Abstract
FIZ1 (Flt-3 Interacting Zinc-finger) interacts and co-purifies with the rod-specific transcription factor NRL (Neural Retina Leucine zipper). We hypothesize that FIZ1 is part of an interface between cell-specific factors, like NRL, and more ubiquitous regulatory networks that vary the absolute expression levels of some rod-specific genes (i.e. Rhodopsin). As part of an ongoing exploration of FIZ1’s role in neural retina, in vivo, we have taken the first look at FIZ1 expression in the developing mouse retina during the retinal maturation period. Using the normal C57/B6 mouse as a model, multiple approaches were used including: immunoblotting, immunohistochemistry, and quantitative real-time PCR. Functional implications of FIZ1/NRL interaction, on NRL-and CRX-mediated activation of the Rhodopsin (Rho) and cGMP-phosphodiesterase β-subunit gene (PDE6B) promoters, were examined by co-transfection assays. Immunoblot analysis revealed that FIZ1 protein levels were lowest in immature mouse neural retina (P0). FIZ1 concentration increased at least ten-fold as the neural retina matured to the adult state (P21 and later). Immunohistochemical comparison of immature post-natal and mature adult retina revealed increasing FIZ1 protein in photoreceptors, the inner plexiform layer, and the ganglion cell layer. Total retinal Fiz1 mRNA content increased as the neural retina matured. The expected increase in Rho mRNA level was also monitored as a genetic marker of photoreceptor maturation. In transient co-transfection assays of CV1 cells, FIZ1 synergized with NRL to activate transcription from the Rho and PDE6B gene promoters with some differences. In the case of the Rho promoter, FIZ1 synergized when both NRL and CRX were present. With the PDE6B promoter, FIZ1 synergized with NRL alone, and the inclusion of CRX decreased this synergy.
Conclusions
These findings support previous evidence that FIZ1 is present in rod-photoreceptors. (Co-immunoprecipitation from nuclear-protein extracts with rod-specific NRL) FIZ1 expression increases in the neural retina during the retinal maturation period. Additionally, in vitro experiments demonstrate that FIZ1 has the potential to significantly increase the NRL-mediated activation of photoreceptor-specific promoters. While CRX is not a strong activator of the PDE6B promoter, alone or with NRL, CRX decreased the synergy of NRL with FIZ1.
Keywords: FIZ1, NRL, CRX, Rhodopsin, cGMP-phosphodiesterase β-subunit, transcription, retinal development, gene, retinal degeneration, mouse
1. Introduction
Throughout retinal development, and in the mature retina, the precise control of photoreceptor gene expression is essential for photoreceptor integrity. Disruptions of these normal gene expression patterns cause photoreceptor degenerations. (Pacione, et al., 2003, Rattner, et al., 1999, Tan, et al., 2001) Transcription factors are integration points for signal transduction networks that orchestrate development. Homeobox transcription-factor genes are expressed in the earliest primordial eye tissue and are essential for vertebrate eye development (i.e. Pax6, Rx, Six3, Six6, and Lhx2). (Carl, et al., 2002, Mathers and Jamrich, 2000, Porter, et al., 1997) The bHLH (basic Helix Loop Helix) gene family of transcription factors also has pivotal roles in the unique patterning of transcription factors that guide retinal development (Mash1, Math3, Ngn2). (Akagi, et al., 2004, Brown, et al., 1998, Brown, et al., 2001, Hojo, et al., 2000, Kanekar, et al., 1997, Marquardt and Gruss, 2002, Moore, et al., 2002, Morrow, et al., 1999, Wang, et al., 2001, Yan and Wang, 1998)
Several transcription factors are clearly essential for photoreceptor development, and their mutations cause retinal degenerations: NRL, CRX, Otx2, Trβ2 (thyroid hormone receptor β2), and NR2E3. (Bessant, et al., 1999, DeAngelis, et al., 2002, Freund, et al., 1997, Haider, et al., 2000, Haider, et al., 2001, Martinez-Gimeno, et al., 2001, Mears, et al., 2001, Ng, et al., 2001, Nishida, et al., 2003, Swain, et al., 1997, Swaroop, et al., 1999) However, our knowledge of the interactions of these cell-specific proteins with each other and upstream signal transduction proteins remains sparse. We do not know how this pool of cell-specific transcription factors interacts with the pool of ubiquitous proteins, which include the general transcription machinery and epigenetic regulators of chromatin/DNA structure. The challenge is to find links between these different functional protein pools: unique and ubiquitous. This requires far more data about, mostly unknown, protein interactions.
Our strategy is to use cell-specific proteins as starting points to discover new interaction networks. We have demonstrated the usefulness of this approach to uncover protein interactions that are functionally relevant to photoreceptor-specific gene expression, including: NRL + CRX, and NR2E3 + NR1D1 (RevErb-alpha). (Cheng, et al., 2004, Mitton, et al., 2000) Peng et al., also using yeast two-hybrid traps, have demonstrated that NR2E3 can bind to CRX in vitro, supporting activation of the Rhodopsin (Rho) promoter, and contributing to repression of cone-opsin promoters. (Peng, et al., 2005) Recently, our use of protein interaction traps discovered FIZ1 binding to the NRL bZip domain. Indeed, native FIZ1 and NRL co-purify from retina nuclear protein extracts, and FIZ1 can alter NRL’s activation of the Rho promoter. (Mitton, et al., 2003) FIZ1’s amino acid sequence is highly conserved in mammals and the human and mouse genes are very similar. (Genbank Accession: NM_032836) FIZ1 mRNA is present in many human and mouse tissues, and FIZ1 protein is present in the photoreceptors and the ganglion cell layer of mature retina. There is evidence that FIZ1 can exist in both the nuclear and cytoplasmic compartments, although its only documented cytoplasmic protein-interaction is with the tyrosine-kinase domain of a receptor essential for myeloid cell development. (Mitton, et al., 2003, Wolf and Rohrschneider, 1999) FIZ1 binds to FLT-3 (FMS-like receptor tyrosine kinase, FLK-2) in myeloid progenitors, and their association increases upon stimulation of cells with FLT-3-ligand. (Wolf and Rohrschneider, 1999) Mutations in the FLT-3 gene constitutively activate Stat-5 and MAP-kinase pathways, promoting myeloid leukemia. (Hayakawa, et al., 2000, Nakao, et al., 1996)
Multiple-Zf proteins, like FIZ1, feature prominently in networks that regulate gene expression, especially those that involve nuclear-receptors. FIZ1 is not homologous to any known Zf-families and we know little of its basic functions in the cell. FIZ1 represents one potential interface between cell-specific transcription factors, like NRL, and the ubiquitous regulatory networks of the cell. Its ability to inhabit both the nuclear and cytoplasmic compartments makes FIZ1 an interesting candidate for this role. As part of our ongoing investigations into the function of this protein, we report that FIZ1’s temporal and spatial expression would permit an interface role in the precise control of photoreceptor-specific gene expression. Retinal FIZ1 protein concentration increases substantially during post-natal maturation of the mouse retina, when maturation-related gene expression is on the rise (i.e. Rho). Furthermore, using co-transfection promoter-activation assays, combinations of specific quantities of expression vectors demonstrate that FIZ1 can promote activation of a minimal Rho, or PDE6B promoters in synergy with NRL and CRX.
2. Materials and methods
The study was designed under approval of Oakland University’s animal care and use committee and complied with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. All eye tissues were obtained at the same time (mid-afternoon) to avoid circadian effects.
2.1. Mouse retinal total RNA extraction
Mouse eyes (C57/B6 strain) were enucleated into PBS. Corneas were punctured with a 30-gauge needle and then removed completely, followed by removal of the lens. Eyecups were trimmed anteriorly to remove the iris and ciliary body, and the sclera/choroid/RPE layers were pealed off the neural retina. The still intact neural retinas were brushed free of isolated RPE-cell patches, and then separated from the optic nerve/disk. These neural retinal cups were fast-frozen to the inside of a 1.5 mL microfuge tube on dry ice, in groups of 4–8.
Total RNA was isolated using the Stratagene mini-Prep RNA kit (Stratagene, La Jolla, California, http://www.stratagene.com). Samples were processed using a 250 µL homogenization volume and DNAase treatment. RNA samples were maintained in mild buffer (RNAse free) with the addition of RNAse inhibitor (Invitrogen Corporation, Carlsbad, California, http://www.invitrogen.com) and stored at −70°C. Depending on the post-natal age of the retinas, 3–4 µg of total RNA per retina were recovered as determined by optical density measurement (260 nm).
2.2. First Strand cDNA Preparation
For quantitative analysis of Fiz1 mRNA using Amplitaq-Gold™ based RT-PCR, the reverse transcriptase reactions were performed using 2 µg of total RNA with Superscript-II™ enzyme as per the manufacturers protocol (Invitrogen). Single reactions resulted in over 40 µL of cDNA. First strand cDNA samples were split into aliquots and kept frozen.
