Highlight
Alternative splicing of the basic chitinase gene PR3b requires genes in the NIC loci of low-nicotine mutants of Burley 21 tobacco.
Key words: Alternative splicing, ethylene, jasmonate, low-nicotine mutant, PR3b, tobacco.
Abstract
Two unlinked semi-dominant loci, A (NIC1) and B (NIC2), control nicotine and related alkaloid biosynthesis in Burley tobaccos. Mutations in either or both loci (nic1 and nic2) lead to low nicotine phenotypes with altered environmental stress responses. Here we show that the transcripts derived from the pathogenesis-related (PR) protein gene PR3b are alternatively spliced to a greater extent in the nic1 and nic2 mutants of Burley 21 tobacco and the nic1nic2 double mutant. The alternative splicing results in a deletion of 65 nucleotides and introduces a premature stop codon into the coding region of PR3b that leads to a significant reduction of PR3b specific chitinase activity. Assays of PR3b splicing in F2 individuals derived from crosses between nic1 and nic2 mutants and wild-type plants showed that the splicing phenotype is controlled by the NIC1 and NIC2 loci, even though NIC1 and NIC2 are unlinked loci. Moreover, the transcriptional analyses showed that the splicing patterns of PR3b in the low-nicotine mutants were differentially regulated by jasmonate (JA) and ethylene (ET). These data suggest that the NIC1 and NIC2 loci display differential roles in regulating the alternative splicing of PR3b in Burley 21. The findings in this study have provided valuable information for extending our understanding of the broader effects of the low-nicotine mutants of Burley 21 and the mechanism by which JA and ET signalling pathways post-transcriptionally regulate the activity of PR3b protein.
Introduction
Nicotine is a natural compound used for defence against attack by insect herbivores in members of the genus Nicotiana and is the predominant alkaloid in most cultivated commercial tobacco (Nicotiana tabacum L.) varieties (Baldwin, 2001; Steppuhn et al., 2008; Kumar et al., 2014; Sears et al., 2014; Shitan et al., 2015). The formation of nicotine begins with ornithine and/or arginine and involves the key catalytic enzymes, PMT (putrescine N-methyltransferase), ODC (ornithine decarboxylase), QPT (quinolinate phosphoribosyltransferase), MPO (N-methylputrescine oxidase), A622 (isoflavone reductase-like), BBL (berberine bridge enzyme-like), and MATE (multidrug and toxic compound extrusion) (Hibi et al., 1994; Sinclair et al., 2000; Heim et al., 2007; Deboer et al., 2009; Kajikawa et al., 2011; Dewey and Xie, 2013; Lewis et al., 2015). The process of nicotine formation takes place in the roots and is regulated by the developmental stage of the plant, phytohormonal signals, and environmental factors (Baldwin, 1998; Xu and Timko, 2004; Shi et al., 2006; Li et al., 2007).
The phytohormone jasmonate (JA) is a major regulator of nicotine synthesis, and an increase in endogenous JA level or an exogenous application of JA or MeJA (methyl jasmonate) rapidly increases the transcript levels of genes encoding enzymes of nicotine biosynthesis (e.g. PMT, QPT), to promote nicotine synthesis (Baldwin, 1998; Imanishi et al., 1998; Shoji et al., 2000b ; Goossens et al., 2003; Saedler and Baldwin, 2004; Xu and Timko, 2004; Cane et al., 2005; Shoji et al., 2008; Zhang et al., 2012). A number of key JA-signalling components have been demonstrated to be involved in the fine-tuning of JA-induced nicotine synthetic genes (Shoji et al., 2008; Shoji and Hashimoto, 2011; Zhang et al., 2012; Woldemariam et al., 2013). Interestingly, several studies also suggested that the gaseous phytohormone ethylene (ET) plays a negative role in nicotine synthesis by suppressing the expression of nicotine synthetic genes (Shoji et al., 2000a ; Winz and Baldwin, 2001; Xu and Timko, 2004). Moreover, members of the plant-specific transcription factor family known as ERFs (ethylene response factors) have been shown to play critical roles in JA/ET signalling and in the regulation of nicotine synthesis (Guo and Ecker, 2004; Gutterson and Reuber, 2004; Shoji et al., 2010; De Boer et al., 2011; Sears et al., 2014).
ERF proteins were first identified as transcription factors regulating plant pathogen resistance in tobacco (Ohme-Takagi and Shinshi, 1995). Thereafter, a large number of ERF transcription factors have been identified as regulators functioning in plant stress tolerance, pathogen resistance, secondary metabolism, and signal transduction of phytohormones [e.g. JA, ET, salicylic acid (SA)] (Chakravarthy et al., 2003; Lorenzo et al., 2003; Gutterson and Reuber, 2004). A major target of ERF proteins is the GCC-box in the promoter regions of pathogenesis-related (PR) protein genes that function in plant pathogen resistance (Ohme-Takagi and Shinshi, 1995; Chakravarthy et al., 2003; Zhang et al., 2004). A number of ERFs were shown to be co-ordinated by JA and ET signalling pathways to regulate pathogen resistance and secondary metabolism (Lorenzo et al., 2003; Gutterson and Reuber, 2004; Zhang et al., 2004; Pre et al., 2008; Shoji et al., 2010). On the other hand, ERF transcription factors could synergistically or antagonistically co-operate with other regulators to co-ordinate the JA and ET signalling (Lorenzo et al., 2003; Pre et al., 2008; Zarei et al., 2011). Therefore, the integrative framework involving ERF transcription factors and their target genes is of great importance for dissecting JA- and/or ET-mediated regulation in plants.
The low-alkaloid mutant of Burley 21 (LA Burley 21) is a genetically stable breeding line that was developed in the early 1930s from Cuban tobacco cigar varieties having very low nicotine content (Valleau, 1949; Legg et al., 1970). Genetic studies in LA Burley 21 demonstrated that nicotine leve1s are controlled by two unlinked semi-dominant loci, A and B (also known as NIC1 and NIC2) (Legg et al., 1969; Legg and Collins, 1971; Hibi et al., 1994). These loci act synergistically in regulating nicotine synthesis (Legg et al., 1969; Hibi et al., 1994; Kidd et al., 2006). The transcript level of PMT was established as a marker widely used in nicotine synthesis studies and subsequent transcriptional analyses identified a set of nicotine synthesis genes that were down-regulated in mutant alleles of NIC1 and NIC2 (Hibi et al., 1994; Reed and Jelesko, 2004; Cane et al., 2005; Shoji et al., 2010). Recent studies have shown that the NIC2 locus contains a cluster of ERF transcription factors (Shoji et al., 2010; Shoji and Hashimoto, 2014). Furthermore, research using a fluorescent differential display (FDD) screen provided evidence that the NIC loci regulated a large number of stress-responsive genes but only a small portion of these genes were involved in nicotine synthesis (Kidd et al., 2006). This may indicate a broad regulatory function of NIC loci.
While the NIC2 locus is comprised of a cluster of ERF-transcription-factor-coding genes, the transcriptional regulation of PR protein genes in the low-nicotine mutants of Burley 21 is less well understood (Lorenzo et al., 2003; Guo and Ecker, 2004; Gutterson and Reuber, 2004; Pre et al., 2008; Shoji et al., 2010; Shoji and Hashimoto, 2014). In this study, the expression patterns of a set of PR protein genes were analysed in the low-nicotine mutants nic1, nic2, and in the double mutant nic1nic2. We identified that PR3b, a basic chitinase gene, is alternatively spliced in plants containing mutant alleles of the NIC1 and NIC2 loci. This splicing resulted in a deletion of 65bp nucleotides and introduced a premature stop codon into the coding region of the PR3b mRNA which, in turn, changed the enzyme-specific activity of PR3b. The genetic linkage between PR3b splicing and the NIC loci and the regulation of PR3b splicing by phytohormone JA and ET were also further investigated. Findings in this study indicate a novel regulatory pattern of PR3b and provide new insights into the genetic basis of the low-nicotine mutants of Burley 21.
