Abstract
Eukaryotic ribosomes, the protein-producing factories of the cell, are composed of four ribosomal RNA molecules and roughly 80 proteins. Their biogenesis is a complex process that involves more than 200 biogenesis factors that facilitate the production, modification, and assembly of ribosomal components and the structural transitions along the maturation pathways of the pre-ribosomal particles. Here, I review recent structural and mechanistic insights into the biogenesis of the large ribosomal subunit that were furthered by cryo-electron microscopy of natively purified pre-60S particles and in vitro reconstituted ribosome assembly factor complexes. Combined with biochemical, genetic, and previous structural data, these structures have provided detailed insights into the assembly and maturation of the central protuberance of the 60S subunit, the network of biogenesis factors near the ribosomal tunnel exit, and the functional activation of the large ribosomal subunit during cytoplasmic maturation.
Keywords: ribosome assembly, structural biology, cytoplasmic maturation, polypeptide exit tunnel, 5S rRNA, proofreading
INTRODUCTION
Ribosomes are giant ribonucleoprotein complexes that synthesize proteins in all living cells. The eukaryotic 80S ribosome is composed of four ribosomal RNA (rRNA) molecules and roughly 80 proteins that form the 40S small and 60S large ribosomal subunits (Klinge et al. 2012; Melnikov et al. 2012). The small subunit binds and decodes the messenger RNA (mRNA) by selection of cognate aminoacyl-transfer RNAs (tRNAs) in its decoding center, whereas the large subunit catalyzes the chemistry of peptide bond formation in its peptidyl transferase center (PTC) active site (Schmeing and Ramakrishnan 2009). The large subunit harbors a number of additional important functional centers (Fig. 1), including (i) the polypeptide exit tunnel (Nissen et al. 2000; Voss et al. 2006), which provides the exit path for the newly synthesized protein; (ii) a binding platform for nascent chain targeting, processing, and folding factors at the exit of the tunnel (Kramer et al. 2009); and (iii) the GTPase-activating center between the P-stalk and the sarcin-ricin loop (SRL), where translational GTPases are recruited and activated (Diaconu et al. 2005; Gao et al. 2009; Voorhees et al. 2010). The L1 stalk and the 5S rRNA-containing central protuberance (CP) are additional landmark features of the 60S subunit (Fig. 1).
FIGURE 1.
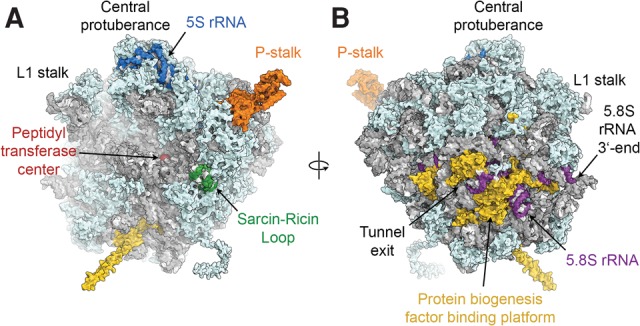
Structure of the yeast 60S ribosomal subunit. (A) View of the 60S subunit from the interface side. (B) View of the 60S subunit from the solvent side. Structural and functional hallmarks of the 60S subunit discussed in the text are colored and labeled. Visualization based on PDB 4V88 and PDB 3J7R (L1 stalk).
While the high-resolution structures of the 40S and 60S subunits as well as the 80S ribosome of both lower and higher eukaryotes have provided detailed insight into the molecular architecture of eukaryotic ribosomes (Ben-Shem et al. 2011; Klinge et al. 2011; Rabl et al. 2011; Voorhees et al. 2014; Khatter et al. 2015), much remains to be learned about how these particles are made. Functional ribosomal subunits are produced in an intricate process termed ribosome assembly or ribosome biogenesis, which is under tight transcriptional control (Warner 1999). Failure of ribosome biogenesis can lead to diseases known as ribosomopathies, while up-regulated ribosome assembly is found in many cancers (Teng et al. 2013). Ribosome assembly has been investigated extensively in the yeast Saccharomyces cerevisiae as well as in the human system. Many aspects of human ribosome biogenesis are conserved in yeast, which has therefore served as a model system for much biochemical, genetic, proteomics, and structural work on this pathway, including the study of human diseases associated with defective ribosome assembly (Woolford and Baserga 2013). The remainder of this section will therefore focus on yeast ribosome biogenesis (Fig. 2).
FIGURE 2.
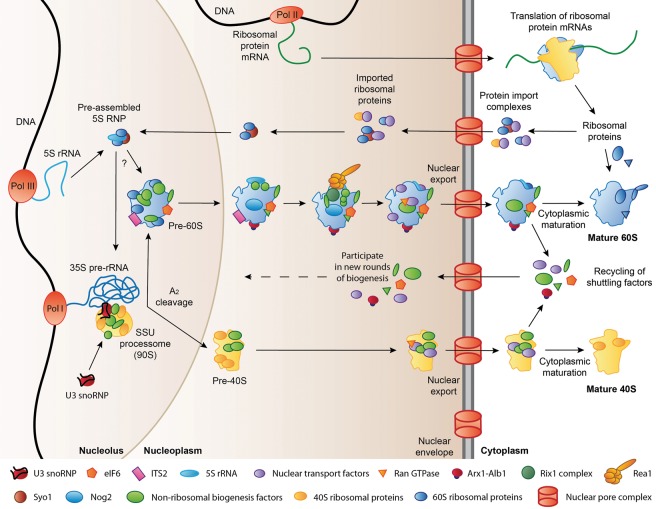
Simplified schematic of ribosome assembly in yeast. Ribosomal protein mRNAs are transcribed by RNA polymerase II (Pol II) and translated in the cytoplasm. A large number of ribosomal proteins are then imported into the nucleus, while a few ribosomal proteins assemble into late cytoplasmic ribosome biogenesis intermediates. In the nucleolus, Pol I and Pol III transcribe the 35S pre-rRNA and 5S rRNA precursors, respectively. The 35S pre-rRNA is incorporated into the SSU processome, where the U3 snoRNP-dependent A2 cleavage separates the nascent large and small subunits, which subsequently undergo maturation and nuclear export independently. The exact timing of the incorporation of the pre-assembled 5S RNP into the nascent large subunit has not yet been determined. Final maturation takes place in the cytoplasm. Shuttling factors that travel from the nucleus to the cytoplasm bound to pre-ribosomal particles are recycled into the nucleus to participate in new rounds of ribosome biogenesis. See text for further details.