For an additional quantitative analysis of Fiz1 mRNA using a different RT-PCR system (Roche Applied Science, Indianapolis, Indiana, http://www.roche-applied-science.com), cDNA samples were prepared from 2 µg of total RNA using the Stratascript™ QPCR cDNA synthesis kit (Stratagene). The cDNA samples were stored at −70° C until further use.
2.3. Quantitative real time PCR
Mouse retinal Fiz1 mRNA levels were measured using Amplitaq Gold™ enzyme (Applied Bio-Systems, Foster City, CA, http://www.appliedbiosystems.com), SYBR Green (Invitrogen) and plasmid cDNA standards in the pGemT-easy vector (Promega, Madison, WI, http://www.promega.com). Primers were first tested by touchdown PCR, using mouse retinal cDNA as template.
For Amplitaq Gold real-time PCR: PCR products were sub-cloned into the pGemT-easy vector as a source of cDNA standards. Reaction efficiencies were the same for templates in pGemTeasy as in retinal cDNA preparations. Expression of Fiz1 mRNA was determined relative to Gapdh mRNA levels using equation-1 for Ct based analysis. Reaction efficiencies are used in this analysis, where Eg was the reaction efficiency for the target (Fiz1 or Rho) and En the reaction efficiency for the normalizer gene (Gapdh). PCR products were confirmed by DNA sequencing.
| Equation-1 |
Thermo cycling conditions and primers are summarized in Table 1. Samples were analyzed with an MX3000P real-time PCR system (Stratagene).
Table 1.
Primers for quantitative PCR analysis of mouse retinal gene expression using Amplitaq Gold Method.
| Fiz1 | TGTAGCGAATGTGGCAAGAGTTTC | AGGGCTTTTCACCAGTGTGGAC | 247bp |
| Gapdh | ACCACAGTCCATGCCATCAC | TCCACCACCCTGTTGCTGTA | 452bp |
| Rho | TCAGAAGGCAGAGAAGGAAGTCAC | TGGTCTTGGTGGATGGATGC | 528bp |
|
Thermo cycling: |
95°C/8min., 40 × (95°C/30sec., 60°C/45sec., 72°C/60sec.), melting curve (90°C – 50°C) |
||
For an additional quantitative PCR analysis, retinal Fiz1 mRNA levels were directly measured using standard curves prepared from QPCR Mouse Reference Total RNA™ (Stratagene). Another reference gene, β-Actin, was also analyzed directly using the same mouse universal reference RNA and a specific hydrolysis-probe (Universal library probe #106, Roche Applied science, Indianapolis, Indiana). This particular strategy with the hydrolysis probe and reference RNA, enabled confirmation of similar β-Actin reference mRNA levels (relative to total RNA) before and after the retinal maturation period. Realtime PCR reactions were performed using Faststart Taqman Probe Master (Rox) mix (Roche Applied Science) with SYBR green dye (for Fiz1) or with Universal Library probe for mouse β-Actin. Primers used, and thermocycling conditions are summarized in Table 2. Samples were analyzed with an MX3000P real-time PCR system (Stratagene).
Table 2.
Primers for quantitative PCR analysis of mouse retinal gene expression using Roche SYBR green and β-actin comparitor.
| β-Actin | TGACAGGATGCAGAAGGAGA | CGCTCAGGAGGAGCAATG | 75bp |
| Fiz1 | TGTAGCGAATGTGGCAAGAGTTTC | AGGGCTTTTCACCAGTGTGGAC | 247bp |
|
Thermo Cycling |
95°C/10 min., 40 × (95°C/30sec., 62°C/45sec., 72°C/30sec.), melting curve (90°C – 50°C) |
||
2.4. Retinal sections and immunohistochemistry
Eyes were obtained from mice (C57/B6) at mid-day. Globes were removed into PBS, and the corneas were punctured a few times with a 30-gauge 1/2 inch needle. Globes were fixed in 0.1 M phosphate buffer, pH 7.2, 4% paraformaldehyde, with gentle agitation for 15 minutes. Cornea and the lens were removed and the retinal cups fixed for 1 hour. Fixed tissues were infused with sucrose using 0.1 M phosphate buffer, pH 7.2, with 10, 15 and 20% sucrose. Tissues were infused with a 2:1 mixture of 0.1 M phosphate buffer, pH 7.2, 20% sucrose and OCT, for 1 hour at room temperature and then frozen in OCT.
Sections were cut (10 µm) in a cryomicrotome and air dried onto positive-charged glass slides. Sections were washed (3 × 5 minutes) in PBS 0.3% Triton X-100 (PBS-Triton), prior to blocking in PBS-Triton + 1.5% goat serum for 30 minutes. Sections were incubated at room temperature with 1/500 dilution of antibody to bovine FIZ1. Control sections were treated with pre-immune rabbit serum. Sections were then washed with PBS-Triton (2 × 2.5 minutes) and incubated with 1/200 dilution of a biotinylated goat-anti-rabbit IgG in PBS-Triton for 30 minutes. Sections were washed (3 × 5 minutes), and incubated with ABC-peroxidase complex (30 minutes) as per the manufacturer’s instructions (Vector Laboratories, Burlingame CA). After a final washing (3 × 5 minutes), the location of FIZ1 protein was visualized by reacting 8 minutes with DAB/hydrogen-peroxide/nickel substrate. Sections were photographed using phase contrast with care to use the same illumination and exposure times for controls and test samples.
2.5. Retinal protein extraction and immunoblotting
Frozen retinas (10 – 14) were homogenized in a 1.5 mL Eppendorf tube with 250 µL of ice-cold 50 mM Tris + 5 mM EDTA + 1 mM DTT, pH 7.4, using a hand held microtube homogenizer. PMSF (final concentration 1mM) was added just at homogenization. Homogenates were centrifuged (20,000g × 10 minutes, 4°C), and the supernatant was frozen before analysis. Protein concentrations were determined using the Bradford assay and aliquots were prepared for DISC-Page by heating (99°C, 5 minutes) with one-volume of 2x-Blue Juice denaturing/reducing buffer (Q-Biogene: Bromophenol blue, urea, SDS, 2-mercapto-ethanol). Denatured samples were centrifuged for 1 minute at 2,000g, prior to loading 25 µg protein onto 12.5 % acrylamide gels in a Bio-Rad Mini-Protein-II apparatus. Proteins were electro-eluted in Towbin’s buffer onto PVDF membranes.
Membranes were blocked for 1-hour in PBS + 5% fat-free milk powder + 0.1% Tween-20, and then washed with PBS-Tween (3 × 5 minutes). Membranes were then incubated with a 1/1000 dilution of primary anti-body (anti-bFiz) in antibody diluent (PBS-Tween + 2.5% fat-free milk powder) for 1-hour, before washing and then incubating with peroxidase-labeled goat anti-rabbit IgG (1/10,000) for 1-hour. Membranes were subjected to chemiluminescent detection using the ECL-Plus Western Blotting reagents (GE-Health /Amersham). Film images were digitized and the relative amounts of FIZ1 protein were determined using Image-J, a Java-based version of the NIH-Image program. (Available at: http://rsb.info.nih.gov/ij/index.html)
In addition to loading equivalent total protein per gel lane, FIZ1 levels were normalized to the total protein actually transferred onto the PVDF membrane. Membranes were stripped of antibodies, washed in PBS-Tween, deionized water, and then stained with Electroblue colloidal protein stain (Q-Biogene) for 15 minutes. After washing in deionized water, the blots were scanned, and the relative amounts of total protein in each lane were determined using Image-J software.
2.6. Tissue protein extraction (non-retinal)
Protein extracts were prepared as for retinal protein extractions; however, tissues were weighed frozen and homogenized with a hand-held polytron generator after addition of PMSF and five volumes of buffer. For electrophoresis, 25 µg of total protein (denatured, reduced) were loaded per lane. Immunoblotting for FIZ1 protein was accomplished as described above.
2.7. Mammalian cell expression of FIZ1 antigen
Monkey kidney cells (line CV1, American Type Culture Collection) were transiently transfected with the pDest26-bFIZ1 plasmid, expressing an N-terminal His-Tag fusion protein of FIZ1 (bovine) in the pDest26 vector (CMV promoter, Invitrogen) . This was also the antigen for production and affinity purification of the FIZ1 antibody as previously described, and shares high homology to the human and mouse FIZ1 proteins. (Mitton, et al., 2003) Cells were transfected with 0, 0.6, or 1.2 ug of pDest26-bFIZ1 plasmid DNA. Plasmid DNA for transfections were prepared using the Qiagen maxi-prep kit (Qiagen, Valencia, CA, http://www.qiagen.com), and CV1 cells were transfected at 70% confluency in 24-well plates using Lipofectamine-plus™ (Invitrogen) as previously described. (Bessant, et al., 1999, Mitton, et al., 2003)
2.8 Transient co-transfection and promoter-activation assays
Monkey kidney cells (line CV1, American Type Culture Collection) were maintained in MEM-alpha (Invitrogen) supplemented with 10% fetal bovine serum, penicillin (50 U/ml) and streptomycin (50 ug/ml, Invitrogen) at 37°C in a humidified incubator with 5% CO2. Cells were not used after more than eight passages. Cells were counted (Beckman Coulter Z1 Particle Counter), and plated in 24-well plates (1.7 × 105 cells/well) the day before transfection. Cells were transfected using Lipofectamine-2000 reagent as per the manufacturers standard recommendations for 24-well format: a DNA (ug) to reagent (uL) ratio of 1:3. (Invitrogen). Briefly, cells were adapted to antibiotic-free and serum-free medium for two hours, and then exposed to the transfection mixes for five hours (antibiotic- and serum-free).