Materials and methods
Plant materials
Plant materials used in this study include wild-type N. tabacum cv. Burley 21 and low-nicotine Burley 21 mutants (nic1, nic2, and nic1nic2). For the transcriptional assay, seeds of the desired tobacco varieties were germinated and grown for 1 week on plates with 1/2 strength Murashige & Skoog (1/2 MS) complete medium for which a commercial product of the MS basal salt mixture (Duchefa, Netherland) was used. Their seedlings were then transferred into sterile hydroponic culture chambers supplied with 200ml liquid 1/2 MS complete medium for four plants each and cultured in a growth room at 25 °C under 2 500 lx light intensity supplied by cool-white fluorescent tubes (Phillips, USA) and a 14h light/10h dark photoperiod. The hydroponic culture medium was changed weekly with fresh medium. For inductive phytohormone treatment, 5-week-old plants were treated using liquid 1/2 MS complete medium containing 50 μM ACC (1-aminocyclopropane-1-carboxylic acid; Sigma-Aldrich), or 50 μM JA (MeJA; Sigma-Aldrich) or the combination of 50 μM ACC and 50 μM JA for 24h. The roots and leaves of tobacco seedlings were collected separately for total RNAs extraction. Roots and leaves from seedlings of control treatment with phytohormone-free medium were collected as controls.
For genetic assay, the low-nicotine nic1 and nic2 mutant plants were fertilized with pollen from wild type Burley 21 to obtain the F1 plants, and the F1 plants were self-pollinated to generate F2 populations. Individual F2 seedlings were initially hydroponically cultured as described above to collect root samples for RNA preparation and then transplanted to the field for the preparation of leaf samples used for the measurement of nicotine content.
Transcriptional analyses using reverse transcription PCR (RT-PCR)
Total RNAs were extracted using the TRIZOL reagent (Invitrogen) according to the manufacturer’s instructions. First-strand cDNAs were synthesized from 5 μg of DNase I-treated total RNAs using a Cloned AMV First-Strand cDNA Synthesis Kit (Invitrogen) with oligo(dT)20. Aliquots containing reverse-transcribed products from 100ng of total RNAs were used as templates for each semi-quantitative RT-PCR or quantitative RT-PCR (qRT-PCR) reaction. Primers used for RT-PCR and qRT-PCR were as follows: 5′-AAAATGGCACTTCTGAACAC-3′ and 5′-CCAGGCTTAATAGAGTTGGA-3′ for PMT1; 5′-ACGAC CAGGTAGCAGCCTAT-3′’ and 5′-TTAGCAGCCGTCATGAA ATC-3′ for PR1a; 5′-TGCAACAATGGGTGGTATTT-3′ and 5′-G GAATCAAAGGGATGTTGCT-3′ for PR1b; 5′-AAGCTGGTTT GGGAAACAAC-3′ and 5′-AAACCACCTAGCATCGTTCC-3′ for PR2b; 5′-AGGAGGTGGAATCAGTGGAC-3′ and 5′-TGACATTA GCACTTGCTTTGG-3′ for PR3b; 5′-GGGTAAACCACCAAAC ACCT-3′ and 5′-GGGAAAGTGATCGGAATGTT-3′ for PR5; 5′-CC ACACAGGTGTGATGGTTG-3′ and 5′-GTGGCTAACACCATCA CCAG-3′ for Actin. 5′-GAGCAATTCAGAAAATTTCAAGAGG-3′ (upstream of the splicing region) was combined, respectively, with 5′-CATTACCCGCGGCTGTCTTGGCTG-3′ (for native PR3b and specific to the fragment to be excised in the splicing) and 5′-CTTGCTTTGGTTTGTGTCCACTG-3′ (for spliced PR3b and specific to the spliced junction) in order to quantify the transcript levels of native and spliced PR3b specifically. Three independent biological replicates were performed for all experiments.
For the semi-quantitative RT-PCR assay, the PCR amplification program was 25 cycles of 1min at 94 °C, 40s at 58 °C, and 40s at 72 °C. The PCR products were separated using a 1% agarose gel, stained with ethidium bromide, and visualized under UV light. qRT-PCR reactions were performed on a Stratagene Mx3000P™ quantitative PCR system (Stratagene, USA) using GoTaq® qPCR Master Mix (Promega). Actin was used as the internal control. The relative transcripts were obtained by calibrating the threshold cycles of genes of interest with that of Actin using the equation 2(–ΔΔCT), as previously described by Zhang et al. (2012), where CT is the cycle number of the threshold point at which fluorescence is detectable.
Rapid amplification of cDNA ends (RACE) of spliced PR3b
Nested PCR method was applied for the RACE PCR using a Smarter RACE kit (Clontech, USA) except that the PCR reagents of a Phire Plant Direct kit (Thermo, USA) were used for the amplification. cDNAs were synthesized with total RNAs from the roots of nic mutants according to the manufacturer’s instruction. 5′-RACE was initially amplified at an annealing temperature of 56 °C with the gene-specific primer 5′-TGACATTAGCACTTGCTTTGG-3′ (downstream of the splicing region) and the universal primer provided by the Smarter RACE kit, and then amplified at an annealing temperature of 60 °C with primer 5′-CTTGCTTTGGTTTGTGTCCACTG-3′ (specific to the spliced junction of PR3b) and the universal primer. 3′-RACE was done in a similar way. The gene-specific primer for the initial amplification is 5′-AGGAGGTGGAATCAGTGGAC-3′ (upstream of the splicing region), and that for the second round amplification is 5′-CAGTGGACACAAACCAAAGCAAG-3′ (specific to the spliced junction of PR3b). The PCR products were ligated into EcoRV-digested pBlueScript II SK+ vector and then sequenced.
Comparison of the RT-PCR and genomic DNA PCR products of PR3b
Genomic DNA was extracted from tobacco roots using the CTAB method, as described by Zhang et al. (2012). 100ng genomic DNA was used as a template for PCR amplification of the PR3b fragment. The PCR amplification program was 30 cycles of 1min at 94 °C, 40s at 58 °C, and 40s at 72 °C.
RT-PCR and genomic DNA PCR amplification products of the PR3b fragments were compared by electrophoresis on a 1.5% (w/v) agarose gel. The amplifications products were visualized by staining with ethidium bromide and exposure under UV light. The 1kb DNA Ladder (Invitrogen, USA) was used as a DNA molecular weight marker.
Gel extraction and abundance estimation of PR3b isoforms
For sequencing of the amplified PR3b DNA or cDNA fragments, the corresponding PCR products were first separated on a 1% agarose gel. The gel fragments containing the amplification products of both native and alternatively spliced PR3b fragments were purified using a Gel Extraction Kit (BBI), ligated into the pBlueScript II SK+ vector (Stratagene), and sequenced using M13 primers. Fifty positive clones of each sample were sequenced, and the abundance of PR3b isoforms estimated.
