Highlight
Alkaline stress disrupts iron deficiency responses in Cucumis species, implicating interference of shoot-to-root iron status signaling.
Key words: Bicarbonate, cucumber, fefe mutant, iron deficiency chlorosis, iron uptake, melon, shoot-to-root signaling.
Abstract
Iron (Fe) is an essential mineral that has low solubility in alkaline soils, where its deficiency results in chlorosis. Whether low Fe supply and alkaline pH stress are equivalent is unclear, as they have not been treated as separate variables in molecular physiological studies. Additionally, molecular responses to these stresses have not been studied in leaf and root tissues simultaneously. We tested how plants with the Strategy I Fe uptake system respond to Fe deficiency at mildly acidic and alkaline pH by measuring root ferric chelate reductase (FCR) activity and expression of selected Fe uptake genes and riboflavin synthesis genes. Alkaline pH increased cucumber (Cucumis sativus L.) root FCR activity at full Fe supply, but alkaline stress abolished FCR response to low Fe supply. Alkaline pH or low Fe supply resulted in increased expression of Fe uptake genes, but riboflavin synthesis genes responded to Fe deficiency but not alkalinity. Iron deficiency increased expression of some common genes in roots and leaves, but alkaline stress blocked up-regulation of these genes in Fe-deficient leaves. In roots of the melon (Cucumis melo L.) fefe mutant, in which Fe uptake responses are blocked upstream of Fe uptake genes, alkaline stress or Fe deficiency up-regulation of certain Fe uptake and riboflavin synthesis genes was inhibited, indicating a central role for the FeFe protein. These results suggest a model implicating shoot-to-root signaling of Fe status to induce Fe uptake gene expression in roots.
Introduction
Iron (Fe) is an important micronutrient that plays crucial roles in plant growth, development, and reproduction (Walker and Waters, 2011; Kobayashi and Nishizawa, 2012; Vigani et al., 2013). Iron uptake into roots of graminaceous plant species (known as Strategy II) is characterized by production and secretion of phytosiderophores that chelate Fe(III) for uptake (Kobayashi and Nishizawa, 2012). Iron uptake by non-graminaceous angiosperm species (known as Strategy I) is characterized by rhizosphere acidification by H+-ATPase proteins, reduction of Fe(III) to Fe(II) by ferric chelate reductase (FCR) proteins, and uptake of Fe(II) by iron transporter proteins (Kobayashi and Nishizawa, 2012). The activity of these Fe uptake proteins is up-regulated in Fe-deficient roots.
Many molecular components of the Strategy I Fe uptake system have been identified. FERRIC REDUCTION OXIDASE 2 (FRO2) encodes the primary root surface FCR in Arabidopsis (Robinson et al., 1999), and the corresponding genes in cucumber (Cucumis sativus L.) and melon (Cucumis melo L.) are called FRO1 (Waters et al., 2007, 2014). Iron (II) transporter genes include IRT1 (Eide et al., 1996; Varotto et al., 2002; Mukherjee et al., 2006) and NRAMP1 (Curie et al., 2000; Thomine et al., 2000; Cailliatte et al., 2010). AtFRO2, AtIRT1, AtNRAMP1, and numerous other genes are up-regulated under Fe deficiency by the basic helix–loop–helix (bHLH) transcription factor AtFIT (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005), a homolog of the tomato FER protein (Ling et al., 2002; Colangelo and Guerinot, 2004). AtFIT gene expression is typically up-regulated by Fe deficiency (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Lucena et al., 2006). A group of four closely related Arabidopsis bHLH genes, AtbHLH38, AtbHLH39, AtbHLH100, and AtbHLH101, are classified in clade Ib of the bHLH superfamily (Wang et al., 2007). The AtFIT protein regulates expression of its target genes as a heterodimer complex of AtFIT and a clade Ib bHLH protein (Colangelo and Guerinot, 2004; Yuan et al., 2008; Wang et al., 2013). The melon C940-fe mutant (fefe) (Nugent and Bhella, 1988, Nugent, 1994) is similar to the fit mutant, in that it does not up-regulate FCR activity and rhizosphere acidification under Fe deficiency, and is chlorotic (Nugent, 1994). Our characterization of the fefe mutant showed that 82 genes that were up-regulated by Fe deficiency in wild-type roots were not regulated by Fe deficiency in the fefe mutant, including key Fe uptake genes (Waters et al., 2014). Thus, both fit and fefe mutants have blocked Fe uptake responses upstream of Fe uptake genes.
Less is known about molecular Fe deficiency responses in leaves than in roots, as only a few studies have profiled genome-wide gene expression in leaves (Waters et al., 2012; Ivanov et al., 2012; Rodríguez-Celma et al., 2013a; Moran Lauter et al., 2014). Several Fe regulated genes respond to Fe deficiency in both leaf and root tissues, whereas some are specific to roots or leaves. While AtFIT is only expressed and regulated by Fe in roots, the transcripts of AtbHLH38, AtbHLH39, AtbHLH100, and AtbHLH101 are up-regulated in both roots and leaves of Fe-deficient Arabidopsis (Wang et al., 2007; Rodríguez-Celma et al., 2013a). In our previous Arabidopsis leaf microarray study (Waters et al., 2012), At1G47400 (named iron responsive protein 1 (AtIRP1) in Rodríguez-Celma et al., 2013a) and AtKCS17 (3-ketoacyl-CoA synthase, At4G34510) were among the most strongly up-regulated genes in Fe-deficient Arabidopsis leaves (10.2-fold for AtIRP1 and 36.0-fold for AtKCS, respectively). A more detailed knowledge of leaf Fe deficiency responses is needed to understand whole-plant adaptations to low Fe conditions, since a leaf-originated signal is thought to be necessary for normal regulation of Fe deficiency responses in roots (De Nisi et al., 2012; García et al., 2013).
Although Fe is abundant in soils, it has low solubility, especially in calcareous, alkaline soils, which occur on 30% of the earth (Chen and Barak, 1982). Plants can show iron deficiency chlorosis (IDC) on alkaline soils (Mengel and Geurtzen, 1986), resulting in reduced growth and yield (Hansen et al., 2004; Rogovska et al., 2007; Briat et al., 2015). Uptake of other metal micronutrients, such as Mn and Zn, can also be inhibited in alkaline soils (George et al., 2012). Soil alkalinity is largely due to bicarbonates (HCO3−) and carbonates (CO32−) (Coulombe et al., 1984; Mengel et al., 1984), and therefore, bicarbonate has been commonly used to induce IDC symptoms in hydroponic Fe nutrition studies (Coulombe et al., 1984; Chaney et al., 1992; Romera et al., 1992a; Lin et al., 1997; Waters and Troupe, 2012). However, whether IDC results from low Fe supply, alkaline stress, or a combination of these factors is still unclear. Most IDC studies to date have not treated Fe supply and alkalinity as separate variables. Studies that applied bicarbonate to both Fe-deficient and Fe-sufficient plants (Fleming et al., 1984; Romera et al., 1992a; Alcántara et al., 2000) were carried out prior to modern molecular methods, and thus it is not clear whether low Fe supply and alkaline stress cause equivalent molecular responses. In Strategy I species, bicarbonate-treated Fe-deficient plants had low root expression of FIT, FRO2, and IRT1, and had inhibited root FCR activity compared with Fe-deficient plants grown without bicarbonate (Romera et al., 1997; Lucena et al., 2007; García et al., 2014). However, we found that cucumber FCR activity was stimulated by bicarbonate treatment in plants supplied with Fe (Waters and Troupe, 2012). Except for Romera et al. (1992a), pH-matched control treatments have not been included in these studies. In pilot studies in our lab with sodium bicarbonate, potassium bicarbonate, and HEPES buffer, the FCR activity response of plants in pH buffered treatments was not distinguishable from the FCR response of bicarbonate-treated plants, regardless of the counter-ion. As such, we use the term alkalinity to refer to bicarbonate treatments. One of our objectives was to determine how pH influences physiological and molecular responses to Fe deficiency in roots and leaves.
Iron deficient Strategy I plant species have long been known to increase efflux of root exudates (Cesco et al., 2010). Some species, such as Arabidopsis thaliana, produce phenolic compounds (Fourcroy et al., 2014; Schmid et al., 2014; Schmidt et al., 2014) while other species, including cucumber and melon, produce flavin compounds (Susin et al., 1993; Welkie, 2000; Rodríguez-Celma et al., 2011b). Although the function of flavin compounds in plant Fe deficiency is not well defined, they may function in reduction or complexation of extracellular Fe to facilitate Fe acquisition (Cesco et al., 2010; Sisó‐Terraza et al., 2016). Proteins involved in riboflavin synthesis increased in abundance in response to Fe deficiency or Fe deficiency in alkaline conditions (Rellán-Álvarez et al., 2010; Rodríguez-Celma et al., 2011b) and genes involved in riboflavin biosynthesis were up-regulated in iron-deficient roots in normal or alkaline conditions (Rellán-Álvarez et al., 2010; Rodríguez-Celma et al., 2013b). However, the expression of riboflavin synthesis genes by alkaline stress separately from Fe deficiency has not been studied. Thus, another objective was to determine how riboflavin synthesis genes respond to Fe deficiency and alkaline stress in roots and leaves.
We addressed the objectives above using molecular and physiological approaches, by measuring leaf chlorosis, root FCR activity, Fe and other metal micronutrient accumulation, and expression of Fe uptake genes and riboflavin synthesis genes in cucumber roots and leaves. Our third objective was to use the melon fefe mutant to determine if alkaline-stimulated root gene expression depends on the fefe Fe deficiency regulatory pathway. The results of this research will lead to increased understanding of how alkaline stress inhibits Fe uptake and causes IDC, and will allow improved design of future studies to develop alkaline stress-tolerant crop varieties.
