ABSTRACT
The structures of RNA Polymerase (Pol) II pre-initiation complexes (PIC) have recently been determined at near-atomic resolution, elucidating unprecedented mechanistic details of promoter opening during transcription initiation. The key structural features of promoter opening are summarized here. Structural knowledge of Pol I and III PIC is also briefly discussed.
Keywords: general transcription factor, pre-initiation complex, promoter opening, RNA polymerase, transcription initiation
Introduction
Expression of genetic information encoded in the duplex DNA genome requires the synthesis of RNA molecules by a family of enzymes called RNA Polymerases (Pol). This essential process, namely transcription, is a key regulatory point for the control of gene expression and mis-regulation of transcription at various steps often leads to human diseases.1 To initiate transcription, Pol and transcription factors (TFs) first need to recognize and bind to promoter regions immediately upstream of the transcription start site (TSS) forming a pre-initiation complex (PIC), followed by the melting of promoter DNA and loading of the template strand into the active site of Pol. Subsequently, Pol, often times with the help of TFs, locates the TSS on the template strand within the transcription bubble and starts the catalysis—formation of the first phosphodiester bond. When the RNA molecule reaches a certain length, Pol will escape the promoter region and enter a processive elongation state, and eventually terminate at the 3′ end of transcribed genes, also in a controlled and coordinated manner.
Bacteria and archaea have only one RNA Pol, while eukaryotes have at least three classes of Pols.2 Pol I synthesizes the large (35S in yeast, 47S in mammals) ribosomal RNA (rRNA) precursor,3 Pol II transcribes all the messenger RNAs (mRNAs),4 and Pol III catalyzes the synthesis of small untranslated RNAs such as the transfer RNAs (tRNAs) and the 5S rRNA.5 The three RNA Pols share a conserved catalytic core, while additional polymerase-specific subunits and general transcription factors (GTFs) are required for regulated gene-specific transcription.6 This is especially true when it comes to promoter opening, as Pol II requires the ATP-dependent translocase activity of the GTF TFIIH for the formation of transcription bubbles.7-10 Recent structural investigations have allowed near-atomic resolution visualization of how transcription bubble is formed in Pol II PIC during transcription initiation.8,11-13 Here, we will summarize the key structural features during promoter opening in both human and yeast systems. We will also briefly review our current structural understanding of transcription initiation mechanisms for TFIIH-independent Pol I and III.
TFIIH-dependent Pol II transcription initiation
The basal level of Pol II transcription on protein-coding genes requires the concerted act of GTFs TFIIA, TFIIB, TFIIE, TFIIF, TFIIH, and TBP (TATA-box binding protein), all of which assemble into a PIC in a stepwise manner.4,14-16 The first step often involves the binding of TBP to the TATA element, and this interaction introduces a ∼90° bend in the promoter.17,18 TBP is required for the transcription of all three Pols in eukaryotes.6 Next, TFIIA and TFIIB are recruited, followed by the recruitment of Pol II/TFIIF. Incorporation of TFIIF into the complex stabilizes the downstream DNA along the cleft of Pol II.8 TFIIE and TFIIH are then recruited to complete the PIC assembly. Promoter melting is achieved by an ATP-dependent translocase activity within TFIIH that threads downstream DNA into the active site of Pol II.7-10 Recent cryo-electron microscopy (cryo-EM) structures of Pol II PICs at near-atomic resolution have elucidated unprecedented mechanistic details of promoter opening during transcription initiation in both human and yeast systems.11-13 Here, we summarize the key structural features from these studies (Fig. 1).
Figure 1.
