Here, Eychenne et al. analyzed how Mediator acts in preinitiation complex (PIC) assembly using in vivo, in vitro, and in silico approaches, and their data suggest a key interaction between Mediator and TFIIB in this process. This study provides novel mechanistic insights into the coordinated function of Mediator and TFIIB in PIC assembly in different chromatin contexts.
Keywords: Mediator, RNA polymerase II transcription, TFIIB, preinitiation complex, Saccharomyces cerevisiae, promoter architecture
Abstract
Mediator is a large coregulator complex conserved from yeast to humans and involved in many human diseases, including cancers. Together with general transcription factors, it stimulates preinitiation complex (PIC) formation and activates RNA polymerase II (Pol II) transcription. In this study, we analyzed how Mediator acts in PIC assembly using in vivo, in vitro, and in silico approaches. We revealed an essential function of the Mediator middle module exerted through its Med10 subunit, implicating a key interaction between Mediator and TFIIB. We showed that this Mediator–TFIIB link has a global role on PIC assembly genome-wide. Moreover, the amplitude of Mediator's effect on PIC formation is gene-dependent and is related to the promoter architecture in terms of TATA elements, nucleosome occupancy, and dynamics. This study thus provides mechanistic insights into the coordinated function of Mediator and TFIIB in PIC assembly in different chromatin contexts.
In eukaryotes, RNA polymerase II (Pol II) transcription machinery is responsible for the synthesis of mRNAs and several classes of noncoding RNAs. This machinery is composed of a huge assembly of nuclear proteins. Regulation of Pol II transcription starts with the binding to gene regulatory elements of specific transcription factors (TFs), which then recruit coregulator complexes (coactivators and corepressors), and proceeds with the binding to promoter DNA of basal components, including Pol II and general TFs (GTFs). A coordinated action of all of these components in the context of promoter chromatin through a complex interaction network is required for regulated transcription of each gene.
Mediator of transcriptional regulation is one of the coregulator complexes serving as a bridge between DNA-binding TFs and the basal Pol II transcription machinery (Kornberg 2005). Together with the GTFs (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH), this large multiprotein complex conserved in all eukaryotes promotes the assembly of preinitiation complexes (PICs) and activates transcription (Flanagan et al. 1991; Kim et al. 1994; Ranish et al. 1999). Mediator is essential for life in yeast and is generally required for Pol II recruitment and transcription (Thompson and Young 1995; Holstege et al. 1998; Soutourina et al. 2011). It also stimulates the phosphorylation of the Pol II C-terminal domain (CTD) by TFIIH (Kim et al. 1994; Jiang et al. 1998). Mediator cooperates with GTFs to stabilize the PIC, in particular through interactions with TFIID, TFIIH, and Pol II (Johnson et al. 2002; Esnault et al. 2008; Soutourina et al. 2011; Eyboulet et al. 2015). A functional interplay between Mediator and TFIIB was also suggested based on in vitro experiments with human Mediator (Baek et al. 2006). In yeast, reconstituted subcomplexes were shown to interact with TFIIB in vitro (Kang et al. 2001). Artificial tethering of TFIIB to the promoter led to Mediator recruitment and PIC assembly in vivo (Lacombe et al. 2013). TFIIB, encoded by the SUA7 gene in yeast, is one of the GTFs important for Pol II recruitment that determines the accuracy of start site selection (Pinto et al. 1992; Liu et al. 2010; Sainsbury et al. 2013).
The Mediator complex is composed of 25 subunits in yeast and up to 30 subunits in mammals (Malik and Roeder 2010). It has a modular organization with head, middle, and tail modules that constitute the core Mediator and the Cdk8 kinase module (Kornberg 2005). In yeast, 10 Mediator subunits are essential for cell viability. The contribution to the function of the complex of most Mediator subunits remains to be elucidated. In addition, how a single complex can be involved in ensuring tight transcriptional regulation of hundreds or thousands of genes through different specific interactions with TFs is largely unknown. The importance of the complex is highlighted by the fact that many Mediator subunits are involved in human diseases like neurodevelopmental pathologies or cancers (Kaufmann et al. 2010; Hashimoto et al. 2011; Spaeth et al. 2011; Schiano et al. 2014).
The function of Mediator middle module subunits has been very poorly studied. Based on protein cross-linking, a model for the Mediator middle module was proposed (Larivière et al. 2013). A cryo-electron microscopic model of a part of the Mediator complex within a partial PIC was also reported recently (Plaschka et al. 2015). Med10 is one of the essential and most conserved Mediator subunits belonging to the middle module. Only one conditional mutant in the yeast Med10 subunit (also known as Nut2), med10-1ts (N2D L61S L64P), was previously isolated (Han et al. 1999). Med10 is engaged in a number of contacts within the complex and interacts with the Med14 subunit that is central in Mediator architecture. The spatial organization of the Mediator modules was revised recently (Tsai et al. 2014; Wang et al. 2014). Med14 links the middle and tail module and, according to a recent model of entire yeast Mediator, makes extensive contacts with subunits from all three core modules (Robinson et al. 2015). A reconstitution of the active human core Mediator complex revealed a critical role of the Med14 subunit in Mediator architecture and function (Cevher et al. 2014).
Regulation of Pol II transcription occurs in the context of promoter chromatin. Most of the Pol II transcribed genes are preceded by a 5′ nucleosome-free promoter region (NFR) on which the PICs assemble. The −1 nucleosome, located on the upstream side of the promoter NFR, can potentially control access to regulatory sequences. The +1 nucleosome is the first nucleosome encountered by the transcription machinery, and its function could differ depending on the nature of the TATA element (TATA box or TATA-like element) present on the promoter (Rhee and Pugh 2012). Promoters containing TATA-like elements are more dependent on the TFIID complex and may rely more on NFR-adjacent nucleosomes for PIC assembly. In addition, histone turnover was reported to be rapid on promoters (Dion et al. 2007), but the function of nucleosome dynamics in PIC assembly remains unknown.
In this study, we selected a conditional mutant in the Med10 Mediator middle subunit and analyzed the effect of med10 mutation on PIC assembly on a genomic scale in vivo and in vitro. We show that this med10 mutation does not have a major effect on Mediator stability but specifically modifies a contact of this subunit with Med14. We demonstrate that Mediator interaction with TFIIB was decreased in the med10 mutant and that this interaction occurs via Med14 and, to a lesser extent, Med10. ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) analysis showed that genome-wide occupancy of PIC components was differentially affected in the med10 mutant. The most pronounced decreases were observed for TFIIB and Pol II. TFIIA and TFIID occupancies were unmodified. Our genome-wide analysis also revealed that the impact of med10 mutation on PIC formation and transcription is gene-dependent and is correlated with the promoter architecture.
Results
The conditional med10 mutant in the Mediator middle module
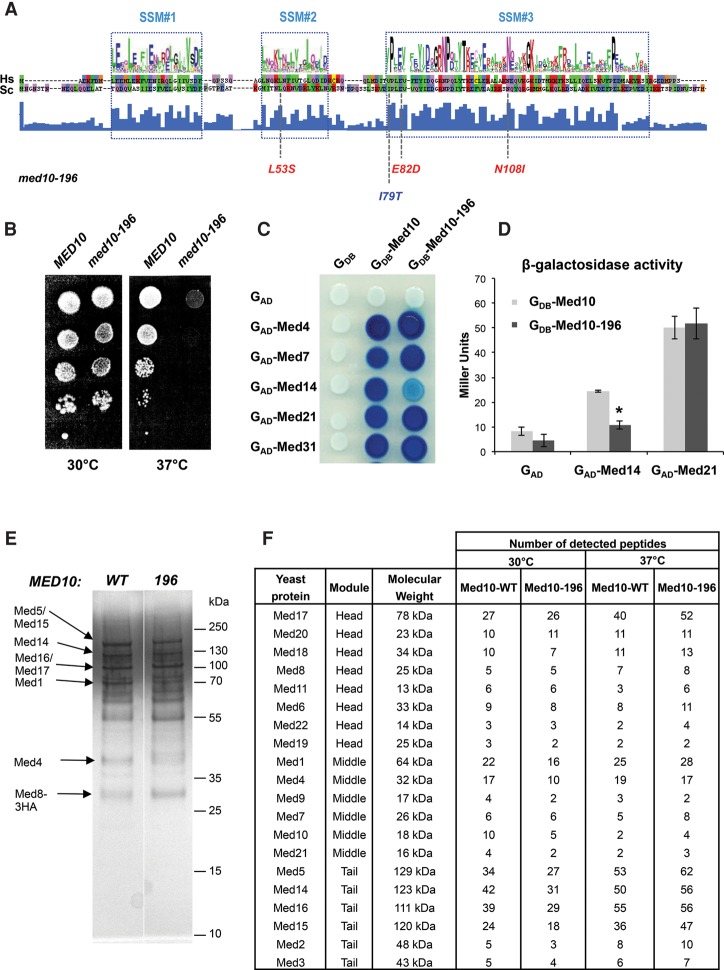
To enhance our understanding of the mechanisms in which Mediator is implicated and to clarify the role of the middle module, we selected a conditional mutant in the Med10 subunit, med10-196, by random mutagenesis using error-prone PCR followed by a screen for a temperature-sensitive growth phenotype as described previously for the Med11 and Med17 subunits (Supplemental Material; Esnault et al. 2008; Soutourina et al. 2011). The four residues mutated in med10-196 (L53S, I79T, E82D, and N108I) were localized within the conserved domains according to multiple sequence alignments and secondary structure features (Bourbon 2008) and corresponded to identical (for L53S, E82D, and N108I) or functionally close (I79T) amino acids in the human Med10 protein (Fig. 1A). med10-196 showed a strong temperature-sensitive phenotype and did not grow at 37°C (Fig. 1B). The individual mutations, when separated by site-directed mutagenesis, displayed normal growth phenotypes (data not shown). Hence, only the med10-196 mutant was used for further analysis.