Several hundred assembly factors contribute to ribosome biogenesis by supporting the processing, modification, and folding of rRNAs, by facilitating the assembly of ribosomal proteins into pre-ribosomes, by enabling the nuclear export of pre-ribosomal particles, and by performing proofreading roles to ensure completion of upstream steps of biogenesis before later processes are initiated (Karbstein 2013; Woolford and Baserga 2013; Gerhardy et al. 2014). Ribosome biogenesis starts in the nucleolus, where RNA polymerase I transcribes the 35S pre-rRNA, which contains the sequences for the 18S, 25S, and 5.8S rRNAs as well as internal and external transcribed spacers (Henras et al. 2008, 2014). The nascent 35S transcript assembles with the U3 small nucleolar ribonucleoprotein (U3 snoRNP), additional box C/D and H/ACA snoRNPs, 40S subunit assembly factors, and early-binding ribosomal proteins to produce the first pre-ribosomal particle, which is termed the SSU processome, and sediments at 90S (Dragon et al. 2002; Grandi et al. 2002; Phipps et al. 2011; Woolford and Baserga 2013; Chaker-Margot et al. 2015; Kornprobst et al. 2016; Zhang et al. 2016). The U3 snoRNP in the SSU processome is important for pre-rRNA cleavage at site A2 in the internal transcribed spacer 1 (ITS1) (Hughes and Ares 1991), which has been suggested to be carried out by the PIN domain nuclease Utp24 (Wells et al. 2016) and can occur co- or post-transcriptionally in yeast (Udem and Warner 1972; Osheim et al. 2004; Kos and Tollervey 2010). A2-cleavage leads to the separation of the 43S and 66S pre-ribosomal particles (Trapman et al. 1975), which subsequently embark on distinct maturation pathways to form the 40S and 60S ribosomal subunits (Woolford and Baserga 2013; Gerhardy et al. 2014). While the nascent 40S subunits are rapidly exported to the cytoplasm, the biogenesis of the 60S ribosomal subunit progresses through several nucleolar and nucleoplasmic assembly and maturation steps that give rise to defined pre-ribosomal particles. These are characterized by the arrival and departure of certain sets of assembly factors and energy-consuming enzymes as well as the accompanying structural transitions of the nascent 60S particle (Bassler et al. 2001; Harnpicharnchai et al. 2001; Saveanu et al. 2001, 2003; Fatica et al. 2002; Nissan et al. 2002; Kressler et al. 2012b; Gerhardy et al. 2014). Additionally, several pre-rRNA cleavage steps, including the removal of ITS2, which is inserted between the sequences for the mature 25S and 5.8S rRNAs in the 27S pre-rRNA species, are required to generate the mature large subunit rRNAs (Henras et al. 2008, 2014; Fernandez-Pevida et al. 2015). Upon completion of the nuclear steps of ribosome assembly, the pre-60S particle recruits nuclear export receptors and is exported to the cytoplasm, where it can enter the pool of translating ribosomes after undergoing final cytoplasmic maturation (Johnson et al. 2002; Zemp and Kutay 2007; Panse and Johnson 2010). Many of the assembly factors released during cytoplasmic maturation are so-called “shuttling factors,” which travel to the cytoplasm bound to the pre-60S subunit during nuclear export and return to the nucleus to participate in new rounds of ribosome biogenesis after their release from the maturing pre-ribosome (Panse and Johnson 2010; Gerhardy et al. 2014).
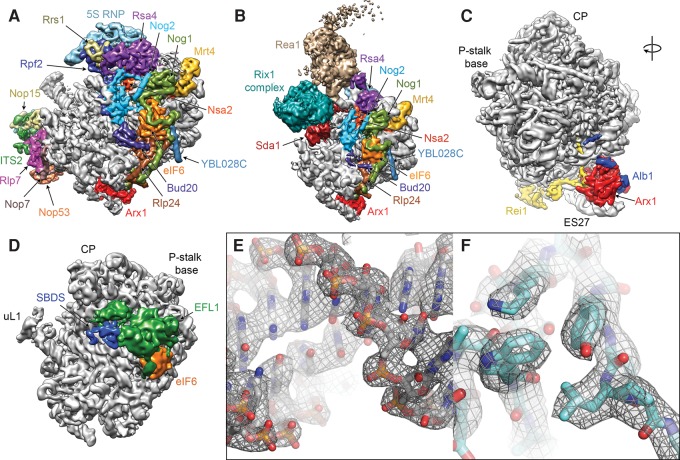
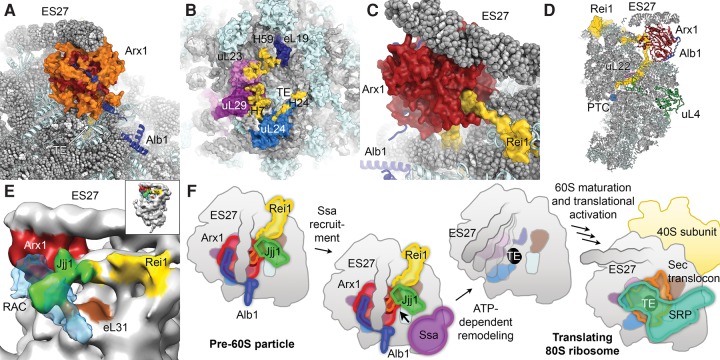
Biochemical, proteomics, cell biological, and genetics experiments as well as structural studies of isolated ribosome assembly factors by X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy have uncovered many aspects of the fascinating and intricate process of eukaryotic ribosome biosynthesis. However, for a complete mechanistic understanding of the 60S assembly process, detailed structural data of pre-60S particles and the interactions of assembly factors on the nascent 60S subunit are crucial. While early cryo-electron microscopic (cryo-EM) visualizations of Nmd3 and eIF6 on the yeast 60S particle (Gartmann et al. 2010; Sengupta et al. 2010) and the X-ray crystal structure of the Tetrahymena thermophila 60S-eIF6 complex (Klinge et al. 2011) provided a first glimpse of the interactions of these assembly factors with the 60S subunit, more detailed insight into the complex interaction networks of assembly factors on the pre-60S particle has only recently started to emerge. Cryo-EM reconstructions of native pre-60S particles at subnanometer and near-atomic resolutions have characterized the architecture of three nuclear biogenesis intermediates, the Nog2, Arx1, and Rix1 particles (Fig. 3A,B; Bradatsch et al. 2012; Leidig et al. 2014; Barrio-Garcia et al. 2016; Wu et al. 2016), which were isolated from native source using tandem affinity purification (TAP) (Puig et al. 2001). Even though they were purified using different bait proteins, the Nog2 and Arx1 particles show a very similar overall architecture. These structures enabled the localization and structural analysis of a large number of biogenesis factors (Wu et al. 2016), revealed a large-scale rearrangement of the 5S RNP during its incorporation at the CP of the nascent 60S subunit (Leidig et al. 2014), and provided insight into the regulation of pre-60S maturation by the giant dynein-like ATPase Rea1 (Barrio-Garcia et al. 2016). Cryo-EM reconstructions of partially or fully in vitro assembled particles also reached near-atomic resolutions and enabled the detailed analysis of the interactions of ribosome assembly factors on the large ribosomal subunit (Fig. 3C–F). The structure of the 60S-Arx1-Alb1-Rei1 complex (Fig. 3C) allowed the building of atomic coordinate models for Arx1 and Rei1 and demonstrated the insertion of Rei1 into the ribosomal polypeptide exit tunnel (Greber et al. 2016), while reconstructions of complexes of the anti-association factor eIF6 (Tif6 in yeast) and its release factors SBDS and EFL1 (Sdo1 and Efl1/Ria1 in yeast, respectively) on the 60S subunit (Fig. 3D) provided important insights into the functional activation of the 60S particle during the final steps of cytoplasmic maturation (Weis et al. 2015).
