathogens (1, 13, 14). Further, elevated concentrations of FIB do not give any indication of the source of fecal contamination, which can hinder remediation efforts. Inability to accurately identify the source of contamination can also lead to inaccurate decisions relating to public health, particularly as different sources of contamination can pose different risks to human health, with human sewage contamination generally posing the greatest risks (15). Other methods to identify the source of contamination, particularly with regard to human sewage, are often necessary to accurately guide remediation efforts.
A variety of methods have been used to indicate human sewage contamination, such as the use of culturable antibiotic-resistant strains of bacteria, which tend to be more prevalent in wastewater (16–20). Additionally, many field studies have utilized microbial source tracking (MST) methods such as host-associated molecular markers to identify multiple sources of contamination (e.g., human, dog, gull, cow) (21–29). Further, in recent years, efforts to characterize the human microbiome have started to reveal the predominant bacteria within the human gut and associated with human skin. Several of these species, such as Bacteroides thetaiotaomicron, B. dorei, Clostridium perfringens, Bifidobacterium adolescentis, and Faecalibacterium prausnitzii, have previously been used as microbial source tracking markers (30–33). However, other prevalent gut-associated bacteria, such as Eubacterium rectale and Ruminococcus bromii, as well as skin-associated bacteria, such as Staphylococcus epidermidis and Propionibacterium acnes, may also be useful markers for human-specific contamination (34–36). Finally, a variety of chemical source tracking (CST) markers have been identified as potential indicators of sewage contamination, including caffeine (37, 38), carbamazepine and other pharmaceuticals (38–40), cotinine (41), and chemical sweeteners (42).
In this study, multiple sampling sites throughout the Humber River watershed (Toronto, Ontario, Canada), including associated tributaries and storm water outfalls, were sampled to identify hot spots of fecal contamination using concentrations of culturable E. coli and ampicillin-resistant (Ampr) E. coli. To attempt to discriminate human and animal sources of the fecal contamination and potentially to identify sewage cross-connections in storm water outfalls, a suite of MST and CST markers were measured at all sampling sites. Additionally, the preliminary application of a quantitative PCR (qPCR) array with a variety of potential MST markers was explored to determine whether these potential markers would be useful in determining the source(s) of fecal contamination. Identification of the source(s) of elevated E. coli concentrations at Sunnyside Beach at the mouth of the Humber River is needed to guide remediation efforts to reduce beach postings and a beach Beneficial Use Impairment within the Toronto Area of Concern (AOC).
MATERIALS AND METHODS
Study area.
This study was conducted within the Humber River watershed in the Toronto AOC. The Humber River is a relatively large river (with the main branch extending 126 km), draining an area of 911 square kilometers into Lake Ontario. Land use in the Humber watershed is 54% rural, 33% urban, 13% urbanizing, and 32% natural cover, and the population in the watershed area in 2014 was 856,200 inhabitants (43). The only sewage treatment plant discharge into the Humber River occurs from two small plants (serving about 7,000 people) that discharge in the central branch of the Humber River above our R2 sampling site. The Humber watershed has been characterized by poor water quality, with contamination in some upper branches of the river historically attributed predominantly to livestock and agricultural contamination (44), while contamination in the lower portion of the river has been attributed to storm water and combined sewer overflows containing raw sewage (45). Sampling sites were selected to represent the major tributaries in the Humber River and those larger storm water outfalls in the lower watershed in proximity to Sunnyside Beach.
Sample collection.
Water samples were taken at both river sites (sites R1 to R7) and storm water outfall sites (S1 to S5) in the Humber River, as well as at the Black Creek tributary (site T1 and associated outfalls T2 and T3), from May to September 2014 (Table 1). Outfalls S1and S5 were not readily accessible and were sampled immediately below the outfall and were thus blended with river water. Two water samples were collected at the same time from each river and outfall site: a 500-ml sample collected in an autoclaved polypropylene bottle for E. coli enumeration and MST assays and an additional 100-ml sample collected in an amber glass bottle for chemical marker analysis (see below). All water samples were placed on ice and transported to the laboratory for processing within 6 h of collection. Additionally, 20 samples each of wastewater influent and effluent were collected from Toronto's four wastewater treatment plants (Ashbridges Bay, Highland Creek, Humber River, and North Toronto) on five separate days (8, 15, and 29 September and 6 and 20 October of 2014). These wastewater samples were used as a reference to compare the concentrations of MST and CST markers in ambient river and storm water samples with concentrations found in wastewater untreated influent and treated effluent.
TABLE 1.
List of sampling site locations and percent detection for endpoint MST markers
| Site name | Type | Locationa | GPS coordinates | Sampling events (n) | % marker |
|||
|---|---|---|---|---|---|---|---|---|
| Human | Ruminant | Gull | Dog | |||||
| R1 | River | Upper Humber (E) | 43°47′51.13″N, 79°34′52.17″W | 16 | 31 | 63 | 38 | 13 |
| R2 | River | Upper Humber (M) | 43°47′27.64″N, 79°35′39.45″W | 16 | 19 | 6 | 38 | 13 |
| R3 | River | Upper Humber (W) | 43°43′9.34″N, 79°32′35.40″W | 16 | 13 | 6 | 44 | 25 |
| R4 | River | Middle Humber | 43°40′43.10″N, 79°30′26.24″W | 16 | 31 | 25 | 81 | 19 |
| T1 | Creek | Black Creek | 43°40′32.25″N, 79°29′48.38″W | 16 | 88 | 31 | 81 | 13 |
| R5b | River | Middle Humber | 43°38′30.39″N, 79°29′25.57″W | 7 | 29 | 29 | 71 | 14 |
| R6 | River | Lower Humber | 43°39′6.80″N, 79°29′29.61″W | 16 | 44 | 25 | 81 | 38 |
| R7 | River | Humber mouth | 43°37′55.32″N, 79°28′15.30″W | 15 | 47 | 13 | 87 | 27 |
| T2 | Outfall | Black Creek | 43°40′30.85″N, 79°29′20.63″W | 14 | 71 | 21 | 29 | 21 |
| T3 | Outfall | Black Creek | 43°40′32.38″N, 79°29′13.65″W | 14 | 57 | 0 | 21 | 7 |
| S1 | Outfall | Middle Humber | 43°39′42.69″N, 79°30′13.69″W | 13 | 69 | 46 | 31 | 15 |
| S2 | Outfall | Lower Humber | 43°39′6.00″N 79°29′28.01″W | 14 | 93 | 21 | 21 | 7 |
| S3 | Outfall | Middle Humber | 43°39′25.63″N, 79°29′58.83″W | 9 | 56 | 0 | 22 | 22 |
| S4 | Outfall | Lower Humber | 43°38′17.49″N, 79°28′39.58″W | 9 | 89 | 22 | 44 | 44 |
| S5 | Outfall | Lower Humber | 43°38′3.18″N, 79°28′28.22″W | 14 | 43 | 14 | 64 | 7 |
E, east; M, middle; W, west.
R5 had a smaller sample size than other sampling sites. Rain events and other seasonal influences were missed at site R5 that were sampled at other sites, precluding simple comparisons among other sites.
E. coli enumeration.
For enumeration of both culturable E. coli and Ampr E. coli, water samples were filtered (using 0.45-μm-pore-size, 47-mm-diameter filters) over a range of dilutions according to standard membrane filtration methods (46). E. coli bacteria were enumerated on differential coliform (DC) media, supplemented with cefsulodin, while Ampr E. coli bacteria were also enumerated on the same DC media additionally supplemented with ampicillin (32 μg/ml). Both culturable E. coli and Ampr E. coli cultures were incubated at 44.5°C for 22 h. Results were reported as CFU counts per 100 milliliters. Filtration blanks were included in every batch of water samples.