Materials and methods
Plant materials and growth conditions
Cucumber seeds of cv Ashley were purchased from Eden Brothers (Asheville, NC, USA) and seeds of Miniature White were purchased from Jonny’s Selected Seeds (Winslow, ME, USA). Melon seeds of cv Edisto were purchased from Victory Seed Company (Molalla, OR, USA), and seeds of C940-fe (fefe; Nugent, 1994) were a gift from Michael A. Grusak (USDA-ARS Children’s Nutrition Research Center, Houston, TX, USA). Seeds were germinated in germination paper soaked with 0.1mM CaSO4 and incubated in the dark at 30 °C for 3 days. Seedlings were transferred to black tubs (four seedlings per tub) with 750mL of nutrient solution made with 1.5mM KNO3, 0.8mM Ca(NO3)2, 0.3mM (NH4)H2PO4, 0.2mM MgSO4, 25 μM CaCl2, 25 μM H3BO3, 2 μM MnCl2, 2 μM ZnSO4, 0.5 μM Na2MoO4, 0.1 μM CuSO4, and 1mM MES buffer (pH 5.5). Iron was supplied as Fe(III)–ethylenediamine-N,N′-bis(2-hydroxyphenylacetic acid) (EDDHA) (Sprint 138, Becker-Underwood) at concentrations indicated below. The Fe(III)–EDDHA chelate is stable at the mildly acidic and alkaline pH used in this study (Chaney et al., 1972, Halvorson and Lindsay, 1972). Plants were grown in a growth chamber at 22 °C with a 16h photoperiod and photosynthetic photon flux density of 300 μmol m−2 s−1 photosynthetically active radiation. Bicarbonate was supplied as potassium bicarbonate (Fisher Scientific) or sodium bicarbonate (Arm & Hammer) with no difference in results. Plants were grown for 4 days in complete solution with 0.5, 1.0, 2.5, or 10 μM Fe (pretreatment), followed by 3 days of treatment with bicarbonate (10mM) at the same Fe concentration as during the pretreatment period. At the end of the treatment, the first true leaf was still growing and the second leaf was emerging (see Supplementary Fig. S1 at JXB online for plant DW). Final nutrient solution pH was determined using a pH meter at the end of the treatment period.
Ferric chelate reductase activity and chlorophyll content
Ferric chelate reductase activity was measured using whole roots of individual cucumber plants after 3 days of treatment. Roots were excised, rinsed in deionized water, and submerged in 20ml assay solution (1mM MES buffer, pH 5.5, 150 μM Fe(III)–EDTA, and 200 μM ferrozine) for 30–60min. Ferrozine–Fe(II) was measured by absorbance at 562nm (subtracting blanks of assay solution with no plants) and reduced Fe was calculated using the extinction coefficient of 28.16mM cm−1. Chlorophyll of the first true leaf was determined using a SPAD-502 chlorophyll meter (Minolta). Significance of differences between Fe treatments at each bicarbonate level, and between bicarbonate treatments at each Fe level were determined by Student’s t-test.
Quantification of dry weight and mineral content
Note that with Fe(III)EDDHA as an Fe source, Fe does not accumulate in the apoplast, in contrast to Fe sources such as Fe(III)EDTA (Longnecker and Welch, 1990; Strasser et al., 1999) or FeSO4 (Waters and Blevins, 2000). Plants were dissected into roots, first true leaf, and the remainder of the plant (stem+cotyledons) after 3d treatments and dried at 70 °C in a drying oven. After measuring dry weight (DW), tissue samples were digested in concentrated nitric acid–hydrogen peroxide with stepwise heating at 100, 125, 150, and 165 °C to dryness, and then resuspended in 5ml 1% HNO3 (Guttieri et al., 2015). Iron, Cu, Zn, and Mn contents for plant parts were quantified by inductively coupled plasma mass spectrometry in the University of Nebraska Redox Biology Center Spectroscopy Facility. Total plant DW or mineral content for each replicate plant was determined by summing plant parts. Significant differences between treatments for DW and each mineral were determined by one-way ANOVA.
RNA preparation, RT-PCR and real-time PCR analyses
Total RNA was extracted from approximately 80mg of frozen tissues of cucumber or melon plants using the RNeasy Plant Mini Kit (Qiagen). RNA samples were treated with DNaseI (Promega, USA) and RNA quality and concentration were determined by A260/A280 ratio. For reverse transcription reactions, 3 μg of total RNA was used with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, CA, USA). Single-stranded cDNA corresponding to 15ng of total RNA was used as a template in 15 μl total volume for real-time PCR assay. Real-time PCRs were carried out with 667nM of gene-specific primers and GoTaq qPCR Master Mix (Promega, USA) using an IQ5 MyiQ detection system (Bio-Rad, Hercules, CA, USA). Coding sequences for cucumber and melon bHLH38, bHLH101, NRAMP1, RIBA1, PYRD, PHS1, DMRLs and F6’H1 (Table 1) were identified from Cucurbit Genomics Database (http://www.icugi.org/cgi-bin/ICuGI/index.cgi) or from the Melonomics database (https://melonomics.net/) by BLAST searches and phylogenetic analysis. Primer sequences (Supplementary Table S1) were designed using the NCBI primer design tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi). The thermal cycler program was one initial cycle of 95 °C, 8min 30s; followed by cycles of 95 °C, 10s; 57 °C, 30s; 72 °C, 15s, with 40 cycles; followed by melt curve analysis for all genes. The relative gene expression considered the 10 μM Fe–0 bicarbonate-treated roots as control, and was calculated using the equation Y= where
Table 1.
Riboflavin synthesis pathway gene IDs and descriptions in roots of cucumber (Cucumis sativa), melon (Cucumis melo), and Arabidopsis thaliana. Normalized read counts for wild-type melon Edisto and the fefe mutant roots at normal Fe supply (10 μM) and under no added Fe conditions are from Waters et al. (2014). Significant fold-changes (FC) in read counts in the Fe-deficient samples are shown in bold.
| Gene name | Cucumber ID | Melon ID | Arabidopsis ID | Description | WT +Fe |
WT –Fe |
WT FC |
fefe +Fe |
fefe –Fe |
fefe FC |
|---|---|---|---|---|---|---|---|---|---|---|