FIGURE 3.
Recent subnanometer and near-atomic resolution cryo-EM structures of 60S subunit biogenesis complexes. (A) Structure of the Nog2-TAP pre-60S particle (EMD-6616). Biogenesis factors and the 5S RNP are colored and labeled. (B) Structure of the Rix1-TAP pre-60S particle (EMD-3199). (C) Structure of the 60S-Arx1-Alb1-Rei1 complex (EMD-3151). (D) Structure of the 60S-SBDS-EFL1-eIF6 complex (EMD-3146). Maps are shown from the subunit interface side, except for C, which is rotated to reveal the tunnel exit area. The high-resolution reconstructions (A,C,D) were low-pass filtered to 6.5 Å for clarity. (E,F) Examples of the high-quality density maps in recent cryo-EM reconstructions of ribosome biogenesis complexes at near-atomic resolution (PDB 5APO, EMD-3151), which allow building of all-atom models of both rRNA (E) and proteins (F).
This rapid progress has been enabled by recent technological breakthroughs in cryo-EM, a structural biology technique that allows the determination of high-resolution structures (Fig. 3E,F) of large molecular assemblies from small amounts of sample and without the need for crystallization (Bai et al. 2015a; Nogales and Scheres 2015). Here, I review these recent structural studies of the biogenesis of the 60S ribosomal subunit in the context of existing functional data and discuss how they have contributed to a more complete mechanistic understanding of 60S ribosomal subunit biogenesis.
The architecture of the immature central protuberance in nucleoplasmic 60S biogenesis intermediates
The most upstream nucleoplasmic 60S biogenesis intermediates for which detailed structural data have been obtained are the Nog2 and Arx1 particles (Bradatsch et al. 2012; Leidig et al. 2014; Wu et al. 2016). Nog2 is a GTPase that has been implicated in the control and coordination of various steps of pre-60S maturation (Saveanu et al. 2001, 2003; Talkish et al. 2012; Matsuo et al. 2014), while Arx1 is a ribosome assembly and nuclear export factor with homology to methionine amino-peptidase (MAP) enzymes (Hung and Johnson 2006; Bradatsch et al. 2007; Kowalinski et al. 2007). Because Arx1 is a shuttling factor that initially binds to early nuclear pre-60S precursors and is released only after export to the cytoplasm, the protein is found associated with a wide range of pre-60S particles (Nissan et al. 2002). The cryo-EM reconstruction of the Arx1 particle (Bradatsch et al. 2012; Leidig et al. 2014) contains the assembly factors Rsa4 and the Rpf2-Rrs1 complex (see below), indicating that this structure represents a relatively early nuclear biogenesis intermediate preceding the Rix1 particle (Leidig et al. 2014). The overall architecture of the Arx1 particle is very similar to the most abundant form of the Nog2-containing particle, the structure of which was determined at near-atomic resolution (Wu et al. 2016). These cryo-EM reconstructions revealed that the solvent side of these pre-ribosomal subunits is already very close to the mature state, while a region that extends from the polypeptide tunnel exit toward the subunit interface side and up to the CP (Fig. 3A), where several ribosomal domains or RNA helices are observed in immature conformations, is covered by a continuous network of biogenesis factors (Bradatsch et al. 2012; Wu et al. 2016). This pattern matches a hierarchical model of 60S subunit formation deduced from the order of ribosomal protein association, according to which the solvent side of the 60S subunit is formed first, while the exit tunnel, the subunit interface, and the CP are formed subsequently (Gamalinda et al. 2014; de la Cruz et al. 2015). Several biogenesis factors are bound near the ribosomal functional centers, including Mrt4 at the P-stalk base, Nog1 between the P-stalk base and the PTC active site, Nog2 at the subunit interface near the PTC, eIF6 near the SRL of the ribosomal GTPase-activating center, and Arx1 and the C-terminal extension of Nog1 at the tunnel exit (Fig. 3A; Bradatsch et al. 2012; Leidig et al. 2014; Wu et al. 2016). This arrangement of biogenesis factors suggests that all functional centers of the 60S subunit, some of them still immature at this point, are protected by assembly factors, some of which may also be involved in proofreading (see section “Probing and proofreading of functional centers of the nascent 60S subunit”).
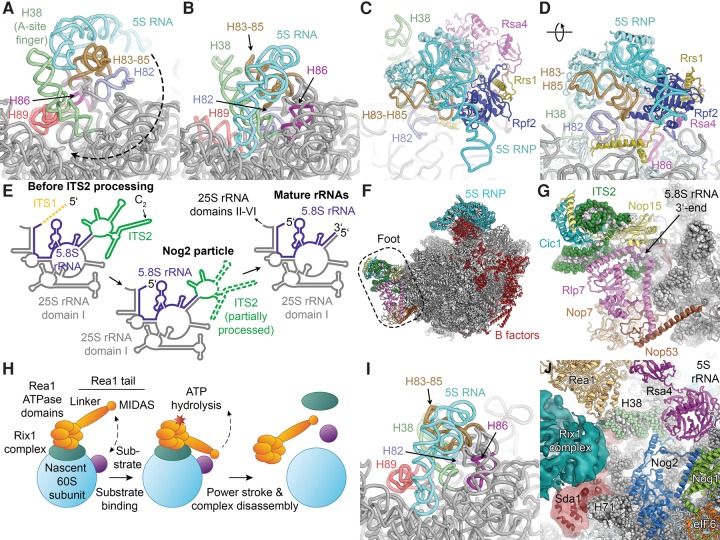
A considerable number of additional biogenesis factors are bound at two hallmark features of the immature pre-60S particles, the foot structure and the strongly rearranged CP (Fig. 4). In the Arx1 particle and the most abundant form of the Nog2 particle, the 5S RNP (consisting of the 5S rRNA and proteins uL5 and uL18; protein names in this review follow the unified nomenclature for ribosomal proteins; Ban et al. 2014) is rotated by roughly 180° relative to its position in the mature subunit (Fig. 4A,B; Ben-Shem et al. 2011; Leidig et al. 2014; Wu et al. 2016). In the immediate vicinity of the rotated 5S RNP in the immature CP, rRNA helix 38 (H38), also termed the A-site finger, as well as helices H82–H88 that eventually form the mature CP, are rearranged, indicating that large structural rearrangements have to occur in this area after the initial incorporation of the 5S RNP (Fig. 4A,B; Leidig et al. 2014). Notably, rRNA H84 engages in a native-like interaction with protein uL5 of the 5S RNP, indicating that it may serve as an initial docking site during assembly (Leidig et al. 2014; Calviño et al. 2015).