The luciferase-reporter construct, pBR-130-luc, provided by Drs Donald Zack (Johns Hopkins) and Shiming Chen (Washington University), contained 130 bp of the bovine Rhodopsin proximal promoter region subcloned into the firefly Luciferase pGL2-basic vector (Kumar, et al., 1996). A similar construct, pGL2-PDE6B (−93/+53), provided by Dr. Debora Farber (UCLA), contained the human PDE6B minimal promoter region (−93/+53). (Lerner and Farber, 2000) The following expression constructs were used for transfection: pED-NRL for NRL (from Dr. Anand Swaroop, University of Michigan), pcDNA-bCRX for CRX (Drs. Zack, Chen) and pDest26-bFIZ1 for FIZ1. (pDest26, CMV-promoter based, from Invitrogen.)
For the Rho-promoter, all transfection mixes contained 555 ng/mL (on cells) of pBR-130luc, and 19 ng/mL of the normalizer Renilla-luciferase construct pRL-CMV (Promega, Madison, WI, USA). Triplicate sample wells were co-transfected with expression vectors for NRL (55 ng/mL), CRX (110 ng/mL), and FIZ1 (165 ng/mL). Mixes contained either the desired expression constructs or corresponding empty plasmids.
For the PDE6B-promoter, all transfection mixes contained 220 ng/mL of pGL2-PDE6B-luc, and 19 ng/mL of the normalizer Renilla-luciferase construct pRL-CMV. (Final concentrations on cells.) Triplicate sample wells were co-transfected with expression DNAs for NRL (55 ng/mL), CRX (110 ng/mL), and FIZ1 (165 ng/mL)
After 48 hours, cells were rinsed in PBS and lysed using Passive Lysis Buffer (Promega). Firefly- and Renilla-luciferase levels were measured simultaneously in a single sample aliquot using Dual-Reporter Luminescence assay reagents and a Tuner Biosystems 20/20n dual-injector luminometer (both from Promega). An experiment included triplicate samples for each DNA combination, and each experiment was repeated at least three times. Individual experiments were analyzed independently. Individual sample activities (Firefly luciferase relative to Renilla luciferase) were normalized to the background activity of the reporter in the presense of empty expression vectors, to control for any unpredictable effects of vector DNA sequences. Data were analyzed for multiple comparisons using ANOVA and post-analysis with the conservative Tukey-Kramer procedure. (GB-Stat software, Dynamic Microsystems, Inc., Silver Spring, MD).
3. Results
3.1. FIZ1 protein is present in several organs
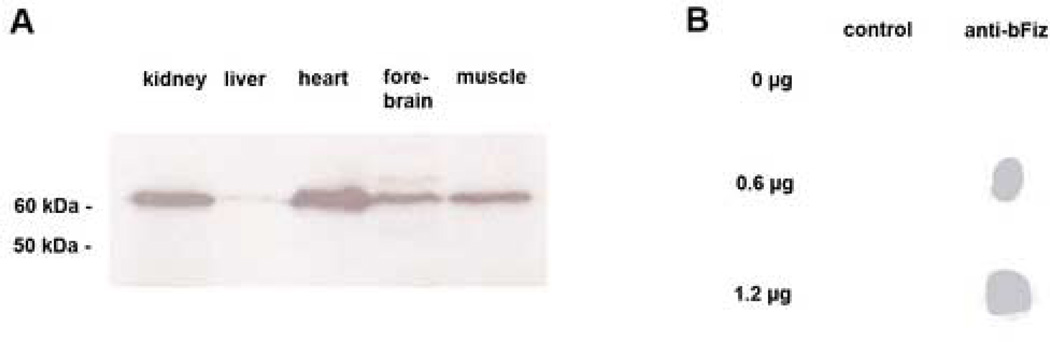
Previous analysis (northern blots) indicated that many mouse and human tissues express Fiz1 mRNA. (Mitton, et al., 2003, Wolf and Rohrschneider, 1999) However, data concerning FIZ1 protein for most non-retinal tissues was lacking. Our analysis shows that several of these tissues do contain FIZ1 protein. (Fig. 1A) Utilizing a polyclonal antibody, that was affinity purified to FIZ1 antigen, we detected a single major band in non-neural tissues and brain (approx. 65 KDa).
Fig. 1.
FIZ1 protein is present in several non-ocular tissues that express Fiz1 mRNA. Expression of FIZ1 protein in mouse tissues was examined by Disc-PAGE and immunoblotting with affinity-purified antibody to the FIZ1 protein. A. FIZ1 protein was detected in all tissues examined. (25 µg total protein per lane) Equivalent loadings were confirmed by direct staining of protein on the blot itself as described in methods. B. Dot blots of protein extracts from CV-1 cells transiently expressing the bovine FIZ1 protein used for production of the FIZ1 antibody. Lysates of cells transfected with 0 µg, 0.6 µg and 1.2 µg of the pDest26-bFiz mammalian expression plasmid were dotted onto nitrocellulose, and then incubated with or without (control) affinity-purified antibody to FIZ1.
The specificity of the antibody to FIZ1 in immunoblotting was confirmed by specific expression of the FIZ1 antigen in transfected CV-1 cells. (Fig. 1B) This same antigen was used to produce the antibody (in rabbits), and for the antigen-specific affinity purification of the final IgG preparation (Invitrogen). Cells with increasing amounts of bovine FIZ1 were extracted for protein (denaturing) and examined by dot-blot. Under the conditions used, the antibody is specific to FIZ1 antigen.
3.2. Neural retina FIZ1 protein expression increases during photoreceptor maturation
Previous investigators have shown that both Rho mRNA and protein levels begin to increase in the mouse retina around postnatal day P5-P6. This pattern holds for other rod visual transduction proteins, such as arrestin and PDE6B. Photoreceptors begin to form inner segments at P5 and are fully functional by P21. Outer-segment length and Rhodopsin protein concentration continue to increase until P30. (Bowes, et al., 1988)
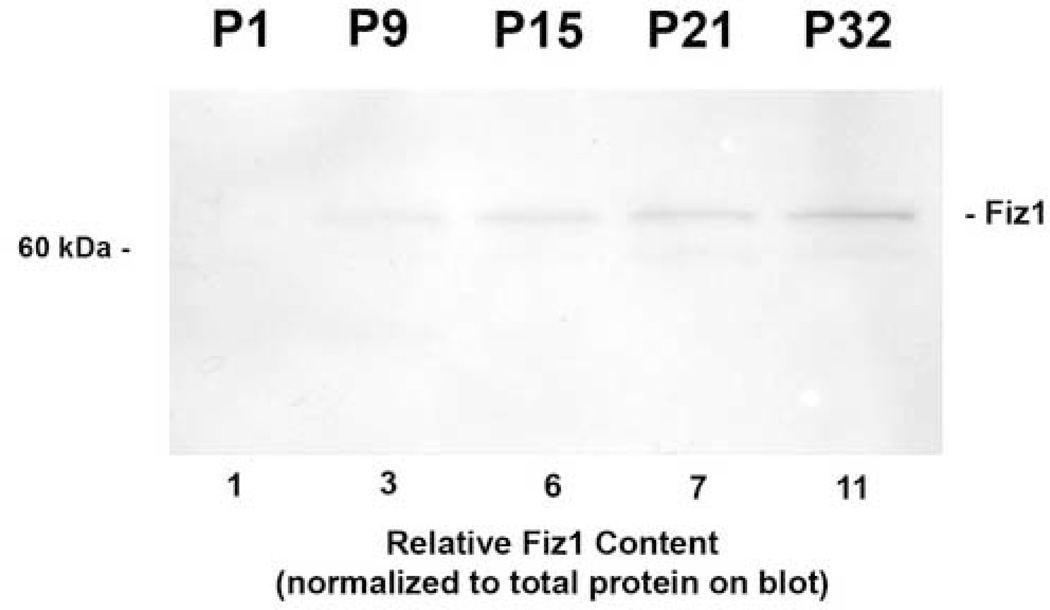
To examine the relative FIZ1 protein content of mouse neural retina throughout the retinal maturation period, samples were compared representing post-natal days P1, P9, P15, P21, and P32. (Fig. 2.) Measurement of the relative FIZ1 protein content revealed a constant increase throughout post-natal retinal development from the earliest time point, when FIZ1 content was relatively low. Image-based analysis of the relative FIZ1 content revealed at least a ten-fold increase from day P1 to day P32.
