Abstract
Sanger sequencing platforms, such as applied biosystems instruments, generate chromatogram files. Generally, for 1 region of a sequence, we use both forward and reverse primers to sequence that area, in that way, we have 2 sequences that need to be aligned and a consensus generated before mutation detection studies. This work is cumbersome and takes time, especially if the gene is large with many exons. Hence, we devised a rapid automated command system to filter, build, and align consensus sequences and also optionally extract exonic regions, translate them in all frames, and perform an amino acid alignment starting from raw sequence data within a very short time. In full capabilities of Automated Mutation Analysis Pipeline (ASAP), it is able to read "*.ab1" chromatogram files through command line interface, convert it to the FASTQ format, trim the low-quality regions, reverse-complement the reverse sequence, create a consensus sequence, extract the exonic regions using a reference exonic sequence, translate the sequence in all frames, and align the nucleic acid and amino acid sequences to reference nucleic acid and amino acid sequences, respectively. All files are created and can be used for further analysis. ASAP is available as Python 3.x executable at https://github.com/aditya-88/ASAP. The version described in this paper is 0.28.
Keywords: alignment, automation, bioinformatics, chromatogram, pipeline, Python, Sanger sequencing, single nucelotide polymorphisms
INTRODUCTION
Automatic Sanger sequencing platforms, such as Applied Biosystems 3130/3500/3730 (Thermo Fisher Scientific, Waltham, MA, USA), generate chromatogram files in the “*.ab1” format. Generally, for each sequence region, we sequence it using both forward and reverse strand-specific primers. The generated files need to be opened in chromatogram viewer software, such as FinchTV (Geospiza, Seattle, WA, USA); then manually, the lower quality sequences are trimmed from both ends, and the remaining good-quality sequences are made into a new file. The reverse sequence is first reverse complemented and then copied to a new file using the same method as for the forward sequence. A consensus sequence is built either using alignment programs, such as ClustalW2,1 or manually finding an overlapping region and then aligning the consensus with the reference sequence. Commercial software, such as SeqMan (DNASTAR Lasergene Suite, DNASTAR, Madison, WI, USA), and free graphic user interface-based applications, such as Clustal X,1 are available for alignment, but still, they require manual intervention, which consumes time and requires skilled manpower. Furthermore, some commercial softwares are also available, but they require a license to be purchased, which generally is annually recurring. Apart from this, ASAP can also extract, translate and align the exonic region from the given sequence. This, when done manually, requires additional hands on time and expertise.
This entire process takes 5–30 min, depending on the experience level and desired work(s) of the user. In labs, such as ours and many others, where sequencing is done in bulk and on a routine basis, this has to be done repeatedly for each sequence. It becomes very cumbersome and takes time to perform this simple task. This gave birth to the idea of developing a simple program that can automate this otherwise simple process; hence, ASAP was developed. ASAP is a very simple code written in Python 3.5 language, which orchestrates different programs to perform all of the steps in an automated method, requiring just 1 primary input from the user and no subsequent intervention.
MATERIALS AND METHODS
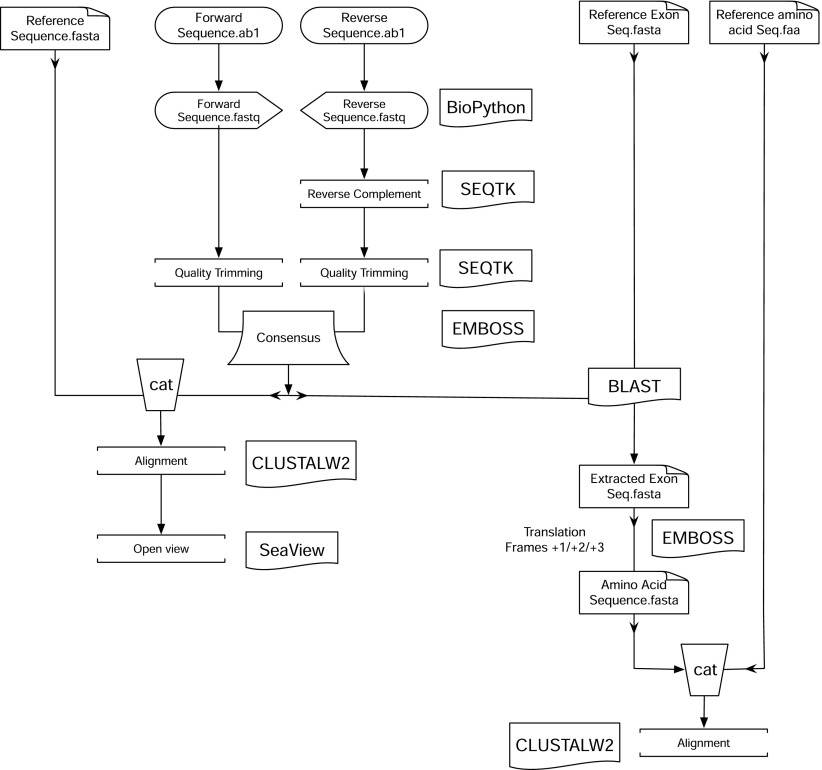
ASAP is a code written in Python language that uses Python components and automatically calls some external programs to achieve its function. In full potential of ASAP, it first converts the input chromatogram sequences (*.ab1) into FASTQ files using BioPython2 and then reverse-complements the reverse sequence(s) using Seqtk (https://github.com/lh3/seqtk), followed by quality trimming of the sequences based on Phred scores by Seqtk. The quality-trimmed files are then aligned, and a consensus sequence is made using the European Molecular Biology Open Software Suite (EMBOSS) merger tool.3 National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST)4 is used to find and extract the exonic region from the entire sequence, based on the given reference exonic sequence. EMBOSS is used to translate the exonic nucleotide sequence in +1/+2/+3 frames. This translated sequence is then aligned with reference amino acid sequence using ClustalW2. A multi-FASTA file is created by adding the previously created nucleotide consensus sequence to the given nucleotide reference sequence using the system’s in-built “cat” command. The sequences of this file are then aligned using ClustalW2, and if ASAP were run for only 1 pair of sequences, then the alignment is automatically opened in the SeaView program.5 The entire pipeline is denoted in Fig. 1 below. ASAP also saves the consensus sequence; alignment file; quality-trimmed forward sequence; reverse-complemented, quality-trimmed reverse sequence; and ClustalW2 alignment report. Optionally, we can provide only 1 sequence to ASAP instead of both forward and reverse, and also, ASAP has an option to skip amino acid alignment too.