| RIBA1 | Csa4M111580 | Melo3C024826 | AT5G64300 | GTP cyclohydrolase II | 656 | 11 118 | 17.0 | 118 | 110 | 0.9 |
| PYRD | Csa6M003430 | MU51870 | AT4G20960 | Diaminohydroxyphosphoribosylamino- pyrimidine deaminase | 369 | 505 | 1.4 | 415 | 394 | 0.9 |
| PHS1 | Csa1M655920 | Melo3C010048 | AT3G47390 | Pyrimidine reductase | 177 | 1670 | 9.4 | 27 | 25 | 1.0 |
| DMRLs | Csa6M366300 | MU59012 | AT2G44050 | Dimethyl-8-ribityllumazine synthase | 1838 | 6234 | 3.4 | 1690 | 1142 | 0.7 |
| RIBC | Csa6M128550 | MU45607 | AT2G20690 | Riboflavin synthase α chain | 220 | 385 | 1.8 | 302 | 233 | 0.8 |
At least three independent RNA extractions (from three different plants) and RT-qPCR reactions with two technical replicates per sample were performed. Significance of differences between Fe treatments at each bicarbonate level, and between bicarbonate treatments at each Fe level were determined by Student’s t-test.
Results
Physiological Fe deficiency and alkaline stress responses in cucumber
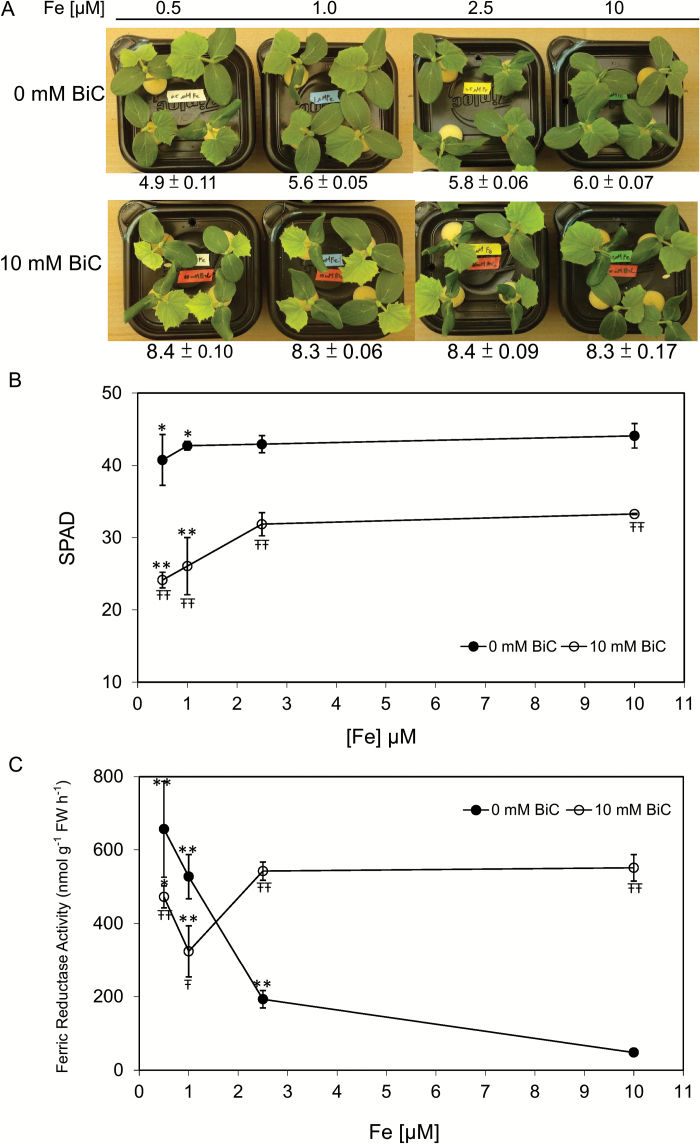
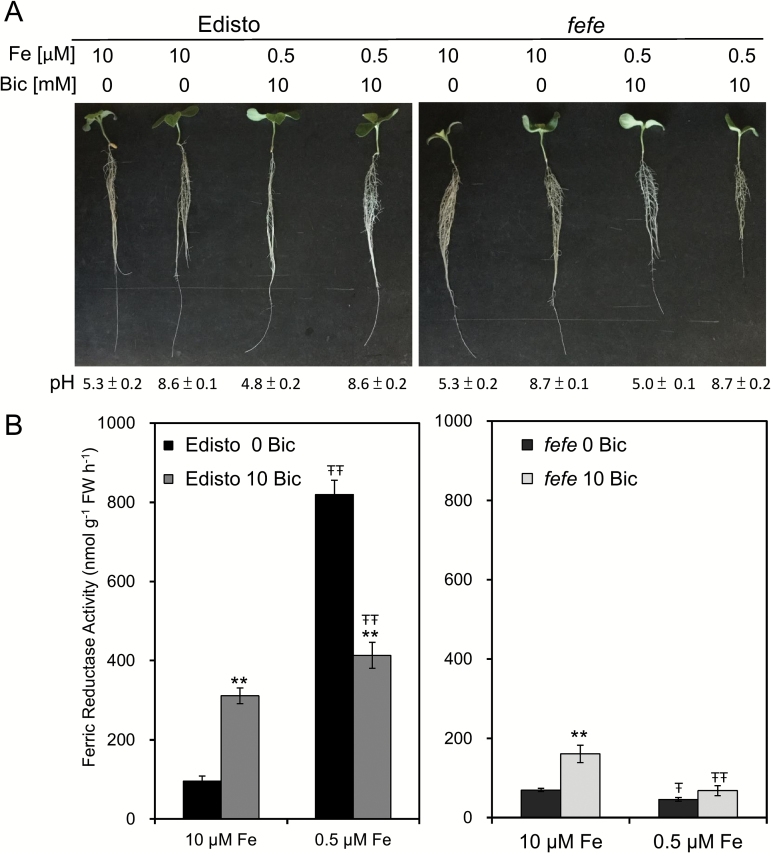
To understand the plant response to Fe supply at normal hydroponic pH and at alkaline pH, we grew plants with or without bicarbonate over a range of Fe supply, using pH-stable Fe(III)–EDDHA to ensure that Fe was soluble at all pH treatments (Chaney et al., 1972; Halvorson and Lindsay, 1972). Without bicarbonate, final solution pH was lower (pH 4.9) at the lowest Fe supply (0.5 μM) compared with final solution pH of the 10 μM Fe control (pH 6.0; Fig. 1A), likely reflecting increased H+-ATPase activity at low Fe supply. With bicarbonate, the final solution pH did not vary by Fe supply, and was 8.3–8.4 in all treatments. The first leaf of cucumber plants was slightly more chlorotic at the lowest Fe supply without bicarbonate, indicating that none of the Fe treatments caused severe Fe deficiency at normal pH. With alkaline stress (10mM bicarbonate), the chlorophyll content of the first leaf was significantly lower at each Fe concentration than in plants without bicarbonate (Fig. 1B). The root FCR activity was highest at 0.5 μM Fe supply without bicarbonate, and was also significantly up-regulated at 1.0 and 2.5 μM Fe (Fig. 1C). However, plants supplied with bicarbonate had similar FCR activity at all Fe concentrations. The FCR activity of plants treated with bicarbonate was significantly higher than that of plants without bicarbonate at 2.5 and 10 μM Fe, but was lower than that of plants without bicarbonate at 0.5 μM Fe. Thus, while low Fe supply and alkaline stress both resulted in elevated FCR activity in cucumber roots, the patterns were quite different. Comparing bicarbonate treatments at specific Fe supplies, bicarbonate caused either inhibition (0.5 μM Fe) or stimulation (10 μM Fe) of FCR activity. We obtained similar chlorophyll and FCR activity results with another variety of cucumber (Miniature White; Supplementary Fig. S2). These results indicated that alkaline stress elevated the root FCR activity relative to Fe-replete plants, but abolished the normal response to Fe supply.
Fig. 1.
Cucumber (cv. Ashley) plant responses to Fe supply in normal pH or alkaline nutrient solution. Cucumber seedlings were pre-treated in hydroponic solution with 0.5, 1.0, 2.5, or 10 μM Fe for 3 days, and then supplied without or with 10mM bicarbonate (BIC) for 4 days. (A) Photograph of the plants in each treatment, with final solution pH (means±SD, n=6) indicated below photograph. (B) Chlorophyll level of first leaf as measured by Minolta SPAD chlorophyll meter. (C) Ferric chelate reductase activities of roots. * and ** indicate statistical significance of P<0.05 and P<0.01, respectively, compared with 10 μM Fe within each curve; Ŧ and ŦŦ indicate statistical significance of P<0.05, P<0.01, respectively, comparing 10mM bicarbonate with 0mM bicarbonate at each Fe supply.
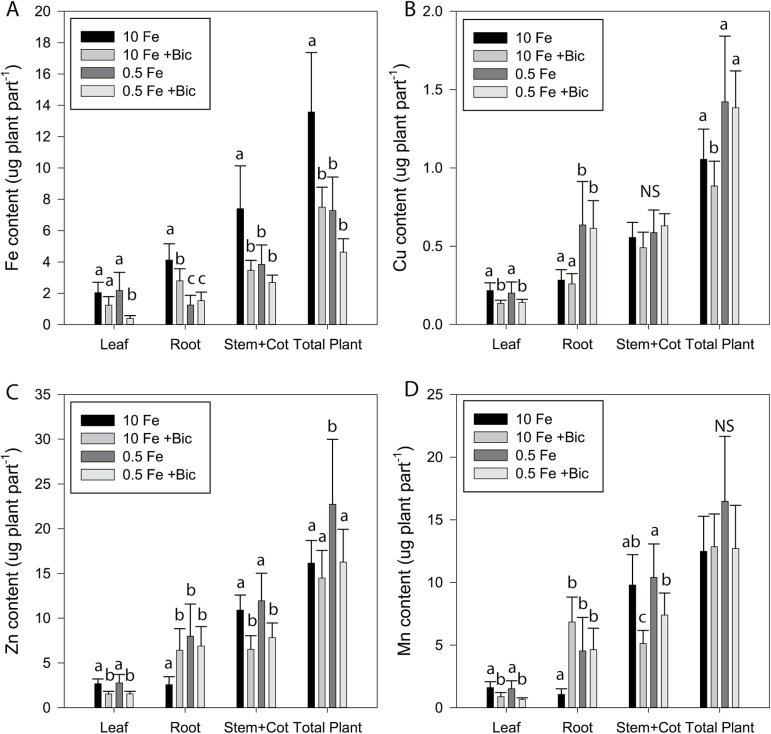
For an additional comparison of plant response to Fe deficiency and alkaline stress, we measured plant biomass and mineral accumulation in plant tissues. Biomass of the total plant or plant parts was not significantly affected by the treatments, except for roots of 0.5 Fe–10mM bicarbonate-treated plants, which were slightly larger than roots of the other treatments (Supplementary Fig. S1). The control plants grown with 10 μM Fe had the highest total Fe content, which decreased by about half under mild Fe deficiency (0.5 μM Fe; Fig. 2A). Bicarbonate treatment at 10 μM Fe supply resulted in an approximately 40% decrease in total Fe content, indicating that alkaline stress inhibited Fe uptake, despite using a pH-stable Fe source. The low Fe, bicarbonate-treated plants had the lowest Fe content in leaves, at 19% of the control value. Iron content decreased in roots and stem+cotyledon with low Fe and/or bicarbonate treatment (Fig. 2A). For other metal micronutrients, Cu, Zn, and Mn contents decreased in leaf tissue of bicarbonate-treated plants in both Fe supply regimes (Fig. 2B–D). In stem+cotyledon, Zn and Mn decreased under bicarbonate treatment, while Cu content was unchanged. In root, Mn and Zn increased in low Fe and bicarbonate treatments, while Cu increased only in low Fe treatments.
Fig. 2.
Mineral content in cucumber (cv. Ashley) plants. Cucumber seedlings were transferred into hydroponics with 0.5 or 10 μM Fe for 4 days pretreatment, and then treated with or without 10mM bicarbonate for 3 days. The roots, first leaf, and stem+cotyledons of each plant were harvested separately. (A) Fe content, (B) Cu content, (C) Zn content, and (D) Mn content in different parts of plants and whole plants. Bars represent mean±SD (n=8). Different letters indicate significant differences (P<0.05) based on ANOVA using the Holm–Sidak method; NS indicates no significant differences between treatments.