Fig. 2.
FIZ1 protein concentration increases during photoreceptor maturation. Relative levels of FIZ1 protein, in total neural retina (mouse), were examined by Disc-PAGE and immunoblotting with affinity-purified antibody to the FIZ1 protein. Relative FIZ1 protein content was determined using the total protein actually transferred to the blot, as described in materials and methods.
3.3. FIZ1 protein expression increases in photoreceptors and ganglion cell layer during retinal maturation
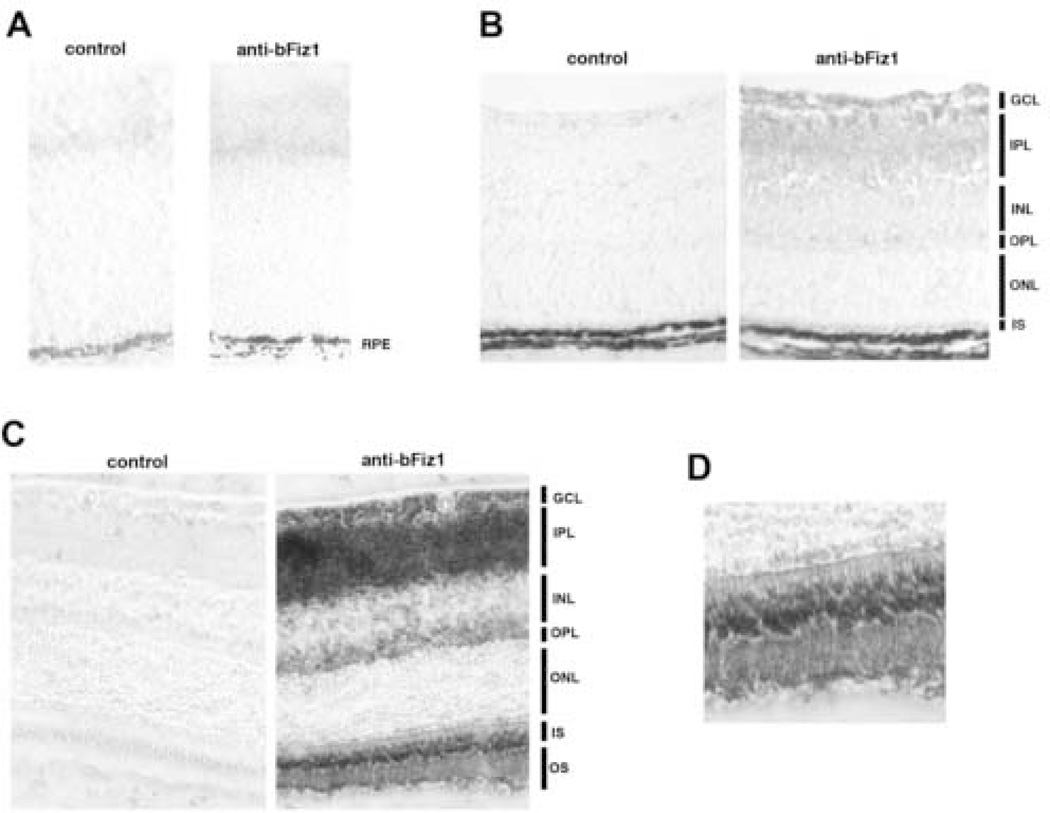
Next, we examined the expression of FIZ1 in fixed-frozen retinal sections at P0 (immature retina), P7 (early photoreceptor maturation) and P45 (fully mature retina). Results were consistent with the immunoblot data. At time P0, even with an extended peroxidase/DAB reaction time, there was no specific FIZ1 staining before the onset of background staining. (Fig. 3A.) FIZ1 protein was detectable at day P7, corresponded to the early stage of photoreceptor/neuronal maturation. Inner-segments were present at the P7 time point with faint immunolabeling for FIZ1. (Fig. 3B.) Low level staining for FIZ1 protein was also present at this time in ganglion cell layer.
Fig. 3.
FIZ1 protein content increases in photoreceptors and the ganglion cell layer during maturation of the mouse retina. A to C. Affinity purified antibody to bFIZ1 (rabbit). Control using pre-immune rabbit serum. Secondary antibody: biotinylated goat anti-rabbit. Detection: avidin-biotin-complex peroxidase, DAB/H2O2 substrate (Vector Labs). Visualized with phase contrast using equivalent exposures. A. Immature retina, day P0; the FIZ1 protein levels are below detection. B. Before outer-segment formation, day P7. The timing of increased FIZ1 protein content is similar in photoreceptors and the ganglion cell layer. Inner segments are just starting to form at P7. C. Mature adult retina. FIZ1 protein was detected in the photoreceptor inner (IS) and outer segments (OS), foot pedicle region in outer plexiform layer (OPL), ganglion cell layer (GCL) and inner plexiform layer (IPL). D. Enlargement of the inner and outer segment region as shown in C.
In adult retina, the immunostaining agreed with our previous immunofluorescent data, detecting FIZ1 protein in photoreceptors and the ganglion cell layer. (Mitton, et al., 2003) (Fig. 3C.) Labeling for FIZ1 was strong in the photoreceptor inner and outer segments of mature neural retina. (Fig. 3D.) FIZ1 is present in photoreceptor nuclei as evident from co-isolation with NRL (rod-specific) from retinal nuclear protein extracts; however, this particular antibody does not intensely label FIZ1 in photoreceptor nuclei of mouse retinal sections. (Mitton, et al., 2003) This may be a result of lower concentration in nuclei (compared to cytoplasm), and some masking of the antigen in vivo.
3.4. Neural retina Fiz1 mRNA content increases during photoreceptor maturation
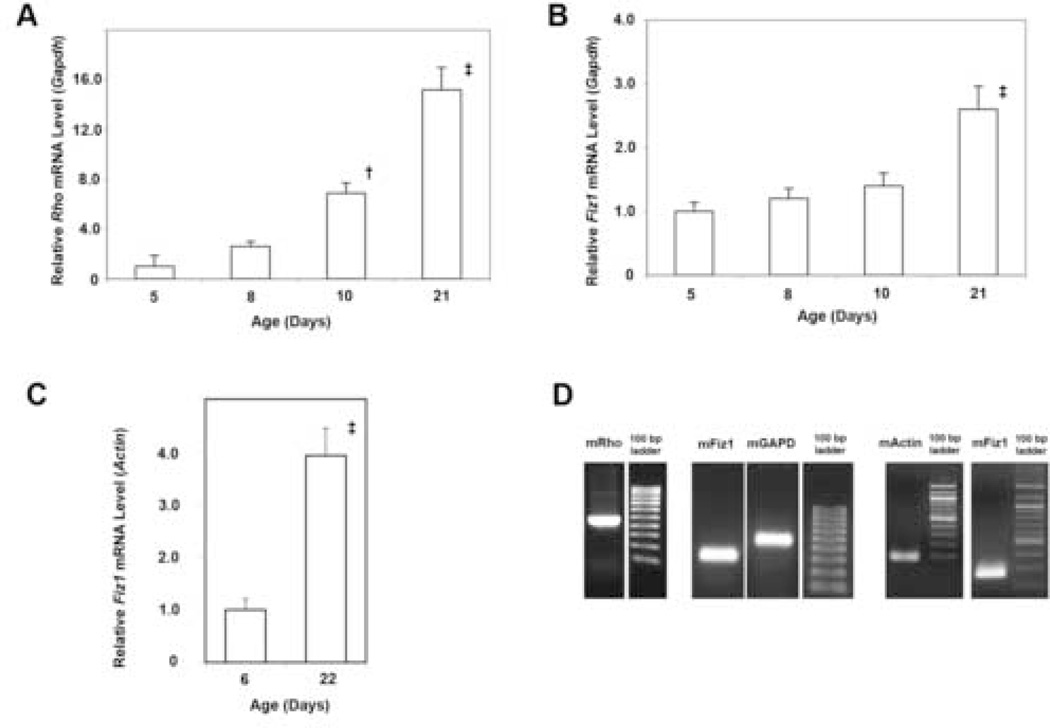
Several key rod-photoreceptor genes, such as Rho and Arrestin, are expressed during the period of photoreceptor maturation. We examined the expression of the Fiz1 gene compared to the Rho gene, since FIZ1 protein levels were increased during this same period. The relative mRNA levels for Rho and Fiz1 were examined at several time points, spanning the maturation period, using quantitative RT-PCR (real time). Rho gene expression increased throughout this period, as expected. (Fig. 4A.) This pattern agreed with previously published data (northern blot), and provided a genetic marker for photoreceptor-specific gene expression during maturation. (Bowes, et al., 1988) Relative Fiz1 mRNA levels also increased in concentration throughout the maturation period when examined relative to Gapdh mRNA levels using the Amplitaq Gold real-time PCR method. (Fig. 4B.)