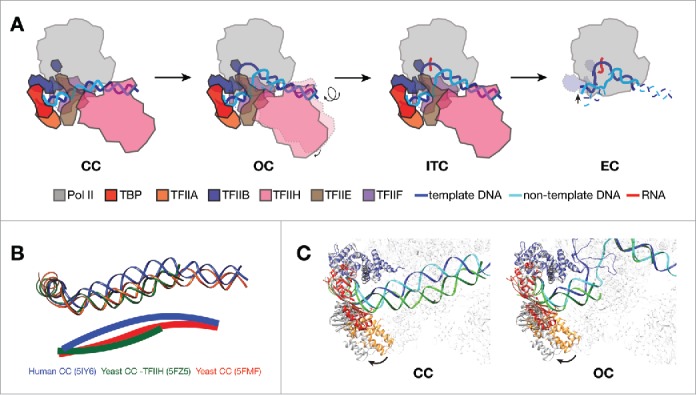
Structural features of Pol II promoter opening. (A) Schematic showing transitions from CC to EC in mammalian transcription system. PIC models are depicted as viewing down from Pol II stalk toward the active site. In CC, promoter DNA is positioned over the cleft of Pol II. The transition into OC involves the rotation and insertion of 12 bp of downstream DNA into the active center cleft by TFIIH. After loading of the template strand into the active center, Pol II PIC starts RNA synthesis and enters the ITC state. When RNA reaches a certain length, displacing the TFIIB (dotted outline in EC) zinc ribbon domain, Pol II clears promoter and turns into a processive elongation mode. (B) DNA paths in CC from three eukaryotic models,11-13 viewing from the same angle as in A. Pol II core subunits were aligned in the three CC structures for the purpose of comparison. DNA paths are also depicted as solid lines through the center of the duplex in the lower panel. Human promoter DNA is bent toward the active site, while yeast structures show clearly different bending properties of promoter DNA. (C) Comparing the positions of TFIIB-TBP-TFIIA between human (colored; PDB ID 5IY611 for CC and 5IY711 for OC) and yeast (gray; 5FZ512 for CC and 5FYW12 for OC) structures by aligning the Pol II core subunits. The TFIIB-TBP-TFIIA lobe rotates away from the rest of PIC in the yeast models. The color scheme for human structures are same as in A, except that the DNA molecule in yeast structures are shown in green (dark green for template strand, and light green for non-template strand).
In the human closed complex (CC) prior to promoter opening, promoter DNA spans over the cleft of Pol II, with its upstream sequence anchored by interactions with TFIIB-TBP-TFIIA, and its downstream sequence contacting Pol II subunit RPB5 and TFIIH (Fig. 1A). The engagement of TFIIH with promoter induces a kink at downstream DNA where it contacts RPB5, bending the DNA toward the Pol II cleft. This likely poses a torsional stress on the DNA that will subsequently be melted by the translocase activity of TFIIH subunit XPB (xeroderma pigmentosum type B).11 In the yeast CC, the promoter DNA does not bend toward the active site of Pol II, but instead, shows a slight outward kink12 (Fig. 1B). Inclusion of TFIIH from a separate study showed that the promoter DNA adopts a slight inward bending conformation over the Pol II cleft13 (Fig. 1B). However, the angle of DNA bending is less than that in the human CC model (Fig. 1B). In addition, the yeast Pol II clamp is in a closed state compared to an open conformation of the human counterpart, possibly due to the different paths of DNA. Interestingly, the extended winged helix (eWH) domain in yeast Tfa1 has a β hairpin structure named “E-wing” that is in close proximity to the promoter DNA at around position −7 (TSS is +1).12 By contrast, the orthologous element in the human CC does not contact DNA, possibly because of the inward bending of the DNA path.11 In comparison with the human structures, the TFIIB-TBP-TFIIA lobe in the yeast structures12,13 rotates around the TFIIB-Pol II interface as the pivot point away from the core PIC in both the CC and the open complex (OC) models (Fig. 1C). These differences might be due to specific mechanisms that different species adopted for transcription initiation, as yeast Pol II is known to scan through promoter regions to locate the TSS that can be as long as 120 base pairs (bp) away from the TATA box.19,20 The configuration of the yeast PIC might present more room over the Pol II cleft for both duplex- and single-stranded DNA during the TSS scanning process. In summary, the CC structures show that the promoter DNA is enclosed over the Pol II active center cleft by Pol II and GTFs, ready for melting by TFIIH.