Figure 1.
Phenotypes of the med10-196 mutant. (A) Location of the mutations in med10-196. The Med10 conserved domains 1, 2, and 3 (signature-specific motif [SSM]) from Saccharomyces cerevisiae (Sc) and Homo sapiens (Hs) with the consensus generated by WebLogo were adapted from Bourbon (2008). The mutated residues corresponding to identical (for L53S, E82D, and N108I) or functionally close (I79T) amino acids in the human Med10 protein are indicated in red and blue, respectively. (B) Thermosensitive growth phenotype of the med10 mutant. Cultures of wild-type and mutant med10 yeast strains were serially diluted, spotted on YPD agar plates, and incubated for 3 d at permissive (30°C) or nonpermissive (37°C) temperatures. (C) Two-hybrid interactions of the Med10-196 protein with its partners. Wild-type or mutant Med10 was fused to the Gal4 DNA-binding domain (GDB-Med10), and Med4, Med7, Med14, Med21, and Med31 were fused to the Gal4 activation domain (GAD-Med4, GAD-Med7, GAD-Med14, GAD-Med21, and GAD-Med31). (D) Quantitative analysis of two-hybrid interactions between Med10 and Med14. Wild-type or mutant Med10 was fused to the Gal4 DNA-binding domain (GDB-Med10), and Med14 or Med21 was fused to the Gal4 activation domain (GAD-Med14 and GAD-Med21). β-Galactosidase was assayed according to the Miller method, as described in the Supplemental Material. The mean values and standard deviation (indicated by error bars) of three independent experiments are shown. Asterisk represents a significant difference between the wild type and the mutant at P-value <0.002 in a Student's t-test. (E) Silver stain SDS-PAGE analysis of purified Mediator complex from wild-type and med10 mutant strains. Cells were grown at 30°C, and core Mediator complex was purified as described in the Supplemental Material. Mediator subunits with low molecular weight were not detectable by silver staining after SDS-PAGE. (F) Mass spectrometry analysis of Mediator integrity in the mutant grown at 30°C or transferred to 37°C. Core Mediator complex containing head, middle, and tail modules was purified from the med10-196 mutant and a wild-type strain. For each Mediator subunit identified by mass spectrometry analysis in wild-type and the med10 mutant, the number of identified peptides is indicated.
Med10 is engaged in multiple contacts within the Mediator complex that can be revealed using the yeast two-hybrid system (Guglielmi et al. 2004). We tested the ability of Med10-196 to interact with Med4, Med7, Med14, Med21, and Med31 (Fig. 1C). All tested interactions were similar between Med10-196 and the wild-type protein except for an interaction with the Med14 subunit that was significantly decreased in the mutant (Fig. 1C,D).
Since Mediator mutations could lead to dissociation of Mediator subunits, impairing Mediator function, we analyzed the effect of med10-196 on the integrity of the core Mediator. Mediator was purified from med10-196 and a wild-type strain (Fig. 1E) and was analyzed by mass spectrometry as described previously (Fig. 1F; Eyboulet et al. 2015). All 20 Mediator subunits detectable in a wild-type strain by this approach were present in the Mediator complex purified from the med10 mutant grown at 30°C (Fig. 1F, 30°C) or after being transferred to 37°C for 90 min (Fig. 1F, 37°C).
med10 mutation affects Mediator–TFIIB interaction
To explore the molecular mechanisms of PIC assembly involving the Med10 Mediator subunit, we analyzed potential interactions between Mediator and PIC components by coimmunoprecipitation (co-IP) experiments. We started by investigating potential Mediator–TFIIB contact with whole-yeast extracts. Mediator was immunoprecipitated via the HA-tagged Med5 Mediator subunit with an anti-HA antibody (Fig. 2A). Using TFIIB-specific antibodies, we showed that TFIIB coimmunoprecipitated with Mediator. We then examined whether this contact was modified in the med10 mutant. The results demonstrated that Mediator interaction with TFIIB was reduced in med10-196 transferred to 37°C compared with the wild type. In contrast, the Mediator interaction with TFIIA remained unchanged by med10 mutation compared with the wild-type strain (Supplemental Fig. S1).
Figure 2.
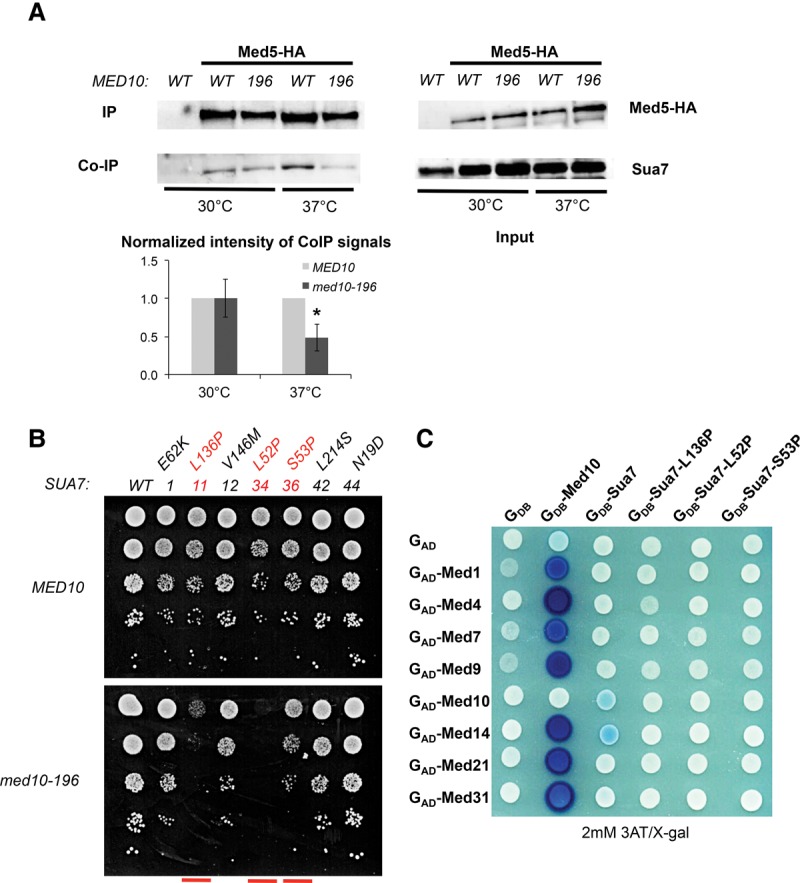
Functional interaction between Mediator and TFIIB. (A) Co-IP between Mediator and TFIIB in the med10-196 mutant compared with the wild-type strain. Wild-type and med10 mutant strains carrying a Med5-HA tag were grown to exponential phase at 30°C or transferred for 90 min to 37°C. Mediator was immunoprecipitated (IP) through Med5-HA from crude extracts (Input) of wild-type and mutant strains using magnetic beads coupled to anti-HA antibodies. MED10 strain carrying a nontagged Mediator subunit was used as a negative control. Coimmunoprecipitated TFIIB was detected by Western blotting using anti-TFIIB antibodies. The intensity of immune staining for coimmunoprecipitated TFIIB signals relative to the wild type was normalized against immunoprecipitation signals and is displayed in the bottom panel. The mean values and standard deviation (indicated by error bars) of three independent experiments are shown. The asterisk represents a significant difference between the wild type and the mutant at P-value <0.05 in a Student's t-test. (B) Specific sua7 mutants have synthetic phenotypes in combination with med10-196. The strain deleted for med10 and complemented by a TRP1 plasmid carrying MED10 or med10-196 and also deleted for sua7 and complemented by a URA3 plasmid carrying SUA7 was transformed by the HIS3 plasmids carrying either a wild-type or mutated version of SUA7. Transformants were serially diluted, spotted on 5-FOA-containing agar plates to counterselect the wild-type SUA7-bearing plasmid (see the Supplemental Material), and incubated for 3 d at 30°C. The sua7 mutants [sua7-11 (L136P), sua7-34 (L52P), and sua7-36 (S53P)] showing synthetic phenotypes with med10-196 are indicated in red. (C) A two-hybrid interaction between Mediator and Sua7 is decreased with Sua7 mutants. Wild-type or mutant Sua7 was fused to the Gal4 DNA-binding domain (GDB-Sua7), and Med1, Med4, Med7, Med9, Med10, Med14, Med21, and Med31 were fused to the Gal4 activation domain (GAD-Med1, GAD-Med4, GAD-Med7, GAD-Med9, GAD-Med10, GAD-Med14, GAD-Med21, and GAD-Med31). Med10 two-hybrid interactions with Mediator subunits are shown as positive controls.