FIGURE 4.
Insights into the maturation of the CP and ITS2 processing from the structures of the Nog2, Arx1, and Rix1 particles. (A) The immature RNA structure in the CP of the Arx1 particle (PDB ID 4V7F; proteins not shown). The rotation of the 5S RNP during CP rearrangement is indicated by a dashed arrow. (B) The RNA structure in the CP of the mature 60S subunit (PDB ID 4V88). (C,D) Architecture of the CP in the Nog2 particle (PDB 3JCT). The 5S RNP sits on top of H83–H85. Rsa4 and Rpf2-Rrs1 bind to the interface between the 5S RNP and the 25S rRNA and stabilize the 5S RNP assembly in the rotated state. (E) Schematic of the location of the ITS2 sequence in the context of the secondary structure diagram of the mature 25S and 5.8S rRNAs (ITS1 shown as dashed line for completeness). ITS2 in the Nog2 particle is partially processed (indicated by dashed lines; approximate regions observed in the structure shown as solid lines). ITS2 secondary structure drawn according to the ring model (Joseph et al. 1999). (F) Overview of the Nog2 particle (PDB 3JCT). B-factors are colored red. (G) Detailed view of the foot structure. RNA and protein components of the foot are color-coded. (H) Schematic of ATP-dependent pre-60S remodeling by Rea1. (I) The RNA structure in the CP of the Rix1 particle (PDB 5FL8). Most of the CP has rearranged toward the mature state, while H89 is still found in an immature conformation. (J) Interactions of the Rix1 complex, Rea1, and Sda1 in the Rix1 particle (PDB 5FL8 and EMD-3199; Nog1/2 from PDB 3JCT).
Together with biochemical, structural, and genetic data that indicate that the 5S RNP is initially assembled away from the ribosome and docked onto the pre-60S particle as a preformed module, these observations reveal a detailed mechanism of the initial assembly pathway of the immature CP. The ribosomal proteins uL5 and uL18, which form the protein component of the 5S RNP, are imported into the nucleus in a concerted manner by the transport adaptor and chaperone Syo1, which recruits the import receptor Kap104 (Kressler et al. 2012a). After binding of the 5S rRNA to the ternary Syo1-uL5-uL18 complex (Kressler et al. 2012a; Calviño et al. 2015), the pre-assembled 5S RNP is incorporated into very early pre-ribosomal particles aided by the assembly factor complex Rpf2-Rrs1 (Zhang et al. 2007). In agreement with a role in 5S RNP recruitment and stabilization of the immature CP, the Rpf2-Rrs1 complex in the Arx1 and Nog2 pre-60S particles is located at the interface between the rotated 5S RNP and the RNA helices that later form the base of the mature CP (Asano et al. 2015; Kharde et al. 2015; Madru et al. 2015; Wu et al. 2016), along with the biogenesis factor Rsa4 (Figs. 3A, 4C,D; Leidig et al. 2014). The high-resolution structure of the Nog2 particle (Wu et al. 2016) showed that helical extensions of both Rrs1 and Rpf2 form interactions at the base of the CP (Fig. 4D) and confirmed that Rrs1 interacts with the protein components of the 5S RNP and the biogenesis factor Rsa4, while Rpf2 forms extensive interactions with the 5S rRNA, uL18, and Rsa4 (Fig. 4C,D; Kharde et al. 2015; Madru et al. 2015; Wu et al. 2016).
Structural insights into pre-rRNA maturation and ITS2 processing
The generation of the mature 25S and 5.8S large subunit rRNAs requires the removal of two internal transcribed spacers, ITS1 and ITS2 (Fig. 4E). In addition to several endo- and exonuclease enzymes, the processing of ITS1 in the 27SA3 rRNA requires a set of assembly factors known as A3-factors, while ITS2 processing at the 27SB rRNA stage requires the so-called B-factors (Woolford and Baserga 2013; Henras et al. 2014). The assembly of the A3-cluster and the B-factors is functionally connected because the recruitment of several B-factors to the pre-60S particle depends on ribosomal proteins uL22, uL29, and eL37 near the tunnel exit (Gamalinda et al. 2013), which in turn rely on the A3-factors for their binding to the pre-60S particle (Sahasranaman et al. 2011). During ITS1 processing, the A3-factors may function to support the folding of the pre-rRNA and the recruitment of ribosomal proteins to enable the formation of the structural scaffold required for pre-rRNA processing (Granneman et al. 2011; Sahasranaman et al. 2011; Jakovljevic et al. 2012; Woolford and Baserga 2013). Biochemical analysis revealed that several A3-factors bind to ITS2 sequences (Granneman et al. 2011; Babiano et al. 2013; Dembowski et al. 2013), suggesting that they function in both ITS1 and ITS2 processing.