To proceed with RNA synthesis after positioning promoter DNA over the cleft, Pol II and GTFs need to melt the promoter DNA and insert the template strand into the cleft, a process that is dependent on TFIIH.21 Comparison of the closed and open complexes reveals that 12 bp of DNA has to be inserted into the cleft of Pol II in order to form the OC (Fig. 1A). The structures also show that TFIIH undergoes a ∼10° rotation along an axis passing through the contact points between TFIIE and TFIIH, as well as between DNA and RPB5, to accommodate the shift in the downstream duplex DNA11 (Fig. 1A). In the human PIC structures, XPB maintains contact with the downstream duplex DNA throughout the transitions from CC to OC to initial transcribing complex (ITC), and this contact position is well preserved throughout the different states11 (Fig. 1A). TFIIH also remains flexibly engaged with the rest of the PIC through the interactions with TFIIE near the RPB4/7 stalk, likely enabling XPB to reach downstream DNA while maintaining the association with the core PIC.11 In a triplex displacement assay, Ssl2 (yeast homolog of human XPB) was shown to be able to track along one strand of DNA backbone with a 5′ to 3′ direction, displacing a triplex-forming oligonucleotide annealed to the complementary strand.10 Interestingly, the processivity of Ssl2 was estimated to be around one turn of duplex DNA,10 which agrees well with the insertion of 12 bp of DNA into the cleft from CC to OC. Taken together, in agreement with a DNA translocase model for TFIIH,7-10 Ssl2/XPB in the PIC binds downstream DNA and probably tracks along the non-template strand DNA upon ATP hydrolysis toward the same direction as RNA synthesis,10 while maintaining contact with the rest of PIC, therefore threading 12 bp of DNA into the cleft passing the RPB5 contact point and unwinding promoter DNA (Fig. 1A).
Upon transitioning into OC, the kink at the DNA-RPB5 contact site disappears as the promoter DNA melts, possibly releasing the tension on the DNA.11 Also observed during the CC-OC transition is the closure of the Pol II clamp. This results in the dissociation of clamp coiled-coil domain with TFIIEβ, and formation of a new interface between the coiled-coil with TFIIEα.11 In OC, density for the TFIIB linker region is clearly visible, stabilized directly between Pol II clamp coiled-coil and rudder on one side and the non-template DNA on the other, whereas its absence in the CC suggests that it is flexible when the promoter DNA is in a closed state.11 The transcription bubble is stabilized by a complex protein–DNA interaction network: the template strand is stabilized as previously suggested by TFIIB and the wall, rudder, and fork loops of Pol II,22,23 while the non-template strand is mainly stabilized by the TFIIB linker region and Pol II fork loops.11 A similar downward movement of TFIIE is observed in the yeast CC-OC transition, whereas the B-reader and B-linker regions remain invisible,12 distinct from the observation in the human complex11 (see above). Binding of the E-ribbon domain triggers a series of structural changes within the yeast PIC that include movement of TFIIB Zinc ribbon domain (B-ribbon) and Pol II fork loop 1 and lid, clearing a path for the entry of the template strand into the cleft.12 Therefore, comparisons of CC and OC have provided mechanistic insight into the process of promoter DNA melting in both human and yeast systems.
ITC structures were captured using a template containing a six-nucleotide RNA molecule mimicking the transcript.11,24 No significant difference was observed between OC and ITC, indicating that PIC stays in the same conformation prior to promoter escaping. To clear the promoter, Pol II has to break the contact with TFIIB. This presumably occurs when the RNA molecule extends to ∼12–13 bp25 that clashes with B-ribbon in the RNA exit channel. Comparison of the mammalian structures of ITC and elongation complex26 (EC) reveals that when entering elongation state, upstream DNA shifts ∼15Å to where the TFIIB N-terminal cyclin fold binds on Pol II in the ITC (Fig. 1A). The transcription bubble collapses from 19 bp in the ITC to 12 bp in the EC. Promoter clearance will trigger Pol II to break up the interactions with the initiation factors, and establish new contacts with elongation factors for a productive and finely regulated transcription elongation.