To investigate the functional interplay between Mediator and TFIIB, we combined 16 specific sua7 (TFIIB) mutations (Wu et al. 1999) with med10-196 and showed that three of them had synthetic phenotypes (Fig. 2B; Supplemental Fig. S2A). sua7-34 (L52P) was colethal with the med10-196 mutation, and sua7-11 (L136P) and sua7-36 (S53P) med10-196 had a slow-growth phenotype at 30°C. On their own, these three synthetic TFIIB mutations do not lead to a start site shift and confer cold- and heat-sensitive phenotypes (Wu et al. 1999). The mutated residues in sua7-34 and sua7-36 localize to the N-terminal zinc ribbon domain (B ribbon), and L136P of sua7-11 is situated in the first part of the core domain. The allele-specific synthetic phenotypes between the med10-196 and sua7 mutations are consistent with the physiological importance of Mediator–TFIIB interaction.
To characterize Mediator middle–TFIIB interaction, Sua7 fused to the Gal4 DNA-binding domain was used in a two-hybrid assay with Mediator middle subunits (Med1, Med4, Med7, Med9, Med10, Med14, Med21, and Med31) fused to the Gal4 activation domain (Fig. 2C; Supplemental Fig. S2B,C). Interactions between Med10 and other Mediator subunits were used as positive controls. We showed that the Med14 Mediator subunit interacts with Sua7 in a two-hybrid system. The Med10 subunit also showed a two-hybrid interaction with TFIIB but with a lower intensity. In addition, these interactions were lost with Sua7-11, Sua7-34, and Sua7-36 mutants, indicating their specificity. Interestingly, the only contact within the Mediator middle module decreased in med10-196 was a two-hybrid interaction with the Med14 subunit (Fig. 1C,D), suggesting that the med10 mutation might alter Med10–Med14 contact, leading to reduced Mediator–TFIIB interaction.
We then investigated the functional interplay between the Mediator Med14 subunit and TFIIB by combining specific sua7 (TFIIB) mutations (Wu et al. 1999) with C-terminal truncations in MED14 that have temperature-sensitive phenotypes (med14-752, med14-686, and med14-483 with a stop codon at position 752, 686, or 483, respectively) (Supplemental Fig. S3A). It should be noted that further C-terminal truncation of MED14 to position 471 led to a lethal phenotype (data not shown). We observed allele-specific synthetic phenotypes between the med14 and sua7 mutations that are consistent with the physiological importance of Mediator–TFIIB interaction (Supplemental Fig. S3B).
Taken together, these results show that Mediator interacts with TFIIB via the Med14 and Med10 Mediator subunits and that this contact is important for a functional interplay between Mediator and the GTF.
med10 mutation differentially affects chromatin occupancy of PIC components
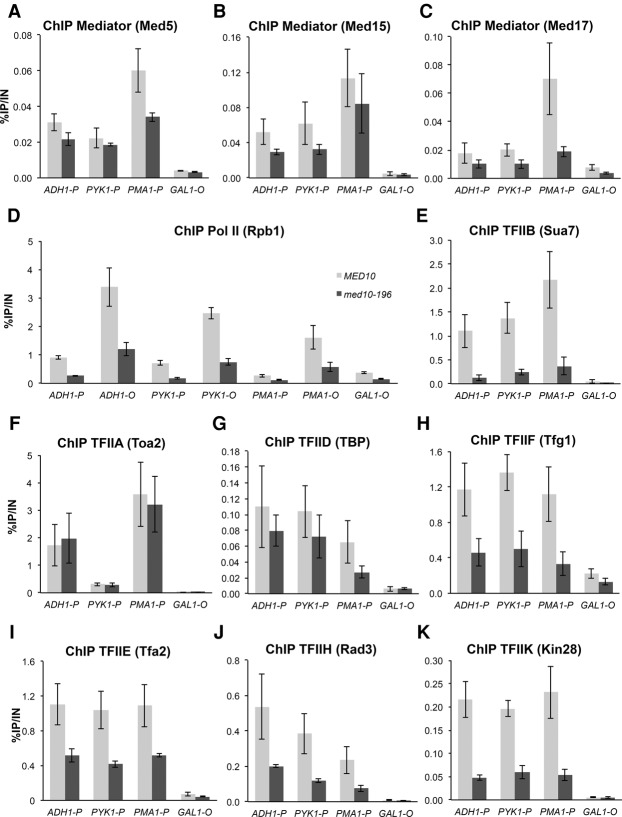
To investigate the molecular consequences of med10 mutation on PIC assembly in vivo, we determined the chromatin occupancy of Mediator, Pol II, and all GTFs on the constitutively expressed genes ADH1, PYK1, and PMA1. The association of Mediator was measured by ChIP experiments with three different Mediator subunits: Med5-HA (tail), Med15-HA (tail), and Med17-HA (head) (Fig. 3A–C). The Mediator occupancy of the promoter regions remained unchanged or decreased slightly in med10-196 except for the PMA1 promoter region showing a decrease in Med5 Mediator subunit occupancy and, to a greater extent, Med17 Mediator subunit occupancy. Pol II (Rpb1) association with the promoter and transcribed regions of class II genes was reduced compared with the wild type (Fig. 3D).
Figure 3.
ChIP analysis of PIC components in med10-196. Cells were grown to exponential phase at 30°C on YPD medium and then transferred for 90 min to 37°C. The immunoprecipitated protein is indicated together with the complex to which it belongs: Mediator tail (Med5) (A), Mediator tail (Med15) (B), Mediator head (Med17) (C), Pol II (Rpb1) (D), TFIIB (Sua7) (E), TFIIA (Toa2) (F), TFIID (TBP) (G), TFIIF (Tfg1) (H), TFIIE (Tfa2) (I), TFIIH core module (Rad3) (J), and TFIIH kinase module (Kin28) (K). All proteins, except Rpb1, were tagged with HA or TAP (see the Supplemental Material). Immunoprecipitated DNA was amplified with primers corresponding to ADH1, PMA1 and PYK1 ORF (O) or promoters (P). P1 primers listed in Supplemental Table S3 located close to upstream regulatory regions were used for Mediator ChIP experiments, and P2 primers located close to core promoters were used for Pol II and GTF ChIP experiments. GAL1 ORF was used as a negative control, since it is repressed in glucose-supplemented rich medium. The mean values and standard deviation (indicated by error bars) of three independent experiments are shown.
To assess the ability of med10 mutation to influence the occupancy of GTFs, ChIP was performed against TFIIA (Toa2 subunit), TFIIB (Sua7), TFIID (TBP), TFIIE (Tfa2 subunit), TFIIF (Tfg1 subunit), and the two TFIIH modules TFIIH core (Rad3 subunit) and TFIIK kinase module (Kin28 subunit) (Fig. 3E–K). Interestingly, the most pronounced effect of med10-196 was observed for TFIIB (Fig. 3E), which is consistent with the decreased Mediator–TFIIB interaction in this mutant. Promoter region occupancy of TFIIA and TBP remained largely unchanged (Fig. 3F,G). In contrast, med10 mutation led to a decrease in the occupancy of all other GTFs, including the TFIIF, TFIIE, and TFIIH modules, to an extent depending on the GTF (Fig. 3H–K). These ChIP results indicated that a mutant in the Mediator middle module has a pronounced effect on the recruitment and/or stability of specific PIC components in vivo.
med10-196 has a global effect on PIC assembly in vivo
To extend the PIC assembly analysis to the whole yeast genome, ChIP-seq experiments were performed for Mediator (Med15 and Med17 subunits), Pol II (Rpb1), and all GTFs (TFIIA [Toa2], TFIIB [Sua7], TFIID [TBP and Taf1], TFIIE [Tfa2], TFIIF [Tfg1], and two TFIIH modules: TFIIH core [Rad3] and TFIIK kinase module [Kin28]). Input DNA and DNA from ChIP with an untagged strain were used as negative controls. Supplemental Figure S4A shows an example of ChIP-seq density distributions for all proteins on a selected class II gene in wild-type strains after subtraction of the normalized control of an untagged strain as described previously (Eyboulet et al. 2013, 2015). As expected, a metagene analysis around the transcription start sites (TSSs) in wild-type strains showed that Pol II is distributed inside transcribed regions and that Mediator (Med15 and Med17 subunits) is located upstream of TSSs on regulatory regions (Supplemental Fig. S4B, top graph). The ChIP-seq distributions of GTFs overlapped with each other close to the TSS with a maximum of peak densities depending on the protein (Supplemental Fig. S4B, bottom graph). A similar analysis performed with the med10 mutant did not indicate any change in Mediator, GTFs, or Pol II peak positions (data not shown).