Fig. 4.
Fiz1 Gene Expression Increases During Photoreceptor Maturation and the Onset of Rhodopsin Gene Expression. A. Total Rho mRNA, normalized to Gapdh, in the mouse neural-retina, measured with the Amplitaq-gold real-time PCR method. (Relative to age P5: † p<0.05; ‡ p<0.01, ANOVA Tukey-Kramer.) B. Total Fiz1 mRNA, normalized to Gapdh, in the mouse neural retina, measured with the Amplitaq-gold real-time PCR method. (Relative to age P5: ‡ p<0.01 .) C. Additional confirmation of total Fiz1 mRNA levels comparing immature and mature mouse neural retina using direct calibration to universal mouse reference RNA, and normalization to β-Actin mRNA. (‡ p<0.01) D. Examples of real-time PCR products for mouse Rho (mRho), Fiz1 (mFiz1), Gapdh (mGAPD) and β-Actin (mActin) using the primers in Tables 1 and 2. PCR products were confirmed by direct DNA sequencing.
As a confirmation, Fiz1 mRNA levels were also examined using a different tissue sample set with normalization to a different reference gene: β-Actin. Both Fiz1 and β-Actin mRNA levels were determined using absolute calibration with a Universal Mouse Reference RNA standard (Stratagene). This analysis method also indicated that neural retina Fiz1 mRNA levels increased 4-fold from the immature (P6) to the mature (P22) state. (Fig. 4C). This analysis was repeated twice. The levels of β-Actin mRNA per total retinal RNA from early (P6) and mature (P22) neural retina were not different. (Data not shown) While the data indicate Fiz1 mRNA levels do not increase as much as the very highly expressed Rho mRNA, a 3–4 fold increase was apparent.
3.5. Synergistic activation of Rho and PDE6B gene promoters by NRL, CRX and FIZ1
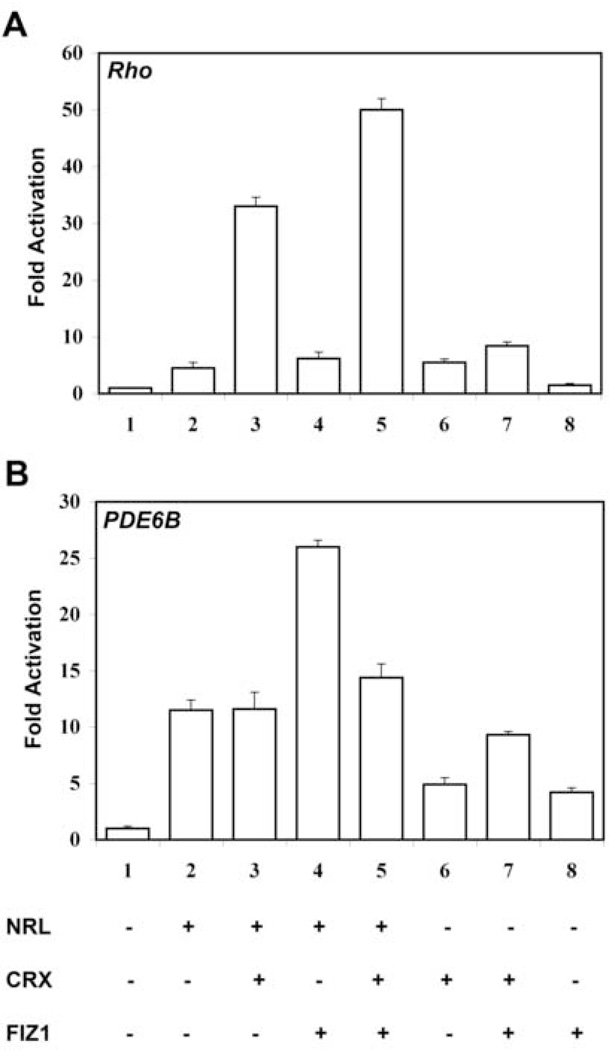
To explore the functional implications of NRL-FIZ1 interaction, we co-transfected CV1 cells with Rho- or PDE6B-promoter reporter plasmids and various combinations of NRL, CRX and FIZ1 expression constructs. FIZ1 synergistically activated NRL-mediated transcription of the Rho, and PDE6B gene promoters with some differences. Representative experiments are shown in Figure 5.
Fig. 5.
FIZ1 activation synergy with NRL and CRX at the Rho and PDE6B promoters.
A. Fold activation of the Rho proximal promoter in co-transfected CV-1 cells. FIZ1 synergy required the presence of both NRL and CRX. All transfection mixes contained the Rho-promoter luciferase-reporter construct, pBR-130luc, and the normalizer Renilla-luciferase construct pRL-CMV. Triplicate sample wells were co-transfected with expression vectors for NRL, CRX, and FIZ1 in the combinations as indicated (expression vector +, corresponding empty vector −). Firefly luciferase activity was normalized to Renilla-luciferase activity for each sample. Fold Activation is shown relative to reporter DNA in the presense of all empty vectors. Bars indicate SD (n = 3).
Fold activation of the PDE6B minimal-promoter in co-transfected CV-1 cells. FIZ1 synergy required the presence of NRL, and was inhibited by CRX. All transfection mixes contained the PDE6B-promoter firefly-luciferase reporter, pGL2-PDE6B-luc, and the normalizer Renilla-luciferase construct pRL-CMV. Triplicate sample wells were co-transfected with expression DNAs for NRL, CRX, and FIZ1 in the combinations as indicated (expression vector +, corresponding empty vector −). Firefly luciferase activity was normalized to Renilla-luciferase activity for each sample. Fold Activation is shown relative to reporter DNA in the presence of all empty vectors. Bars indicate SD (n = 3).
In Rho promoter assays (Figure 5A), FIZ1 alone had no effect on promoter activation. NRL, or CRX, could activate the promoter about 5-fold compared to empty vectors (p<0.01, ANOVA, Tukey-Kramer post analysis). NRL combined with CRX resulted in synergistic activation of the Rho promoter: 33-fold relative to empty vectors (p<0.01, Tukey-Kramer). These experiments utilized relatively more NRL and less Fiz1 DNA (more similar amounts) than previously found to inhibit NRL-mediated activation of the Rho promoter. Here, FIZ1 had little effect on NRL- or CRX-mediated activation of Rho promoter. However, with both NRL and CRX, FIZ1 increased the total activation synergy by over 30% relative to NRL plus CRX (p<0.01, Tukey-Kramer).
Using the same relative expression DNA concentrations for the PDE6B promoter (Figure 5B), CRX alone had no significant effect compared to empty vectors (N.D., Tukey-Kramer). The same was true for FIZ1 alone. CRX combined with FIZ1 did result in a 10-fold activation of the promoter relative to empty vectors (p<0.01, Tukey-Kramer). NRL alone was a strong activator of the PDE6B reporter: 12-fold relative to empty vectors (p<0.01, Tukey-Kramer). While CRX had no effect when added to NRL, FIZ1 was synergistic with NRL and doubled promoter activation to 26-fold (NRL versus NRL/FIZ1, p<0.01, Tukey-Kramer). Furthermore, when CRX was combined with NRL and FIZ1, there was a 40-percent decrease in the total promoter activity compared to NRL/FIZ1 (p<0.01, Tukey-Kramer). These results indicate that FIZ1 can promote NRL-mediated activation of this minimal PDE6B gene promoter.
4. Discussion
The Rhodopsin (Rho) gene is an excellent model to explore the pathways that precisely regulate rod-specific gene expression. Rho expression is rod-specific and the amount of Rhodopsin protein is regulated at the level of transcription. (Treisman, et al., 1988) Transcription factors that regulate the Rho gene also regulate other rod-specific genes that share a similar expression pattern. (Kumar, et al., 1996, Lerner, et al., 2001, Mears, et al., 2001, Rehemtulla, et al., 1996) Rho expression varies during development, dramatically increasing during maturation of rod outer-segments. (Bowes, et al., 1988) In mature retina, Rho expression continues to be regulated to follow the circadian patterns of rod outer-segment formation. (Bowes, et al., 1988, Matsumoto and Bok, 1984, von Schantz, et al., 1999)
The Rho proximal promoter region is sufficient for rod-specific expression and includes several DNA sequence elements that bind to proteins in vitro. These include the NRL response element (NRE). NRL binds to NRE elements in several rod-specific genes: Rho, Pde6b, Pde6a and Gnat-1. (Ahmad, 1995, Chen, et al., 1997, Chen and Zack, 1996, DesJardin and Hauswirth, 1996, Kumar, et al., 1996, Lerner, et al., 2001, Mears, et al., 2001, Morabito, et al., 1991, Nie, et al., 1996, Rehemtulla, et al., 1996) NRL can bind directly with CRX to synergistically co-activate the Rho promoter. (Mitton, et al., 2000) FIZ1 represents a model to study an interface between these crucial transcription factors and the more ubiquitous regulatory machinery of the cell.