TFIIH-independent Pol I and III transcription initiation
Besides the conserved catalytic core, Pol I and III also contain subunits that show structural and functional homology to TFIIE and TFIIF, suggesting an evolutionarily conserved mechanism of transcription initiation.6 Meanwhile, unique mechanisms also exist, as Pol I and III do not depend on TFIIH for promoter opening, which is indispensable for Pol II transcription.6 However, TFIIH was shown to be an elongation factor for Pol I transcription,27 suggesting that it might be a general factor for all eukaryotic Pols. Recent structural studies utilizing hybrid techniques28-33 have provided much insight into the specialized transcription initiation of Pol I and III machineries.
Recruitment of Pol I to the rDNA promoter requires the TF Rrn3 (TIF-IA in mammals).3 Yeast Pol I in complex with Rrn3 was recently solved to sub-nanometer resolution using cryo-EM.31,32 Rrn3 associates with Pol I through contacts with stalk A43/A14 and subunits A190 and AC40, consistent with the positioning of Rrn3 on Pol I based on chemical crosslinking and mass spectrometry (CXMS) data.34 Association with Rrn3 stabilizes Pol I in its monomeric and active form, and results in a narrower active center cleft than that was observed in previous crystallography studies.29,30 Pol I PIC formation also requires a TFIIB-like factor, Rrn7 in the yeast core factor (CF) or TAF1B in the human selectivity factor 1 (SL1).35,36 CXMS analysis has revealed the molecular architecture of the yeast CF and its interaction with TBP.28 Based on the crosslinking data and PIC models for Pol II and III, the authors proposed a model for the minimal Pol I PIC containing Pol I, Rrn3, CF, and TBP.28 However, structures of Pol I PIC in its various functional states and/or in association with its regulatory factors have yet to be determined.
Pol III is more resistant to structural biology approaches than Pol I and II, as its structure has just recently been determined at near-atomic resolution using cryo-EM.33 Pol III transcription initiation requires different sets of general factors depending on the specific gene context. At type I and II promoters, binding of TFIIIC to the gene-internal promoter elements either by direct (type II promoters, e.g., tRNA genes) or TFIIIA-mediated (type I promoters, e.g., 5S rRNA gene) interactions recruits TFIIIB, which in turn recruits Pol III to initiate transcription.5 In Saccharomyces cerevisiae, the N-terminal domain of Brf1 (TFIIB-related factor 1) participates in a network of protein–protein interactions resembling that of TFIIB in Pol II PIC, whereas its cyclin repeats domain contacts the Pol III-specific subunit C34.37 The N-terminal region of yeast Bdp1 was positioned within the active center cleft of Pol III PIC, contacting Pol III subunits C128, C37, and Brf1.38 TFIIIC is composed of two subcomplexes that are connected by the interaction between Tfc4/τ131 and Tfc3/τ138. This interface overlaps with Bdp1 binding sites, suggesting a role in Pol III PIC assembly.39 SNAPc (small nuclear RNA-activating protein complex) is required for transcription by human Pol III at type III genes (e.g., U6 snRNA and 7SK RNA genes), by recruiting a different version of TFIIIB that contains BRF2.5 Structure of Pol III PIC will be essential for interpreting many past results, as well as providing new insight into mechanisms of eukaryotic transcription, and cryo-EM holds great promise for achieving this in the future.