To investigate how general the effects of med10-196 on PIC assembly in vivo were, the genome-wide occupancy of Mediator, Pol II, and all GTFs was analyzed using ChIP-seq experiments in the mutant to compare them with the wild-type strains. To detect the potential global effects of Mediator mutations on PIC component occupancy, we performed the normalization step relative to quantitative PCR (qPCR) data on a set of selected regions as described previously (Eyboulet et al. 2015). Regression analysis of PIC component binding in med10-196 versus wild type was systematically performed.
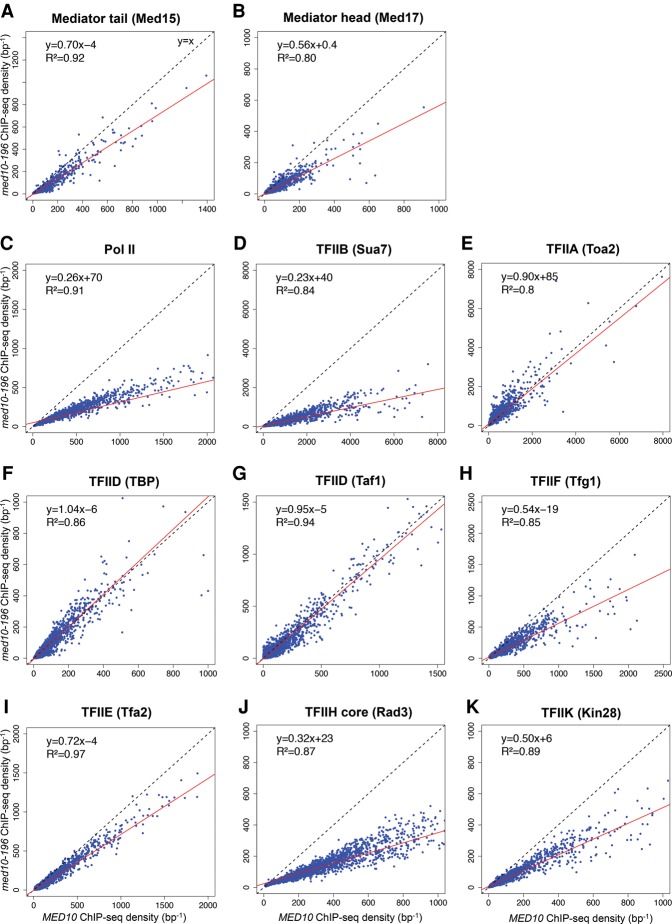
Genome-wide Mediator occupancy was analyzed for the Med15 tail subunit and the Med17 head subunit. As described above, ChIP results on selected class II gene promoters showed no effect or a slight decrease in Mediator occupancy in the med10-196 mutant, except for the PMA1 promoter region with a decrease in Med5 Mediator occupancy and, to a greater extent, Med17 Mediator occupancy. Genome-wide analysis of the med10-196 mutant revealed a global 1.4-fold decrease (the slope of regression line, shown in red in Fig. 4A, is equal to 0.70) in Med15 Mediator occupancy with a high correlation coefficient (R2 is equal to 0.92) (Fig. 4A). The effect of med10 mutation on Med17 Mediator occupancy was more pronounced, with a global 1.8-fold decrease suggesting some differences in the chromatin association of Mediator head and tail modules in med10-196 (Fig. 4B).
Figure 4.
Genome-wide effects of the med10-196 mutation on PIC component distribution. Cells were grown at 30°C in YPD medium and then transferred for 90 min to 37°C. The ChIP-seq densities of sequence tags in Mediator tail (Med15) (A), Mediator head (Med17) (B), TFIIB (Sua7) (D), TFIIA (Toa2) (E), TFIID (TBP) (F), TFIID (Taf1) (G), TFIIF (Tfg1) (H), TFIIE (Tfa2) (I), TFIIH core (Rad3) (J), and TFIIK (Kin28) (K) were calculated for promoter regions of Pol II transcribed genes. (C) The densities of sequence tags in the Pol II ChIP-seq experiments were calculated for the Pol II transcribed genes. Tag densities were normalized relative to qPCR data on a set of selected genes. Each point on the plot corresponds to one promoter region or one ORF. Promoter regions correspond to intergenic regions in tandem or in divergent orientation, excluding intergenic regions encompassing Pol III transcribed genes. In all, 2694 intergenic regions corresponding to 3303 Pol II-enriched genes were used for these analyses. A linear regression (red line) for ChIP-seq density in the mutant versus ChIP-seq density in wild type and an R2 correlation coefficient are indicated.
Similar to the ChIP results on selected gene promoters, Pol II and TFIIB occupancies were the most affected by the med10 mutation. A large global decrease was observed for genome-wide binding of Pol II (3.8-fold) and TFIIB (4.3-fold) in med10-196 (Fig. 4C,D). In contrast, genome-wide occupancy of two other PIC components, TFIIA and TFIID (TBP and Taf1 subunits), remained largely unchanged compared with the wild type (slopes are equal to 0.90, 1.04, and 0.95, and R2 is equal to 0.8, 0.86, and 0.94, respectively) (Fig. 4E–G). A global decrease in med10-196 was also observed for TFIIF, TFIIE, TFIIH core, and TFIIK occupancy to varying extents depending on the GTF (1.8-fold, 1.4-fold, 3.1-fold, and twofold decrease, respectively) (Fig. 4H–K). The genome-wide analysis of the med10 mutant showed a key role of the Mediator middle module in PIC assembly in vivo and demonstrated that med10 mutation can lead to a moderate effect on Mediator occupancy accompanied by a large decrease in Pol II and TFIIB occupancy and a decrease in several PIC components. TFIIA and TFIID behaved in a radically different manner without any change in med10-196 compared with the wild type. Our results also indicate an independent behavior for PIC components, including TFIIE, core and kinase TFIIH modules, or TFIIF and Pol II.
Gene-specific effects of med10-196 mutation on PIC assembly in vivo
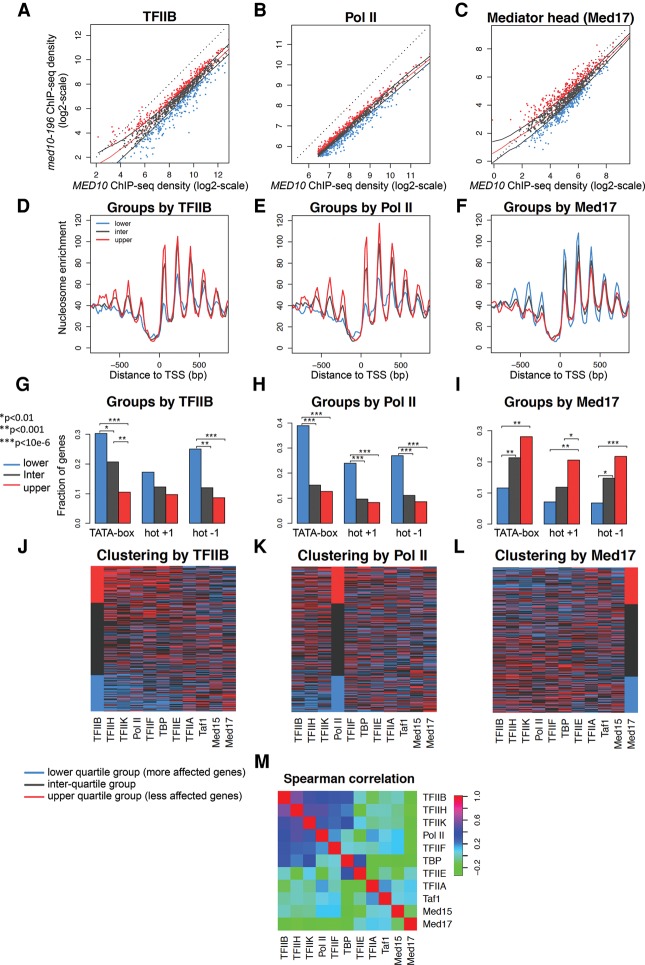
Our previous analysis of the impact of conditional mutations in the Med17 Mediator subunit on genome-wide PIC assembly showed a global nature of the effects but also suggested that some effects could still be gene-specific (Eyboulet et al. 2015). The high correlation coefficients obtained when comparing the genome-wide occupancy of the different PIC components between med10-196 and the wild type highlight the global effects of the mutation (Fig. 4). To address the question of the gene-specific effects, we analyzed the variations of the ratio across genes between the normalized ChIP-seq tag densities computed in the mutant and the wild type. For each measured GTF and Mediator subunit, tag densities in the mutant versus the wild type were plotted in log2 scale, such as a constant ratio translates into an alignment of the points along a line parallel to the diagonal (Y = X) whose intercept corresponds to the log2 ratio (Fig. 5A–C).