Molecular responses to Fe deficiency and alkaline stress in cucumber
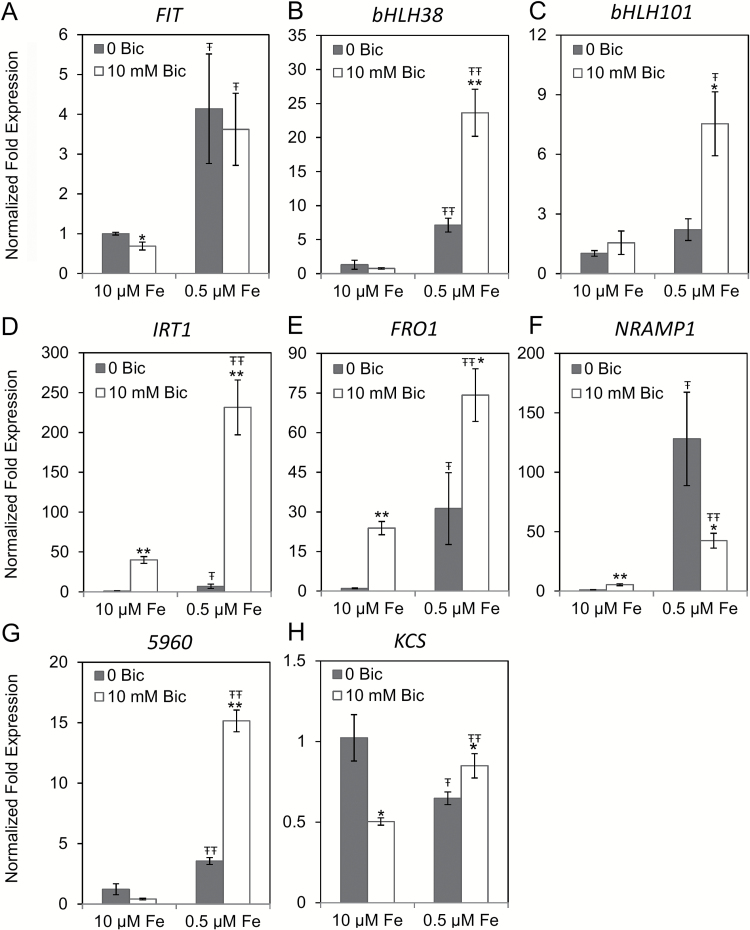
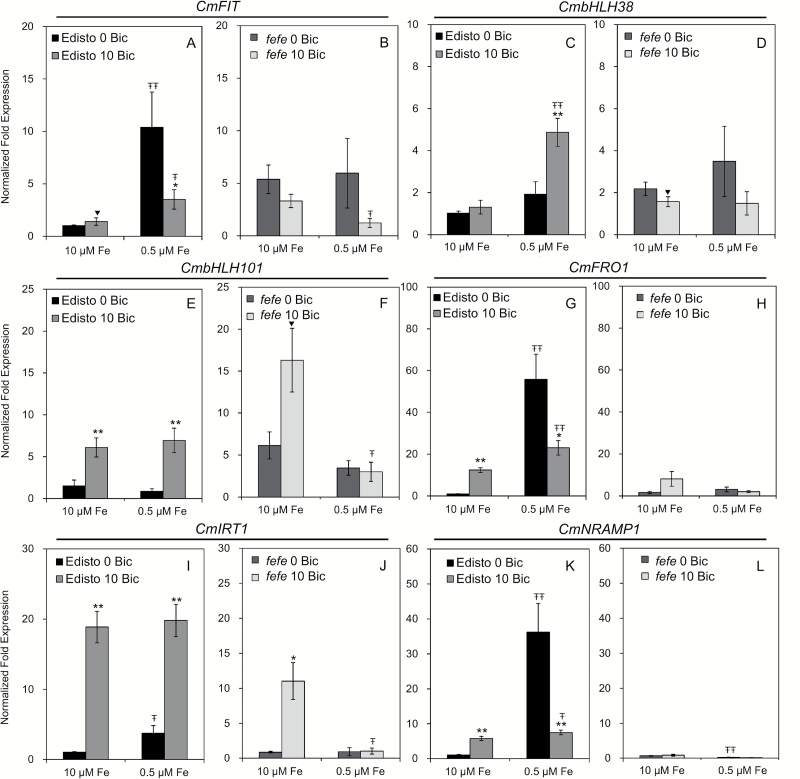
To gain a molecular understanding of similarities and differences between molecular responses to Fe deficiency and alkaline stress, we measured gene expression of known Fe deficiency responsive genes in root and first leaf tissue of cucumber plants. The Fe uptake transcription factor genes CsFIT and CsbHLH38 were up-regulated by Fe deficiency in roots (Fig. 3A, B), but CsFIT was not further induced by bicarbonate treatment at low Fe and was not induced by bicarbonate at replete Fe. Transcripts of CsbHLH38 and CsbHLH101 were greatly induced by bicarbonate in roots with low Fe supply, but there was no stimulation by bicarbonate in roots with normal Fe supply (Fig. 3B, C). The expression of CsIRT1 and CsFRO1 was increased under Fe deficiency (Fig. 3D, E), and their expression was increased further by bicarbonate at low Fe concentration. Bicarbonate also stimulated expression of these genes at normal Fe supply. In contrast, while the expression of CsNRAMP1 was strongly induced by Fe deficiency (Fig. 3F), bicarbonate treatment at low Fe supply resulted in much lower induction compared with the treatment without bicarbonate. Two additional genes based on previous leaf microarray results from Fe-deficient Arabidopsis (Waters et al., 2012) were studied. The 5960 gene (Csa2M005960, homologous to AtIRP1) had an expression pattern similar to bHLH38. The CsKCS gene (Csa2M361630) was not consistently regulated by Fe or bicarbonate in roots, although some treatments were statistically significant.
Fig. 3.
Quantitative RT-PCR analyses of transcript levels of Fe uptake-related or Fe regulated genes in cucumber (cv. Ashley) roots after 3 days of treatment. The results were normalized to ubiquitin as an internal reference. The transcript abundance of each gene (means±SE, n=3) was normalized to roots of the control treatment (10 μM Fe and 0mM bicarbonate). * and ** indicate statistical significance at P<0.05 and P<0.01, respectively, comparing bicarbonate treatments within the same Fe supply; Ŧ and ŦŦ indicate statistical significance at P<0.05 and P<0.01, respectively, comparing different Fe supply within the same bicarbonate treatment.
In leaf tissue, two patterns of gene expression were apparent, both of which were substantially different from root gene expression patterns. CsFIT, CsFRO2, or CsNRAMP1 transcripts were not detected in leaf tissue. None of the other genes’ expression increased in leaf in response to bicarbonate at full Fe supply (Fig. 4). The transcripts of CsbHLH38 and Cs5960 were highly induced in leaf tissue under Fe deficiency, but this induction was completely abolished in the leaf of bicarbonate-treated plants (Fig. 4A, D). However, these transcripts were synergistically increased by Fe deficiency and alkaline stress in roots (Fig. 3B, G). CsbHLH101 and CsIRT1 were induced by Fe deficiency, to a lower extent than in roots, but the addition of bicarbonate diminished their expression in the Fe-deficient leaf to control levels (Fig. 4B, C), whereas their expression was stimulated in roots (Fig. 3C, D). The expression of CsKCS was induced by Fe deficiency in leaf, but not in root (Figs. 3H and 4E). Thus, alkaline stress interfered with the molecular leaf response to low Fe supply.
Fig. 4.
Quantitative RT-PCR analyses of transcript levels of Fe uptake-related or Fe regulated genes in cucumber (cv. Ashley) first leaf tissue after 3 days of treatment. The results were normalized to ubiquitin as an internal reference. The transcript abundance of each gene (means±SE, n=3) was normalized to transcript abundance in roots of the control treatment (10 μM Fe and 0mM bicarbonate). * and ** indicate statistical significance at P<0.05 and P<0.01, respectively, comparing bicarbonate treatments within the same Fe supply; Ŧ and ŦŦ indicate statistical significance at P<0.05 and P<0.01, respectively, comparing different Fe supply within the same bicarbonate treatment.
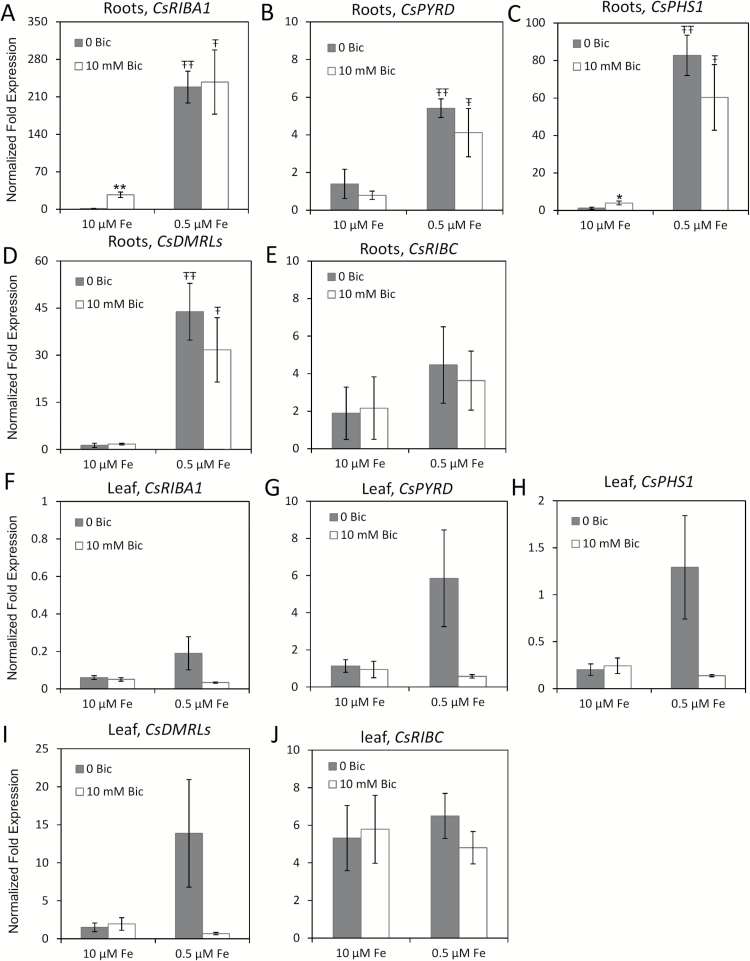
Expression of riboflavin synthesis pathway genes
We identified homologs of genes known to be involved in the riboflavin synthesis pathway (Table 1) and measured their gene expression in response to Fe deficiency and alkaline stress. The riboflavin synthesis genes were up-regulated by Fe deficiency in roots, except for CsRIBC (Fig. 5). The addition of bicarbonate to the low Fe treatment did not greatly change the transcript levels, but CsRIBA1 and CsPHS1 riboflavin synthesis genes were significantly induced by bicarbonate under 10 μM Fe supply. Thus, plant Fe status seems to be a more important factor than pH for induction of riboflavin synthesis genes, in contrast to the Fe uptake genes. In the leaf, none of the riboflavin synthesis genes were up-regulated by bicarbonate treatment at full Fe supply. Three of the genes were up-regulated by Fe deficiency, but this up-regulation was abolished when bicarbonate treatment was also applied (Fig. 5F–J), similar to what we observed for other leaf genes (Fig. 4).
Fig. 5.
Quantitative RT-PCR analyses of riboflavin synthesis genes in cucumber (cv. Ashley) roots (A–E) and first leaf (F–J) after 3 days of treatment. The results were normalized to ubiquitin as an internal reference. The transcript abundance of each gene (means±SE, n=3) was normalized to transcript abundance in roots of the control treatment (10 μM Fe and 0mM bicarbonate). * and ** indicate statistical significance at P<0.05 and P<0.01, respectively, comparing bicarbonate treatments within the same Fe supply; Ŧ and ŦŦ indicate statistical significance at P<0.05 and P<0.01, respectively, comparing different Fe supply within the same bicarbonate treatment.