The foot structure (Bradatsch et al. 2012), which is the second hallmark of the immature Arx1 and Nog2 particles—the first one being the immature structure of the CP—is a large mass of density observed near the 3′-end of the 5.8S rRNA and the 5′-end of the 25S rRNA (Figs. 1B, 3A, 4F) and harbors the partially processed ITS2 (Leidig et al. 2014; Wu et al. 2016). In agreement with a functional role of A3-factors in ITS2 processing, the cryo-EM structure of the Arx1 particle and the near-atomic resolution structure of the Nog2 particle revealed that the A3-factors Rlp7, Nop7, Nop15, and Cic1, along with the biogenesis factor Nop53, assemble around ITS2 segments in the foot that extend from the 5′-end of the 25S rRNA and the 3′-end of the 5.8S rRNA (Fig. 4G; Leidig et al. 2014; Wu et al. 2016). During ITS2 processing, the components of the four-subunit Las1 complex perform the Las1-mediated initial incision at site C2 as well as the Rat1-dependent trimming of the 5′-end of the resulting 25.5S rRNA (Geerlings et al. 2000; Gasse et al. 2015). The A3-factors in the foot may regulate ITS2 processing by controlling the access of these nucleases to the packaged ITS2 RNA (Wu et al. 2016), in agreement with the finding that the Rat1 exonuclease, one of the subunits of the Las1 complex (Gasse et al. 2015), degrades rather than properly processes pre-rRNAs in A3-factor mutants (Sahasranaman et al. 2011). The biogenesis factor Nop53 found in the pre-60S foot binds the exosome-associated helicase Mtr4 (Thoms et al. 2015), thereby mediating the pre-ribosomal recruitment of the nuclear exosome, which participates in the conversion of the 7S pre-rRNA produced by C2 cleavage to the mature 5.8S rRNA (Mitchell et al. 1996, 1997; Henras et al. 2014; Fernandez-Pevida et al. 2015).
In addition to the processing enzyme complexes themselves and the constituents of the foot structure, processing of ITS2 requires the B-factors, a hierarchically assembling network of 60S maturation factors (Saveanu et al. 2001, 2003; Talkish et al. 2012). The B-factors include Rlp24, eIF6, and Nog1 (Basu et al. 2001; Jensen et al. 2003; Saveanu et al. 2003), which are located near the GTPase-activating center of the 60S subunit, as well as Rpf2-Rrs1 and Nsa2 (Morita et al. 2002; Lebreton et al. 2006a; Zhang et al. 2007), which have been localized near the CP (Figs. 3A, 4F; Bassler et al. 2014; Leidig et al. 2014; Wu et al. 2016). The two branches of the B-factor recruitment pathway near the CP and near the GTPase-activating center converge at the GTPase Nog2, whose binding to the interface side of the pre-60S particle depends on the presence of the other B-factors (Talkish et al. 2012). All B-factors that have been localized in the existing cryo-EM structures of pre-60S particles (Bradatsch et al. 2012; Leidig et al. 2014; Wu et al. 2016) are relatively far removed from the site of ITS2 processing (Fig. 4F), indicating that they may function in the ITS2 maturation pathway by coordinating pre-rRNA processing with maturation events at other sites of the pre-60S particle, such as the CP, rather than by participating in the processing reactions directly (see next section).
Maturation of the central protuberance and checkpoint control of nuclear export factor recruitment
The first detailed structural insights into the maturation steps from the Arx1 and Nog2 particles toward nuclear export competence came from the cryo-EM reconstruction of the Rix1 particle (Barrio-Garcia et al. 2016). The Rix1 particle contains the Rix1-Ipi1-Ipi3 subcomplex (Rix1 complex), which is likely a pentameric assembly containing Rix1, Ipi1, and Ipi3 in 2:1:2 stoichiometry (Barrio-Garcia et al. 2016) and the giant (550 kDa) dynein-like AAA+-type ATPase Rea1 (Nissan et al. 2002, 2004). Depletion of the Rix1 complex or Rea1 in yeast leads to similar 60S biogenesis defects, including impairment of ITS2 processing and nuclear export (Galani et al. 2004; Krogan et al. 2004). In addition to the AAA ATPase ring, Rea1 contains a highly mobile tail domain (Ulbrich et al. 2009), which harbors a metal ion-dependent adhesion site (MIDAS) (Garbarino and Gibbons 2002). The MIDAS interacts with the Rea1 substrates Ytm1 in the nucleolus (Bassler et al. 2010) and Rsa4 in the nucleoplasm (Ulbrich et al. 2009) to allow the ATP-driven mechanochemical removal of these biogenesis factors from the maturing pre-60S particle (Fig. 4H; Ulbrich et al. 2009; Kressler et al. 2012b). Ytm1 forms part of a larger complex with the biogenesis factors Erb1 and Nop7 (Miles et al. 2005; Tang et al. 2008; Wegrecki et al. 2015; Thoms et al. 2016), which are therefore also affected by the remodeling activity of Rea1. Two-dimensional negative stain EM analysis of pre-60S particles containing Rea1 revealed a tadpole-like particle, with a globular body formed by the main mass of the pre-60S subunit and a highly mobile tail harboring the Rea1 tail domain (Nissan et al. 2004). The AAA motor domains of Rea1 and the Rix1 complex localize to the connection between tail and body of the “tadpole” (schematic in Fig. 4H; Nissan et al. 2004; Ulbrich et al. 2009). A direct physical interaction between Rix1 and Rea1 that depends on the C-terminal segment of Rix1 and a sequence insertion in the second ATPase module of Rea1 is important for Rea1 recruitment to the pre-60S particle and functional ribosome biogenesis (Barrio-Garcia et al. 2016).
In overall agreement with these assignments based on low-resolution negative stain analysis, the cryo-EM reconstruction of the Rix1 particle revealed large additional densities for Rea1 and the Rix1 complexes near the CP (Fig. 3B; Barrio-Garcia et al. 2016). Furthermore, the hallmarks of the Arx1 and Nog2 particles located upstream in the 60S maturation pathway (Bradatsch et al. 2012; Leidig et al. 2014; Wu et al. 2016) are no longer present in this complex: The 5S RNP in the Rix1 particle has assumed a nonrotated and near-mature conformation (Fig. 4I), and the foot structure harboring the partially processed ITS2 with its associated biogenesis factors has been removed (Fig. 3B; Barrio-Garcia et al. 2016). Interestingly, both the foot removal and the remodeling of the CP fail in the presence of Rix1 mutations that impair the recruitment of Rea1, while ATPase-deficient Rea1 mutants do not lead to retention of the foot or the rotated 5S RNP, indicating that these two maturation events depend on the binding of Rea1 but not its ATPase activity (Barrio-Garcia et al. 2016). Consistent with these observations, the Rea1 substrate Rsa4 is still present in the CP of the Rix1 particle, indicating that the ATP-dependent remodeling step that removes Rsa4 occurs further downstream in the assembly pathway. Although these data suggest that foot removal and CP rotation occur before removal of Rsa4, a subpopulation of particles isolated by Nog2-TAP shows a near-mature conformation of the CP and lacks Rsa4 but retains the foot (Wu et al. 2016). Therefore, the extent of coordination and the relative timing of these two maturation events require further investigation.