Figure 5.
Clustering analysis of genome-wide PIC occupancy ratios between med10-196 and the wild type. (A–C) ChIP-seq tag densities in the mutant versus the wild type were plotted as in Figure 4 but with log2 scales. To aggregate data at the gene level, divergent genes with double peaks for GTFs were excluded from the analysis as well as intergenic regions encompassing Pol III transcribed genes and centromeric regions. The genes with the lowest Pol II occupancy (lowest 25%) were also excluded from the analysis. For each PIC component, three groups of genes were defined according to the occupancy ratios between the med10-196 mutant and the wild type: the lowest 25% (lower quartile group in blue), the highest 25% (upper quartile group in red), and the ratios between 25% and 75% (interquartile group in gray). The red line corresponds to the median trend, and black lines correspond to the first and third quartiles of the data. Blue points correspond to the lowest 25% of mutant/wild-type values, and gray points correspond to values between 25% and 75% of the mutant/wild type. The red points correspond to the 25% of the genes that have the highest mutant/wild-type ratio. Tag densities were calculated as described in the legend for Figure 4 and were analyzed for the TFIIB (A,D,G,J), Pol II (B,E,H,K), and Mediator (Med17) groups (C,F,I,L). (D,E,F) The groups determined by TFIIB (A), Pol II (B), and the Mediator Med17 subunit (C) were analyzed for nucleosome occupancy in a 1600-base-pair (bp) window centered on the TSS. P-values determined by Wilcox test for the differences between the gene groups for the maximum values of nucleosome occupancy on the region between 0 and 100 bp relative to the TSS were as follows: For TFIIB groups, lower quartile versus interquartile was 3 × 10−6, interquartile versus upper quartile was <10 × 10−12, and lower quartile versus upper quartile was <10 × 10−12; for Pol II groups, lower quartile versus interquartile was 6 × 10−4, interquartile versus upper quartile was 6 × 10−3, and lower quartile versus upper quartile was 7 × 10−6; and for Med17 groups, interquartile versus upper quartile was 0.002, and lower quartile versus upper quartile was 6 × 10−5. (G–I) The groups determined by TFIIB (A), Pol II (B), and the Mediator Med17 subunit (C) were analyzed for the presence of the TATA box for dynamic (hot) nucleosomes −1 and +1. P-values determined by Fisher test are indicated by asterisks with a value key in the left panel. (J–L) The heat maps clustered by the TFIIB groups (J), Pol II groups (K), and Med17 groups (L) summarize the group distribution according to the occupancy ratios between the mutant and the wild type for each GTF, Mediator subunit, and Pol II. For each PIC component, lower quartile group genes are colored in blue, interquartile group genes are gray, and upper quartile group genes are red. The genes are ordered in the heat maps inside each group by interquartile range (IQR) score. (M) Pair-wise Spearman correlations between mutant and wild-type ratios for each PIC component were calculated. The correlated PIC components (>0.48) were TFIIB, TFIIH, TFIIK, and Pol II.
We first investigated the amplitude of the med10 mutation effects on the different PIC component occupancy. For each PIC component, we classified the genes into three groups based on the amplitude of the med10 mutation effects: the upper quartile group (ratios in the highest 25%) (Fig. 5, in red), the interquartile group (general trend) (Fig. 5, in gray), and the lower quartile group (ratios in the lowest 25%) (Fig. 5, in blue) as described in the Materials and Methods (Fig. 5; Supplemental Fig. S5). Since the general effect on occupancy was negative for most PIC elements, the lower quartile group corresponded to the most impacted genes. However, for TFIIA and TFIID (TBP and Taf1), the lower and upper quartiles distinguished genes that are impacted negatively and positively.
Next, to investigate the functional interplay between promoter chromatin architecture and PIC assembly, we compared the nucleosome occupancy profiles from Rhee and Pugh (2012) between the three groups of genes built to reflect the impact of med10-196 on each PIC component (Fig. 5D–F; Supplemental Fig. S5). These groups did not correlate with changes in nucleosome positioning. However, for GTFs such as TFIIB, TFIIH, TFIIK, and Pol II, the groups correlated with the occupancy for nucleosomes on TSS-surrounding regions; namely, the most affected genes (lower quartile groups) showed low nucleosome occupancy, whereas the least affected genes (upper quartile groups) displayed high nucleosome occupancy (Fig. 5D,E; Supplemental Fig. S5). An opposite trend was observed for changes in Mediator occupancy between the mutant and the wild type: The least affected genes (upper quartile groups) showed lower nucleosome occupancy than the most affected genes (lower quartile groups) (Fig. 5F).
Next, we determined how TATA-box-containing genes (Rhee and Pugh 2012) were distributed between the groups (Fig. 5G–I; Supplemental Fig. S5). The genes classified as the most affected by med10-196 for TFIIB and Pol II occupancy contained a TATA box significantly more often than those classified as the least affected (Fig. 5G,H). Again, the situation was opposite for the impact of the mutation on Mediator occupancy: The genes classified as the least affected contained a TATA box more often than the genes classified as the most affected (Fig. 5I). The dynamics of nucleosome −1 and +1 as defined by Dion et al. (2007) follow the TATA-box enrichment pattern (Fig. 5G–I; Supplemental Fig. S5). Both the most affected genes for TFIIB and Pol II occupancy and the least affected genes for Mediator occupancy exhibited promoter regions with more dynamic NFR-adjacent nucleosomes (hot nucleosome −1 and +1).
Patterns of nucleosome occupancy, TATA-box presence, and nucleosome dynamics suggest that some level of correlation exists between the impacts of med10-196 mutation on the different PIC components measured at the gene level. To describe these correlations more directly, the three groups of genes reflecting the impact of med10-196 on each PIC component were visualized in heat maps with genes ordered by TFIIB groups (Fig. 5J), Pol II groups (Fig. 5K), and Med17 groups (Fig. 5L). For each PIC component, the group of more affected genes is colored in blue, the general trend group genes are in gray, and the group of less affected genes is in red. These heat maps allow the observation of a positive correlation between the impact of the mutation on TFIIB, TFIIH, TFIIK, and Pol II occupancy and a negative correlation between the impact on these GTFs and on Mediator subunits. To quantify the observed relationships, we performed a Spearman correlation analysis (Fig. 5M). The impacts of med10-196 were the most intercorrelated for TFIIB, TFIIH, TFIIK, and Pol II. The impact on TFIIF also showed some correlation with this set of four PIC components. Although TBP occupancy was globally unchanged in the med10 mutant, slight gene-specific variations also tended to follow the variations observed for the four most correlated PIC elements but simultaneously showed some positive correlation with the impact on TFIIE. In contrast, the measured impact of med10 mutation on Mediator occupancy exhibited slight negative correlations with most other PIC components.
In conclusion, the impact of Mediator med10 mutation on PIC formation and transcription has amplitude, which is gene-dependent and correlated with the promoter architecture in terms of TATA elements, nucleosome occupancy, and dynamics.
med10 mutation affects PIC assembly in vitro
To investigate the role of the essential Med10 subunit of Mediator on PIC assembly in vitro, we performed biochemical experiments of PIC formation on immobilized DNA templates (Ranish et al. 1999). Nuclear extracts were prepared from the med10-196 mutant and the wild-type strain as described previously (Ranish et al. 1999). The amounts of PIC components, including Mediator (Med14 and Med17 subunits), Pol II (Rpb3 subunit), TFIIB (Sua7), and TFIIA (Toa1 subunit), were analyzed by Western blotting with med10-196 and wild-type nuclear extracts (Fig. 6, lanes 1,2, Input). Histone H3 was used as a loading control. All PIC components tested were unchanged in the med10 mutant compared with the wild-type extract. We then investigated the impact of med10-196 mutation on the PIC formation in vitro using an immobilized template system. The PICs were assembled from Med10 wild-type or Med10-196 nuclear extracts in the absence or presence of Gal4-Gcn4 activator and analyzed by Western blotting (Fig. 6, lanes 3–10, PIC). As expected, the activator stimulated the recruitment of the PIC components tested, including Mediator, Pol II, TFIIB, and TFIIA, using nuclear extract prepared from the wild-type strain (Fig. 6, lanes 3,4). Under the conditions of the assay, TFIIH bound to the immobilized template to a similar extent in the presence and absence of the activator (data not shown), which likely reflects the nonspecific DNA-binding activities of this factor, consistent with previous observations (Baek et al. 2006). This situation has precluded the analysis of the med10 mutation's effect on TFIIH recruitment in vitro. In agreement with our ChIP-seq results, the recruitment of Mediator, Pol II, and TFIIB was defective in the med10-196 nuclear extract (Fig. 6, lane 8) compared with the wild-type extract (Fig. 6, lane 4). Similar to the situation that we observed for TFIIA genome-wide binding, the level of this factor in PIC assembled in vitro was unchanged in the med10 mutant extract. Addition of increasing amounts of purified wild-type core Mediator stimulated the recruitment of each of the affected PIC components (Fig. 6, lanes 9,10). The results show that a functional Med10 Mediator subunit is required for the stable recruitment of Mediator, Pol II, and TFIIB, suggesting a role for this Mediator subunit in efficient PIC assembly and/or stability.