4.1. FIZ1 protein expression in various tissues
Western blots of protein extracts from several tissues, previously shown to contain Fiz1 mRNA by northern blot, confirmed the presence of FIZ1 protein. Previous analysis of FIZ1 protein content in several hematopoietic progenitor and fibroblast cell-lines seemed to suggest that endogenous FIZ1 protein is easier to detect in more differentiated cells (i.e. NIH-3T3). (Wolf and Rohrschneider, 1999) We previously found that proliferating COS and CV-1 cells contain very low levels of FIZ1. (Mitton, et al., 2003) One could speculate that the function of FIZ1 is associated with processes during and after maturation of cell-type.
4.2. FIZ1 protein expression in retinal development
Examination of the mouse neural retina at several points of postnatal development revealed that retinal FIZ1 expression is developmentally regulated. Previous work had only examined mature retina. FIZ1 protein levels were minimal in neonatal mouse retina, when about half of the retinal precursor cells have not completed a final mitosis (P0 in the mouse.) A clear and dramatic increase, in FIZ1 protein content was seen through the period of neuronal maturation (approximately P5 to P21).
While 50% of the cells destined to the rod-photoreceptor fate are born by P0, expression of the Rho gene does not occur until P4-P5. This expression pattern is similar for other rod-specific visual transduction genes, including Pde6b. (Bowes, et al., 1988) If FIZ1 has influence on the expression level of photoreceptor specific genes, such as Rho, it is expressed during the correct time period.
In frozen sections of neural retina, immunolabeling for FIZ1 protein was quite intense in the inner segment region. The antibody used can detect FIZ1 protein in retinal nuclear protein extracts, but does not label FIZ1 well in nuclei on sections. (Mitton, et al., 2003) This could reflect sensitivity limitations on sections, or a masking of antigen. Several nuclear proteins have been difficult to detect in retinal sections, possibly due to masking of antigen within the large protein complexes involved in gene regulation. Different antibodies to NRL, for example, have displayed varied utility on retinal sections and blots. (Rehemtulla, et al., 1996, Swain, et al., 2001) The location of FIZ1 in photoreceptor nuclei is supported by co-immunoprecipitation with NRL from retinal nuclear extracts. (Mitton, et al., 2003) Intense labeling of FIZ1 in photoreceptor inner segments, suggests that this protein is also present in the photoreceptor cytoplasmic compartment.
The presence and appearance of FIZ1 in other regions of the retina, such as ganglion cells, suggests that FIZ1 may have a maturation-related function in these cells as well. During this time of development, retinal neurons begin to express genes that are functionally cell-specific (i.e. Rho), and synaptogenesis progresses in bipolar, photoreceptor, and ganglion cells. (Bachman and Balkema, 1993, Rich, et al., 1997) Could FIZ1 have different interactions with other transcription factors involved in maturation-related gene expression in these other retinal cell-types? At this time, we can only speculate.
4.3. FIZ1 can modulate NRL and CRX-mediated activation of rod specific gene promoters, Rho and PDE6B
Previously we demonstrated that FIZ1 interacts with NRL using yeast two-hybrid screening, GST-pulldown and co-immunoprecipitation from retinal nuclear-protein extracts. To test the functional implications of FIZ1/NRL interaction, we analyzed its role in NRL/CRX-mediated activation of the Rho, and PDE6B gene promoters in vitro. Previous tests had demonstrated an inhibition of NRL-mediated activation of the Rho promoter with greater quantities of FIZ1 expression DNA and lower quantity of NRL expression DNA. Continuing our explorations with this assay system, we included a second rod-specific promoter (PDE6B). We reduced the differences in quantities amoung the different expression plasmids and decreased the total DNA concentration, to minimize the over expression required for data collection. Under these parameters it is apparent that FIZ1 can contribute to the activation synergy at the minimal Rho, and PDE6B gene promoters. Lerner et al., (2005), reported a similar experience with the Sp4 transcription factor (Zn-finger protein). Excessive amounts of Sp4 expression vector had an inhibitory effect on promoter reporters, while a reduced amount synergized with CRX on the Rhodopsin promoter.
FIZ1 alone had little effect on the Rho proximal-promoter, and a small additive effect with NRL or CRX. When combined with NRL and CRX, FIZ1 synergized to give the maximum activation of all the combinations tested. With the PDE6B minimal-promoter, maximum activation resulted from FIZ1 in synergy with NRL alone. Addition of CRX to NRL and FIZ1 resulted in a decrease of NRL-FIZ1 synergy. We also observed slight activation of PDE6B promoter by CRX and FIZ1.
Different effects of FIZ1 at these promoters could be attributed, in part, to the different effects of NRL and CRX at these same promoters. Our results with NRL and CRX in co-transfection assays are consistent with previous reports. NRL and CRX clearly synergize on the Rho proximal-promoter, but CRX does not produce this strong effect on the PDE6B minimal-promoter, in spite of the presence of a CRX binding element (Chau, et al., 2000, Lerner, et al., 2002, Mitton, et al., 2000). Our results here, comparing the two promoters, show that CRX lacks synergy with NRL on the PDE6B promoter. However, recent Chromatin immunoprecipitation studies by Peng at al., (2005), indicate that CRX co-occupies the mouse Pde6b gene promoter with NRL and NR2E3 in vivo. Lerner et al., have recently shown that CRX can synergize with Sp4 (a zinc-finger transcription factor) to activate the Rho promoter, and may slightly inhibit Sp4-mediated activation of the PDE6B promoter. (Lerner, et al., 2005) Our results here indicate that CRX can decrease the activation synergy of FIZ1 with NRL at the PDE6B promoter.
At the molecular level, the different effects of NRL, CRX, and FIZ1 on these two promoters could result from the different combinations and spacing of DNA-binding elements in the promoters. It is also possible that an unknown factor may associate at one promoter and not the other; it is wise to keep in mind that we do not possess a complete inventory of all proteins present on these promoters in vivo. (Peng, et al., 2005) Furthermore, NRL and CRX interaction does not require interaction with DNA. (Mitton et al., 2000) Therefore, the same protein combinations could be present on different promoters, yet use a different balance of protein and DNA associations. These alternative structures could propagate different effects on the transcriptional apparatus.
Same-site reorientation is another mechanism, acting on bZIP proteins, that may have implications for the NRL-mediated promoters used in this study. Erlasnon, et al. (1996) have shown that the orientation of the AP-1 (Fos/Jun) DNA binding surface can be reversed, on the same DNA element, due to interaction of its leucine zipper with NFAT. This ability of bZIP proteins to reverse orientation, on the same DNA-binding element, would reverse the orientation of their N-terminal activation-domains. It is possible to envision similar effects on NRL.
Interaction of a bZIP and Homeodomain (HD) were discovered through studies of NRL/CRX association. (Mitton, et al., 2000) Likewise, CRX and NR2E3, interact via their DNA-binding domains. (Peng et al., 2005) Studies of Sp4 indicate that the Sp4 DNA-binding domain is also sufficient and necessary for interaction with the CRX HD. (Lerner, et al., 2005) Currently we do not know if FIZ1 has DNA-binding activity; however, the FIZ1-binding domain of NRL is the bZIP domain, including the leucine zipper. (Mitton et al., 2003) We would note that FIZ1 binding to NRL’s bZIP domain is compatible with a synergy effect. While the basic portion of a bZIP dimer interfaces with dsDNA, the leucine-zipper portion of the complex extends away from the DNA normal axis. This leucine-zipper dimer is free to interact with other proteins along its entire length. X-ray crystallographic models of NFAT and AP1, bound to DNA, have illustrated this clearly. (Chen et al., 1998)
The human FIZ1 gene is located near several other genes important to retinal development, such as the CRX gene, on Chromosome-19. This chromosome is particularly rich in Zf-proteins. Analysis of the human and mouse genome sequences does not reveal any significant homology for FIZ1 with other known or theoretical genes. This includes other Zf-proteins, some of which fall into closely related families based on overall amino-acid sequence. FIZ1 represents a unique Zf-protein in its overall structure. FIZ1 has eleven Zn-fingers organized into groups of four, two, two and three Zn-fingers. The N-terminal (four fingers) and C-terminal (three fingers) groups are capable of interacting with the kinase domain of the RTK, Flt-3/Flk-2. (Wolf and Rohrschneider, 1999)
Zn-finger domains typically provide for DNA-protein and protein-protein interactions. Zf-proteins feature prominently as regulators of gene repression, activation and chromatin reorganization (supporting both repression and activation). Several Zf-proteins (EKLF, SP1, GATA-1) utilize motifs of two or three Zn-fingers to recruit the chromatin remodeling complex SWI/SNF to the β-Globin gene promoter. (Armstrong, et al., 1998, Emerson, 2002, Kadam, et al., 2000) At this time we cannot rule out the possibility that FIZ1 may have the ability to interact directly with DNA, in light of the fact that FIZ1 has multiple Zn-finger domains. Promoter activation studies here cannot answer this particular question; however, it is possible for a protein to alter the activation context with or without DNA association. For example, current evidence suggests that NR2E3 exerts its effects through interaction with CRX without binding to DNA directly. (Peng et al., 2005)
NR2E3 (a nuclear receptor) is required for normal photoreceptor development, and recent studies have demonstrated NR2E3 can interact with CRX and regulate transcription at Opsin promoters. (Cheng, et al., 2004, Haider, et al., 2000, Peng, et al., 2005) Nuclear receptors, as a family, are noted for both repression and activation effects that are mediated through Zn-finger proteins. (Belandia, et al., 2002) Recent studies also indicate that the interaction of NR2E3 with NRL or CRX may not be direct and additional transcriptional regulatory proteins might be necessary for the formation of a stable complex in vivo. Appearance of FIZ1 during retinal maturation is consistent with our hypothesis that FIZ1 is required for the precise regulation of genes imparting cell-specific functionality. FIZ1 may be one additional regulatory protein, which forms a complex with NRL, CRX, Sp4 and NR2E3, and is necessary for regulating the precise control of photoreceptor-specific genes such as Rho and PDE6B.