Alkaline stress responses in fefe melon plants
Since our results with cucumber indicated that plant Fe uptake genes were up-regulated by both alkaline stress and Fe deficiency, we tested whether up-regulation of their melon counterparts requires a functional FeFe gene. The WT line Edisto was not strongly chlorotic under low Fe (0.5 μM Fe) or alkaline pH (10mM bicarbonate) stress. The fefe mutant was more sensitive to low Fe supply, and fefe roots had a yellow coloration under low Fe with bicarbonate (Fig. 6A, right). Edisto FCR activity was stimulated by bicarbonate at 10 μM Fe supply. FCR activity was stimulated by Fe deficiency (0.5 μM), but this activity was repressed by adding bicarbonate to the low Fe treatment (Fig. 6B, left). FCR activity was not up-regulated in fefe melon at low Fe supply, but FCR was significantly stimulated by bicarbonate at 10 μM Fe, but to a lower extent than in Edisto (Fig. 6B, right). This indicated that alkaline stress stimulation of FCR activity is not entirely dependent on the FeFe protein.
Fig. 6.
Melon plant growth and ferric chelate reductase activity with Fe deficiency and bicarbonate treatments. Wild-type (Edisto) and fefe mutant seedlings were grown in hydroponics with 0.5 or 10 μM Fe for pretreatment for 4 days, and then supplied without or with 10mM bicarbonate for 3 days. (A) Photograph of the plants. The final nutrient solution pH of each treatment (means±SE, n=6) is indicated below each plant. (B) Ferric chelate reductase activities of roots after 4 days. * and ** indicate statistical significance at P<0.05 and P<0.01, respectively, comparing bicarbonate treatments within the same Fe supply; Ŧ and ŦŦ indicate statistical significance at P<0.05 and P<0.01, respectively, comparing Fe supply treatments within the same bicarbonate treatment.
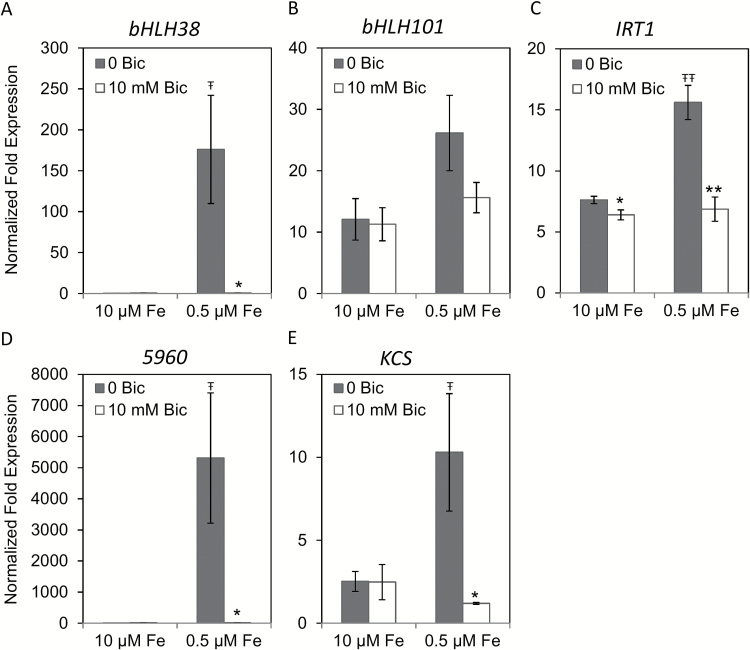
In the WT roots, the CmFIT gene was up-regulated by Fe deficiency, but its expression was greatly reduced by bicarbonate treatment (Fig. 7A). CmbHLH38 was induced by bicarbonate in WT roots at low Fe supply (Fig. 7C), and CmbHLH101 was induced by bicarbonate at both low Fe and high Fe supply (Fig. 7E). The expression pattern of CmFRO1 corresponded to FCR activity in WT melon (Figs. 6B and 7G). The transcript patterns of both CmIRT1 and CmNRAMP1 in Edisto were consistent with expression in cucumber (Figs 2D, F and 7I, K). In fefe roots, CmFIT, CmbHLH38, CmFRO1, and CmNRAMP1 did not respond to Fe deficiency or to bicarbonate treatment. However, both CmbHLH101 and CmIRT1 had higher expression in bicarbonate-treated, Fe-sufficient roots. These results suggested that the alkaline stress response was mostly, but not entirely, dependent on fefe.
Fig. 7.
Quantitative RT-PCR analyses of Fe uptake related genes in roots of wild-type Edisto (A, C, E, G, I, and K) and fefe mutant (B, D, F, H, J, and L) after 3 days of treatment. The results were normalized to ubiquitin as an internal reference. The transcript abundance of each gene (means±SE, n=3) of both genotypes was normalized to Edisto root samples of the control treatment (10 μM Fe and 0mM bicarbonate). * and ** indicate statistical significance at P<0.05 and P<0.01, respectively, comparing bicarbonate treatments within the same Fe supply; Ŧ and ŦŦ indicate statistical significance at P<0.05 and P<0.01, respectively, comparing different Fe supply within the same bicarbonate treatment.
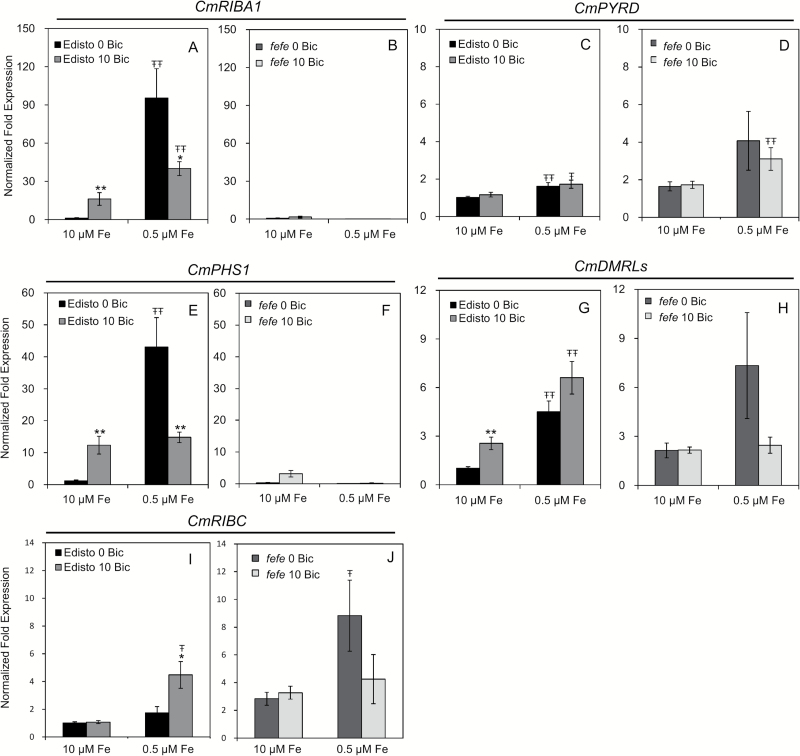
Expression of riboflavin synthesis pathway genes in fefe melon
To test whether Fe deficiency regulation of riboflavin synthesis genes requires the FeFe gene, we gathered RNAseq expression data from our previous study (Waters et al., 2014) (Table 1). In the WT, three (RIBA1, PHS1, and DMRLs) of the five genes were significantly up-regulated under Fe deficiency, but none of these genes were up-regulated by Fe deficiency in the fefe mutant. To test whether up-regulation of these genes in response to alkaline stress depends on the FeFe gene, we measured their expression in WT and fefe mutant roots by RT-qPCR. Consistent with the RNAseq results, CmRIBA1, CmPHS1, and CmDMRLs were induced by Fe deficiency in WT. Transcript levels of CmRIBA1 and CmPHS1 were lower in bicarbonate-treated Fe-deficient Edisto roots (Fig. 8A, C), similar to CmFIT expression, but CmDMRLs expression was increased (Fig. 8G). In the fefe mutant, expression of CmPYRD was somewhat higher in low Fe, both with or without bicarbonate treatment, and CmDMRLs was up-regulated under low Fe without bicarbonate (Fig. 8). However, expression of CmRIBA1 and CmPHS1 was almost completely abolished in the fefe mutant, suggesting that expression of these genes strongly depends on the FeFe regulatory pathway.
Fig. 8.
Quantitative RT-PCR analyses of riboflavin synthesis genes in roots of wild-type Edisto (A, C, E, G, and I) and fefe mutant (B, D, F, H, and J) after 3 days of treatment. The results were normalized to ubiquitin as an internal reference. The transcript abundance of each gene (means±SE, n=3) of both genotypes was normalized to Edisto root samples of the control treatment (10 μM Fe and 0mM bicarbonate). * and ** indicate statistical significance at P<0.05 and P<0.01, respectively, comparing bicarbonate treatments within the same Fe supply; Ŧ and ŦŦ indicate statistical significance at P<0.05 and P<0.01, respectively, comparing different Fe supply within the same bicarbonate treatment.
Discussion
Although it is well known that IDC occurs in alkaline soils with low Fe availability, the physiological and molecular basis for this phenomenon is not well understood. In this study, we treated Fe supply and pH as separate variables. Using a range of Fe supply with two nutrient solution pHs, we found that root FCR activity responded to Fe supply only at normal pH, not at alkaline pH (Fig. 1C). We then used low and normal Fe supply, with or without bicarbonate, to determine whether gene expression in roots and leaves responded to Fe supply at each pH, and whether gene expression responded to pH at each Fe supply. Whether root FCR activity is inhibited or induced by alkaline pH depends on the point of reference. At low Fe supply, bicarbonate treatment inhibited FCR activity, relative to the treatment without bicarbonate. However, at alkaline pH the FCR activity at 10 and 2.5 μM Fe was elevated, relative to activity at normal pH. We found similar results for melon FCR activity (Fig. 6B), in that bicarbonate treatment inhibited FCR activity in Fe-deficient plants, while in Fe-supplied plants, bicarbonate stimulated FCR activity. These results are consistent with a previous study in cucumber and sunflower (Romera et al., 1992a). Together, these results explain conflicting reports in the literature, where bicarbonate treatment decreased FCR activity (Romera et al., 1997; Lucena et al., 2007; García et al., 2014) or stimulated FCR activity (Waters and Troupe, 2012), depending on Fe supply.