Figure 6.
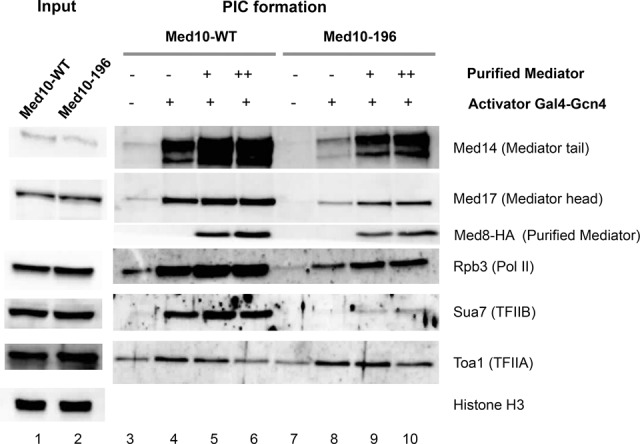
In vitro PIC formation in the med10-196 mutant. (Lanes 1,2) Nuclear extracts were prepared from the wild-type and med10-196 mutant strains and analyzed by Western blotting (Input). Histone H3 was used as a loading control. PIC assembly assay was performed by incubating the indicated nuclear extracts with the HIS4 immobilized template for 40 min in the absence (lanes 3,7) or presence (lanes 4–6,8–10) of Gal4-Gcn4 activator as described in the Supplemental Material. Increasing amounts of purified core Mediator were added as indicated (lanes 5,6 for Med10 wild-type extract; lanes 9,10 for Med10-196 extract). Western blotting for Med8-HA was used as a control for the purified Mediator addition.
Discussion
In this study, an integrative analysis of a conditional mutant in the Med10 Mediator middle module subunit was performed combining in vivo, in vitro, and in silico approaches, giving mechanistic insights into our understanding of Mediator function. We show that the Mediator middle module has a general role in PIC formation and that a functional interplay between Mediator and TFIIB is important for PIC component assembly and Pol II transcription. (1) We demonstrated an essential role of Mediator in TFIIB recruitment and/or stabilization during PIC formation in vivo and in vitro. We identified Mediator interactions with this GTF via Med14 and Med10 subunits using a two-hybrid approach. med10 mutation specifically modified a contact of this subunit with Med14 and led to a decreased Mediator–TFIIB interaction, demonstrating a functional link between Mediator and TFIIB in vivo. (2) Our results reveal a general function of the Med10 Mediator subunit and the Mediator middle module in PIC assembly and/or stability in vivo and in vitro. med10 mutation has a differential effect on PIC component occupancy, with a very large global decrease in Pol II and TFIIB. (3) Our genome-wide analysis revealed that the range of the effects of med10 mutation on PIC formation and transcription is gene-specific and is correlated with the promoter architecture, including the presence of TATA elements, the level of nucleosome occupancy, and nucleosome dynamics.
Functional link of Mediator with TFIIB
Previously, a functional cooperation between Mediator and TFIIB was suggested in yeast and humans (Kang et al. 2001; Baek et al. 2006; Lacombe et al. 2013). Our present work supports the existence of a strong functional link between Mediator and TFIIB through contacts between the middle module and the GTF. We identified Med14 and Med10 as the subunits that interact with TFIIB, as indicated by two-hybrid interactions and their loss in specific TFIIB mutants. Moreover, Mediator–TFIIB co-IP decreased in med10-196, and allele-specific synthetic phenotypes were identified between sua7 and med10-196 or med14 mutations. These observations are in line with the physiological importance of Mediator–TFIIB interaction. Interestingly, the sua7-36 (S53P) mutant that had a synthetic phenotype with med10-196 was shown previously to be defective for activation of specific genes (Wu and Hampsey 1999). The only contact within the Mediator middle decreased in med10-196 was a two-hybrid interaction with the Med14 subunit, but all Mediator core subunits, including Med14, were identified by mass spectrometry analysis of purified Med10-196 Mediator. We noted that TFIIB was the PIC component whose recruitment was the most affected by the med10 mutation in biochemical experiments with immobilized templates and in our genome-wide location analysis of PIC formation. Based on our results, we propose that the Med10 mutation specifically alters the Med10–Med14 contact, introducing conformational or functional changes in the Mediator middle module that lead to reduced Mediator interaction with TFIIB, destabilization of the GTFs within the PIC, and reduced Pol II recruitment and transcription.
Although our data show that Med14 and Med10 interact with TFIIB, we do not exclude the possibility that other Mediator subunits might be contacting this GTF. A recent cryo-electron microscopic model of a partial PIC, including a part of Mediator, suggests that Med18–Med20 subunits of the Mediator head bind the TFIIB B-ribbon domain (Plaschka et al. 2015). However, this model does not precisely define the positions of the Med10 and Med14 subunits, and the PIC complex is partial and stabilized by a cross-link with BS3, precluding any conclusion about the positioning of Med14–TFIIB and Med10–TFIIB contacts. In addition, an earlier biochemical study with reconstituted Mediator modules showed a pull-down between TFIIB and the Med9/10 subcomplex (Kang et al. 2001). It should also be noted that in vivo PIC assembly is a dynamic process involving different contacts acting at specific steps and that Mediator is particularly dynamic and flexible in this process.
Both Mediator and TFIIB are interacting with Pol II and are important for the enzyme recruitment. The PIC assembly culminates in Pol II recruitment, and therefore it is not surprising that many PIC components contact Pol II, the main player of transcription. We could not exclude that, in addition to Mediator–TFIIB contact, other interactions within the PIC could be involved in med10 mutation effects.
Global role of the Mediator middle module in PIC formation
This study demonstrates a general function of the Med10 Mediator subunit and Mediator middle module in PIC assembly and/or stability in vivo and in vitro. We showed that the behaviors of TFIIB, TFIIH, TFIIK, and Pol II were correlated on individual promoters when Med10 was mutated. TFIID and TFIIA were essentially unaffected, suggesting that their recruitment can be independent from that of the other GTFs or that these components are stabilized in the med10 mutant.
Based on in vitro reconstitution studies, a model for PIC assembly in a linear sequence was initially proposed with TFIID recruitment as a first component, followed by TFIIA and TFIIB arrival, Pol II recruitment in association with TFIIF, and completion of PIC assembly by TFIIH and TFIIE (Buratowski et al. 1989; Ranish and Hahn 1996). This model, which does not include Mediator, cannot explain our in vivo observations that suggest an independent behavior for PIC components that are supposed to be incorporated together such as TFIIE, core and kinase TFIIH modules, or TFIIF and Pol II. The Med10 mutation led to a large decrease in Pol II occupancy, but the effect on TFIIF, which is thought to be incorporated to the PIC together with Pol II, was more moderate. According to a linear model, TFIIE and TFIIH finalize the PIC assembly and depend on the incorporation of Pol II. However, the ranges of the med10 mutation effects differ considerably. Recent electron microscopy-based structural models of human and yeast PIC have provided important information on transcription initiation mechanisms (He et al. 2013, 2016; Murakami et al. 2013, 2015; Plaschka et al. 2015, 2016). However, they are currently incomplete (Plaschka et al. 2015), and the dynamics of PIC assembly are unavailable. Our functional in vivo study thus complements structural studies to investigate different aspects of PIC assembly on promoters.
Previously, our analysis of mutants in the Med11 and Med17 Mediator head subunits showed that Mediator stabilizes TFIIK kinase and TFIIH core modules independently and selectively contributes to TBP recruitment or stabilization on the chromatin and that a direct Mediator–Pol II interaction is generally required for Pol II recruitment and transcription (Esnault et al. 2008; Soutourina et al. 2011; Eyboulet et al. 2015). Based on our results, we proposed a model for PIC assembly through multiple pathways (Esnault et al. 2008) and suggested that Mediator independently orchestrates multiple steps of PIC assembly in vivo (Eyboulet et al. 2015). In the present work, we analyzed genome-wide occupancy of all PIC components and showed that mutations in the Med10 Mediator middle module that affect Mediator–TFIIB contact led to a pronounced decrease in TFIIB (coordinated with some but not all PIC components) and a global impact on Pol II recruitment and transcription, demonstrating a role of Mediator in PIC assembly on a genomic scale at the step of TFIIB binding.