DNA extraction and PCR.
An additional 300 ml was filtered (using 0.45-μm-pore-size, 47-mm-diameter filters), as described above, for DNA extraction. Filters were frozen, for no more than 1 week, at −80°C until ready for DNA extraction. Filters were then folded and placed into Powerbead tubes, and the filter contents were extracted using Powersoil DNA isolation kits (Mo Bio Laboratories, Inc., Carlsbad, CA, USA) according to the manufacturer's instructions. Extraction blanks were included in every batch of DNA extractions.
PCR and quantitative PCR (qPCR) assays were performed on all extracted DNA samples (including filtration and extraction blanks). PCR assays included general (Bac32), human (HF183), ruminant (CF128), and dog (DG37) Bacteroidales assays and gull Catellicoccus (Gull2) assays, performed with previously published primer sets (47–49). MST assays for qPCR included general (GenBactF3), human (HF183), cow (CowM2), and dog (DG37) Bacteroidales assays and gull Catellicoccus (qGull4) assays, performed with previously published primer and probe sets (49–53).
Each PCR consisted of 2.5 μl 10× IDTE buffer (Integrated DNA Technologies, Coralville, IA, USA), 0.2 μl 100 mM deoxynucleoside triphosphate (dNTP) mixture, 0.16 μl 10% bovine serum albumin (BSA), 0.5 μl each of forward and reverse primers (78 pM), 0.25 μl HotMaster Taq DNA polymerase (5Prime GmbH, Hilden, Germany), 19.89 μl nuclease-free water, and 1 μl of extracted DNA. Reactions were carried out in 96-well plates using an Eppendorf Mastercycler (Hamburg, Germany). Each 96-well plate included a negative control consisting of nuclease-free water and a positive control of DNA extracted from a known fecal source. For all plates, the negative control produced no band on the subsequent gel, while the positive control produced a band of the correct molecular weight for the corresponding target. Cycler conditions were consistent with previously published assays (47–49).
Each of the qPCRs consisted of 2 μl of an internal amplification control (IAC), 2.5 μl 2 mg/ml BSA, 3 μl nuclease-free water, 12.5 μl TaqMan universal master mix 2.0 (Applied Biosystems, Carlsbad, CA, USA), 3 μl of a primer/probe mixture (100 μM for both primers and probe), and 2 μl of extracted DNA. Reactions were carried out in 96-well plates using a CFX96 cycler (Bio-Rad, Hercules, CA, USA). All reactions were carried out in duplicate, including no-template controls (NTC), negative controls consisting of 2 μl salmon testes DNA, and positive controls consisting of 2 μl of DNA extracted from a known fecal source. Standard curves were run on every qPCR plate (13 total standard curves were run for each target). For all qPCR runs, NTC and negative-control samples never showed amplification, while amplification was observed in all positive controls. Samples that amplified at cycle 30 ± 3 cycles in the IAC were considered uninhibited. No samples were deemed inhibited. Thermocycler settings were 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min for all targets except gull. Thermocycler settings for the gull qPCR assay were 95°C for 5 min and 45 cycles of 95°C for 15 s and 60°C for 30 s. All qPCR results were reported as copy numbers (CN) per 100 milliliters.
Standard curves for all qPCR assays were constructed using synthesized plasmid DNA (pIDTSMART with ampicillin resistance; Integrated DNA Technologies, Coralville, IA, USA). DNA used for the standard curve was serially diluted using AE buffer (Qiagen, Valencia, CA, USA) to concentrations ranging from 102 to 105 gene copies/reaction. DNA used for the IAC was similarly constructed using synthesized plasmid DNA (pIDTSMART with ampicillin resistance; Integrated DNA Technologies, Coralville, IA, USA) with complementary primer sites included in each assay and every reaction to verify that there was no inhibition from the ambient water matrices. All qPCR runs had an efficiency level of between 90% and 110%, with an R2 of >0.95, and results were normalized to reaction efficiency.
qPCR arrays.
One wastewater influent sample and 36 water samples were selected for exploratory testing using Qiagen microbial DNA qPCR arrays (Qiagen, Valencia, CA). The water samples were collected on one rain event sampling day in late July and one dry weather sampling day in early August. The qPCR array design was customized in a 96-well format to include DNA markers for B. thetaiotaomicron, B. dorei, Bif. adolescentis, Catellicoccus marimammalium, Clostridium perfringens, Eu. rectale, F. prausnitzii, P. acnes, R. bromii, S. epidermidis, and Turicibacter sanguinis. The qPCR arrays were run according to the manufacturer's instructions. As these arrays are not quantitative without standard curves, the results were reported in a semiquantitative form using an inverse cycle threshold: this value was obtained by subtracting the cycle at which a sample amplified from the maximum number of 40 cycles. Therefore, the greater the inverse cycle, the more copies of a given target detected within that sample.
CST marker analysis.
CST marker analysis for all samples was performed by Environment Canada's National Laboratory for Environmental Testing (Burlington, Ontario, Canada). The full methods are presented in “Additional methods” in the supplemental material. For comparison, chemical analysis was performed on wastewater samples (influent and effluent collected from Toronto's four wastewater treatment plants; n = 20 for both influent and effluent samples). Concentrations were measured for the compounds caffeine, carbamazepine, codeine, cotinine, and acetaminophen and the artificial sweetener acesulfame. For quantitative analyses, if a sampling site had detectable levels of a chemical in greater than 50% of water samples, water samples with values below the detection limit (“nondetects”) were adjusted to values of one-half the detection limit for that chemical. For sites where a chemical was detected less than 50% of the time, nondetects were assigned a value of 0. The results of the statistical analyses were robust regardless of whether data corresponding to one-half the detection limit or untransformed data were used.
Statistical analysis.
Data from both measures of E. coli (culturable and Ampr; CFU counts per 100 milliliters) and all MST markers (CN per 100 milliliters) were log transformed prior to analysis. t tests were used to assess differences in E. coli and MST marker concentrations between river sites and outfalls. Multivariate analysis of variance (MANOVA) was used to determine the main effect of sampling site among river sites or among outfalls, where response variables were E. coli concentrations. The main effect of sampling site (including wastewater treatment plant influent and effluent samples) was similarly assessed via MANOVA where response variables were chemical marker concentrations. Tukey's post hoc test was performed if a significant effect was detected. Chi square tests were used to determine differences in endpoint MST marker detection. Spearman correlations were used to assess relationships among E. coli, qPCR, and MST and CST marker concentrations. All analyses were performed in Statistica v.12, and results were considered significant at the α level of 0.05.