Bioactivity assay of spliced PR3b variant
To obtain proteins for the enzymatic assay, the coding sequences of native PR3b and alternatively spliced PR3b were amplified with the same 5′-end primer 5′-AAAGGATCCATGAGCATTAAGCTATCTT-3′ (restriction site is italicized) and specific 3′-end primers 5′-AACAGCACCCCTGATAGC-3′ (for native PR3b) and 5′-TTCCAAAGCATGACACCTC-3′ (for spliced PR3b), and cloned in-frame with the glutathione S-transferase (GST) tag coding region in pGEX-4T-2 vector (Novagen) through restriction sites BamHI and SmaI. Then, the protein expression vectors were transformed into E. coli BL21 cells to induce prokaryotic protein expression by treating cells with 1mM IPTG for 3h at 37 °C. The recombinant proteins were purified using GST affinity column chromatography according to the manufacturer’s protocol (Invitrogen) and dialysed against the dialysing buffer (50mM sodium phosphate, 10% glycerol, pH 6.5). The empty pGEX-4T-2 vector was used to produce control GST protein in the same procedure. The enzyme-specific activity was determined with a fluorimetric Chitinase Assay Kit (Sigma CS1030) with 4-methylumbelliferyl β-d-N, N ′, N ′-triacetylchitotriose [4MU-GlcNAc3] as substrate (Brotman et al., 2012) according to the manufacturer’s introduction. After incubation for 1h at 37 °C, fluorescence of the reaction mixture was measured by a fluorescence spectrophotometer (excitation at 360nm, emission at 450nm).
Leaf nicotine measurement
Dry leaf samples were subjected to alkaloids extraction as previously described previously (Goossens et al., 2003; Zhang et al., 2012) with minor modification. Briefly, 10mg of homogenized dry leaf sample was soaked in 1ml of 10% NaOH (w/v) for 20min and then extracted by vortexing with an equivalent volume of dichloromethane. The organic layer was collected after centrifugation. The nicotine content was measured on an Agilent Technologies 7890A Chromatograph equipped with a DB 5 MS column and Agilent Technologies 5975C inert MSD detector with helium as the carrier gas. The column temperature was held at 100 °C for 5min, increased to 210 °C at an increment of 50 °C min–1, and then held at 210 °C for 4min. The ion source temperature was 230 °C and the quadrupole temperature was 150 °C. Nicotine from Sigma–Aldrich was used as the standard control.
Gene accessions
The NCBI accession numbers for the N. tabacum genes mentioned in this article are as follows: PMT1 (AF126810), PR1a (X12737), PR1b (X66942), PR2b (M59442), PR3b (Z11564), PR5 (M29279), and Actin (X63603).
Results
JA/ET-induced expression patterns of a set of PR protein genes in the roots of Burley 21 tobacco
It is well documented that PR protein genes functioning in plant pathogen resistance are regulatory targets of ERF transcription factors (Chakravarthy et al., 2003; Guo and Ecker, 2004; Gutterson and Reuber, 2004). The NIC1 and NIC2 loci integrate the regulation of nicotine biosynthetic genes and stress-responsive genes (Hibi et al., 1994; Cane et al., 2005; Kidd et al., 2006), and the NIC2 locus contains a cluster of ERF transcription factors (Shoji et al., 2010). Therefore, we hypothesized that mutations in NIC1 or NIC2 are likely to alter the PR protein gene expression patterns. To explore this possibility, we first selected a set of PR protein genes known to be regulated by the JA and/or ET pathways and ERF transcription factors (Ohme-Takagi and Shinshi, 1995; Chakravarthy et al., 2003; Lorenzo et al., 2003; Zhang et al., 2004; van Loon et al., 2006), and tested their induction by JA and/or ACC (1-aminocyclopropane-1-carboxylic acid; the immediate precursor of ethylene) in the roots of wild-type Burley 21 tobacco. The PR protein genes selected included PR1a (acidic PR1 gene), PR1b (basic PR1 gene), PR2b (basic beta-1,3-glucanase gene), PR3b (basic chitinase III gene), and PR5 (osmotin gene). The expression level of PMT1, a well characterized gene involved in nicotine biosynthesis (Riechers and Timko, 1999), was also analysed as a control.
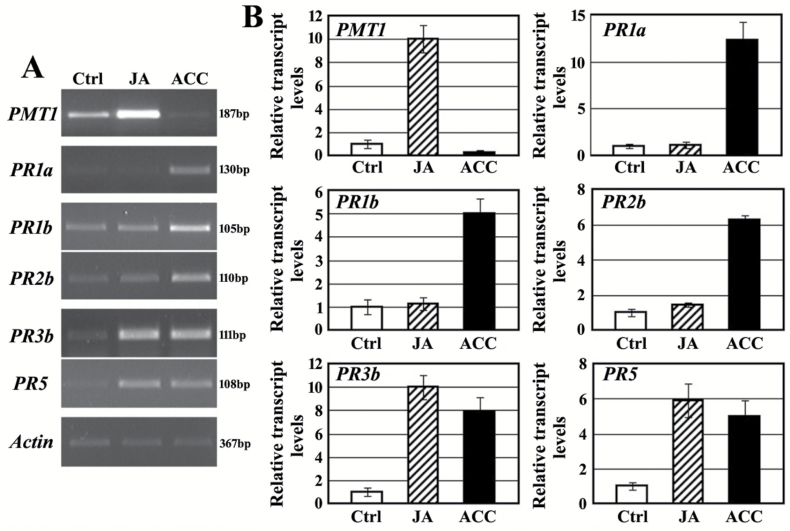
We analysed the expression patterns of the selected PR protein genes in the roots of wild-type tobacco treated for 24h with JA or ACC. As shown in Fig. 1A, the designed primers could specifically amplify the target fragments of PR protein genes in the roots of wild-type Burley 21. The expression of PMT1 was dramatically induced by JA but inhibited by ACC; the expression of PR1a, PR1b, and PR2b could only be induced by ACC treatment and the expression of PR3b and PR5 was induced by both JA and ACC treatments (Fig. 1A, B). These findings established a preliminary regulatory relationship between the nicotine biosynthetic pathway and the regulation of PR protein genes by the JA and ET signalling pathways in Burley 21 tobacco.
Fig. 1.
Induced expression patterns of the nicotine synthetic gene PMT1 and PR protein genes in the roots of wild-type tobacco Burley 21. (A) Expression pattern assay by semi-quantitative RT-PCR. The representative results of three independent replicates are shown. The sizes of the amplified products are indicated on the right. (B) Transcript levels of PMT1 and PR protein genes based on qRT-PCR analysis. Ctrl indicates untreated control. JA and ACC indicate treatment with JA and ACC, respectively. The transcription level of each gene in the Ctrl is set as ‘1’. Actin was used as an internal control for both semi-quantitative RT-PCR and qRT-PCR.