Root molecular response to alkalinity and Fe supply
Alkalinity up-regulated many of the Fe deficiency responses even when plants were grown with normal Fe supply. While the cucumber transcription factor genes did not respond to alkaline pH at 10 μM Fe, the Fe uptake genes were up-regulated (Fig. 3). In Fe-deficient Arabidopsis, AtbHLH38 and AtbHLH39 proteins interact with AtFIT, and the resulting heterodimer regulates transcription of downstream genes like AtFRO2 and AtIRT1 (Yuan et al., 2008). The difference in expression between transcription factors and their targets suggests that there could be regulators in addition to FIT/bHLH that respond to alkaline pH to up-regulate the expression Fe uptake genes in Fe replete, alkaline stressed plants. Alternatively, FIT or subgroup Ib bHLH protein activity (Meiser et al., 2011; Hindt and Guerinot, 2012), rather than transcriptional regulation, may respond to alkaline pH at normal Fe supply.
While melon and cucumber FCR activity responded to Fe supply only at normal pH (Figs 1 and 6), cucumber Fe uptake gene expression responded to Fe deficiency at both normal and alkaline pH (Fig. 3). At low Fe supply, most of the cucumber Fe uptake genes (except CsFIT and CsNRAMP1) responded to alkaline treatment by a further increase in transcript abundance (Fig. 3). This was not the case in melon, which had diminished Fe uptake gene expression in response to Fe deficiency at alkaline pH (Fig. 6), similar to previous results in Arabidopsis (García et al., 2014). In a field setting in which plants suffer from IDC, both low Fe supply and alkaline pH occur. In a prior study, low levels of bicarbonate increased expression of CsFRO1 and CsIRT1 in Fe-deficient roots (as seen here), while higher levels decreased expression of these Fe uptake genes (Lucena et al., 2007; García et al., 2014). The bicarbonate concentration we chose is in the stimulatory range for cucumber, but is in the inhibitory range for melon. In Fe-deficient plants higher concentrations of bicarbonate were required to inhibit Fe uptake gene expression in Arabidopsis than in cucumber (Lucena et al., 2007). Extending this difference in response between Arabidopsis and cucumber, and cucumber and melon to other dicot species, these results provide insight into why some species of plants are more sensitive to IDC than others.
Fe deficiency and alkaline stress regulation of riboflavin synthesis genes
Cucumber and melon roots produce and release flavin compounds in response to Fe deficiency (Welkie, 2000; Rodríguez-Celma et al., 2011a). In cucumber and WT melon roots, most of the riboflavin synthesis genes were up-regulated by Fe deficiency in both normal pH and alkaline media (Figs 5 and 8, and Table 1). Bicarbonate treatment slightly increased expression of two or three (depending on species) of the riboflavin synthesis genes, but only at full Fe supply, and there was no synergistic up-regulation at the combined low Fe and alkaline treatment as was seen for Fe uptake genes. These results indicate that although alkaline stress induces some of the same gene expression responses as Fe deficiency, alkaline stress is not precisely equivalent to Fe deficiency stress. The overlapping but not equivalent effect of Fe deficiency and alkaline stress was seen in results of a recent metabolomics study that showed that abundance of some common metabolites changed in roots treated with low Fe or with alkaline solution, while abundance of other metabolites changed only in one of these conditions (Schmidt et al., 2014). In Medicago truncatula, several genes and proteins of the riboflavin synthesis pathway were up-regulated in Fe-deficient roots (Rodríguez-Celma et al., 2011a, b, 2013b). Iron-deficient Medicago combined with alkaline stress had increased abundance for one riboflavin synthesis protein and decreased abundance for another riboflavin synthesis protein, relative to Fe-deficient plants at normal pH (Rodríguez-Celma et al., 2011a), and accumulated more flavins than Fe-deficient plants at normal pH, although release of flavins from roots was decreased at alkaline pH (Rodríguez-Celma et al., 2011b). When the Arabidopsis bHLH38 and bHLH39 genes were overexpressed in tobacco, the plants produced more riboflavin (Vorwieger et al., 2007). Our results provide a mechanism for increased riboflavin synthesis in melon and cucumber under Fe deficiency, and suggest that plants that are under alkaline stress but with normal Fe supply may also produce more riboflavin.
Alkaline stress stimulation of Fe uptake gene expression depends on the FeFe regulatory pathway
Most of the Fe uptake-related genes were not regulated by Fe deficiency in the fefe mutant, as expected, and most of these genes also were not responsive to alkaline stress (Fig. 7B, D, H, L), suggesting that the alkaline stress signal depends mainly on the FeFe regulatory pathway. However, CmHLH101 and CmIRT1 were up-regulated in the fefe mutant by alkaline stress under full Fe supply (Fig. 7F, J), suggesting that alkaline stress can at least partially regulate these genes independently of the FeFe regulatory pathway. These genes were not induced in fefe mutants by alkaline stress under low Fe supply, which suggests that, as for other genes, the combination of Fe deficiency and alkaline stress may result in inhibition of Fe uptake gene expression.
Many of the Fe uptake genes that were up-regulated by alkaline stress in melon and cucumber have homologs in Arabidopsis that are targets of the primary Fe homeostasis transcription factor FIT (Colangelo and Guerinot, 2004). The expression of the F6’H1 gene that is required for synthesis of phenolic root exudates also depends on FIT (Schmid et al., 2014). The melon FeFe gene has not been identified, but is predicted to be a transcription factor that is functionally upstream of CmFIT (Waters et al., 2014). Since melon is a flavin producer rather than a phenolics producer like Arabidopsis, we were able to test whether this primary Fe homeostasis regulator is required for expression of riboflavin synthesis genes, and whether alkaline stress responses require the FeFe gene. The three riboflavin synthesis genes that were up-regulated by Fe deficiency in wild-type melon (CmRIBA1, CmPHS1, and CmDMRLs) were dependent on FeFe for their up-regulation by Fe deficiency (Fig. 8). Combined with results for Fe uptake genes, these results show that FeFe is a master regulator of both Fe uptake genes and Fe deficiency-induced riboflavin synthesis. These results are consistent with an early characterization of the fefe mutant, where riboflavin efflux into the nutrient solution was increased by Fe deficiency in the wild-type, but was not increased in the fefe mutant (Welkie, 1996), and with our previous RNAseq results (Waters et al., 2014). Thus, both flavin (melon) and phenolic (Arabidopsis) producing Strategy I species depend on master Fe uptake regulators for increased synthesis and efflux of root exudates. Up-regulation of riboflavin synthesis genes in melon roots by alkaline stress in Fe-sufficient plants also was abolished in the fefe mutant (Table 1 and Fig. 8), demonstrating that, like the Fe deficiency signal, alkaline stress regulation of riboflavin synthesis genes depends on the FeFe regulatory pathway. There is evidence for a role for both phenolics and flavins in Fe uptake under alkaline stress (Rodríguez-Celma et al., 2013b; Schmidt et al., 2014; Schmid et al., 2014), which may explain why the phenolic and riboflavin synthesis genes are regulated by Fe uptake transcription factors.
Whole plant responses to Fe supply and alkaline stress: implications for intraplant signaling and IDC
Alkaline stress may have increased expression of Fe deficiency up-regulated genes because alkaline treatment resulted in lower plant Fe accumulation. Despite increased expression of CsFRO1 and CsIRT1 (Fig. 3D, E) and FCR activity in roots of plants treated with full Fe supply and bicarbonate, whole plant Fe content was lower (Fig. 2A) and was similar to that of Fe-deficient plants, indicating that Fe uptake was inhibited by alkaline stress, consistent with previous studies (Fleming et al., 1984; Alhendawi et al., 1997). Leaf chlorophyll was also lower at all Fe levels in alkaline stressed plants (Figs. 1B and 2A) and so was Fe in stem+cotyledon and roots, while root Zn and Mn increased. These results suggest that normal Strategy I root Fe uptake processes (e.g. FCR activity, Fe(II) transporter function) do not function properly when plants are growing at alkaline pH. FCR activity of sugar beet was inhibited by alkaline pH of the assay medium (Susin et al., 1996). While our FCR assay at pH 5.5 indicated that FCR activity was increased, the alkaline pH of the growth medium may disrupt the ability of the FCR protein to reduce Fe, and may inhibit other aspects of root Fe uptake, such as specificity of Fe over Zn and Mn (George et al., 2012). These results extend the previous evidence that alkaline stress blocks Fe uptake and translocation.
The results discussed so far show that cucumber roots up-regulated certain Fe uptake and riboflavin synthesis genes in response to Fe limitation and alkaline stress in roots. In contrast, in cucumber leaf none of the Fe uptake genes or riboflavin synthesis genes were up-regulated in response to alkaline stress. The expression of CsIRT1, CsbHLH101, and CsbHLH38 was up-regulated by Fe deficiency in leaf tissue, but this up-regulation was completely inhibited by bicarbonate (Fig. 4A–C), indicating that alkaline stress blocks Fe deficiency responses in leaves. It has long been suggested that alkaline stress inhibits Fe uptake into leaf cells (Mengel, 1994). Our results suggest that this lack of uptake could be caused by a lack of Fe uptake gene expression rather than simply by physical effects, such as alkalization of xylem sap pH. This abolition of gene up-regulation by Fe deficiency under alkaline stress was also true for homologs of Arabidopsis genes that are not known to be involved in Fe uptake but were up-regulated by Fe deficiency in leaves (5960 and KCS; Fig. 4D, E). Thus, this inhibition of Fe deficiency up-regulated genes by alkaline stress occurs more generally than only for Fe uptake genes. These results show that the leaf responded to alkaline stress quite differently from the root, and this may have important implications for whole-plant Fe sensing.