RESULTS
E. coli enumeration.
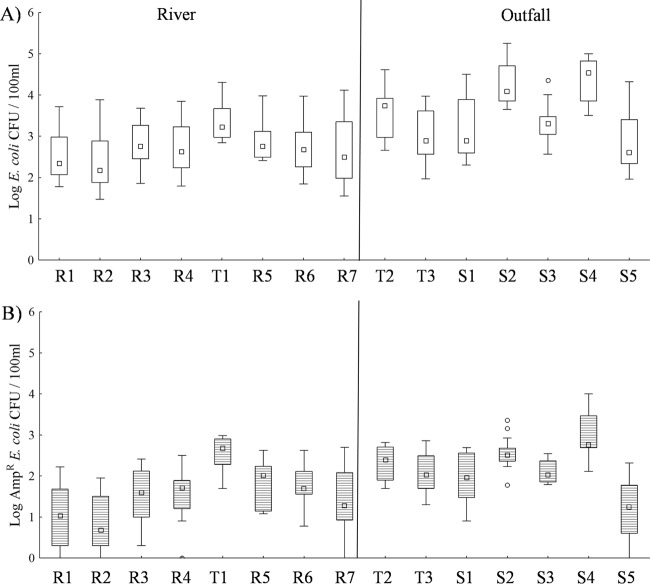
Culturable E. coli concentrations were significantly correlated (P < 0.05) with Ampr E. coli concentrations (rs = 0.87) for the pooled data set. t tests revealed significantly greater concentrations of E. coli by both measures (culturable and Ampr) in outfall sites than in river sites (P < 0.001 for both measures). Among the river sites, sampling site had a significant effect on both measures of E. coli (F21,259 = 3.06, P < 0.001; Fig. 1). Post hoc analyses revealed that site T1 had significantly greater concentrations of culturable E. coli than sites R2 (P = 0.003 for both measures) and R1 (P = 0.021 and 0.025, respectively). Ampr E. coli concentrations were also significantly greater in site T1 than in sites R1 to R4 and R7 (P ≤ 0.010). Among outfall sites, sampling site also had a significant effect on both measures of E. coli (F18,190 = 5.26, P < 0.001; Fig. 1). Post hoc analysis revealed that outfall S2 had significantly greater culturable E. coli concentrations than all other outfalls, except S4 (P ≤ 0.009), and that S4 had significantly greater culturable E. coli concentrations than outfalls T3, S5, and S1 (P ≤ 0.002).
FIG 1.
Box plots of (A) culturable E. coli and (B) Ampr E. coli at each sampling site. Box plots show the median E. coli concentration between the 25th and 75th data quartiles; whiskers extend to the outermost data point within ±1.5 data points of this interquartile range. Open circles depict outlier values.
Microbial source tracking.
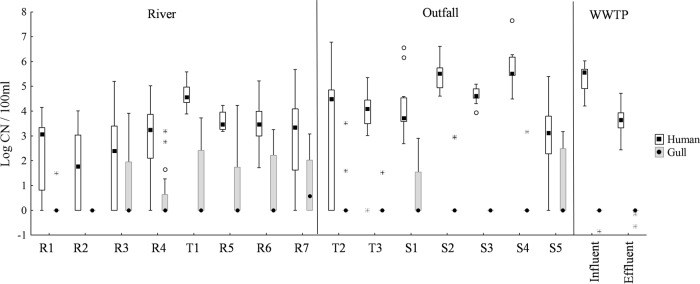
The general Bacteroidales marker (Bac32) was detected in all but one sample and was therefore not included in further analyses. A chi square test revealed significant differences in the levels of detection of host-associated endpoint MST markers between river sites and outfalls (Table 1); the human marker was detected significantly more frequently in outfalls than in river sites (χ2 = 16.32, P < 0.001), while the gull marker was detected significantly more frequently in river sites (χ2 = 19.35, P < 0.001). As the qPCR cow marker was never detected in any river or outfall samples, and the qPCR dog marker was detected only rarely in outfalls, they were excluded from further analyses. t tests revealed that outfalls had significantly higher concentrations of the human Bacteroidales qPCR marker than river sites (P < 0.001; Fig. 2).
FIG 2.
Box plots of human and gull qPCR marker concentrations for each site. Box plots show the median E. coli concentration between the 25th and 75th data quartiles; whiskers extend to the outermost data point within ±1.5 data points of this interquartile range. Open circles depict outlier values. WWTP, wastewater treatment plant.
Among river sites, sampling location had a significant effect on endpoint MST marker detection (Table 1). The human Bacteroidales marker was detected significantly more frequently at site T1 than at other river sites (χ2 = 25.5, P = 0.001), the ruminant Bacteroidales marker was detected significantly more often at site R1 than at other river sites (χ2 = 20.0, P = 0.006), and the gull marker was detected significantly less often at sites R1 and R2 than at other river sites (χ2 = 22.4, P = 0.002). Sampling location also had a significant effect on qPCR marker concentrations among river sites (F21,311 = 3.62, P < 0.001; Fig. 2). Post hoc analyses determined that site T1 had significantly greater concentrations of human-specific Bacteroidales than all river sites except R5 and R6 (P ≤ 0.009; Fig. 2).
Among outfall sites, no significant differences were observed for detection of any endpoint marker among sites. However, MANOVA detected a significant effect of outfall location on qPCR marker concentrations (F18,221 = 4.75, P < 0.001; Fig. 2). Post hoc analyses determined that outfall S2 had significantly greater concentrations of the human Bacteroidales marker than outfalls T2, T3, and S5 (P = 0.007, 0.037, and < 0.001, respectively) and that outfall S4 had significantly greater concentrations than outfalls T2, T3, and S5 (P = 0.004, 0.020, and < 0.001, respectively).
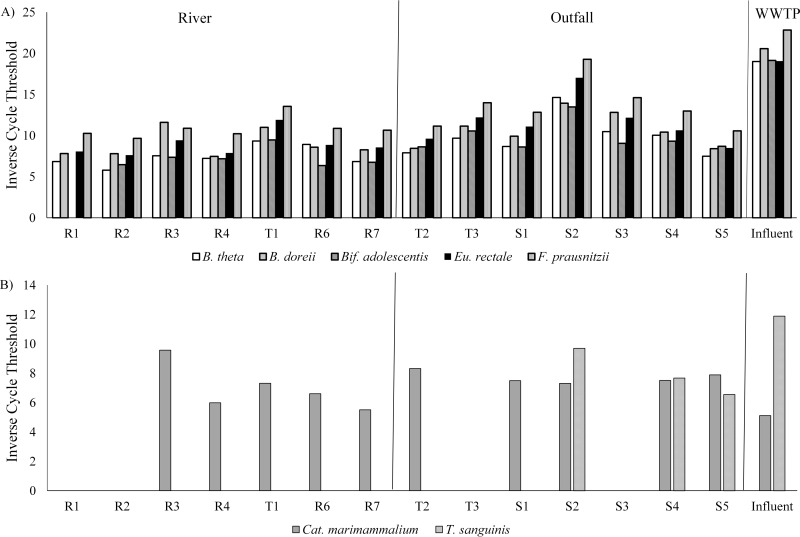
The inverse cycle thresholds obtained for select qPCR array markers are displayed in Fig. 3. Generally, C. perfringens was more prevalent in outfalls than in river sites (data not shown). B. thetaiotaomicron and B. dorei were present in all sites, with the highest levels in S2. Bif. adolescentis was present at all sites except for R1, with the highest levels in S2. F. prausnitzii and Eu. rectale were ubiquitous, and concentrations in S2 approached the levels found in wastewater influent. R. bromii was commonly found, with higher concentrations in the outfall and middle-to-lower Humber River sites (data not shown). P. acnes was found at higher concentrations in river than in storm water outfalls, while S. epidermidis was detected at only one storm water outfall (S2) and in wastewater influent (data not shown). C. marimammalium was present at the majority of sites but less often in the upper watershed away from Lake Ontario. T. sanguinis was not detected in the river and was detected only in outfalls from the lower Humber River.
FIG 3.
Mean inverse cycle thresholds obtained using qPCR arrays with targets for (A) human and (B) gull and Canada goose.
CST markers.