PR3b is alternatively spliced in low-nicotine mutants of Burley 21
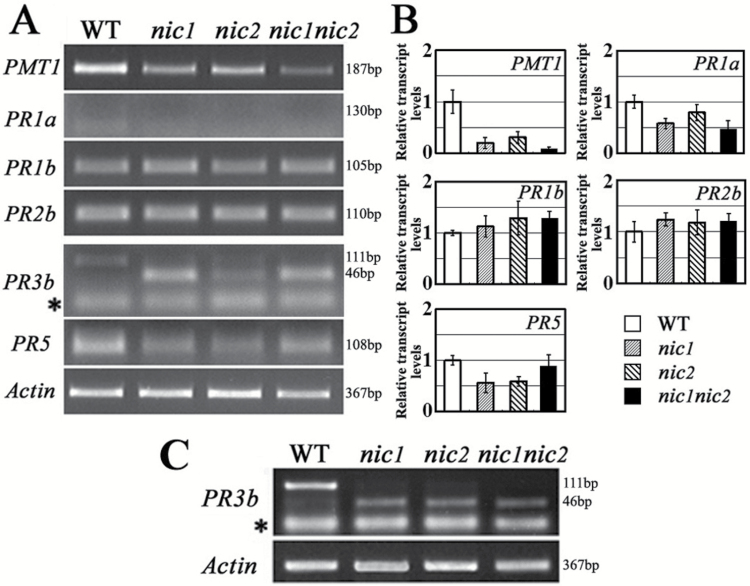
To investigate the potential roles of NIC loci in regulating PR protein genes, transcript levels of the selected PR protein genes were comparatively analysed in the untreated low-nicotine mutants (nic1, nic2, and nic1nic2) as well as the wild-type control. RT-PCR assays revealed that the transcript level of PMT1 was down-regulated to different extents in the mutants (Fig. 2A, B). No obvious transcriptional differences were observed for PR1b and PR2b in the wild type or low-nicotine mutants of Burley 21, whereas the transcript levels of PR1a and PR5 were slightly down-regulated in the low-nicotine mutants (Fig. 2A, B). While all the above amplifications gave the expected specific products, the amplification products of PR3b from the low-nicotine mutants showed two distinct bands in the gel: a faint band of the same size amplified from wild-type Burley 21 and a smaller, more intense band which is hard to visualize in the wild-type control (Fig. 2A). The amplification of PR3b transcripts from leaf tissue gave a similar result to the roots (Fig. 2C). To establish whether the smaller band was an alternative transcript of PR3b or a non-specifically amplified fragment, the PR3b RT-PCR amplification products were sequenced. This revealed that the larger product is the amplification of native PR3b transcript while the smaller one is an amplification of an alternatively spliced transcript of PR3b (Fig. 3B). These data implied a difference in the post-transcriptional regulation of PR3b transcripts between the wild type and the low-nicotine mutants of Burley 21.
Fig. 2.
Transcription patterns of PMT1 and PR protein genes in the wild type and in low-nicotine mutants of Burley 21. (A) Expression patterns of PMT1 and PR protein genes in roots by semi-quantitative RT-PCR assay. (B) Expression levels of PMT1 and PR protein genes determined by qRT-PCR. The transcription level of each gene in the Ctrl is set as ‘1’. (C) Transcripts of PR3b gene in leaves. (A, C) The representative results of three independent replicates and the sizes of the amplified products are indicated on the right. WT indicates wild type Burley 21. nic1, and nic2 indicate low-nicotine mutant alleles of NIC1 and NIC2, and nic1nic2 indicates the double mutant. The asterisk indicates primer dimers in the amplification products of the PR3b gene. Actin was used as an internal control.
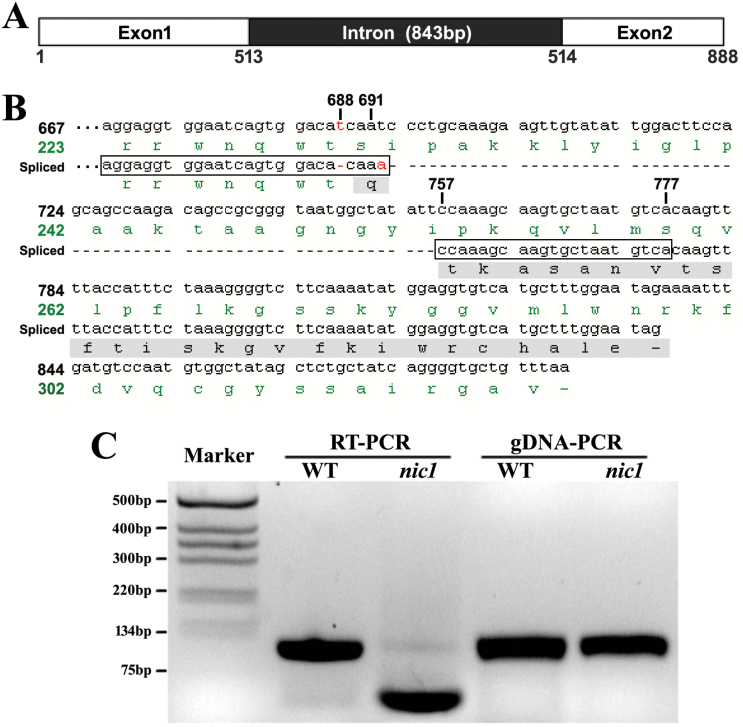
Fig. 3.
Characterization of the alternative splicing of PR3b. (A) Schematic gene structure of PR3b. The details are shown in Supplementary Fig. S1. (B) Alignment of the alternatively spliced PR3b fragment (Spliced) against the native PR3b cDNA (sequence with labelled nucleotide numbers). The numbers indicate the position of the indicated nucleotide at the native PR3b coding region. The alternative spliced region is highlighted by rectangles and dashes between the rectangles indicate the excised region by the alternative splicing. Positions corresponding to the single-nucleotide mutation sites are indicated by red characters. The amino acid sequences in green are deduced from the native PR3b cDNA. The amino acid sequence highlighted by the grey bar shows the coding change region caused by alternatively splicing of PR3b; the dash in the amino acid sequence indicates the position of the stop codon. (C) Comparison of RT-PCR and genomic DNA PCR (gDNA-PCR) products amplified using the same primers. Marker indicates the DNA molecular marker. WT, wild type; nic1, low-nicotine mutant nic1.
Characterization of the alternatively spliced transcripts of PR3b
In order to reveal further information about the alternatively spliced transcript of PR3b, we compared its nucleotide sequence with the native PR3b cDNA sequence using DNA sequence alignment analysis. The genomic sequence of PR3b contains two exons and one intron (Fig. 3A; see Supplementary Fig. S1 at JXB online), and the alternative splicing occurs in the region of exon 2. The amplified fragment corresponding to native PR3b cDNA is 111bp in length (nt. 667–777 of the 888bp coding sequence), while that of the alternatively spliced transcript is 46bp in length (nt. 667–712 of the 771bp coding sequence; Fig. 3B). Therefore, a fragment of 65bp in length was deleted through the alternative splicing. To confirm whether the smaller fragment was a real spliced transcript of PR3b or an occasionally non-specific amplification product, we designed primers to amplify a longer fragment overlapping the previously amplified region and analysed the sequence details. The results revealed a consensus in the sequence observed to be alternatively spliced (Supplementary Fig. S2). As 65 is a non-integral multiple of three, the alternative splicing altered the reading frame as well as the amino acid sequence of PR3b and introduced a premature stop codon in the coding region of PR3b (Fig. 3B). This implies a potential functional alteration of PR3b caused by the alternative splicing. Intriguingly, two single-nucleotide changes were also observed, i.e. one missing T at position 688 and one extra A behind position 691 of the native PR3b (Fig. 3B). A following RACE (rapid amplification of cDNA ends) PCR was applied to determine whether other splicing events are present in the coding sequence of PR3b, using primers specific to the spliced junction of the spliced PR3b variant. Yet, no other splicing was observed (Supplementary Fig. S3).
To determine whether this was a consequence of transcription shifts of unidentified genes with homologous sequence to the splicing region of PR3b in tobacco, we amplified the corresponding genomic fragment using the same primers used for RT-PCR amplification. Figure 3C shows results of the comparison of RT-PCR and genomic DNA PCR amplification products of the wild type and the nic1 mutant of Burley 21. This revealed that the genomic DNA PCR products from all of the plants were the same size as the native PR3b fragment and that no products corresponding to the alternatively spliced PR3b fragment were observed (Fig. 3C). This indicated that the 46bp fragment was an alternatively spliced product of PR3b. Furthermore, we observed that the truncated PR3b fragment was also present in wild-type tobacco at trace amounts (Fig. 3C) and that the amplification products of the native PR3b fragment were observed in the RT-PCR amplification products of low-nicotine mutants.