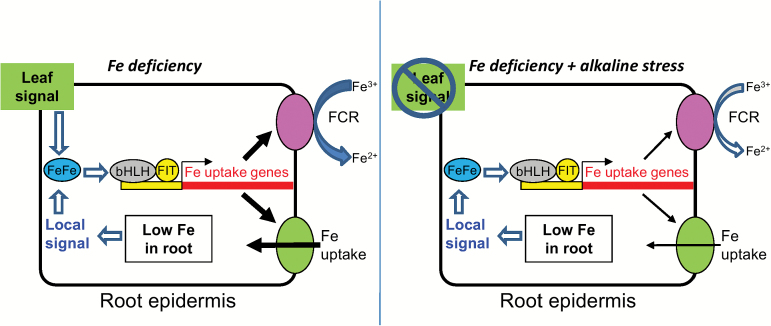
Intraplant signaling of Fe status is not well understood, but both local sensing of root Fe status and shoot-to-root signaling of leaf Fe status are important for up-regulation of root Fe uptake responses (Romera et al., 1992b; Vert et al., 2003; Lucena et al., 2006; Gayomba et al., 2015) through the FIT regulatory pathway. Our results give new clues into why alkaline stress may stimulate root Fe uptake responses in Fe-sufficient plants, but inhibit these responses in Fe-deficient plants. In bicarbonate-treated cucumber plants with full Fe supply, lower root Fe concentration may stimulate a local signal to increase root FCR activity and Fe uptake gene expression. The lack of response to Fe deficiency in bicarbonate-treated cucumber leaf (Fig. 4) suggests that alkaline stress interferes with leaf Fe sensing, which would be expected to inhibit shoot-to-root signaling. Since leaf signaling may be a major factor in up-regulation of root Fe uptake responses (Enomoto and Goto, 2008; García et al., 2013), a lack of signal from alkaline-stressed, Fe-deficient leaves may explain why plants grown at alkaline pH did not increase root FCR activity at low Fe supply beyond the activity at full Fe supply. That is, the local Fe status root signal was already fully activated by decreased root Fe concentration resulting from alkaline stress, but the leaf signal to further increase root FCR activity was absent. This model (Fig. 9) would also explain why both alkaline and Fe deficiency signals rely mainly on the FeFe regulatory pathway upstream of FIT (Figs. 7 and 8).
Fig. 9.
Model of responses to Fe deficiency and Fe deficiency plus alkaline stress. A local root signal for Fe deficiency acts through the FeFe regulatory pathway upstream of FIT and a bHLH partner, which activate Fe uptake genes and subsequent Fe uptake activity. An Fe deficiency signal from the leaf up-regulates the same pathway in an additive manner (left). When alkaline stress is combined with Fe deficiency (right), it blocks the normal Fe deficiency response in leaf, resulting in a loss of shoot-to-root signal. Alkaline stress combined with Fe deficiency also weakens root Fe uptake responses at the transcript and protein activity levels.
What is not clear from our current knowledge is why leaves exposed to alkaline stress fail to respond appropriately to low Fe supply, or why Fe-deficient roots cannot maintain up-regulated FCR activity and Fe uptake gene expression (Figs. 3, 4 and 7, and Lucena et al., 2007) under alkaline stress. Because a slight induction of FCR activity (Fig. 6) and CmbHLH101 and CmIRT1 up-regulation occurred in fefe mutant plants treated with full Fe supply and bicarbonate, there may be a direct pH signal that interacts with Fe status signaling. Further research into mechanisms of leaf Fe sensing and shoot-to-root signaling, and plant sensing of rhizosphere pH will be necessary to fully understand the IDC phenomenon that occurs in low Fe availability, alkaline soils. However, since plants respond to Fe deficiency differently when also exposed to alkaline stress, this study indicates that knowledge of plant responses to low Fe supply alone, or alkaline stress alone, will not be adequate to understand IDC. Future molecular physiological IDC studies will need to include Fe supply and pH treatments singly and in combination to fully understand the IDC phenomenon, and will need to incorporate leaf and root measurements simultaneously.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. DW of cucumber plants.
Figure S2. Cucumber (cv Miniature White) leaf chlorophyll and ferric chelate reductase activity in response to Fe supply in normal pH or alkaline nutrient solution.
Table S1. Primers used in this study.
Acknowledgements
The authors thank Tori Hinrichs, Erin Kinley, and Laura Armbrust for technical support and Mary Guttieri and Raghuprakash Kastoori Ramamurthy for helpful discussions. This work was funded in part by grants NSF-IOS-1257568 and USDA-2014-67013-21658 to BMW.
References
- Alcántara E, Romera FJ, Cañete M, de la Guardia MD. 2000. Effects of bicarbonate and iron supply on Fe(III) reducing capacity of roots and leaf chlorosis of the susceptible peach root stock “Nemaguard”. Journal of Plant Nutrition 23, 1607–1617. [Google Scholar]
- Alhendawi RA, Römheld V, Kirkby EA, Marschner H. 1997. Influence of increasing bicarbonate concentrations on plant growth, organic acid accumulation in roots and iron uptake by barley, sorghum, and maize. Journal of Plant Nutrition 20, 1731–1753. [Google Scholar]
- Briat J, Dubos C, Gaymard F. 2015. Iron nutrition, biomass production, and plant product quality. Trends in Plant Science 20, 33–40. [DOI] [PubMed] [Google Scholar]
- Cailliatte R, Schikora A, Briat JF, Mari S, Curie C. 2010. High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. The Plant Cell 22, 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesco S, Neumann G, Tomasi N, Pinton R, Weisskopf L. 2010. Release of plant-borne flavonoids into the rhizosphere and their role in plant nutrition. Plant and Soil 329, 1–25. [Google Scholar]
- Chaney RL, Brown JC, Tiffin LO. 1972. Obligatory reduction of ferric chelates in iron uptake by soybeans. Plant Physiology 50, 208–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaney RL, Coulombe BA, Bell PF, Angle JS. 1992. Detailed method to screen dicot cultivars for resistance to Fe-chlorosis using FeDTPA and bicarbonate in nutrient solutions. Journal of Plant Nutrition 15, 2063–2083. [Google Scholar]
- Chen Y, Barak P. 1982. Iron nutrition of plants in calcareous soils. Advances in Agronomy 35, 217–240. [Google Scholar]
- Colangelo EP, Guerinot ML. 2004. The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. The Plant Cell 16, 3400–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe B, Chaney RL, Wiebold W. 1984. Bicarbonate directly induces iron chlorosis in susceptible soybean cultivars. Soil Science Society of America Journal 48, 1297–1301. [Google Scholar]
- Curie C, Alonso J, Ecker J, Briat J. 2000. Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochemical Journal 347, 749–755. [PMC free article] [PubMed] [Google Scholar]
- De Nisi P, Vigani G, Dell’Orto M, Zocchi G. 2012. Application of the split root technique to study iron uptake in cucumber plants. Plant Physiology and Biochemistry 57, 168–174. [DOI] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML. 1996. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proceedings of the National Academy of Sciences of the United States of America 93, 5624–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto Y, Goto F. 2008. Long-distance signaling of iron deficiency in plants. Plant Signaling & Behavior 3, 396–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A, Chaney R, Coulombe B. 1984. Bicarbonate inhibits Fe‐stress response and Fe uptake‐translocation of chlorosis‐susceptible soybean cultivars. Journal of Plant Nutrition 7, 699–714. [Google Scholar]
- Fourcroy P, Sisó‐Terraza P, Sudre D, Savirón M, Reyt G, Gaymard F, Abadía A, Abadia J, Álvarez‐Fernández A, Briat J. 2014. Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytologist 201, 155–167. [DOI] [PubMed] [Google Scholar]
- García MJ, García‐Mateo MJ, Lucena C, Romera FJ, Rojas CL, Alcántara E, Pérez‐Vicente R. 2014. Hypoxia and bicarbonate could limit the expression of iron acquisition genes in Strategy I plants by affecting ethylene synthesis and signaling in different ways. Physiologia Plantarum 150, 95–106. [DOI] [PubMed] [Google Scholar]
- García MJ, Romera FJ, Stacey MG, Stacey G, Villar E, Alcántara E, Pérez-Vicente R. 2013. Shoot to root communication is necessary to control the expression of iron-acquisition genes in Strategy I plants. Planta 237, 65–75. [DOI] [PubMed] [Google Scholar]
- Gayomba SR, Zhai Z, Jung H, Vatamaniuk OK. 2015. Local and systemic signaling of iron status and its interactions with homeostasis of other essential elements. Frontiers in Plant Science 6, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George E, Horst W, Neumann E. 2012. Adaptation of plants to adverse chemical soil conditions. Marschner’s Mineral Nutrition of Higher Plants 3, 409–472. [Google Scholar]
- Guttieri MJ, Baenziger PS, Frels K, Carver B, Arnall B, Waters BM. 2015. Variation for grain mineral concentration in a diversity panel of current and historical Great Plains hard winter wheat germplasm. Crop Science 55, 1035–1052. [Google Scholar]
- Halvorson A, Lindsay W. 1972. Equilibrium relationships of metal chelates in hydroponic solutions. Soil Science Society of America Journal 36, 755–761. [Google Scholar]
- Hansen N, Jolley V, Naeve S, Goos R. 2004. Iron deficiency of soybean in the North Central US and associated soil properties. Soil Science and Plant Nutrition 50, 983–987. [Google Scholar]
- Hindt MN, Guerinot ML. 2012. Getting a sense for signals: regulation of the plant iron deficiency response. Biochimica et Biophysica Acta 1823, 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov R, Brumbarova T, Bauer P. 2012. Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Molecular Plant 5, 27–42. [DOI] [PubMed] [Google Scholar]
- Jakoby M, Wang H, Reidt W, Weisshaar B, Bauer P. 2004. FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Letters 577, 528–534. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nishizawa NK. 2012. Iron uptake, translocation, and regulation in higher plants. Annual Review of Plant Biology 63, 131–152. [DOI] [PubMed] [Google Scholar]
- Lin S, Cianzio S, Shoemaker R. 1997. Mapping genetic loci for iron deficiency chlorosis in soybean. Molecular Breeding 3, 219–229. [Google Scholar]
- Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M. 2002. The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proceedings of the National Academy of Sciences of the United States of America 99, 13938–13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker N, Welch RM. 1990. Accumulation of apoplastic iron in plant roots: a factor in the resistance of soybeans to iron-deficiency induced chlorosis? Plant Physiology 92, 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucena C, Romera FJ, Rojas CL, García MJ, Alcántara E, Pérez-Vicente R. 2007. Bicarbonate blocks the expression of several genes involved in the physiological responses to Fe deficiency of Strategy I plants. Functional Plant Biology 34, 1002–1009. [DOI] [PubMed] [Google Scholar]
- Lucena C, Waters BM, Romera FJ, Garcia MJ, Morales M, Alcantara E, Perez-Vicente R. 2006. Ethylene could influence ferric reductase, iron transporter, and H+-ATPase gene expression by affecting FER (or FER-like) gene activity. Journal of Experimental Botany 57, 4145–4154. [DOI] [PubMed] [Google Scholar]
- Meiser J, Lingam S, Bauer P. 2011. Posttranslational regulation of the iron deficiency basic helix-loop-helix transcription factor FIT is affected by iron and nitric oxide. Plant Physiology 157, 2154–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengel K. 1994. Iron availability in plant tissues-iron chlorosis on calcareous soils. Plant and Soil 165, 275–283. [Google Scholar]
- Mengel K, Breininger MT, Bübl W. 1984. Bicarbonate, the most important factor inducing iron chlorosis in vine grapes on calcareous soil. Plant and Soil 81, 333–344. [Google Scholar]
- Mengel K, Geurtzen G. 1986. Iron chlorosis on calcareous soils. Alkaline nutritional condition as the cause for the chlorosis. Journal of Plant Nutrition 9, 161–173. [Google Scholar]
- Moran Lauter AN, Peiffer GA, Yin T, Whitham SA, Cook D, Shoemaker RC, Graham MA. 2014. Identification of candidate genes involved in early iron deficiency chlorosis signaling in soybean (Glycine max) roots and leaves. BMC Genomics 15, 702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee I, Campbell NH, Ash JS, Connolly EL. 2006. Expression profiling of the Arabidopsis ferric chelate reductase (FRO) gene family reveals differential regulation by iron and copper. Planta 223, 1178–1190. [DOI] [PubMed] [Google Scholar]
- Nugent PE. 1994. Iron chlorotic melon germplasm C940-fe. HortScience 29, 50–51. [Google Scholar]
- Nugent PE, Bhella HS. 1988. A new chlorotic mutant of muskmelon. HortScience 23, 379–381. [Google Scholar]
- Rellán-Álvarez R, Andaluz S, Rodríguez-Celma J, Wohlgemuth G, Zocchi G, Álvarez-Fernández A, Fiehn O, López-Millán A, Abadía J. 2010. Changes in the proteomic and metabolic profiles of Beta vulgaris root tips in response to iron deficiency and resupply. BMC Plant Biology 10, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML. 1999. A ferric-chelate reductase for iron uptake from soils. Nature 397, 694–697. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Celma J, Lattanzio G, Grusak MA, Abadía A, Abadía J, López-Millán A. 2011a Root responses of Medicago truncatula plants grown in two different iron deficiency conditions: changes in root protein profile and riboflavin biosynthesis. Journal of Proteome Research 10, 2590–2601. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Celma J, Lin WD, Fu GM, Abadia J, Lopez-Millan AF, Schmidt W. 2013b Mutually exclusive alterations in secondary metabolism are critical for the uptake of insoluble iron compounds by Arabidopsis and Medicago truncatula. Plant Physiology 162, 1473–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Celma J, Pan IC, Li W, Lan P, Buckhout TJ, Schmidt W. 2013a The transcriptional response of Arabidopsis leaves to Fe deficiency. Frontiers in Plant Science 4, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Celma J, Vazquez-Reina S, Orduna J, Abadia A, Abadia J, Alvarez-Fernandez A, Lopez-Millan AF. 2011b Characterization of flavins in roots of Fe-deficient strategy I plants, with a focus on Medicago truncatula. Plant & Cell Physiology 52, 2173–2189. [DOI] [PubMed] [Google Scholar]
- Rogovska NP, Blackmer AM, Mallarino AP. 2007. Relationships between soybean yield, soil pH, and soil carbonate concentration. Soil Science Society of America Journal 71, 1251–1256. [Google Scholar]
- Romera FJ, Alcantara E, de la Guardia MD. 1992a Effects of bicarbonate, phosphate and high pH on the reducing capacity of Fe‐deficient sunflower and cucumber plants. Journal of Plant Nutrition 15, 1519–1530. [Google Scholar]
- Romera FJ, Alcántara E, de la Guardia MD. 1992b Role of roots and shoots in the regulation of the Fe efficiency responses in sunflower and cucumber. Physiologia Plantarum 85, 141–146. [Google Scholar]
- Romera FJ, Alcántara E, de la Guardia MD. 1997. Influence of bicarbonate and metal ions on the development of root Fe (III) reducing capacity by Fe‐deficient cucumber (Cucumis sativus) plants. Physiologia Plantarum 101, 143–148. [Google Scholar]
- Schmid NB, Giehl RF, Doll S, Mock HP, Strehmel N, Scheel D, Kong X, Hider RC, von Wiren N. 2014. Feruloyl-CoA 6’-hydroxylase1-dependent coumarins mediate iron acquisition from alkaline substrates in Arabidopsis. Plant Physiology 164, 160–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Günther C, Weber M, Spörlein C, Loscher S, Böttcher C, Schobert R, Clemens S. 2014. Metabolome analysis of Arabidopsis thaliana roots identifies a key metabolic pathway for iron acquisition. PloS ONE 9, e102444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisó‐Terraza P, Rios JJ, Abadía J, Abadía A, Álvarez‐Fernández A. 2016. Flavins secreted by roots of iron‐deficient Beta vulgaris enable mining of ferric oxide via reductive mechanisms. New Phytologist 209, 733–745. [DOI] [PubMed] [Google Scholar]
- Strasser O, Köhl K, Römheld V. 1999. Overestimation of apoplastic Fe in roots of soil grown plants. Plant and Soil 210, 179–189. [Google Scholar]
- Susin S, Abadia A, Gonzalez-Reyes JA, Lucena JJ, Abadia J. 1996. The pH requirement for in vivo activity of the iron-deficiency-induced “turbo” ferric chelate reductase. Plant Physiology 110, 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susin S, Abian J, Sanchez-Baeza F, Peleato ML, Abadia A, Gelpi E, Abadia J. 1993. Riboflavin 3’- and 5’-sulfate, two novel flavins accumulating in the roots of iron-deficient sugar beet (Beta vulgaris). Journal of Biological Chemistry 268, 20958–20965. [PubMed] [Google Scholar]
- Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI. 2000. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proceedings of the National Academy of Sciences of the United States of America 97, 4991–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto C, Maiwald D, Pesaresi P, Jahns P, Salamini F, Leister D. 2002. The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. The Plant Journal 31, 589–599. [DOI] [PubMed] [Google Scholar]
- Vert GA, Briat JF, Curie C. 2003. Dual regulation of the Arabidopsis high-affinity root iron uptake system by local and long-distance signals. Plant Physiology 132, 796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigani G, Zocchi G, Bashir K, Philippar K, Briat JF. 2013. Cellular iron homeostasis and metabolism in plant. Frontiers in Plant Science 4, 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorwieger A, Gryczka C, Czihal A, Douchkov D, Tiedemann J, Mock H, Jakoby M, Weisshaar B, Saalbach I, Bäumlein H. 2007. Iron assimilation and transcription factor controlled synthesis of riboflavin in plants. Planta 226, 147–158. [DOI] [PubMed] [Google Scholar]
- Walker EL, Waters BM. 2011. The role of transition metal homeostasis in plant seed development. Current Opinion in Plant Biology 14, 318–324. [DOI] [PubMed] [Google Scholar]
- Wang H, Klatte M, Jakoby M, Bäumlein H, Weisshaar B, Bauer P. 2007. Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226, 897–908. [DOI] [PubMed] [Google Scholar]
- Wang N, Cui Y, Liu Y, Fan H, Du J, Huang Z, Yuan Y, Wu H, Ling H. 2013. Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Molecular Plant 6, 503–513. [DOI] [PubMed] [Google Scholar]
- Waters BM, Blevins DG. 2000. Ethylene production, cluster root formation, and localization of iron (III) reducing capacity in Fe deficient squash roots. Plant and Soil 225, 21–31. [Google Scholar]
- Waters BM, Lucena C, Romera FJ, Jester GG, Wynn AN, Rojas CL, Alcántara E, Pérez-Vicente R. 2007. Ethylene involvement in the regulation of the H -ATPase CsHA1 gene and of the new isolated ferric reductase CsFRO1 and iron transporter CsIRT1 genes in cucumber plants. Plant Physiology and Biochemistry 45, 293–301. [DOI] [PubMed] [Google Scholar]
- Waters BM, McInturf SA, Amundsen K. 2014. Transcriptomic and physiological characterization of the fefe mutant of melon (Cucumis melo) reveals new aspects of iron–copper crosstalk. New Phytologist 203, 1128–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters BM, McInturf SA, Stein RJ. 2012. Rosette iron deficiency transcript and microRNA profiling reveals links between copper and iron homeostasis in Arabidopsis thaliana. Journal of Experimental Botany 63, 5903–5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters BM, Troupe GC. 2012. Natural variation in iron use efficiency and mineral remobilization in cucumber (Cucumis sativus). Plant and Soil 352, 185–197. [Google Scholar]
- Welkie GW. 1996. Iron‐deficiency stress responses of a chlorosis‐susceptible and a chlorosis‐resistant cultivar of muskmelon as related to root riboflavin excretion. Journal of Plant Nutrition 19, 1157–1169. [Google Scholar]
- Welkie GW. 2000. Taxonomic distribution of dicotyledonous species capable of root excretion of riboflavin under iron deficiency. Journal of Plant Nutrition 23, 1819–1831. [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling H. 2008. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Research 18, 385–397. [DOI] [PubMed] [Google Scholar]
- Yuan YX, Zhang J, Wang DW, Ling HQ. 2005. AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Research 15, 613–621. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.