Mediator impact on PIC assembly related to promoter architecture
Mediator serves as a bridge between specific TFs and the general Pol II transcriptional machinery, acting in gene-specific transcriptional regulation. Despite the global nature of the Mediator effects on PIC assembly, the range of these effects could be gene-specific, as suggested by previous Med17 mutant analysis (Eyboulet et al. 2015). Here, we directly addressed the question of a global versus a gene-specific nature of Mediator impact on PIC formation by performing in-depth analysis of our ChIP-seq data of all PIC components in the Med10 mutant compared with the wild type. We determined how the genes most or least affected for one PIC component are distributed for Mediator, GTFs, and Pol II. The range of the effects of Med10 mutation is not simply due to the experimental variability. The genes the most and the least affected by Mediator mutation were, in general, specific for each PIC component but show a common tendency (correlation) for several factors like TFIIB, TFIIH, TFIIK, and Pol II.
Since the PIC assembly occurs in a chromatin context, we examined the nucleosomal organization of the promoter regions in different groups for Med10 mutation impact. This analysis revealed a connection between the nucleosome occupancy around the TSS and the impact of Med10 Mediator mutation on TFIIB, TFIIH, TFIIK, and Pol II occupancy, with the most affected genes showing significantly lower nucleosome occupancy on TSS-surrounding regions. It should be noted that it was not the case for TFIIA, TFIID (Taf1), TFIIE, and TFIIF (Supplemental Fig. S5). Interestingly, the opposite tendency was observed for the Mediator subunits, since the most impacted genes tended to have higher nucleosome occupancy. Previously, in the wild-type context, a mode of PIC and nucleosome assembly on promoter regions was proposed to be TATA element-dependent with a competitive assembly for TATA-box-containing promoters and a cooperative assembly for promoters with TATA-like elements (Rhee and Pugh 2012). Our work demonstrates that the Med10 Mediator subunit mutation has an impact on PIC assembly in a TATA element-dependent and nucleosome-dependent manner. It should be noted that the recruitment or stability of TBP that recognizes the TATA elements remained globally unchanged in the med10 Mediator mutant as well as the Taf1 TFIID-specific subunit. Interestingly, the dynamics of NFR-adjacent nucleosomes −1 and +1 (Dion et al. 2007) follow the tendency similar to that of the TATA-box presence. For the mutant to wild-type occupancy ratios of Mediator subunits, the opposite situation occurs. A generally modest effect on Mediator subunit occupancy is accompanied by a higher nucleosome occupancy, depletion for TATA-box-containing promoters, and low dynamics of −1 and +1 nucleosomes for the most affected groups. It should be noted that Mediator is recruited by specific TFs on upstream regulatory elements that could have chromatin organization different from that of core promoter elements. Our results suggest that changes in Mediator occupancy that are the consequence of the Med10 Mediator mutation have a reverse relationship with promoter architecture compared with the core PIC components (TFIIB, TFIIH, TFIIK, and Pol II). We also suggest that the transcriptional effects of Med10 Mediator mutation are related to the function of Mediator in coordination with TFIIB and other GTFs, leading to coordinated changes in Pol II occupancy and transcription. We envision that Mediator occupancy does not completely reflect its in vivo function, suggesting the importance of evaluating the dynamics of Mediator binding in future studies.
Taken together, our results suggest a functional interplay between the PIC assembly mechanisms coordinated by Mediator and the promoter architecture. The TATA-box-containing genes are thought to be characterized by a greater plasticity and flexibility in their expression. Here we show that Mediator has a global impact on the PIC assembly and transcription and that the TATA-box-containing genes with lower nucleosome occupancy and dynamic −1 and +1 nucleosomes are the most influenced by Med10 Mediator mutation. From a mechanistic point of view, this work suggests how functional interplay between Mediator, TFIIB, other GTFs, and the promoter architecture leads to gene-specific transcription. It will help to understand the global to gene-specific rules governing transcription regulation. An integrative analysis of PIC assembly mechanisms using mutants in different Mediator subunits will likely permit future modeling of the PIC assembly pathways at each promoter and prediction of the Mediator-related regulation rules.
Materials and methods
Strains and plasmids
All Saccharomyces cerevisiae strains are described in Supplemental Table S1. All plasmids are listed in Supplemental Table S2. The oligonucleotides used in this study are in Supplemental Table S3.
ChIP and ChIP-seq
ChIP experiments were performed as described previously (Ghavi-Helm et al. 2008). Cell cultures (100 mL) were grown to exponential phase in YPD medium at 30°C, transferred for 90 min to a shaking incubator set at 37°C to allow gradual warming, and cross-linked with 1% formaldehyde for 10 min. The 3HA-tagged proteins were immunoprecipitated with 12CA5 antibody and Pol II with 8WG16 anti-CTD antibody (Covance) bound to IgG magnetic beads (Dynabead). TAP-tagged proteins were immunoprecipitated with IgG magnetic beads (Dynabeads). ChIP-seq experiments were performed as described previously (Eyboulet et al. 2013). Chromatin preparation for ChIP-seq experiments was performed as described previously for conventional ChIP, except that an additional sonication step with a Bioruptor (Diagenode) (six cycles of 40 sec with medium intensity setting) was included to generate DNA fragments of ∼200-base-pair (bp) mean size. DNA sequencing of 40-nucleotide tags was performed on a GA-IIx, Hi-Seq, or Next-Seq sequencer using the procedures recommended by the manufacturer (Illumina). Input DNA and DNA from ChIP with an untagged strain were used as negative controls. The ChIP-seq data have been deposited to Array Express under accession number E-MTAB-4607.
Data analysis
ChIP-seq data were analyzed as described previously (Eyboulet et al. 2013, 2015). The number of mappable tags for each ChIP-seq experiment is in Supplemental Table S4. To compare read counts in wild-type and med10 mutant ChIP-seq data, a count of reads was determined on promoter regions of Pol II transcribed genes for all GTFs and Mediator and on Pol II transcribed gene ORFs for Pol II. For ChIP-seq data analysis, promoter regions were defined as corresponding intergenic regions in tandem or in divergent orientation on the yeast genome. To consider only Pol II transcribed gene promoters, intergenic regions encompassing Pol III transcribed genes were excluded. Read numbers were normalized relative to qPCR data on a set of selected regions (Supplemental Table S5) as described previously (Eyboulet et al. 2015). The median of normalization coefficients between ChIP-qPCR and read ratios used for ChIP-seq data normalization is in Supplemental Table S6.
To aggregate data at the gene level (Fig. 5; Supplemental Fig. S5), tag density (per 1-bp bin) was averaged across whole intergenic regions, corresponding to the regions between tandem genes, except for Pol II, where the averaging was performed over the ORFs. Divergent genes with double peaks for GTFs were excluded from the analysis as well as centromeric regions. R software version 3.2.2 was used for computational analysis. In order to avoid potential biases that could arise from low occupancy values associated with low transcription levels, the genes showing the lowest Pol II occupancy (the first quartile) were excluded from the analyses shown in Figure 5 and Supplemental Figure S5,leading to 1065 remaining genes.
To investigate the amplitude of the med10 mutation effects on the binding of a particular GTF, Mediator subunit, or Pol II for each of our measurements, the genes were divided into three groups: The lower quartile group with the lowest 25% of mutant versus wild-type ratios comprises 267 genes, and the upper quartile group with the highest 25% comprises another 267 genes, while the interquartile group with the values between 25% and 75% comprises 531 genes. These groups for TFIIB, Pol II, and Mediator (Med17) are illustrated in Figure 5, A–C. As the trend of mutant versus wild-type values was not always perfectly linear and in order to correct for this nonlinearity, we developed a script that estimates locally (i.e., for a given x) the median and the interquartile range (IQR) to compute a score obtained as [y-median(x)]/IQR(x), where x and y denote the values for a particular gene in the wild type and the mutant, respectively. In practice, for each x, we applied the linear quantile regression implemented in the Quantreg package (version 5.19) to obtain the local median and IQR after weighting the points according to their distance to x with a normal kernel and a bandwidth equal to one-fifth of the total span of the data on the X-axis. The collections of the genes most influenced by the mutation and the least influenced by mutation were defined with this score.
The promoter architecture was determined for each of the three groups. The presence of the TATA boxes and nucleosome enrichment profiles in a 1600-bp window centered on the TSS were taken from Rhee and Pugh (2012). The dynamics of the nucleosomes (Dion et al. 2007) were determined for +1 and −1 nucleosomes, and only the significantly dynamic (hot) nucleosomes were included in the analysis. To determine the significance of the results for the genes containing the TATA box and hot −1 and hot +1 nucleosomes, we performed a Fisher test, while, for the difference between nucleosome enrichment, the P-values were obtained by a Wilcox test applied to the maximum value obtained in the 100 bp downstream from the TSS for each gene. Both Fisher test and Wilcox test are from the R base package stats.
Spearman correlations between two sets of mutant versus wild-type ratio values on the genes in the GTFs, Mediator subunits, or Pol II were computed and visualized using the heat maps produced with heatmap3 package version 1.1.1.