MANOVA revealed a significant effect of sample type (river, outfall, influent, or effluent) on the concentrations of CST markers (F18,620 = 172.17, P < 0.001; select CST markers are shown in Fig. 4). Post hoc analysis revealed that wastewater influent had significantly higher concentrations of caffeine, cotinine, and acetaminophen than any other site type (P < 0.001 for all analyses). While there was no significant difference in carbamazepine concentrations between wastewater influent and effluent, both wastewater sample types had significantly higher concentrations of the chemical than samples from river or outfall sites (P < 0.001 for all analyses). Codeine and acesulfame concentrations were significantly higher in wastewater influent than in any other site type (P < 0.001 for all analyses), and effluent had significantly greater concentrations of the chemicals than samples from river or outfall sites (P < 0.001 for all analyses). Excluding wastewater samples from the analysis, t tests revealed that outfall sites had significantly greater concentrations of caffeine, acetaminophen, and acesulfame than river sites (P = 0.030, <0.001, and <0.001, respectively) and that river sites had significantly greater concentrations of codeine than outfalls (P = 0.009).
FIG 4.
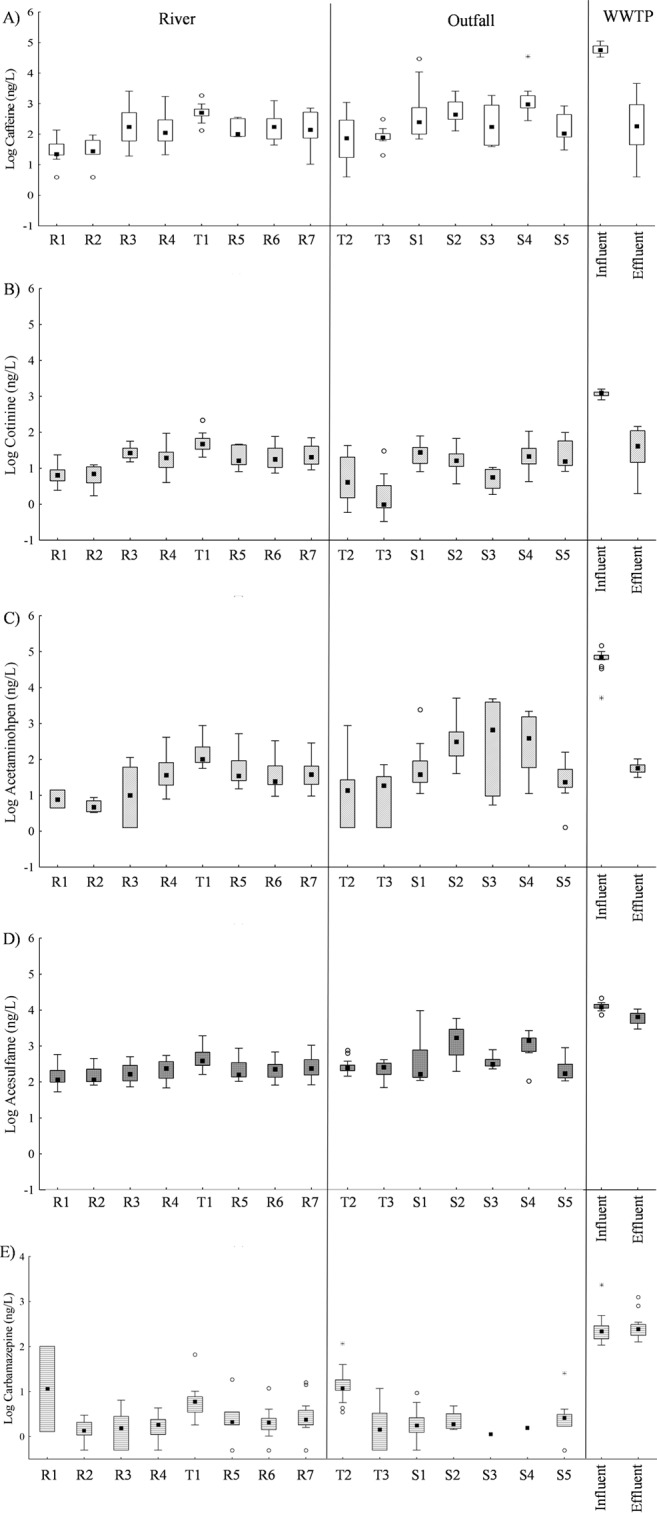
Box plots of (A) caffeine, (B) cotinine, (C) acetaminophen, (D) acesulfame, and (E) carbamazepine for each site. Box plots show the median E. coli concentration between the 25th and 75th data quartiles; whiskers extend to the outermost data point within ±1.5 data points of this interquartile range. Open circles depict outlier values, and asterisks depict extreme values.
Among river sites, MANOVA detected a significant effect of sampling location on chemical marker concentrations (F42,454 = 2.90, P < 0.001), with site T1 having significantly higher concentrations of the CST markers than many river sites (Fig. 4). Post hoc analyses revealed that site T1 had significantly greater concentrations of caffeine than site R2 (P = 0.003), significantly greater concentrations of carbamazepine than sites R1 to R4 (P = 0.026, 0.003, 0.033, and 0.017, respectively), and significantly greater concentrations of cotinine than all other river sites (P ≤ 0.033).
Among outfalls, MANOVA detected a significant effect of outfall on CST marker concentrations (F36,301 = 3.87, P < 0.001; Fig. 4), although post hoc analyses revealed no significant differences among outfalls for caffeine or acetaminophen concentrations. Outfall T2 had significantly greater concentrations of carbamazepine than all other outfalls (P ≤ 0.006). Outfall S2 had significantly greater concentrations of codeine than all other outfalls (P ≤ 0.013) and significantly greater concentrations of acesulfame than outfalls T2, T3, and S5 (P = 0.031, 0.024, and 0.028, respectively).
Correlations between E. coli, qPCR, and CST markers.
Spearman correlations between E. coli and qPCR marker concentrations are presented in Table 2 for river sites and Table 3 for outfall sites and wastewater samples. Among river sites, both measures of E. coli were significantly associated with human and gull qPCR markers, although the correlations were considerably higher with the human marker (Table 2). Among outfall sites, significant positive correlations were observed between concentrations of the human qPCR markers and both measures of E. coli, although no significant correlations were observed between either measure of E. coli and the qPCR gull marker (Table 3).
TABLE 2.