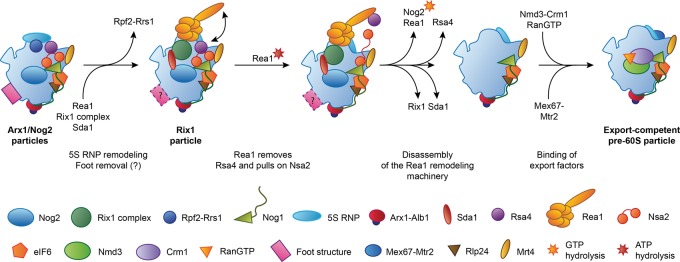
Rea1 and the Rix1 complex form an intricate network of interactions with the remainder of the pre-60S particle, including interactions at the CP, with H38, and with the biogenesis factor Sda1 at the subunit interface (Fig. 4J). These interactions may enable the Rix1 complex and Rea1 to sense the maturation stage of the pre-60S subunit and induce further maturation, such as release of Rsa4, only upon completion of the large-scale rearrangement of the CP (Barrio-Garcia et al. 2016). The model for maturation of the CP that emerges from the available data (Fig. 5) is that initial binding of the Rix1 complex triggers destabilization of the contacts between the rotated 5S RNP, Rpf2-Rrs1, and Rsa4, enabling the coordinated rotation of the 5S RNP and H38 toward their mature positions (Fig. 4A,B,I; Barrio-Garcia et al. 2016). Completion of these structural transitions allows the stable binding of the Rix1-Rea1 remodeling machinery, which then uses the ATPase activity of Rea1 to remove Rsa4 (Barrio-Garcia et al. 2016). Because Rsa4 is linked to the assembly factor Nsa2, this step induces a cascade of structural rearrangements relayed through Nsa2, the removal of which eventually allows accommodation of rRNA H89 in its mature conformation near the PTC (Fig. 4B,I; Bassler et al. 2014). Subsequent acquisition of export competence requires GTP hydrolysis by Nog2 (Fig. 5), whose presence in the pre-60S particle inhibits recruitment of the export receptor Nmd3 because of an extensive overlap between the binding sites of these two maturation factors, as determined by biochemical (Matsuo et al. 2014) and structural analysis (Sengupta et al. 2010; Wu et al. 2016). Additionally, Nog2 likely stabilizes an immature, splayed-out conformation of H71 at the subunit interface, as observed in the Nog2, Arx1, and Rix1 particles (Fig. 4J; Leidig et al. 2014; Barrio-Garcia et al. 2016; Wu et al. 2016), which may further impede Nmd3 association. The remodeling activity of Rea1 and the requirement for Nog2 GTPase activation may act in concert to establish a checkpoint that allows export factor recruitment and subsequent nuclear export only after completion of the maturation of the CP (Matsuo et al. 2014; Barrio-Garcia et al. 2016). It is interesting to note that the initial recruitment of the checkpoint GTPase Nog2 occurs in the context of the B-factors, which are required for processing of ITS2 (see above). This indicates that ITS2 processing and maturation of the CP may be governed, monitored, and coordinated by an integrated network of assembly factors, a hypothesis that also agrees with the observation that defective recruitment of Rea1 impacts both removal of the ITS2-containing foot structure and rotation of the 5S RNP (Barrio-Garcia et al. 2016).
FIGURE 5.
Schematic of the transition from the Arx1 and Nog2 particles to an export competent pre-60S subunit. The relative timing of foot removal and CP rotation cannot be unambiguously established based on the available structures.
Overview of nuclear export and cytoplasmic maturation of the pre-60S particle
The nuclear pore complex (NPC) is a giant molecular assembly embedded in the nuclear envelope that provides a path for exchange of macromolecules between the nucleus and the cytoplasm (Grossman et al. 2012; Knockenhauer and Schwartz 2016). While small molecules can diffuse through the pore freely, large macromolecules need to recruit export factors that help them pass through a meshwork formed by FG-rich repeats of nucleoporins that fill the channel at the center of the NPC (Grossman et al. 2012). The nascent 60S particle acquires export competence by recruiting its full set of nuclear export factors, which include Nmd3, Mex67-Mtr2, Arx1, Bud20, and Ecm1 in yeast (Ho et al. 2000b; Gadal et al. 2001; Bradatsch et al. 2007; Yao et al. 2007, 2010; Hung et al. 2008; Altvater et al. 2012; Bassler et al. 2012). The requirement for a large number of export factors may originate from the large size of the pre-60S subunit, which at more than 2 MDa molecular weight is among the largest cargoes transported through the NPC (Knockenhauer and Schwartz 2016). Nuclear export factors use a variety of mechanisms to facilitate passage of pre-ribosomal particles through the NPC. Arx1 and the Mex67-Mtr2 complex can interact directly with the FG-rich repeats of nucleoporins that line the channel through the NPC (Bradatsch et al. 2007; Yao et al. 2007). Nmd3 contains a nuclear export sequence (NES) that is recognized by the exportin Crm1 (Xpo1 in humans), which binds to FG-repeat nucleoporins and mediates nuclear export in cooperation with the small GTPase Ran (Hurt et al. 1999; Ho et al. 2000b; Gadal et al. 2001; Thomas and Kutay 2003; Cook et al. 2007). The mechanism by which Ecm1 and Bud20 contribute to nuclear export has not been fully clarified yet. Bud20 has been reported to bind to FG-rich nucleoporins directly (Altvater et al. 2012) or to harbor a NES-like element (Bassler et al. 2012). However, because Bud20 does not recruit Crm1 in vitro (Altvater et al. 2012), this NES-like element may not be functionally active. Besides these export factors, the protein Gle2 ensures efficient nuclear export of the pre-60S particle by engaging in non-FG interactions with the nucleoporin Nup116 (Occhipinti et al. 2013). Additionally, the HEAT-repeat containing proteins Sda1 (Dez et al. 2006) and Rrp12 (Oeffinger et al. 2004) have been reported to contribute to pre-60S nuclear export. The structural results from the cryo-EM analysis of the Rix1 pre-60S particle raise the possibility that the export defect observed in Sda1-deficient cells (Dez et al. 2006) might originate not from a defect in nuclear export as such, but from the failure to complete the Rea1- and Nog2-dependent maturation checkpoint (see above), which would preclude recruitment of bona fide export factors, such as Nmd3 (Barrio-Garcia et al. 2016).