FIGURE 1.
Flow chart of ASAP pipeline. External programs are in boxes on right. “cat” is a *nix command to concoct multiple files into 1.
ASAP accepts *.ab1 chromatogram files, 2 files per sequence, 1 forward and 1 reverse, or just any one of these sequences. The program can be multiplexed per reference sequence. For example, if there are 100 pairs of sequences to be aligned for 1 single gene region, then all of the sequencing chromatograms can be given to the program at once with 1 reference sequence. ASAP will produce 1 alignment file with a reference per pair of sequence.
Requirements
Requirements include the Linux/Unix System with EMBOSS, NCBI BLAST+, Seqtk, Python 3.x, BioPython, and ClustalW2 installed. ASAP comes as a Python executable file and requires no installation. One can also download the ASAP zip archive, which has a simplified installer for ASAP. Furthermore, a test data, including 18 pairs of chromatogram files, along with the reference sequences for testing the pipeline, can also be downloaded from the same GitHub page for testing purposes. The data are of mixed quality required for testing the pipeline. They can easily run on personal computers and require no purchasing or registration, and all used external programs are freely available. Simple use instructions can be found in Supplemental information S1. To ease the process of installing dependencies, we recommend that users, especially new users who are not acquainted with Linux, install Bio-Linux,6 which has the majority of the dependencies preinstalled. Furthermore, with the use of any flavor of Ubuntu Linux, one can add Bio-Linux repositories into them and simply install most of the dependencies using simple commands.7
RESULTS AND DISCUSSION
Time taken by ASAP was compared with time taken by a manual method. Ten sequence pairs with 1 reference sequence, 1 amino acid sequence, and 1 exon nucleotide sequence were selected, and the analysis was performed by both of the methods by colleagues. ASAP took 39.406 s (timed with Python’s cProfiler) to perform analysis of these 10 pairs of chromatogram sequences on a personal computer with a quad-core Intel Core i7 processor, 8GB RAM, and Mac OS X 10.11.4 installed. This, when done manually, takes 10–20 min per sequence pair, which when added for 10 sequences, ranges between 1 hr 40 min and 3 h 20 min. The program takes even less time (6–7 s) if the exon extraction part is skipped. The results of ASAP were validated with the results achieved through a manual method, and it was found to be accurate. Sanger sequencing sometimes generates data that aren’t clean, and as ASAP automates the otherwise manual scrutiny of the data, it is required to check how it behaves in case of the failed/impure sequencing run. It was found that when the chromatogram file contained bad reads or only “NNNN,” which means a bad or completely failed sequencing run, respectively, ASAP was able to identify and skip all of those sequences and log them into a text file, titled “Failed_Report.txt.” The user can then go back and check those sequences for possible causes of failure.
Conclusion
ASAP greatly reduces the time and expertise required for achieving the process of getting raw sequencing files to generating alignment with a control/desired sequence using a simple code written in Python language. The said program can run on any present day desktop/ laptop system, running Linux/Unix/Mac OS ×. The described pipeline and all required external programs are freely available to the public, which greatly increases its accessibility and further development.
Supplementary Material
ACKNOWLEDGMENTS
A.S. thanks his mentor, P.B., for encouragement and support and colleagues Dr. Meenu Singh, Mr. Rajendra Marathe, Ms. Madhulika Sharma, and Mr. Sandeep Negi from the same institute and fellow Mr. Ravinder Sirohi from the Institute of Microbial Technology (Chandigarh, India) for installing and testing the software. There are no conflicts of interest.
REFERENCES
- 1.Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007;23:2947–2948. [DOI] [PubMed] [Google Scholar]
- 2.Cock PJ, Antao T, Chang JT, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009;25:1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 2000;16:276–277. [DOI] [PubMed] [Google Scholar]
- 4.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990;215:403–410. [DOI] [PubMed] [Google Scholar]
- 5.Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 2010;27:221–224. [DOI] [PubMed] [Google Scholar]
- 6.EOS Environmental Omics. Bio-Linus overview. 2015. Available at: http://environmentalomics.org/bio-linux/. Accessed September 23, 2016.
- 7.Natural Environment Research Council Environmental Bioinformatics CentreNatural Environment Research Council. Install Bio-Linus software. 2016. Available at: http://nebc.nerc.ac.uk/nebc_website_frozen/nebc.nerc.ac.uk//tools/bio-linux-7/other-bl-docs/package-repository. Accessed September 23, 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.