In summary, FIZ1 protein content is developmentally regulated in post-natal retinal development. Total FIZ1 protein and mRNA content increase during the photoreceptor maturation period in mouse neural retina. The timing of this increase in Fiz1 expression corresponded to the increasing expression of photoreceptor-specific genes, such as Rho and PDE6B. Transient co-transfection studies indicate that FIZ1 can work with NRL and CRX, to alter the activation of rod-specific gene promoters. Further studies will examine the interaction of FIZ1 with CRX and NR2E3.
Acknowledgments
The authors wish to thank: Dr. Anand Swaroop (The University of Michigan), Dr. Debora Farber (ULCA), Dr. Donald Zack (Johns Hopkins Medical School) and Dr. Shiming Chen (Washington University) for providing reagents as noted in methods. Also, Richard Schlaf (Oakland University cytotechnology student) for technical assistance with co-transfection and luciferase assays. Funding support for this work: NEI/NIH grant R01EY014626 to K.M.; NEI/NIH research-support grant EY014803 to the Oakland University Eye Research Institute.
References
- Ahmad I. Mash-1 is expressed during ROD photoreceptor differentiation and binds an E-box, E(opsin)-1 in the rat opsin gene. Brain Res Dev Brain Res. 1995;90:184–189. doi: 10.1016/0165-3806(96)83500-0. [DOI] [PubMed] [Google Scholar]
- Akagi T, Inoue T, Miyoshi G, Bessho Y, Takahashi M, Lee JE, Guillemot F, Kageyama R. Requirement of multiple basic helix-loop-helix genes for retinal neuronal subtype specification. J Biol Chem. 2004;279:28492–28498. doi: 10.1074/jbc.M400871200. [DOI] [PubMed] [Google Scholar]
- Armstrong JA, Bieker JJ, Emerson BM. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- Bachman KM, Balkema GW. Developmental expression of a synaptic ribbon antigen (B16) in mouse retina. J Comp Neurol. 1993;333:109–117. doi: 10.1002/cne.903330109. [DOI] [PubMed] [Google Scholar]
- Belandia B, Orford RL, Hurst HC, Parker MG. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. Embo J. 2002;21:4094–4103. doi: 10.1093/emboj/cdf412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessant DA, Payne AM, Mitton KP, Wang QL, Swain PK, Plant C, Bird AC, Zack DJ, Swaroop A, Bhattacharya SS. A mutation in NRL is associated with autosomal dominant retinitis pigmentosa. Nat Genet. 1999;21:355–356. doi: 10.1038/7678. [DOI] [PubMed] [Google Scholar]
- Bowes C, Van Veen T, Farber DB. Opsin, G-protein and 48-kDa protein in normal and rd mouse retinas: developmental expression of mRNAs and proteins and light/dark cycling of mRNAs. Exp Eye Res. 1988;47:369–390. doi: 10.1016/0014-4835(88)90049-8. [DOI] [PubMed] [Google Scholar]
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carl M, Loosli F, Wittbrodt J. Six3 inactivation reveals its essential role for the formation and patterning of the vertebrate eye. Development. 2002;129:4057–4063. doi: 10.1242/dev.129.17.4057. [DOI] [PubMed] [Google Scholar]
- Chau KY, Chen S, Zack DJ, Ono SJ. Functional domains of the cone-rod homeobox (CRX) transcription factor. J Biol Chem. 2000;275:37264–37270. doi: 10.1074/jbc.M002763200. [DOI] [PubMed] [Google Scholar]
- Chen L, Glover JN, Hogan PG, Rao A, Harrison SC. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- Chen S, Zack DJ. Ret 4, a positive acting rhodopsin regulatory element identified using a bovine retina in vitro transcription system. J Biol Chem. 1996;271:28549–28557. doi: 10.1074/jbc.271.45.28549. [DOI] [PubMed] [Google Scholar]
- Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- Cheng H, Khanna H, Oh EC, Hicks D, Mitton KP, Swaroop A. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum Mol Genet. 2004;13:1563–1575. doi: 10.1093/hmg/ddh173. [DOI] [PubMed] [Google Scholar]
- Deangelis MM, Grimsby JL, Sandberg MA, Berson EL, Dryja TP. Novel Mutations in the NRL Gene and Associated Clinical Findings in Patients With Dominant Retinitis Pigmentosa. Arch Ophthalmol. 2002;120:369–375. doi: 10.1001/archopht.120.3.369. [DOI] [PubMed] [Google Scholar]
- Desjardin LE, Hauswirth WW. Developmentally important DNA elements within the bovine opsin upstream region. Invest Ophthalmol Vis Sci. 1996;37:154–165. [PubMed] [Google Scholar]
- Emerson BM. Specificity of gene regulation. Cell. 2002;109:267–270. doi: 10.1016/s0092-8674(02)00740-7. [DOI] [PubMed] [Google Scholar]
- Erlanson DA, Chytil M, Verdine GL. The leucine zipper domain controls the orientation of AP-1 in the NFAT. AP-l.DNA complex. Chem Biol. 1996;3:981–991. doi: 10.1016/s1074-5521(96)90165-9. [DOI] [PubMed] [Google Scholar]
- Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick JA, Duncan A, Scherer SW, Tsui LC, Loutradis-Anagnostou A, Jacobson SG, Cepko CL, Bhattacharya SS, Mcinnes RR. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91:543–553. doi: 10.1016/s0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- Haider NB, Jacobson SG, Cideciyan AV, Swiderski R, Streb LM, Searby C, Beck G, Hockey R, Hanna DB, Gorman S, Duhl D, Carmi R, Bennett J, Weleber RG, Fishman GA, Wright AF, Stone EM, Sheffield VC. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat Genet. 2000;24:127–131. doi: 10.1038/72777. [DOI] [PubMed] [Google Scholar]
- Haider NB, Naggert JK, Nishina PM. Excess cone cell proliferation due to lack of a functional NR2E3 causes retinal dysplasia and degeneration in rd7/rd7 mice. Hum Mol Genet. 2001;10:1619–1626. doi: 10.1093/hmg/10.16.1619. [DOI] [PubMed] [Google Scholar]
- Hayakawa F, Towatari M, Kiyoi H, Tanimoto M, Kitamura T, Saito H, Naoe T. Tandem-duplicated Flt3 constitutively activates STAT5 and MAP kinase and introduces autonomous cell growth in IL-3-dependent cell lines. Oncogene. 2000;19:624–631. doi: 10.1038/sj.onc.1203354. [DOI] [PubMed] [Google Scholar]
- Hojo M, Ohtsuka T, Hashimoto N, Gradwohl G, Guillemot F, Kageyama R. Glial cell fate specification modulated by the bHLH gene Hes5 in mouse retina. Development. 2000;127:2515–2522. doi: 10.1242/dev.127.12.2515. [DOI] [PubMed] [Google Scholar]
- Kadam S, Mcalpine GS, Phelan ML, Kingston RE, Jones KA, Emerson BM. Functional selectivity of recombinant mammalian SWI/SNF subunits Genes. Dev. 2000;14:2441–2451. doi: 10.1101/gad.828000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekar S, Perron M, Dorsky R, Harris WA, Jan LY, Jan YN, Vetter ML. Xath5 participates in a network of bHLH genes in the developing Xenopus retina. Neuron. 1997;19:981–994. doi: 10.1016/s0896-6273(00)80391-8. [DOI] [PubMed] [Google Scholar]
- Kumar R, Chen S, Scheurer D, Wang QL, Duh E, Sung CH, Rehemtulla A, Swaroop A, Adler R, Zack DJ. The bZIP transcription factor Nrl stimulates rhodopsin promoter activity in primary retinal cell cultures. J Biol Chem. 1996;271:29612–29618. doi: 10.1074/jbc.271.47.29612. [DOI] [PubMed] [Google Scholar]
- Lerner LE, Farber D. Transcriptional Regulation of the cGMP Phosphodiesterase β-Subunit Gene. Meth Enzymol. 2000;315:617–635. doi: 10.1016/s0076-6879(00)15871-9. [DOI] [PubMed] [Google Scholar]