Supplementary Material
Acknowledgments
We thank M. Hampsey for sua7 mutant plasmids; S. Hahn for immobilized template plasmids; F.J. Asturias for the CA001 strain; C. Cibot for Med14 truncation mutants; M.-C. Nevers and H. Volland for anti-Med14 and anti-Med17 antibodies; C. Thermes, E. Van Dijk, and Y. Jaszczyszyn for performing the high-throughput sequencing of ChIP samples; the Service de Pharmacologie et Immunoanalyse (Commissariat à l’Énergie Atomique/Saclay) for monoclonal antibodies; and A. Goldar, C. Gazin, and V. Fromion for fruitful discussions. This work has benefited from the facilities and expertise of the high-throughput sequencing and the Service d'Identification et de Caractérisation des Protéines par Spectrométrie de Masse platforms of the Institute for Integrative Biology of the Cell, Centre National de la Recherche Scientifique. This work was supported by the Agence Nationale de la Recherche (ANR-08-BLAN-0229, ANR 11 BSV8 020 01, and ANR-14-CE10-0012-01) and the Fondation ARC (SL 2201130607079). T.E. was supported by the Fondation pour la Recherche Médicale (FDT20150531985).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.285775.116.
References
- Baek HJ, Kang YK, Roeder RG. 2006. Human Mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J Biol Chem 281: 15172–15181. [DOI] [PubMed] [Google Scholar]
- Bourbon HM. 2008. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res 36: 3993–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S, Hahn S, Guarente L, Sharp PA. 1989. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56: 549–561. [DOI] [PubMed] [Google Scholar]
- Cevher MA, Shi Y, Li D, Chait BT, Malik S, Roeder RG. 2014. Reconstitution of active human core Mediator complex reveals a critical role of the MED14 subunit. Nat Struct Mol Biol 21: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315: 1405–1408. [DOI] [PubMed] [Google Scholar]
- Esnault C, Ghavi-Helm Y, Brun S, Soutourina J, Van Berkum N, Boschiero C, Holstege F, Werner M. 2008. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol Cell 31: 337–346. [DOI] [PubMed] [Google Scholar]
- Eyboulet F, Cibot C, Eychenne T, Neil H, Alibert O, Werner M, Soutourina J. 2013. Mediator links transcription and DNA repair by facilitating Rad2/XPG recruitment. Genes Dev 27: 2549–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyboulet F, Wydau-Dematteis S, Eychenne T, Alibert O, Neil H, Boschiero C, Nevers MC, Volland H, Cornu D, Redeker V, et al. 2015. Mediator independently orchestrates multiple steps of preinitiation complex assembly in vivo. Nucleic Acids Res 43: 9214–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan PM, Kelleher RJ III, Sayre MH, Tschochner H, Kornberg RD. 1991. A mediator required for activation of RNA polymerase II transcription in vitro. Nature 350: 436–438. [DOI] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Michaut M, Acker J, Aude JC, Thuriaux P, Werner M, Soutourina J. 2008. Genome-wide location analysis reveals a role of TFIIS in RNA polymerase III transcription. Genes Dev 22: 1934–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi B, van Berkum NL, Klapholz B, Bijma T, Boube M, Boschiero C, Bourbon HM, Holstege FC, Werner M. 2004. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res 32: 5379–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SJ, Lee YC, Gim BS, Ryu GH, Park SJ, Lane WS, Kim YJ. 1999. Activator-specific requirement of yeast mediator proteins for RNA polymerase II transcriptional activation. Mol Cell Biol 19: 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto S, Boissel S, Zarhrate M, Rio M, Munnich A, Egly JM, Colleaux L. 2011. MED23 mutation links intellectual disability to dysregulation of immediate early gene expression. Science 333: 1161–1163. [DOI] [PubMed] [Google Scholar]
- He Y, Fang J, Taatjes DJ, Nogales E. 2013. Structural visualization of key steps in human transcription initiation. Nature 495: 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Yan C, Fang J, Inouye C, Tjian R, Ivanov I, Nogales E. 2016. Near-atomic resolution visualization of human transcription promoter opening. Nature 533: 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728. [DOI] [PubMed] [Google Scholar]
- Jiang YW, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway JW, Conaway RC, Kornberg RD. 1998. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc Natl Acad Sci 95: 8538–8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KM, Wang J, Smallwood A, Arayata C, Carey M. 2002. TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev 16: 1852–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JS, Kim SH, Hwang MS, Han SJ, Lee YC, Kim YJ. 2001. The structural and functional organization of the yeast mediator complex. J Biol Chem 276: 42003–42010. [DOI] [PubMed] [Google Scholar]
- Kaufmann R, Straussberg R, Mandel H, Fattal-Valevski A, Ben-Zeev B, Naamati A, Shaag A, Zenvirt S, Konen O, Mimouni-Bloch A, et al. 2010. Infantile cerebral and cerebellar atrophy is associated with a mutation in the MED17 subunit of the transcription preinitiation mediator complex. Am J Hum Genet 87: 667–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. 1994. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77: 599–608. [DOI] [PubMed] [Google Scholar]
- Kornberg RD. 2005. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci 30: 235–239. [DOI] [PubMed] [Google Scholar]
- Lacombe T, Poh SL, Barbey R, Kuras L. 2013. Mediator is an intrinsic component of the basal RNA polymerase II machinery in vivo. Nucleic Acids Res 41: 9651–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larivière L, Plaschka C, Seizl M, Petrotchenko EV, Wenzeck L, Borchers CH, Cramer P. 2013. Model of the Mediator middle module based on protein cross-linking. Nucleic Acids Res 41: 9266–9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. 2010. Structure of an RNA polymerase II–TFIIB complex and the transcription initiation mechanism. Science 327: 206–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. 2010. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 11: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K, Elmlund H, Kalisman N, Bushnell DA, Adams CM, Azubel M, Elmlund D, Levi-Kalisman Y, Liu X, Gibbons BJ, et al. 2013. Architecture of an RNA polymerase II transcription pre-initiation complex. Science 342: 1238724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K, Tsai KL, Kalisman N, Bushnell DA, Asturias FJ, Kornberg RD. 2015. Structure of an RNA polymerase II preinitiation complex. Proc Natl Acad Sci 112: 13543–13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto I, Ware DE, Hampsey M. 1992. The yeast SUA7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site selection in vivo. Cell 68: 977–988. [DOI] [PubMed] [Google Scholar]
- Plaschka C, Lariviere L, Wenzeck L, Seizl M, Hemann M, Tegunov D, Petrotchenko EV, Borchers CH, Baumeister W, Herzog F, et al. 2015. Architecture of the RNA polymerase II–Mediator core initiation complex. Nature 518: 376–380. [DOI] [PubMed] [Google Scholar]
- Plaschka C, Hantsche M, Dienemann C, Burzinski C, Plitzko J, Cramer P. 2016. Transcription initiation complex structures elucidate DNA opening. Nature 533: 353–358. [DOI] [PubMed] [Google Scholar]
- Ranish JA, Hahn S. 1996. Transcription: basal factors and activation. Curr Opin Genet Dev 6: 151–158. [DOI] [PubMed] [Google Scholar]
- Ranish JA, Yudkovsky N, Hahn S. 1999. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev 13: 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. 2012. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483: 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ, Trnka MJ, Pellarin R, Greenberg CH, Bushnell DA, Davis R, Burlingame AL, Sali A, Kornberg RD. 2015. Molecular architecture of the yeast Mediator complex. Elife 4: e08719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury S, Niesser J, Cramer P. 2013. Structure and function of the initially transcribing RNA polymerase II–TFIIB complex. Nature 493: 437–440. [DOI] [PubMed] [Google Scholar]
- Schiano C, Casamassimi A, Rienzo M, de Nigris F, Sommese L, Napoli C. 2014. Involvement of Mediator complex in malignancy. Biochim Biophys Acta 1845: 66–83. [DOI] [PubMed] [Google Scholar]
- Soutourina J, Wydau S, Ambroise Y, Boschiero C, Werner M. 2011. Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science 331: 1451–1454. [DOI] [PubMed] [Google Scholar]
- Spaeth JM, Kim NH, Boyer TG. 2011. Mediator and human disease. Semin Cell Dev Biol 22: 776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CM, Young RA. 1995. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci 92: 4587–4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KL, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Asturias FJ. 2014. Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 157: 1430–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sun Q, Ding Z, Ji J, Wang J, Kong X, Yang J, Cai G. 2014. Redefining the modular organization of the core Mediator complex. Cell Res 24: 796–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WH, Hampsey M. 1999. An activation-specific role for transcription factor TFIIB in vivo. Proc Natl Acad Sci 96: 2764–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WH, Pinto I, Chen BS, Hampsey M. 1999. Mutational analysis of yeast TFIIB. A functional relationship between Ssu72 and Sub1/Tsp1 defined by allele-specific interactions with TFIIB. Genetics 153: 643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.