Enzyme-specific activity of the spliced PR3b variant
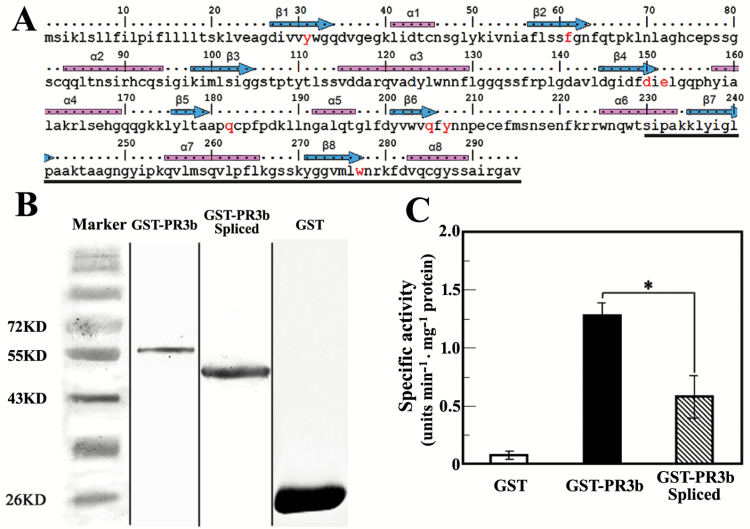
Tobacco PR3b is a class III plant chitinase, having a GH18 (glycosyl hydrolase family 18) domain with a pronounced active-site cleft at the C-terminal end of its beta-barrel (Hurtado-Guerrero and van Aalten, 2007; Tyler et al., 2010). The alternative splicing of PR3b altered the amino acid sequence after Thr229 at its C-terminus which contains secondary structure regions (α6/7/8, β7/8) and an amino acid (Trp277) that are conserved across the GH18 chitinase family (Fig. 4A; Hurtado-Guerrero and van Aalten, 2007; Tyler et al., 2010). Thus, the observed structural alterations suggested that a change in PR3b activity might have occurred. To determine this hypothesis, we expressed the wild-type PR3b protein and its spliced variant as GST-tagged fusions in E. coli BL21 cells, and the purified proteins (Fig. 4B) were tested for their chinolytic activity in a specific bioassay with GST as the control. Results showed that the enzyme-specific activity of native PR3b is about 2-fold higher than that of the spliced PR3b variant (Fig. 4C), suggesting that the alternative splicing of PR3b results in a significant reduction in the enzyme-specific activity of PR3b.
Fig. 4.
Specific chitinase activity of native and alternatively spliced PR3b. (A) Conserved α-helixes (blue arrows), β-sheets (pink bars), and amino acids (red characters) of PR3b compared with the catalytic cores of GH18 family chitinases (Hurtado-Guerrero and van Aalten, 2007). The region affected by alternative splicing is underlined with a black line. (B) Purified proteins of GST-tagged PR3b and GST separated by SDS-PAGE gel. ‘GST-PR3b’ is the GST-tagged protein of native PR3b; ‘GST-PR3b Spliced’ is the GST-tagged protein of spliced PR3b. Marker is the protein standard ruler. The picture shown is a combination of representative lanes from gels with serial elutions of the purified proteins. (C) Enzyme-specific activity of GST-tagged native and alternatively spliced PR3b. One unit equals 1 μmol of released 4-MU. GST is used for the control reaction. Values are the average of three replicates. Error bar, mean ±SD. The asterisk indicates a significant difference between the paired data sets (P <0.05, Student’s t test).
Genetic linkage between enhanced PR3b splicing and the NIC loci in Burley 21
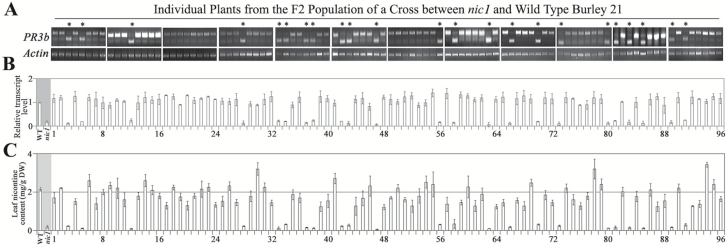
Low-nicotine mutants nic1 and nic2 of Burley 21 were derived from the double mutant nic1nic2 of Burley 21 (Legg and Collins, 1971), thus the coincident PR3b splicing in all these low-nicotine varieties somehow implies a linkage to the NIC loci. To explore the relationship between enhanced PR3b splicing and the NIC loci in Burley 21 further, we performed genetic analyses with an F2 population of the cross between the nic1 mutant and wild-type Burley 21. In ~25% of the F2 plants (23/96), PR3b is alternatively spliced as it is in the nic mutants of Burley 21 (Fig. 5A), and the transcript levels of PMT1 in these lines were also considerably lowered (Fig. 5B). Furthermore, the leaf nicotine content of the corresponding lines is very low (less than 0.35mg g–1 dry weight) compared with the other lines (Fig. 5C). We also analysed PR3b splicing in a small group of F2 plants of the cross between the nic2 mutant and wild-type Burley 21 and obtained a similar result (Supplementary Fig. S4). These findings support a positive link between enhanced PR3b splicing and the NIC loci in Burley 21. On the other hand, the NIC1 and NIC2 loci are two unlinked loci (Legg et al., 1969; Legg and Collins, 1971). The correlation between PR3b splicing and the NIC loci in Burley 21 implies the involvement of both NIC1 and NIC2 loci in regulating PR3b splicing.
Fig. 5.
Genetic linkage between alternative splicing of PR3b and the NIC1 locus in the Burley 21 background. (A) Alternative splicing of PR3b in individual F2 plants of a cross between nic1 and wild-type Burley 21. (B) Transcript levels of PMT1 in the roots of individual F2 plants. Transcript level of PMT1 in the roots of wild-type Burley 21 was set as ’1’. Actin was used as an internal control. (C) Leaf nicotine content of individual F2 plants. The values shown are the means of three technical replicates. Error bar, mean ±SD.
Regulation of PR3b splicing in the low-nicotine mutants of Burley 21 by phytohormones JA and ET
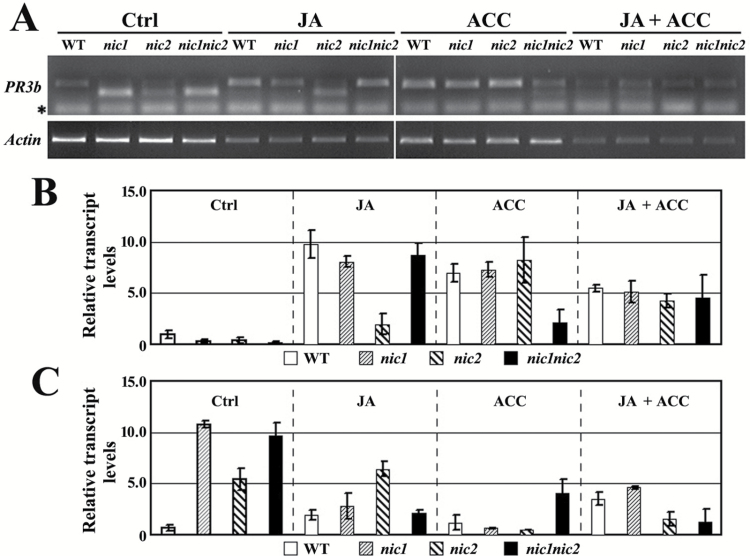
NIC1 and NIC2 are unlinked loci and they may display differential roles in regulating PR3b splicing. We noticed the induction of PR3b expression by JA and the precursor of ET which might be able to alter the regulatory effects of NIC loci on PR3b splicing. We then investigated the potentials of these two hormones on the alternative splicing of PR3b transcripts. The results showed that the alternative splicing patterns of PR3b were altered by treatments with JA, ACC, or the combination of JA and ACC (Fig. 6). JA treatment suppressed the alternative splicing of PR3b in nic1 and nic1nic2 mutants but not in nic2. By contrast, treatment with ACC or the combination of JA and ACC suppressed splicing in all nic mutants to different extents. Interestingly, the alternatively spliced fragment of PR3b was faintly visible in the amplification products of wild-type tobacco treated with JA or the combination of JA and ACC (Fig. 6A). The abundance of native and alternatively spliced PR3b was quantified by qRT-PCR amplification using specific primers. Our results showed that JA and/or ACC treatment could accentuate the transcript levels of native PR3b and attenuate the transcript levels of spliced PR3b in the low-nicotine mutants (Fig. 6B, C). These findings were similar to those observed in semi-quantitative RT-PCR. Taken together, our results suggest that the alternative splicing of PR3b is differentially regulated by JA and ET in the wild type and low-nicotine mutants of Burley 21 tobacco.