Perspectives
Much of our understanding of transcription initiation by eukaryotic RNA Pol comes from studies of Pol II. Various structural biology means including, but not limited to, X-ray crystallography, cryo-EM, and CXMS, have provided molecular details of PIC assembly, promoter opening, transcription initiation, and elongation. Meanwhile, there are many questions that remain unanswered. For instance, as stated above, Pol I and III PIC structures are largely lacking. Future work will continue to elucidate the architectures of these large multi-protein assemblies that are responsible for the majority of the RNA synthesis within a living cell. Another unanswered question is how PIC is regulated and assembled by transcriptional co-activator complexes, such as Mediator and TFIID. A partial Pol II PIC containing only Pol II, TBP, TFIIB, and TFIIF was captured with a core Mediator,24 but the tail module of Mediator and TFIIE and TFIIH are missing in this structure. TFIID bound to promoter DNA has recently been visualized using cryo-EM and docked with the human CC to generate a model for TFIID-dependent PIC.40 Directly visualizing these more complete complexes will provide even richer details for the regulation of transcription. Last but not least, the current PIC structures were assembled on naked DNA templates, while genomic DNA wraps around histones forming a more compact chromatin structure in vivo. How Pols initiate and elongate transcription on chromatin template is another interesting topic for future investigations.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
Y. Han is a recipient of the Chicago Biomedical Consortium Postdoctoral Award. The authors would also like to thank Susan Fishbain for critical reading of the manuscript.
Funding
Y. He is supported by the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust and an Institutional Research Grant, IRG-15-173-21, from the American Cancer Society.
ORCID
References
- [1].Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell 2013; 152:1237-1251; PMID:23498934; http://dx.doi.org/ 10.1016/j.cell.2013.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Roeder RG, Rutter WJ. Multiple forms of DNA–dependent RNA polymerase in eukaryotic organisms. Nature 1969; 224:234-237; PMID:5344598; http://dx.doi.org/ 10.1038/224234a0 [DOI] [PubMed] [Google Scholar]
- [3].Schneider DA. RNA polymerase I activity is regulated at multiple steps in the transcription cycle: recent insights into factors that influence transcription elongation. Gene 2012; 493:176-184 PMID:21893173; http://dx.doi.org/ 10.1016/j.gene.2011.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Roeder R. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci 1996; 21:327-335; PMID:8870495; http://dx.doi.org/ 10.1016/S0968-0004(96)10050-5 [DOI] [PubMed] [Google Scholar]
- [5].Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev 2002; 16:2593-2620; PMID:12381659; http://dx.doi.org/ 10.1101/gad.1018902 [DOI] [PubMed] [Google Scholar]
- [6].Vannini A, Cramer P. Conservation between the RNA Polymerase I, II, and III transcription initiation machineries. Mole Cell 2012; 45:439-446; http://dx.doi.org/ 10.1016/j.molcel.2012.01.023 [DOI] [PubMed] [Google Scholar]
- [7].Grünberg S, Warfield L, Hahn S. Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP–dependent promoter opening. Nat Struct Mol Biol 2012; 19:788-796; PMID:22751016; http://dx.doi.org/ 10.1038/nsmb.2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].He Y, Fang J, Taatjes DJ, Nogales E. Structural visualization of key steps in human transcription initiation. Nature 2013; 495:481-486; PMID:23446344; http://dx.doi.org/ 10.1038/nature11991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kim TK, Ebright RH, Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science 2000; 288:1418-1422; PMID:10827951; http://dx.doi.org/ 10.1126/science.288.5470.1418 [DOI] [PubMed] [Google Scholar]