- Lerner LE, Gribanova YE, Ji M, Knox BE, Farber DB. Nrl and Sp nuclear proteins mediate transcription of rod-specific cGMP-phosphodiesterase beta-subunit gene: involvement of multiple response elements. J Biol Chem. 2001;276:34999–35007. doi: 10.1074/jbc.M103301200. [DOI] [PubMed] [Google Scholar]
- Lerner LE, Gribanova YE, Whitaker L, Knox BE, Farber D. The Rod cGMP-phosphodiesterase B-Subunit Promoter is a Specific Target for Sp4 and is not Activated by Other Sp Proteins or CRX. J Biol Chem. 2002;227:25877–25889. doi: 10.1074/jbc.M201407200. [DOI] [PubMed] [Google Scholar]
- Lerner LE, Peng GH, Gribanova YE, Chen S, Farber DB. Sp4 is expressed in retinal neurons, activates transcription of photoreceptor-specific genes, and synergizes with Crx. J Biol Chem. 2005;280:20642–20650. doi: 10.1074/jbc.M500957200. [DOI] [PubMed] [Google Scholar]
- Marquardt T, Gruss P. Generating neuronal diversity in the retina: one for nearly all Trends. Neurosci. 2002;25:32–38. doi: 10.1016/s0166-2236(00)02028-2. [DOI] [PubMed] [Google Scholar]
- Martinez-Gimeno M, Maseras M, Baiget M, Beneito M, Antinolo G, Ayuso C, Carballo M. Mutations P51U and G122E in retinal transcription factor NRL associated with autosomal dominant and sporadic retinitis pigmentosa. Hum Mutat. 2001;17:520. doi: 10.1002/humu.1135. [DOI] [PubMed] [Google Scholar]
- Mathers PH, Jamrich M. Regulation of eye formation by the Rx and pax6 homeobox genes. Cell Mol Life Sci. 2000;57:186–194. doi: 10.1007/PL00000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto B, Bok D. Diurnal variations in amino acid incorporation into inner segment opsin. Invest Ophthalmol Vis Sci. 1984;25:1–9. [PubMed] [Google Scholar]
- Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- Mitton KP, Swain PK, Chen S, Xu S, Zack DJ, Swaroop A. The leucine zipper of NRL interacts with the CRX homeodomain. A possible mechanism of transcriptional synergy in rhodopsin regulation. J Biol Chem. 2000;275:29794–29799. doi: 10.1074/jbc.M003658200. [DOI] [PubMed] [Google Scholar]
- Mitton KP, Swain PK, Khanna H, Dowd M, Apel IJ, Swaroop A. Interaction of retinal bZIP transcription factor NRL with Flt3-interacting zinc-finger protein Fiz1: possible role of Fiz1 as a transcriptional repressor. Hum Mol Genet. 2003;12:365–373. doi: 10.1093/hmg/ddg035. [DOI] [PubMed] [Google Scholar]
- Moore KB, Schneider ML, Vetter ML. Posttranslational mechanisms control the timing of bHLH function and regulate retinal cell fate. Neuron. 2002;34:183–195. doi: 10.1016/s0896-6273(02)00666-9. [DOI] [PubMed] [Google Scholar]
- Morabito MA, Yu X, Barnstable CJ. Characterization of developmentally regulated and retina-specific nuclear protein binding to a site in the upstream region of the rat opsin gene. J Biol Chem. 1991;266:9667–9672. [PubMed] [Google Scholar]
- Morrow EM, Furukawa T, Lee JE, Cepko CL. NeuroD regulates multiple functions in the developing neural retina in rodent. Development. 1999;126:23–36. doi: 10.1242/dev.126.1.23. [DOI] [PubMed] [Google Scholar]
- Nakao M, Yokota S, Iwai T, Kaneko H, Horiike S, Kashima K, Sonoda Y, Fujimoto T, Misawa S. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996;10:1911–1918. [PubMed] [Google Scholar]
- Ng L, Hurley JB, Dierks B, Srinivas M, Salto C, Vennstrom B, Reh TA, Forrest D. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat Genet. 2001;27:94–98. doi: 10.1038/83829. [DOI] [PubMed] [Google Scholar]
- Nie Z, Chen S, Kumar R, Zack DJ. RER, an evolutionarily conserved sequence upstream of the rhodopsin gene, has enhancer activity. J Biol Chem. 1996;271:2667–2675. doi: 10.1074/jbc.271.5.2667. [DOI] [PubMed] [Google Scholar]
- Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, Furukawa T. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- Pacione LR, Szego MJ, Ikeda S, Nishina PM, Mcinnes RR. Progress toward understanding the genetic and biochemical mechanisms of inherited photoreceptor degenerations. Annu Rev Neurosci. 2003;26:657–700. doi: 10.1146/annurev.neuro.26.041002.131416. [DOI] [PubMed] [Google Scholar]
- Peng GH, Ahmad O, Ahmad F, Liu J, Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum Mol Genet. 2005;14:747–764. doi: 10.1093/hmg/ddi070. [DOI] [PubMed] [Google Scholar]
- Porter FD, Drago J, Xu Y, Cheema SS, Wassif C, Huang SP, Lee E, Grinberg A, Massalas JS, Bodine D, Alt F, Westphal H. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development. 1997;124:2935–2944. doi: 10.1242/dev.124.15.2935. [DOI] [PubMed] [Google Scholar]
- Rattner A, Sun H, Nathans J. Molecular genetics of human retinal disease. Annu Rev Genet. 1999;33:89–131. doi: 10.1146/annurev.genet.33.1.89. [DOI] [PubMed] [Google Scholar]
- Rehemtulla A, Warwar R, Kumar R, Ji X, Zack DJ, Swaroop A. The basic motif-leucine zipper transcription factor Nrl can positively regulate rhodopsin gene expression. Proc Natl Acad Sci U S A. 1996;93:191–195. doi: 10.1073/pnas.93.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich KA, Zhan Y, Blanks JC. Migration and synaptogenesis of cone photoreceptors in the developing mouse retina. J Comp Neurol. 1997;388:47–63. [PubMed] [Google Scholar]
- Swain PK, Chen S, Wang QL, Affatigato LM, Coats CL, Brady KD, Fishman GA, Jacobson SG, Swaroop A, Stone E, Sieving PA, Zack DJ. Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron. 1997;19:1329–1336. doi: 10.1016/s0896-6273(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Swain PK, Hicks D, Mears AJ, Apel IJ, Smith JE, John SK, Hendrickson A, Milam AH, Swaroop A. Multiple phosphorylated isoforms of NRL are expressed in rod photoreceptors. J Biol Chem. 2001;276:36824–36830. doi: 10.1074/jbc.M105855200. [DOI] [PubMed] [Google Scholar]
- Swaroop A, Wang QL, Wu W, Cook J, Coats C, Xu S, Chen S, Zack DJ, Sieving PA. Leber congenital amaurosis caused by a homozygous mutation (R90W) in the homeodomain of the retinal transcription factor CRX: direct evidence for the involvement of CRX in the development of photoreceptor function. Hum Mol Genet. 1999;8:299–305. doi: 10.1093/hmg/8.2.299. [DOI] [PubMed] [Google Scholar]
- Tan E, Wang Q, Quiambao AB, Xu X, Qtaishat NM, Peachey NS, Lem J, Fliesler SJ, Pepperberg DR, Naash MI, A1-Ubaidi MR. The relationship between opsin overexpression and photoreceptor degeneration. Invest Ophthalmol Vis Sci. 2001;42:589–600. [PubMed] [Google Scholar]
- Treisman JE, Morabito MA, Barnstable CJ. Opsin expression in the rat retina is developmentally regulated by transcriptional activation. Mol Cell Biol. 1988;8:1570–1579. doi: 10.1128/mcb.8.4.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Schantz M, Lucas RJ, Foster RG. Circadian oscillation of photopigment transcript levels in the mouse retina. Brain Res Mol Brain Res. 1999;72:108–114. doi: 10.1016/s0169-328x(99)00209-0. [DOI] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf I, Rohrschneider LR. Fiz1, a novel zinc finger protein interacting with the receptor tyrosine kinase Flt3. J Biol Chem. 1999;274:21478–21484. doi: 10.1074/jbc.274.30.21478. [DOI] [PubMed] [Google Scholar]
- Yan RT, Wang SZ. neuroD induces photoreceptor cell overproduction in vivo and de novo generation in vitro. J Neurobiol. 1998;36:485–496. [PMC free article] [PubMed] [Google Scholar]