Spearman correlation coefficients between E. coli concentrations, qPCR, and chemical assays for river sitesa
| Sampling site | qPCR assay | Spearman correlation coefficient |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Culturable E. coli | Ampr E. coli | Caffeine | Carbamazepine | Codeine | Cotinine | Acetaminophen | Acesulfame | ||
| All river samples | Human | 0.75* | 0.83* | 0.72* | 0.23* | 0.51* | 0.65* | 0.60* | 0.23* |
| Gull | 0.28* | 0.44* | 0.38* | 0.08 | 0.13 | 0.38* | 0.37* | 0.18 | |
| R1 | Human | 0.62* | 0.79* | 0.43 | 0.55* | 0.50 | −0.16 | ||
| Gull | −0.25 | 0.24 | −0.31 | 0.12 | 0.31 | ||||
| R2 | Human | 0.52* | 0.68* | 0.48 | 0 | −0.52 | 0.41 | 0.80 | 0.01 |
| Gull | |||||||||
| R3 | Human | 0.54* | 0.74* | 0.57* | 0.04 | 0.26 | 0.67* | 0.60* | 0.23 |
| Gull | 0.50* | 0.70* | 0.57* | 0.23 | 0.22 | 0.47 | 0.60* | 0.18 | |
| R4 | Human | 0.78* | 0.90* | 0.78* | 0.33 | 0.11 | 0.72* | 0.32 | −0.05 |
| Gull | 0.35 | 0.08 | 0.57* | 0.38 | 0.42 | 0.57* | 0.77* | 0.40 | |
| T1 | Human | 0.49 | 0.70* | 0.46 | −0.18 | 0.31 | 0.22 | 0.5 | −0.14 |
| Gull | 0.64* | 0.46 | 0.27 | −0.65* | −0.36 | 0.43 | 0.04 | −0.55* | |
| R5b | Human | 0.43 | 0.59 | 0.14 | −0.54 | 0.09 | −0.09 | 0.67 | 0.12 |
| Gull | −0.45 | −0.18 | −0.68 | −0.68 | 0.27 | −0.03 | −0.67 | −0.26 | |
| R6 | Human | 0.77* | 0.78* | 0.76* | −0.31 | 0.57* | 0.60* | 0.64* | −0.01 |
| Gull | 0.39 | 0.45 | 0.43 | 0.19 | 0.42 | 0.29 | 0.55* | 0.06 | |
| R7 | Human | 0.70* | 0.62* | 0.65* | −0.39 | 0.64* | 0.56* | 0.64* | −0.27 |
| Gull | −0.09 | 0.46 | −0.35 | −0.24 | −0.33 | −0.25 | −0.12 | 0.15 | |
Blank cells represent situations where nondetects limited the ability to obtain a correlation. *, significant correlation (P < 0.05).
R5 had a smaller sample size than other sampling sites. Rain events and other seasonal influences were missed at site R5 that were sampled at other sites, precluding simple comparisons among other sites.
TABLE 3.
Spearman correlation coefficients between E. coli concentrations, qPCR, and chemical assays for outfall and wastewater (influent and effluent) samplesa
| Sampling site | qPCR assay | Spearman correlation coefficient |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Culturable E. coli | Ampr E. coli | Caffeine | Carbamazepine | Codeine | Cotinine | Acetaminophen | Acesulfame | ||
| All outfall samples | Human | 0.77* | 0.66* | 0.66* | −0.06 | 0.52* | 0.31* | 0.61* | 0.44* |
| Gull | 0.16 | 0.06 | 0.19 | −0.04 | −0.07 | 0.42* | 0.10 | −0.28* | |
| T2 | Human | 0.86* | 0.71* | 0.90* | 0.44 | 0.49 | 0.87* | 0.82* | −0.31 |
| Gull | 0.50 | 0.40 | 0.62* | −0.20 | −0.04 | 0.52 | 0.64* | −0.29 | |
| T3 | Human | 0.40 | 0.31 | 0.35 | −0.32 | 0.03 | 0.45 | 0.39 | 0.10 |
| Gull | 0.31 | 0.31 | 0.23 | 0.48 | 0.46 | 0.31 | −0.08 | −0.46 | |
| S1 | Human | 0.60* | 0.22 | 0.92* | −0.40 | 0.61* | 0.62* | 0.72* | 0.45 |
| Gull | 0.69* | 0.74* | 0.19 | 0.08 | −0.20 | 0.06 | 0.14 | −0.30 | |
| S2 | Human | −0.04 | −0.13 | −0.09 | −0.40 | 0.45 | −0.15 | 0.37 | 0.23 |
| Gull | −0.01 | 0.05 | −0.05 | −0.26 | −0.63* | 0.40 | −0.45 | −0.56* | |
| S3 | Human | 0.10 | 0.25 | −0.18 | −0.37 | 0.21 | −0.39 | −0.57 | |
| Gull | |||||||||
| S4 | Human | 0.62 | 0.64 | −0.07 | 0.37 | 0.20 | 0.15 | −0.42 | |
| Gull | 0.55 | 0.55 | 0.55 | 0.27 | 0.14 | ||||
| S5 | Human | 0.61* | 0.50 | 0.67* | −0.01 | 0.10 | 0.70* | 0.41 | −0.01 |
| Gull | 0.25 | 0.48 | 0.19 | −0.17 | −0.27 | 0.41 | 0.11 | 0.04 | |
| Wastewater | Human | NSb | NS | 0.78* | −0.22 | 0.76* | 0.68* | 0.70* | 0.68* |
| Gull | NS | NS | 0.13 | 0.07 | 0.11 | 0.19 | 0 | 0.27 | |
Blank cells represent situations where nondetects limited the ability to obtain a correlation. *, significant correlation (P < 0.05).
NS, the parameter was not sampled.
Analysis of river sites for CST markers showed significant positive correlations between culturable and Ampr E. coli concentrations and caffeine (rs = 0.69 and 0.72, respectively), codeine (rs = 0.36 and 0.32, respectively), cotinine (rs = 0.67 and 0.71, respectively), and acetaminophen (rs = 0.41 and 0.53, respectively). Analysis of outfalls showed significant positive correlations between culturable and Ampr E. coli concentrations and caffeine (rs = 0.63 and 0.42, respectively), codeine (rs = 0.47 and 0.36, respectively), and acetaminophen (rs = 0.61 and 0.40, respectively). Significant positive correlations were also observed in outfall samples between concentrations of culturable E. coli and cotinine (rs = 0.40) and acesulfame (rs = 0.27).
DISCUSSION
This study found that all Humber river sites were usually in excess of the limit specified in Ontario provincial guidelines for recreational water of 100 E. coli CFU/100 ml (54) and had some level of human sewage contamination. The river hot spot for E. coli was site T1. It had the highest concentrations of E. coli and the human qPCR marker, which is consistent with a previous study which found high levels of an alternate HF183 marker in Black Creek (21). Our results identify the importance of reducing sewage contamination in Black Creek for reducing E. coli levels and the potential for health risks downstream. The source of the HF183 human marker in the upper branches of the Humber River could be two small wastewater treatment plants upstream of site R2, sewage cross-connects within storm water systems in smaller rural communities, or leaking septic systems. An MST study using the HF183 marker in Michigan in the United States recently detected widespread septic system impacts on surface water quality in rural areas (55). The increases in both E. coli and human marker concentrations in the middle and lower Humber River are likely due to increased impacts from combined sewer overflows and storm water systems with sewage cross-connections.
River sites were also frequently impacted by gull fecal contamination, although at relatively low concentrations, which is consistent with a previous study that found widespread gull contamination along coastal and riverine systems in southern Ontario (56). Within the upper watershed, particularly at site R1, there also appeared to be high levels of ruminant contamination, as detected by endpoint PCR. While this is consistent with past research showing this area of the Humber River watershed to have a history of livestock operations (44), the qPCR CowM2 marker was never detected at this (or any other) site. It is possible that ruminant contamination was present at this site, albeit at low levels. Additionally, the concentrations of the CowM2 marker have been previously reported to vary based upon the age and diet of cows, which may have affected detection of this marker (57). However, CF128 marker detection may have been the result of false positives, as previous host specificity testing of this CF128 marker (58) revealed that it had only a 43.89% probability of correctly detecting a true positive. Detection of the CF128 marker could, therefore, have been indicative of another fecal pollution source upstream of site R1.