After nuclear export, the pre-60S particle is subjected to cytoplasmic maturation (Fig. 6). The Drg1-dependent removal of Rlp24 from the pre-60S particle, which appears to be linked to nuclear export (Kappel et al. 2012), initiates a cascade of biogenesis factor release events and ribosomal protein exchanges that ultimately result in the acquisition of functionality and release of the 60S subunit into the pool of translating ribosomes (Pertschy et al. 2007; Zemp and Kutay 2007; Lo et al. 2010; Panse and Johnson 2010). Importantly, these maturation events occur in a well-defined order (Lo et al. 2010), where completion of upstream events is required for initiation of downstream maturation. More specifically, the replacement of Rlp24 by eL24 (Pertschy et al. 2007; Kappel et al. 2012) enables the recruitment of Rei1, which cooperates with the maturation factors Jjj1 and Ssa to release Arx1 from the tunnel exit (Hung and Johnson 2006; Lebreton et al. 2006b; Demoinet et al. 2007; Meyer et al. 2007, 2010). In a parallel branch of the pathway, the Yvh1-dependent exchange of the stalk protein uL10 for the placeholder Mrt4 leads to formation of the P-stalk (Kemmler et al. 2009; Lo et al. 2009; Rodríguez-Mateos et al. 2009). Even though Yvh1 was initially found to be a shuttling protein (Kemmler et al. 2009; Lo et al. 2009), raising the possibility that it might associate with pre-60S subunits in the nucleus, more recent data obtained by innovative mass spectrometry techniques show that Yvh1 binding is cytoplasmic and occurs downstream from the Drg1-dependent release of Rlp24 (Altvater et al. 2012). The final steps of maturation, which include the release of Nmd3 and eIF6 from the subunit interface side of the 60S particle, depend on the completion of both Arx1 release and P-stalk assembly (Lebreton et al. 2006b; Lo et al. 2010). While the coupling between the release of Arx1 and the final maturation events at the subunit interface is not well understood, the need to recruit the elongation factor like GTPase EFL1 in order to release eIF6 provides a mechanistic explanation for the dependence of eIF6 release on P-stalk assembly (Senger et al. 2001; Lo et al. 2010).
FIGURE 6.
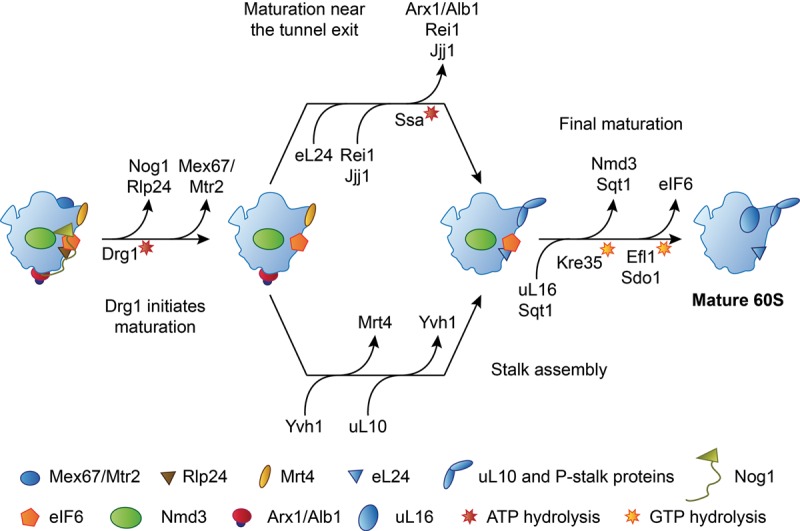
The pathway of the cytoplasmic maturation of the 60S subunit. Only binding and release events discussed in the text are shown. For an exhaustive review, see Gerhardy et al. (2014). The release reactions of Nmd3 and eIF6 are ordered according to Weis et al. (2015); see text for details.
The release of eIF6 by Sdo1 (SBDS) and EFL1 (Bécam et al. 2001; Senger et al. 2001; Menne et al. 2007) and the Kre35 (Lsg1)-dependent removal of the nuclear export adaptor Nmd3 (Hedges et al. 2005; West et al. 2005; Sengupta et al. 2010) are both candidates for being the final step of cytoplasmic 60S maturation before the subunit enters the pool of translating ribosomes (Fig. 6). Both of these release reactions depend on the previous incorporation of ribosomal protein uL16 (Hedges et al. 2005; West et al. 2005; Bussiere et al. 2012; De Keersmaecker et al. 2013; Sulima et al. 2014a; Weis et al. 2015), which is cotranslationally captured by its dedicated chaperone Sqt1 and then assembled into the maturing pre-60S particle (West et al. 2005; Pausch et al. 2015). Because Sqt1 can be trapped on pre-60S particles by certain GTPase-deficient mutants of Kre35, Sqt1 itself may also be transiently incorporated into the pre-60S particle during uL16 insertion and may be released together with Nmd3 in a Kre35-dependent manner (West et al. 2005; Pausch et al. 2015). The presence of Sqt1 and Nmd3 may preclude full accommodation of uL16 into its binding site, suggesting that stable binding of uL16 may be coupled to Nmd3 and Sqt1 release (West et al. 2005; Pausch et al. 2015).
Because uL16 has been found to be required for the recruitment of SBDS and the release of eIF6 (Bussiere et al. 2012; Sulima et al. 2014a; Weis et al. 2015), and because eIF6-bound D. discoideum late pre-60S subunits contain uL16 and lack Nmd3 (Weis et al. 2015), the release of eIF6 has recently been proposed to occur after removal of Nmd3 (Weis et al. 2015). Furthermore, available structural data (Pausch et al. 2015; Weis et al. 2015) and the mapping of the binding sites of Nmd3 on the 60S subunit (Sengupta et al. 2010; Matsuo et al. 2014) raise the possibility that the presence of Sqt1 and Nmd3 might interfere with the binding of the eIF6-releasing factors due to steric hindrance or overlap of binding sites. In contrast, previous yeast genetics data indicated that Nmd3 release requires previous removal of eIF6, placing Nmd3 downstream from eIF6 release. More specifically, interfering with the release of eIF6 by depletion of EFL1 or mutations in Sdo1 (SBDS) or uL16 causes retention of Nmd3 on the pre-60S particle (Lo et al. 2010; Bussiere et al. 2012). Nmd3 release can be restored by alleles of eIF6 that bind the pre-60S particle less tightly and allow eIF6 to dissociate without support from release factors, indicating that the presence of eIF6 itself inhibits the release of Nmd3 (Lo et al. 2010; Bussiere et al. 2012). Further experimental data will be required to unambiguously determine the last step of cytoplasmic pre-60S maturation.