Fig. 6.
Phytohormone-induced splicing patterns of PR3b. (A) Splicing patterns of PR3b in the wild type (WT) and in low-nicotine mutants (nic1, nic2, and nic1nic2). The representative results of three independent replicates are shown. The asterisk indicates the primer dimers in the PCR amplification products. (B) Quantification of specific transcript of native PR3b. (C) Quantification of specific transcript of spliced PR3b. Ctrl indicates the untreated control; JA, ACC, and JA+ACC (the combination of JA and ACC) indicate different phytohormone treatments. The PR3b transcript level in the WT of the Ctrl treatment is set as ‘1’ (B, C). Actin was used as an internal control.
We also cloned the RT-PCR amplification products from each plant into the pBluescript II SK+ vector and sequenced them to determine the presence of the native and alternatively spliced transcripts. The sequencing analyses revealed that the major amplification products in the wild-type plants were the 111bp native fragments and that there were quite a few transcripts of the 46bp spliced fragment (Table 1). The major amplification products in the low-nicotine mutants were the 46bp spliced fragment, however, there were also considerable transcripts of the 111bp native fragment and their abundances were affected by phytohormone treatments (Table 1). These results are consistent with the results of the qRT-PCR assays and showed the presence of the spliced fragment in wild-type plants. This evidence indicates that mutations of the NIC loci altered the abundance of native and alternatively spliced PR3b transcripts in the low-nicotine mutants, i.e. mutation in nic1 or nic2 specifically enhanced the abundance of alternatively spliced PR3b transcripts.
Table 1.
Abundance of native and alternatively spliced transcripts of PR3b in the RT-PCR amplification products of the roots of the wild type (WT) and in low-nicotine mutants of Burley 21
| Treatment | Ctrl | JA | ACC | JA+ACC | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | WT | nic1 | nic2 | nic1nic2 | WT | nic1 | nic2 | nic1nic2 | WT | nic1 | nic2 | nic1nic2 | WT | nic1 | nic2 | nic1nic2 |
| Native | 49 | 12 | 22 | 10 | 46 | 44 | 17 | 48 | 50 | 50 | 50 | 37 | 41 | 36 | 49 | 50 |
| Spliced | 1 | 38 | 28 | 40 | 4 | 6 | 33 | 2 | 0 | 0 | 0 | 13 | 9 | 14 | 1 | 0 |
Data were collected from a representative RT-PCR amplification of three independent replicates. Ctrl indicates the untreated controls. JA, ACC, and JA+ACC indicate the different phytohormone treatments. Native indicates the number of colonies containing native PR3b fragments. Spliced indicates the number of colonies containing alternatively spliced PR3b fragments.
Discussion
By investigating the expression patterns of a set of PR protein genes in the low-nicotine mutants (nic1, nic2, and nic1nic2) of Burley 21 tobacco, we identified the basic chitinase gene PR3b as being alternatively spliced and then characterized this phenomenon in this study.
Tobacco PR3b belongs to the class III chitinase which has a conserved GH18 domain with eight-stranded β/α-barrel (Hurtado-Guerrero and van Aalten, 2007). The alternative splicing of PR3b mRNA caused a reading frame change and introduced a premature stop codon. Therefore, it changed the C-terminal amino acid sequence of PR3b and resulted in the loss of conserved domains and amino acid. And, this splicing could result in a reduction of the enzyme-specific activity of PR3b by half. PR3b has a functional role in tobacco resistance against fungal pathogens (Lawton et al., 1992; van Loon et al., 2006). Presumably, the enhanced alternative splicing of PR3b in the low-nicotine mutants might cause an alteration in the susceptibility to fungal pathogens. However, the transcription patterns of other PR protein genes were also altered in the nic mutants (Supplementary Fig. S5), which resulted in a certain difficulty in determining the specific change in anti-fungal capability caused by PR3b splicing. Two single-nucleotide changes (a missing T and an extra A) were also observed in the alternative spliced sequence of PR3b. Presumably, this was caused by nucleotide-deletion/insertion during mRNA splicing (Gott and Emeson, 2000). Yet, the cause of reading frame shift is the deletion of the 65bp fragment but not the single-nucleotide change.
In general pre-mRNA splicing, introns mostly start from GU, end with AG, and contain a so-called ‘branch site’ with the sequence CU(A/G)A(C/U) at about 20–50 bases upstream of the AG-end (Meyer et al., 2015). Apparently, the PR3b splicing found in this study does not meet this rule. Alternative splicing is a complicated regulatory mechanism (Meyer et al., 2015), and has been reported for several genes functioning in the plant response to pathogen attack (Dinesh-Kumar and Baker, 2000; Zhang and Gassmann, 2007). A previous finding that shares some similarities with this study is that of the alternative splicing of acidic chitinase II in Citrus clementina. This introduced a premature stop codon but the chitinase activity could be induced following MeJA treatment (Del Carratore et al., 2011). Similarly, the enhanced alternative splicing of PR3b in the low-nicotine mutants could be suppressed by JA and/or ACC (the precursor of ET) treatments. Although the acidic chitinase II of C. clementina does not share any similarity with the basic chitinase PR3b in N. tabacum, they both provide evidence for JA-induced alternative splicing of chitinase, implying a common regulatory mechanism. These findings also provided evidence of JA/ET-signalling pathways in co-ordinating the alternative splicing of PR protein genes.
The low-nicotine mutants nic1 and nic2 were derived from LA Burley 21, i.e. the low-nicotine mutant nic1nic2 of Burley 21 (Legg et al., 1970; Legg and Collins, 1971). Hence, the coincident PR3b splicing patterns in all these low-nicotine varieties, to some extent, imply a linkage to the NIC loci. Consistently, the genetic analyses of F2 populations of crosses between wild-type Burley 21 and the nic1 mutant suggested a positive link between PR3b splicing and the NIC loci in Burley 21. On the other hand, findings in this study showed that PR3b was spliced in both the wild type and the low-nicotine mutants of Burley 21, except that the spliced PR3b transcripts were enhanced to higher levels in the low-nicotine mutants of Burley 21. Thus, observation of enhanced PR3b splicing in these low-nicotine varieties suggests that PR3b splicing is co-ordinately regulated by the NIC1 and NIC2 loci, even though they are two unlinked loci (Legg et al., 1970; Legg and Collins, 1971). Furthermore, the alternative splicing of PR3b in the low-nicotine mutants is differentially regulated by the phytohormones JA and ET. The alternative splicing of PR3b could be suppressed by ACC in all of the low-nicotine mutants to different extents but was only repressed by JA in nic1 and nic1nic2. The difference in phytohormone-induced splicing patterns of PR3b in the low-nicotine mutants suggests that the NIC1 and NIC2 loci display differential roles in regulating PR3b splicing.