- [10].Fishburn J, Tomko E, Galburt E, Hahn S. Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. PNAS 2015; 112:3961-3966; PMID:25775526; http://dx.doi.org/ 10.1073/pnas.1417709112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].He Y, Yan C, Fang J, Inouye C, Tjian R, Ivanov I, Nogales E. Near-atomic resolution visualization of human transcription promoter opening. Nature 2016; 533:359-365; PMID:27193682; http://dx.doi.org/ 10.1038/nature17970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Plaschka C, Hantsche M, Dienemann C, Burzinski C, Plitzko J, Cramer P. Transcription initiation complex structures elucidate DNA opening. Nature 2016; 533:353-358; PMID:27193681; http://dx.doi.org/ 10.1038/nature17990 [DOI] [PubMed] [Google Scholar]
- [13].Murakami K, Tsai KL, Kalisman N, Bushnell DA, Asturias FJ, Kornberg RD. Structure of an RNA polymerase II preinitiation complex. PNAS 2015; 112:13543-13548; PMID:26483468; http://dx.doi.org/ 10.1073/pnas.1518255112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Goodrich JA, Cutler G, Tjian R. Contacts in Context: promoter specificity and macromolecular interactions in transcription. Cell 1996; 84:825-830; PMID:8601306; http://dx.doi.org/ 10.1016/S0092-8674(00)81061-2 [DOI] [PubMed] [Google Scholar]
- [15].Boeger H, Bushnell DA, Davis R, Griesenbeck J, Lorch Y, Strattan JS, Westover KD, Kornberg RD. Structural basis of eukaryotic gene transcription. FEBS Letters 2004; 579:899-903; http://dx.doi.org/ 10.1016/j.febslet.2004.11.027 [DOI] [PubMed] [Google Scholar]
- [16].Buratowski S, Hahn S, Guarente L, Sharp PA. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 1989; 56:549-561; PMID:2917366; http://dx.doi.org/ 10.1016/0092-8674(89)90578-3 [DOI] [PubMed] [Google Scholar]
- [17].Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature 1993; 365:520-527; PMID:8413605; http://dx.doi.org/ 10.1038/365520a0 [DOI] [PubMed] [Google Scholar]
- [18].Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature 1993; 365:512-520; PMID:8413604; http://dx.doi.org/ 10.1038/365512a0 [DOI] [PubMed] [Google Scholar]
- [19].Buratowski S, Sopta M, Greenblatt J, Sharp PA. RNA polymerase II-associated proteins are required for a DNA conformation change in the transcription initiation complex. Proc Natl Acad Sci USA 1991; 88:7509-7513; PMID:1881889; http://dx.doi.org/ 10.1073/pnas.88.17.7509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell 1987; 49:295-297; PMID:2882858; http://dx.doi.org/ 10.1016/0092-8674(87)90277-7 [DOI] [PubMed] [Google Scholar]
- [21].Goodrich JA, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell 1994; 77:145-156; PMID:8156590; http://dx.doi.org/ 10.1016/0092-8674(94)90242-9 [DOI] [PubMed] [Google Scholar]
- [22].Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science 2010; 327:206-209; PMID:19965383; http://dx.doi.org/ 10.1126/science.1182015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, Cramer P. RNA polymerase II–TFIIB structure and mechanism of transcription initiation. Nature 2009; 462:323-330; PMID:19820686; http://dx.doi.org/ 10.1038/nature08548 [DOI] [PubMed] [Google Scholar]
- [24].Plaschka C, Larivière L, Wenzeck L, Seizl M, Hemann M, Tegunov D, Petrotchenko EV, Borchers CH, Baumeister W, Herzog F et al.. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature 2015; 518:376-380; PMID:25652824; http://dx.doi.org/ 10.1038/nature14229 [DOI] [PubMed] [Google Scholar]
- [25].Čabart P, ∨jvári A, Pal M, Luse DS. Transcription factor TFIIF is not required for initiation by RNA polymerase II, but it is essential to stabilize transcription factor TFIIB in early elongation complexes. PNAS 2011; 108:15786-15791; PMID:21896726; http://dx.doi.org/ 10.1073/pnas.1104591108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bernecky C, Herzog F, Baumeister W, Plitzko JM, Cramer P. Structure of transcribing mammalian RNA polymerase II. Nature 2016; 529:551-554; PMID:26789250; http://dx.doi.org/ 10.1038/nature16482 [DOI] [PubMed] [Google Scholar]