This study also found that all storm water outfalls had some level of human sewage contamination. Among storm water outfalls, S2 and S4 had the highest levels of E. coli as well as the highest frequency of the human PCR marker and the highest concentrations of the human qPCR marker. Consequently, these outfalls are likely impacted by sewage cross-connections and represent important targets for remediation. In contrast, outfalls S1 and S5 had higher levels of gull contamination than other outfalls. However, outfall S1 was sampled in the river slightly downstream of the outfall due to dangerous conditions encountered in accessing the outfall directly and results from that site therefore likely reflect the combined impacts from upstream river water quality and the outfall. Similarly, outfall S5 likely reflects the combined impacts from upstream river quality and this submerged outfall.
Aside from analysis of E. coli concentrations and employment of conventional PCR/qPCR techniques, this study also examined the utility of a variety of alternate markers of human sewage contamination, including the use of ampicillin-resistant E. coli, which has previously been shown to be present in high concentrations in treated and untreated wastewater (19). At river sites in this study, Ampr E. coli was found to be more highly correlated with concentrations of the human qPCR marker than culturable E. coli, suggesting that Ampr E. coli might be a more useful indicator of human sewage contamination in the Humber River. However, this did not extend to storm water outfalls, where a significant correlation between Ampr E. coli and the human qPCR marker was observed at only one outfall site. Caution should therefore be used in using Ampr E. coli as an indicator of sewage contamination, as these organisms can be ubiquitous in a given watershed.
The data obtained from the qPCR array were relatively consistent with the standard qPCR results. The human gut-associated bacteria B. thetaiotaomicron, B. dorei, Bif. adolescentis, F. prausnitzii, Eu. rectale, and R. bromii appeared more abundant in outfalls than in river sites. There were higher concentrations of these markers in outfall S2 and at river site T1, consistent with our HF183 qPCR results, suggesting that these markers could be good indicators of human sewage contamination. In contrast, the skin-associated bacterium P. acnes showed an inverse pattern and was detected more frequently in river sites than in outfalls. As this bacterial species is skin associated, this may reflect sources such as gray water rather than sewage discharge into the river. The qPCR array markers for Catellicoccus marimammalium (gull) and T. sanguinis (possibly Canada geese) were also consistent with our gull qPCR results and were found where we might expect to find impacts from Canada geese closer to the lake. However, it should be noted that, as the qPCR arrays are not rigorously quantitative, and as only two sampling events were assessed by this method, these conclusions are only preliminary. Further study utilizing these arrays is necessary to evaluate the efficacy of these markers.
Differences were also detected among the CST markers regarding their efficacy in assessing sewage contamination sources. Comparison of wastewater influent and effluent samples revealed that CST markers had different levels of persistence. Caffeine, cotinine, and acetaminophen had significantly higher concentrations in sewage influent than in the associated effluent (Fig. 4), which suggests that these CST markers are more likely to degrade quickly and, therefore, might be more indicative of recent raw sewage contamination. Caffeine, cotinine, and acetaminophen were the CST markers most often correlated with the human qPCR marker (Tables 2 and 3), supporting the idea of their utility as predictors of raw sewage contamination. Other studies have also found that the presence of caffeine tends to be indicative of more-recent sewage contamination (59). Carbamazepine has previously been observed to be a useful indicator of sewage contamination, although perhaps from less-recent contamination events, or treated sewage, due to its greater potential to resist degradation (60, 61).
However, caution should be used when interpreting CST markers. While the most contaminated river site (based on E. coli concentrations, human PCR marker detection, and human qPCR marker concentrations), T1, tended to have higher concentrations of most CST markers, in particular, of cotinine, codeine, acetaminophen, and acesulfame, than most of the other river sites, this was not true of the most contaminated outfalls. Only outfalls S2 and S4 had concentrations of codeine and acetaminophen that were significantly greater than those at other outfalls. Further, none of the most sewage-contaminated sites (T1, S2, and S4) had significant correlations between any chemical marker and the human qPCR marker. The lack of significant correlations with the human qPCR marker at these sites may indicate a site-specific utility for CST markers. Storm water outfalls can exhibit unique “upstream” storm watershed conditions which may render the relationships between chemical and human qPCR markers unreliable depending upon aspects such as different human consumption patterns, the scale and mechanism of sewage input into a storm water system, and different attenuation mechanisms of markers.
MST methods were able to identify human sewage as the likely source of fecal contamination at key Humber watershed E. coli hot spots, with Ampr E. coli concentrations showing some potential for an incremental benefit in detecting human sewage contamination in Humber River sites. Some CST markers were also consistent in identifying human sewage contamination at key E. coli hot spots. The CST markers caffeine, cotinine, and acetaminophen showed promise as indicators of recent raw sewage contamination. However, these CST markers present potential challenges for source determination, as significant correlations with human qPCR markers were not observed at most sites. Consequently, the results of this study indicate that while CST markers can be helpful in identifying raw sewage contamination, the additional use of MST methodologies can provide more-reliable identification of the source(s) of fecal contamination and alleviate potential confounding factors related to use of CST methods alone.
Supplementary Material
ACKNOWLEDGMENTS
We thank students Dennis Chuong, Teagan Maier-Downing, Phoenix Shum, and Caroline Tom for assistance.
We thank Environment and Climate Change Canada's Great Lakes Action Plan and Strategic Applications of Genomics in the Environment (STAGE) Programs for support.
All primers and probes from previously reported studies were used solely for research purposes.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01675-16.
REFERENCES
- 1.Sidhu JPS, Ahmed W, Gernjak W, Aryal R, McCarthy D, Palmer A, Kolotelo P, Toze S. 2013. Sewage pollution in urban stormwater runoff as evident from the widespread presence of multiple microbial and chemical source tracking markers. Sci Total Environ 463–464:488–496. [DOI] [PubMed] [Google Scholar]
- 2.Parker JK, McIntyre D, Noble RT. 2010. Characterizing fecal contamination in stormwater runoff in coastal North Carolina, USA. Water Res 44:4186–4194. doi: 10.1016/j.watres.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 3.Brownell MJ, Harwood VJ, Kurz RC, McQuaig SM, Lukasik J, Scott TM. 2007. Confirmation of putative stormwater impact on water quality at a Florida beach by microbial source tracking methods and structure of indicator organism populations. Water Res 41:3747–3757. doi: 10.1016/j.watres.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Dorfman M. 2006. Testing the waters: a guide to water quality at vacation beaches. National Resource Defense Council, Washington, DC. [Google Scholar]
- 5.Gaffield SJ, Goo RL, Roberts LA, Jackson RJ. 2003. Public health effects of inadequately managed stormwater runoff. Am J Public Health 93:1527–1533. doi: 10.2105/AJPH.93.9.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curriero FC, Patz JA, Rose JB, Lele S. 2001. The association between extreme precipitation and waterborne disease outbreaks in the United States, 1948–1994. Am J Public Health 91:1194–1199. doi: 10.2105/AJPH.91.8.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noble RT, Griffith JF, Blackwood AD, Fuhrman JA, Gregory JB, Hernandez X, Liang X, Bera AA, Schiff K. 2006. Multitiered approach using quantitative PCR to track sources of fecal pollution affecting Santa Monica Bay, California. Appl Environ Microbiol 72:1604–1612. doi: 10.1128/AEM.72.2.1604-1612.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajal VB, McSwain BS, Thompson DE, Leutenegger CM, Wuertz S. 2007. Molecular quantitative analysis of human viruses in California stormwater. Water Res 41:4287–4298. doi: 10.1016/j.watres.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Pongmala K, Autixier L, Madoux-Humery A-S, Fuamba M, Galarneau M, Sauvé S, Prévost M, Dorner S. 2015. Modelling total suspended solids, E. coli and carbamazepine, a tracer of wastewater contamination from combined sewer overflows. J Hydrol 531:830–839. doi: 10.1016/j.jhydrol.2015.10.042. [DOI] [Google Scholar]