The regulation of alternative RNA splicing is an important part of the gene regulation network (Black, 2003; Reddy, 2007; Syed et al., 2012; Yang et al., 2014) which involves the regulation of stress and phytohormone responses (Palusa et al., 2007; Reddy and Shad Ali, 2011). The finding of PR3b splicing regulation by JA/ET and NIC loci in Burley 21 is valuable to the genetic studies of low-nicotine mutants and could provide clues to unravel the mechanism by which JA/ET-signalling pathways regulate PR protein gene splicing.
Supplementary data
Supplementary data can be found at JXB online.
Figure S1. Alignment of PR3b genomic sequence and CDS (coding sequence) amplified from tobacco Burley 21.
Figure S2. Sequence analysis of PR3b splicing region amplified with different primer sets.
Figure S3. Rapid amplification of cDNA ends (RACE) of alternatively spliced PR3b.
Figure S4. Alternative splicing of PR3b in the F2 population of a cross between nic2 and wild-type Burley 21.
Figure S5. Phytohormone-induced transcription patterns of PR protein genes.
Acknowledgements
This work was financially supported by the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (Elite youth program to HZ, ASTIP-TRIC05), the Program of Chongqing Tobacco Company (NY20140403030022), the Key Special Program of China National Tobacco Corporation [110201301009 (JY-09), 110201301005 (JY-05)], and Guizhou Science and Technology Fund (J[2010]2088). MPT was supported in part by funds from 22nd Century LLC.
References
- Baldwin IT. 1998. Jasmonate-induced responses are costly but benefit plants under attack in native populations. Proceedings of the National Academy of Sciences, USA 95, 8113–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin IT. 2001. An ecologically motivated analysis of plant-herbivore interactions in native tobacco. Plant Physiology 127, 1449–1458. [PMC free article] [PubMed] [Google Scholar]
- Black DL. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annual Review of Biochemistry 72, 291–336. [DOI] [PubMed] [Google Scholar]
- Brotman Y, Landau U, Pnini S, Lisec J, Balazadeh S, Mueller-Roeber B, Zilberstein A, Willmitzer L, Chet I, Viterbo A. 2012. The LysM receptor-like kinase LysM RLK1 is required to activate defense and abiotic-stress responses induced by overexpression of fungal chitinases in Arabidopsis plants. Molecular Plant 5, 1113–1124. [DOI] [PubMed] [Google Scholar]
- Cane KA, Mayer M, Lidgett AJ, Michael AJ, Hamill JD. 2005. Molecular analysis of alkaloid metabolism in AABB v. aabb genotype Nicotiana tabacum in response to wounding of aerial tissues and methyl jasmonate treatment of cultured roots. Functional Plant Biology 32, 305–320. [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Tuori RP, D’Ascenzo MD, Fobert PR, Despres C, Martin GB. 2003. The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. The Plant Cell 15, 3033–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer K, Tilleman S, Pauwels L, Vanden Bossche R, De Sutter V, Vanderhaeghen R, Hilson P, Hamill JD, Goossens A. 2011. APETALA2/ETHYLENE RESPONSE FACTOR and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. The Plant Journal 66, 1053–1065. [DOI] [PubMed] [Google Scholar]
- Dewey RE, Xie J. 2013. Molecular genetics of alkaloid biosynthesis in Nicotiana tabacum . Phytochemistry 94, 10–27. [DOI] [PubMed] [Google Scholar]
- Deboer KD, Lye JC, Aitken CD, Su AK, Hamill JD. 2009. The A622 gene in Nicotiana glauca (tree tobacco): evidence for a functional role in pyridine alkaloid synthesis. Plant Molecular Biology 69, 299–312. [DOI] [PubMed] [Google Scholar]
- Del Carratore R, Magaldi E, Podda A, Beffy P, Migheli Q, Maserti BE. 2011. A stress responsive alternative splicing mechanism in Citrus clementina leaves. Journal of Plant Physiology 168, 952–959. [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Baker BJ. 2000. Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proceedings of the National Academy of Sciences, USA 97, 1908–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens A, Hakkinen ST, Laakso I, et al. 2003. A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proceedings of the National Academy of Sciences, USA 100, 8595–8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gott JM, Emeson RB. 2000. Functions and mechanisms of RNA editing. Annual Review of Genetics 34, 499–531. [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR. 2004. The ethylene signaling pathway: new insights. Current Opinion in Plant Biology 7, 40–49. [DOI] [PubMed] [Google Scholar]
- Gutterson N, Reuber TL. 2004. Regulation of disease resistance pathways by AP2/ERF transcription factors. Current Opinion in Plant Biology 7, 465–471. [DOI] [PubMed] [Google Scholar]
- Heim WG, Sykes KA, Hildreth SB, Sun J, Lu RH, Jelesko JG. 2007. Cloning and characterization of a Nicotiana tabacum methylputrescine oxidase transcript. Phytochemistry 68, 454–463. [DOI] [PubMed] [Google Scholar]
- Hibi N, Higashiguchi S, Hashimoto T, Yamada Y. 1994. Gene expression in tobacco low-nicotine mutants. The Plant Cell 6, 723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado-Guerrero R, van Aalten DM. 2007. Structure of Saccharomyces cerevisiae chitinase 1 and screening-based discovery of potent inhibitors. Chemistry & Biology 14, 589–599. [DOI] [PubMed] [Google Scholar]
- Imanishi S, Hashizume K, Nakakita M, Kojima H, Matsubayashi Y, Hashimoto T, Sakagami Y, Yamada Y, Nakamura K. 1998. Differential induction by methyl jasmonate of genes encoding ornithine decarboxylase and other enzymes involved in nicotine biosynthesis in tobacco cell cultures. Plant Molecular Biology 38, 1101–1111. [DOI] [PubMed] [Google Scholar]
- Kajikawa M, Shoji T, Kato A, Hashimoto T. 2011. Vacuole-localized berberine bridge enzyme-like proteins are required for a late step of nicotine biosynthesis in tobacco. Plant Physiology 155, 2010–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd SK, Melillo AA, Lu RH, Reed DG, Kuno N, Uchida K, Furuya M, Jelesko JG. 2006. The A and B loci in tobacco regulate a network of stress response genes, few of which are associated with nicotine biosynthesis. Plant Molecular Biology 60, 699–716. [DOI] [PubMed] [Google Scholar]
- Kumar P, Pandit SS, Steppuhn A, Baldwin IT. 2014. Natural history-driven, plant-mediated RNAi-based study reveals CYP6B46’s role in a nicotine-mediated antipredator herbivore defense. Proceedings of the National Academy of Sciences, USA 111, 1245–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton K, Ward E, Payne G, Moyer M, Ryals J. 1992. Acidic and basic class III chitinase mRNA accumulation in response to TMV infection of tobacco. Plant Molecular Biology 19, 735–743. [DOI] [PubMed] [Google Scholar]
- Legg PD, Chaplin JF, Collins GB. 1969. Inheritance of percent total alkaloids in Nicotiana tabacum L.: Populations derived from crosses of low alkaloid lines with burley and flue-cured varieties. Journal of Heredity 60, 213–217. [Google Scholar]
- Legg PD, Collins GB. 1971. Inheritance of percent total alkaloids in Nicotiana tabacum L. II. Genetic effects of two loci in Burley 21 Í LA Burley 21 populations. Canadian Journal of Genetics and Cytology 13, 287. [Google Scholar]
- Legg PD, Colons GB, Litton CC. 1970. Registration of LA Burley 21 tobacco germplasm. Crop Science 10, 212. [Google Scholar]