- [27].Assfalg R, Lebedev A, Gonzalez OG, Schelling A, Koch S, Iben S. TFIIH is an elongation factor of RNA polymerase I. Nucleic Acids Res 2012; 40:650-659; PMID:21965540; http://dx.doi.org/ 10.1093/nar/gkr746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Knutson BA, Knutson BA, Luo J, Ranish J, Hahn S. Architecture of the Saccharomyces cerevisiae RNA polymerase I core factor complex. Nat Struct Mol Biol 2014; 21:810-816; PMID:25132180; http://dx.doi.org/ 10.1038/nsmb.2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fernández-Tornero C, Moreno-Morcillo M, Rashid UJ, Taylor NMI, Ruiz FM, Gruene T, Legrand P, Steuerwald U, Müller CW. Crystal structure of the 14-subunit RNA polymerase I. Nature 2013; 502:644-649; http://dx.doi.org/ 10.1038/nature12636 [DOI] [PubMed] [Google Scholar]
- [30].Engel C, Sainsbury S, Cheung AC, Kostrewa D, Cramer P. RNA polymerase I structure and transcription regulation. Nature 2013; 502:650-655; PMID:24153182; http://dx.doi.org/ 10.1038/nature12712 [DOI] [PubMed] [Google Scholar]
- [31].Pilsl M, Crucifix C, Papai G, Krupp F, Steinbauer R, Griesenbeck J, Milkereit P, Tschochner H, Schultz P. Structure of the initiation-competent RNA polymerase I and its implication for transcription. Nat Commun 2016; 7:12126; PMID:27418187; http://dx.doi.org/ 10.1038/ncomms12126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Engel C, Plitzko J, Cramer P. RNA polymerase I–Rrn3 complex at 4.8 Å resolution. Nat Commun 2016; 7:12129; PMID:27418309; http://dx.doi.org/ 10.1038/ncomms12129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hoffmann NA, Jakobi AJ, Moreno-Morcillo M, Glatt S, Kosinski J, Hagen WJH, Sachse C, Müller CW. Molecular structures of unbound and transcribing RNA polymerase III. Nature 2015; 528:231-236; PMID:26605533; http://dx.doi.org/ 10.1038/nature16143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Blattner C, Jennebach S, Herzog F, Mayer A, Cheung ACM, Witte G, Lorenzen K, Hopfner KP, Heck AJR, Aebersold R et al.. Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth. Genes Dev 2011; 25:2093-2105; PMID:21940764; http://dx.doi.org/ 10.1101/gad.17363311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Knutson BA, Hahn S. Yeast Rrn7 and human TAF1B Are TFIIB-related RNA polymerase I general transcription factors. Science 2011; 333:1637-1640; PMID:21921198; http://dx.doi.org/ 10.1126/science.1207699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Naidu S, Friedrich JK, Russell J, Zomerdijk JCBM. TAF1B is a TFIIB-like component of the basal transcription machinery for RNA polymerase I. Science 2011; 333:1640-1642; PMID:21921199; http://dx.doi.org/ 10.1126/science.1207656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Khoo SK, Wu CC, Lin YC, Lee JC, Chen HT. Mapping the protein interaction network for TFIIB-related factor Brf1 in the RNA polymerase III preinitiation complex. Mol Cell Biol 2014; 34:551-559; PMID:24277937; http://dx.doi.org/ 10.1128/MCB.00910-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hu HL, Wu CC, Lee JC, Chen HT. A Region of Bdp1 necessary for transcription initiation that is located within the RNA polymerase III active site cleft. Mol Cell Biol 2015; 35:2831-2840; PMID:26055328; http://dx.doi.org/ 10.1128/MCB.00263-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Male G, Appen von A, Glatt S, Taylor NMI, Cristovao M, Groetsch H, Beck M, Müller CW. Architecture of TFIIIC and its role in RNA polymerase III pre-initiation complex assembly. Nat Commun 2015; 6:7387; PMID:26060179; http://dx.doi.org/ 10.1038/ncomms8387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Louder RK, He Y, López-Blanco JR, Fang J, Chacón P, Nogales E. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 2016; 531:604-609; PMID:27007846; http://dx.doi.org/ 10.1038/nature17394 [DOI] [PMC free article] [PubMed] [Google Scholar]