- 10.Panasiuk O, Hedström A, Marsalek J, Ashley RM, Viklander M. 2015. Contamination of stormwater by wastewater: a review of detection methods. J Environ Manage 152:241–250. doi: 10.1016/j.jenvman.2015.01.050. [DOI] [PubMed] [Google Scholar]
- 11.US Environmental Protection Agency. 2012. Recreational water quality criteria. EPA-820-F-12-058. US Environmental Protection Agency, Washington, DC. [Google Scholar]
- 12.Health Canada. 2012. Guidelines for Canadian recreational water quality. Health Canada, Toronto, Ontario, Canada. [Google Scholar]
- 13.McQuaig SM, Scott TM, Lukasik JO, Paul JH, Harwood VJ. 2009. Quantification of human polyomaviruses JC virus and BK virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Appl Environ Microbiol 75:3379–3388. doi: 10.1128/AEM.02302-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selvakumar A, Borst M. 2006. Variation of microorganism concentrations in urban stormwater runoff with land use and seasons. J Water Health 4:109–124. [PubMed] [Google Scholar]
- 15.Soller JA, Schoen ME, Bartrand T, Ravenscropt JE, Ashbolt NJ. 2010. Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res 44:4674–4691. doi: 10.1016/j.watres.2010.06.049. [DOI] [PubMed] [Google Scholar]
- 16.Parveen S, Murphree RL, Edmiston L, Kaspar CW, Portier KM. 1997. Association of multiple-antibiotic-resistance profiles with point and nonpoint sources of Escherichia coli in Apalachicola Bay. Appl Environ Microbiol 63:2607–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooke MD. 1976. Antibiotic resistance among coliform and fecal coliform bacteria isolated from sewage, seawater, and marine shellfish. Antimicrob Agents Chemother 9:879–884. doi: 10.1128/AAC.9.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reinthaler FF, Posch J, Feierl G, Wust G, Haas D, Ruckenbauer G, Mascher F, Marth E. 2003. Antibiotic resistance of E. coli in sewage and sludge. Water Res 37:1685–1690. doi: 10.1016/S0043-1354(02)00569-9. [DOI] [PubMed] [Google Scholar]
- 19.Bouki C, Venieri D, Diamadopoulos E. 2013. Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: a review. Ecotoxicol Environ Saf 91:1–9. doi: 10.1016/j.ecoenv.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 20.Kotlarska E, Łuczkiewicz A, Pisowacka M, Burzyński A. 2015. Antibiotic resistance and prevalence of class 1 and 2 integrons in Escherichia coli isolated from two wastewater treatment plants, and their receiving waters (Gulf of Gdansk, Baltic Sea, Poland). Environ Sci Pollut Res Int 22:2018–2030. doi: 10.1007/s11356-014-3474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edge TA, Hill S, Seto P, Marsalek J. 2010. Library-dependent and library-independent microbial source tracking to identify spatial variation in faecal contamination sources along a Lake Ontario beach (Ontario, Canada). Water Sci Technol 62:719–727. doi: 10.2166/wst.2010.335. [DOI] [PubMed] [Google Scholar]
- 22.Reischer GH, Ebdon JE, Bauer JM, Schuster N, Ahmed W, Aström J, Blanch AR, Blöschl G, Byamukama D, Coakley T, Ferguson C, Goshu G, Ko G, de Roda Husman AM, Mushi D, Poma R, Pradhan B, Rajal V, Schade MA, Sommer R, Taylor H, Toth EM, Vrajmasu V, Wuertz S, Mach RL, Farnleitner AH. 2013. Performance characteristics of qPCR assays targeting human- and ruminant-associated Bacteroidetes for microbial source tracking across sixteen countries on six continents. Environ Sci Technol 47:8548–8556. doi: 10.1021/es304367t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staley ZR, Chase E, Mitraki C, Crisman TL, Harwood VJ. 2013. Microbial water quality in freshwater lakes with different land use. J Appl Microbiol 115:1240–1250. doi: 10.1111/jam.12312. [DOI] [PubMed] [Google Scholar]
- 24.Hagedorn C, Blanch AR, Harwood VJ. 2011. Microbial source tracking: methods, applications, and case studies. Springer-Verlag, New York, NY. [Google Scholar]
- 25.Harwood VJ, Staley C, Badgley BD, Borges K, Korajkic A. 2014. Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol Rev 38:1–40. doi: 10.1111/1574-6976.12031. [DOI] [PubMed] [Google Scholar]
- 26.Korajkic A, Badgley BD, Brownell MJ, Harwood VJ. 2009. Application of microbial source tracking methods in a Gulf of Mexico field setting. J Appl Microbiol 107:1518–1527. doi: 10.1111/j.1365-2672.2009.04351.x. [DOI] [PubMed] [Google Scholar]
- 27.Staley C, Gordon KV, Schoen ME, Harwood VJ. 2012. Performance of two human-associated microbial source tracking qPCR methods in various Florida water types and implications for microbial risk assessments. Appl Environ Microbiol 78:7317–7326. doi: 10.1128/AEM.01430-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoeckel DM, Harwood VJ. 2007. Performance, design, and analysis in microbial source tracking studies. Appl Environ Microbiol 73:2405–2415. doi: 10.1128/AEM.02473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.US Environmental Protection Agency. 2005. Microbial source tracking guide document. EPA/600-R-05-064. US Environmental Protection Agency, Washington, DC. [Google Scholar]
- 30.Abdelzaher AM, Wright ME, Ortega C, Solo-Gabriele HM, Miller G, Elmir S, Newman X, Shih P, Bonilla JA, Bonilla TD, Palmer CJ, Scott TM, Lukasik JO, Harwood VJ, McQuaig S, Sinigalliano CD, Gidley M, Plano LR, Zhu X, Wang JD, Fleming LE. 2010. Presence of pathogens and indicator microbes at a non-point source subtropical recreational marine beach. Appl Environ Microbiol 76:724–732. doi: 10.1128/AEM.02127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mieszkin S, Yala J-F, Joubrel R, Gourmelon M. 2010. Phylogenetic analysis of Bacteroidales 16S rRNA gene sequences from human and animal effluents and assessment of ruminant faecal pollution by real-time PCR. J Appl Microbiol 108:974–984. [DOI] [PubMed] [Google Scholar]
- 32.Gómez-Doñate M, Ballesté E, Muniesa M, Blanch AR. 2012. New molecular quantitative PCR assay for detection of host-specific Bifidobacteriaceae suitable for microbial source tracking. Appl Environ Microbiol 78:5788–5795. doi: 10.1128/AEM.00895-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng G, Yampara-Iquise H, Jones JE, Carson CA. 2009. Development of Faecalibacterium 16S rRNA gene marker for identification of human faeces. J Appl Microbiol 106:634–641. doi: 10.1111/j.1365-2672.2008.04037.x. [DOI] [PubMed] [Google Scholar]
- 34.Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet J-P, Ugarte E, Muñoz-Tamayo R, Paslier DLE, Nalin R, Dore J, Leclerc M. 2009. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol 11:2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- 35.Grice EA, Segre JA. 2011. The skin microbiome. Nat Rev Microbiol 9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, Cantarel BL, Coutinho PM, Henrissay B, Crock LW, Russell A, Verberkmoes NC, Hettich RL, Gordon JI. 2009. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci U S A 106:5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buerge IJ, Poiger T, Müller MD, Buser H-R. 2003. Caffeine, an anthropogenic marker for wastewater contamination of surface waters. Environ Sci Technol 37:691–700. doi: 10.1021/es020125z. [DOI] [PubMed] [Google Scholar]
- 38.Daneshvar A, Aboulfadl K, Viglino L, Broséus R, Sauvé S, Madoux-Humery A-S, Weyhenmeyer GA, Prévost M. 2012. Evaluating pharmaceuticals and caffeine as indicators of fecal contamination in drinking water sources of the Greater Montreal region. Chemosphere 88:131–139. doi: 10.1016/j.chemosphere.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 39.Clara M, Strenn B, Kreuzinger N. 2004. Carbamazepine as a possible anthropogenic marker in the aquatic environment: investigations on the behaviour of Carbamazepine in wastewater treatment during groundwater infiltration. Water Res 38:947–954. doi: 10.1016/j.watres.2003.10.058. [DOI] [PubMed] [Google Scholar]
- 40.Nikolaou A, Meric S, Fatta D. 2007. Occurrence patterns of pharmaceuticals in water and wastewater environments. Anal Bioanal Chem 387:1225–1234. doi: 10.1007/s00216-006-1035-8. [DOI] [PubMed] [Google Scholar]
- 41.Bradley PM, Barber LB, Kolpin DW, McMahon PB, Chapelle FH. 2007. Biotransformation of caffeine, cotinine, and nicotine in stream sediments: implications for use as wastewater indicators. Environ Toxicol Chem 26:1116–1121. doi: 10.1897/06-483R.1. [DOI] [PubMed] [Google Scholar]
- 42.Oppenheimer J, Eaton A, Badruzzaman M, Haghani AW, Jacangelo JG. 2011. Occurrence and suitability of sucralose as an indicator compound of wastewater loading to surface waters in urbanized regions. Water Res 45:4019–4027. doi: 10.1016/j.watres.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 43.Toronto and Region Conservation. 2016. Etobicoke Creek and Mimico Creek watersheds. Toronto and Region Conservation for the Living City. http://www.trca.on.ca/the-living-city/watersheds/humber-river/ Accessed 23 March 2016.