- Lewis RS, Lopez HO, Bowen SW, Andres KR, Steede WT, Dewey RE. 2015. Transgenic and mutation-based suppression of a berberine bridge enzyme-like (BBL) gene family reduces alkaloid content in field-grown tobacco. PLoS One 10, e0117273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Teng W, Shi Q, Zhang F. 2007. Multiple signals regulate nicotine synthesis in tobacco plant. Plant Signaling & Behavior 2, 280–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sanchez-Serrano JJ, Solano R. 2003. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. The Plant Cell 15, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Koester T, Staiger D. 2015. Pre-mRNA splicing in plants: in vivo functions of RNA-binding proteins implicated in the splicing process. Biomolecules 5, 1717–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H. 1995. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. The Plant Cell 7, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palusa SG, Ali GS, Reddy AS. 2007. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. The Plant Journal 49, 1091–1107. [DOI] [PubMed] [Google Scholar]
- Pre M, Atallah M, Champion A, De Vos M, Pieterse CM, Memelink J. 2008. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiology 147, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS. 2007. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annual Review of Plant Biology 58, 267–294. [DOI] [PubMed] [Google Scholar]
- Reddy AS, Shad Ali G. 2011. Plant serine/arginine-rich proteins: roles in precursor messenger RNA splicing, plant development, and stress responses. Wiley Interdisciplinary Reviews: RNA 2, 875–889. [DOI] [PubMed] [Google Scholar]
- Reed DG, Jelesko JG. 2004. The A and B loci of Nicotiana tabacum have non-equivalent effects on the mRNA levels of four alkaloid biosynthetic genes. Plant Science 167, 1123–1130. [Google Scholar]
- Riechers DE, Timko MP. 1999. Structure and expression of the gene family encoding putrescine N-methyltransferase in Nicotiana tabacum: new clues to the evolutionary origin of cultivated tobacco. Plant Molecular Biology 41, 387–401. [DOI] [PubMed] [Google Scholar]
- Saedler R, Baldwin IT. 2004. Virus-induced gene silencing of jasmonate-induced direct defences, nicotine and trypsin proteinase-inhibitors in Nicotiana attenuata . Journal of Experimental Botany 55, 151–157. [DOI] [PubMed] [Google Scholar]
- Sears MT, Zhang H, Rushton PJ, Wu M, Han S, Spano AJ, Timko MP. 2014. NtERF32: a non-NIC2 locus AP2/ERF transcription factor required in jasmonate-inducible nicotine biosynthesis in tobacco. Plant Molecular Biology 84, 49–66. [DOI] [PubMed] [Google Scholar]
- Shi Q, Li C, Zhang F. 2006. Nicotine synthesis in Nicotiana tabacum L. induced by mechanical wounding is regulated by auxin. Journal of Experimental Botany 57, 2899–2907. [DOI] [PubMed] [Google Scholar]
- Shitan N, Hayashida M, Yazaki K. 2015. Translocation and accumulation of nicotine via distinct spatio-temporal regulation of nicotine transporters in Nicotiana tabacum . Plant Signaling & Behavior 10, e1035852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T, Hashimoto T. 2011. Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant & Cell Physiology 52, 1117–1130. [DOI] [PubMed] [Google Scholar]
- Shoji T, Hashimoto T. 2014. Stress-induced expression of NICOTINE2-locus genes and their homologs encoding Ethylene Response Factor transcription factors in tobacco. Phytochemistry 113, 41–49. [DOI] [PubMed] [Google Scholar]
- Shoji T, Kajikawa M, Hashimoto T. 2010. Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. The Plant Cell 22, 3390–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T, Nakajima K, Hashimoto T. 2000. a Ethylene suppresses jasmonate-induced gene expression in nicotine biosynthesis. Plant & Cell Physiology 41, 1072–1076. [DOI] [PubMed] [Google Scholar]
- Shoji T, Ogawa T, Hashimoto T. 2008. Jasmonate-induced nicotine formation in tobacco is mediated by tobacco COI1 and JAZ genes. Plant & Cell Physiology 49, 1003–1012. [DOI] [PubMed] [Google Scholar]
- Shoji T, Yamada Y, Hashimoto T. 2000. b Jasmonate induction of putrescine N-methyltransferase genes in the root of Nicotiana sylvestris . Plant & Cell Physiology 41, 831–839. [DOI] [PubMed] [Google Scholar]
- Sinclair SJ, Murphy KJ, Birch CD, Hamill JD. 2000. Molecular characterization of quinolinate phosphoribosyltransferase (QPRTase) in Nicotiana . Plant Molecular Biology 44, 603–617. [DOI] [PubMed] [Google Scholar]
- Steppuhn A, Schuman MC, Baldwin IT. 2008. Silencing jasmonate signalling and jasmonate-mediated defences reveals different survival strategies between two Nicotiana attenuata accessions. Molecular Ecology 17, 3717–3732. [DOI] [PubMed] [Google Scholar]
- Syed NH, Kalyna M, Marquez Y, Barta A, Brown JW. 2012. Alternative splicing in plants--coming of age. Trends in Plant Science 17, 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler L, Bragg JN, Wu J, Yang X, Tuskan GA, Vogel JP. 2010. Annotation and comparative analysis of the glycoside hydrolase genes in Brachypodium distachyon . BMC Genomics 11, 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valleau WD. 1949. Breeding low-nicotine tobacco. Journal of Agricultural Research 78, 171–181. [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CM. 2006. Significance of inducible defense-related proteins in infected plants. Annual Review of Phytopathology 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Winz RA, Baldwin IT. 2001. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. IV. Insect-induced ethylene reduces jasmonate-induced nicotine accumulation by regulating putrescine N-methyltransferase transcripts. Plant Physiology 125, 2189–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldemariam MG, Dinh ST, Oh Y, Gaquerel E, Baldwin IT, Galis I. 2013. NaMYC2 transcription factor regulates a subset of plant defense responses in Nicotiana attenuata . BMC Plant Biology 13, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Timko M. 2004. Methyl jasmonate induced expression of the tobacco putrescine N -methyltransferase genes requires both G-box and GCC-motif elements. Plant Molecular Biology 55, 743–761. [DOI] [PubMed] [Google Scholar]
- Yang S, Tang F, Zhu H. 2014. Alternative splicing in plant immunity. International Journal of Molecular Sciences 15, 10424–10445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarei A, Korbes AP, Younessi P, Montiel G, Champion A, Memelink J. 2011. Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Molecular Biology 75, 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang D, Chen J, Yang Y, Huang Z, Huang D, Wang XC, Huang R. 2004. Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum . Plant Molecular Biology 55, 825–834. [DOI] [PubMed] [Google Scholar]
- Zhang HB, Bokowiec MT, Rushton PJ, Han SC, Timko MP. 2012. Tobacco transcription factors NtMYC2a and NtMYC2b form nuclear complexes with the NtJAZ1 repressor and regulate multiple jasmonate-inducible steps in nicotine biosynthesis. Molecular Plant 5, 73–84. [DOI] [PubMed] [Google Scholar]
- Zhang XC, Gassmann W. 2007. Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiology 145, 1577–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.