- 44.Mar P. 1991. Clean up rural beaches (CURB): plan for the east Humber River, Centreville Creek, and Bruce Creek watersheds. SBN 0-7729-8651-7. Metropolitan Toronto and Region Conservation Authority, Toronto, Ontario, Canada. [Google Scholar]
- 45.Environment Canada and the Ontario Ministry of the Environment. 2011. Toronto and region area of concern status of beneficial use impairments September 2010. En164-22/4-2011E-PDF. Environment Canada and the Ontario Ministry of the Environment, Gatineau, Quebec, Canada, and Toronto, Ontario, Canada. [Google Scholar]
- 46.American Public Health Association. 1999. Standard methods for the examination of water and wastewater, 20th ed American Public Health Association, Washington, DC. [Google Scholar]
- 47.Bernhard AE, Field KG. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl Environ Microbiol 66:4571–4574. doi: 10.1128/AEM.66.10.4571-4574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu J, Santo Domingo JW, Lamendella R, Edge T, Hill S. 2008. Phylogenetic diversity and molecular detection of bacteria in gull feces. Appl Environ Microbiol 74:3969–3976. doi: 10.1128/AEM.00019-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Green HC, White KM, Kelty CA, Shanks OC. 2014. Development of rapid canine fecal source identification PCR-based assays. Environ Sci Technol 48:11453–11461. doi: 10.1021/es502637b. [DOI] [PubMed] [Google Scholar]
- 50.Green HC, Haugland RA, Varma M, Millen HT, Borchardt MA, Field KG, Walters WA, Knight R, Sivaganesan M, Kelty CA, Shanks OC. 2014. Improved HF183 quantitative real-time PCR assay for characterization of human fecal pollution in ambient surface water samples. Appl Environ Microbiol 80:3086–3094. doi: 10.1128/AEM.04137-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryu H, Griffith JF, Khan IUH, Hill S, Edge TA, Toledo-Hernandez C, Gonzalez-Nieves J, Santo Domingo J. 2012. Comparison of gull feces-specific assays targeting the 16S rRNA genes of Catellicoccus marimammalium and Streptococcus spp. Appl Environ Microbiol 78:1909–1916. doi: 10.1128/AEM.07192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shanks OC, Atikovic E, Blackwood AD, Lu J, Noble RT, Santo Domingo J, Seifring S, Sivaganesan M, Haugland RA. 2008. Quantitative PCR for detection and enumeration of genetic markers of bovine fecal pollution. Appl Environ Microbiol 74:745–752. doi: 10.1128/AEM.01843-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shanks OC, Sivaganesan M, Peed L, Kelty CA, Blackwood AD, Greene MR, Noble RT, Bushon RN, Stelzer EA, Kinzelman J, Anan'eva T, Sinigalliano C, Wanless D, Griffith J, Cao Y, Weisberg S, Harwood VJ, Staley C, Oshima KH, Varma M, Haugland RA. 2012. Interlaboratory comparison of real-time PCR protocols for quantification of general fecal indicator bacteria. Environ Sci Technol 46:945–953. [DOI] [PubMed] [Google Scholar]
- 54.Ontario Ministry of Health. 1998. Beach management protocol—safe water program. Ontario Ministry of Health, Toronto, Ontario, Canada. [Google Scholar]
- 55.Verhougstraete MP, Martin SL, Kendall AD, Hyndman DW, Rose JB. 2015. Linking fecal bacteria in rivers to landscape, geochemical, and hydrologic factors and sources at the basin. Proc Natl Acad Sci U S A 112:10419–10424. doi: 10.1073/pnas.1415836112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu J, Ryu H, Hill S, Schoen M, Ashbolt N, Edge TA, Santo Domingo J. 2011. Distribution and potential significance of a gull fecal marker in urban coastal riverine areas of southern Ontario, Canada. Water Res 45:3960–3968. doi: 10.1016/j.watres.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 57.Shanks OC, Kelty CA, Peed L, Sivaganesan M, Mooney T, Jenkins M. 2014. Age-related shifts in density and distribution of genetic marker water quality indicators in cow and calf feces. Appl Environ Microbiol 80:1588–1594. doi: 10.1128/AEM.03581-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Staley ZR, Edge TA. 2016. Comparative microbial source tracking methods for identification of fecal contamination sources at Sunnyside Beach in the Toronto region area of concern. J Water Health 14. doi: 10.2166/wh.2016.296. [DOI] [PubMed] [Google Scholar]
- 59.Sauvé S, Aboulfadl K, Dorner S, Payment P, Deschamps G, Pévost M. 2012. Fecal coliforms, caffeine and carbamazepine in stormwater collection systems in a large urban area. Chemosphere 86:118–123. doi: 10.1016/j.chemosphere.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 60.Madoux-Humery A-S, Dorner S, Sauvé S, Aboulfadl K, Galarneau M, Servais P, Prévost M. 2013. Temporal variability of combined sewer overflow contaminants: Evaluation of wastewater micropollutants as tracers of fecal contamination. Water Res 47:4370–4382. doi: 10.1016/j.watres.2013.04.030. [DOI] [PubMed] [Google Scholar]
- 61.Guérineau H, Dorner S, Carrière A, McQuaid N, Sauvé S, Aboulfadl K, Hajj-Mohamad M, Prévost M. 2014. Source tracking of leaky sewers: a novel approach combining fecal indicators in water and sediments. Water Res 58:50–61. doi: 10.1016/j.watres.2014.03.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.