Arx1, Rei1, Jjj1, and Alb1: biogenesis factors near the polypeptide tunnel exit
Currently, no structures of natively purified export-competent or newly exported 60S biogenesis intermediates are available. However, in vitro reconstitution experiments using purified mature 60S subunits and recombinantly expressed biogenesis factors Arx1, Alb1, Rei1, and Jjj1 have provided detailed insights into the interplay of cytoplasmic maturation factors near the tunnel exit of the large ribosomal subunit (Fig. 3C). As mentioned previously, Arx1 functions as a nuclear export factor for the pre-60S particle in yeast (Bradatsch et al. 2007; Hung et al. 2008). A central cavity in the fold of Arx1, which corresponds to the catalytic pocket of homologous MAP enzymes, has been suggested to serve as a binding site for FG-repeat nucleoporins of the NPC, thereby mediating nuclear export (Bradatsch et al. 2007). However, the structures of the natively purified Arx1 particle (Bradatsch et al. 2012; Leidig et al. 2014), as well as the higher-resolution structures of in vitro reconstituted complexes of the yeast 60S subunit and Arx1 (Greber et al. 2012, 2016), showed that this pocket faces the ribosomal subunit surface near the polypeptide tunnel exit, and that access to it is restricted by several Arx1-specific extensions to the conserved MAP-like core fold that are involved in interactions with the 60S subunit (Fig. 7A; Greber et al. 2016). Even though these observations do not exclude that highly flexible FG-repeats of nucleoporins can access the Arx1 cavity, it is possible that structural elements of Arx1 other than the putative nucleoporin-binding pocket are involved in mediating nuclear export. Notably, Ebp1, the human homolog of Arx1, does not show affinity for FG-repeat nucleoporins (Bradatsch et al. 2007), suggesting that the nuclear export function of Arx1 might be limited to certain organisms, while the structural roles of Arx1 (see below) may be conserved between yeast and humans.
FIGURE 7.
Structural analysis of 60S-bound assembly factors near the polypeptide tunnel exit. (A) Structure of Arx1 and Alb1 on the 60S subunit (PDB ID 5APO). Extensions (orange) of the Arx1 core fold (red) shield access to the tunnel exit (TE). (B) Arx1-binding sites (yellow) map to ribosomal proteins eL19, uL23, uL29, and uL24 as well as to rRNA (H7, H24, and H59 indicated). (C) Rei1 interacts with Arx1 and enters the tunnel. (D) Rei1 occupies almost the entire tunnel, passing the constriction formed by uL4 and uL22, and approaching the PTC active site. (E) The low-resolution reconstruction of the 60S-Arx1-Rei1-Jjj1 complex (EMD-2167) with the cryo-EM volume of RAC (EMD-6105) superposed in semitransparent blue reveals similar binding sites for Jjj1 and RAC. (F) Schematic of the maturation events controlled by the network of assembly factors near the polypeptide tunnel exit of the pre-60S particle (only tunnel-associated factors are shown). After maturation, protein biogenesis factors, such as SRP or the Sec translocon, can use the binding sites near the tunnel exit of the translating 80S ribosome.
The ribosomal binding site of Arx1 includes ribosomal proteins uL23, uL24, uL29, and eL19, as well as rRNA helix 59 (Bradatsch et al. 2012; Greber et al. 2012, 2016), which are important binding sites for nascent chain processing, folding, and targeting factors, such as the signal recognition particle (SRP) or the Sec translocon (Fig. 7B; Halic et al. 2004; Becker et al. 2009; Kramer et al. 2009). Binding of Arx1 near the tunnel exit may serve to block the premature binding of these protein biogenesis factors to the nascent 60S subunit (Bradatsch et al. 2012; Greber et al. 2012). Additionally, Arx1 may be unable to associate with the pre-60S particle if its binding sites are not properly formed, allowing it to proofread the correct assembly of the protein biogenesis factor-binding platform near the tunnel exit (Greber et al. 2016). Tunnel exit assembly defects due to depletion of ribosomal proteins in that area have been shown to lead to impaired or delayed ITS2 processing (Pöll et al. 2009; Babiano and de la Cruz 2010; Babiano et al. 2012; Gamalinda et al. 2013); however, there is currently no evidence for a direct involvement of Arx1 in this pathway. Interestingly, Arx1 also binds ES27, an essential and highly mobile expansion segment (Sweeney et al. 1994; Jeeninga et al. 1997) that occurs in two main conformations, pointing toward either the polypeptide tunnel exit or the L1 stalk (Beckmann et al. 2001). Arx1 stabilizes ES27 in the tunnel conformation by binding to its tip (Bradatsch et al. 2012; Greber et al. 2012). The functional importance of this interaction has not been fully clarified yet, but arresting the expansion segment may aid in compaction of the pre-60S particle for nuclear export.
In its native cellular environment, Arx1 exists in a stable complex with the small highly basic protein Alb1 (Lebreton et al. 2006b; Bradatsch et al. 2007), which is recruited to the pre-60S particle by this interaction. Genetics evidence indicates that the deletion of Alb1 can rescue the growth defects of cells that harbor mutations in Arx1 release factors (Lebreton et al. 2006b; Demoinet et al. 2007; Meyer et al. 2007), suggesting that Alb1 may be involved in modulating the affinity of Arx1 to the pre-60S particle. In agreement with this notion, the structural assignment of Alb1 in complex with Arx1 and the 60S subunit by cryo-EM and chemical cross-linking-mass spectrometry (CX-MS) experiments shows that Alb1 interacts with both Arx1 and the 60S subunit (Fig. 7A; Greber et al. 2016).
After nuclear export of the pre-60S particle, the Arx1-Alb1 complex needs to be released and recycled back to the nucleus to participate in a new round of 60S subunit assembly. The cytoplasmic 60S maturation factor Rei1, which joins the pre-60S particle after the Drg1-dependent removal of Rlp24 and incorporation of eL24 (Fig. 6; Pertschy et al. 2007; Lo et al. 2010), plays an important role in this process. Rei1 was initially suggested to mediate the nuclear recycling of Arx1, which was found to accumulate in the cytoplasm in yeast cells lacking Rei1 (Lebreton et al. 2006b; Demoinet et al. 2007). Arx1 remains associated with the pre-60S subunit in the absence of Rei1, indicating that Rei1 is required for the release of Arx1 from the pre-60S particle (Hung and Johnson 2006; Meyer et al. 2010). The structure of the in vitro reconstituted 60S-Arx1-Rei1-Alb1 complex at 3.4 Å resolution visualized a physical interaction between Rei1 and Arx1 (Fig. 7C; Greber et al. 2016), rationalizing the increased stability of the 60S-Arx1 interaction in Rei1-containing complexes observed previously (Greber et al. 2012). These findings suggest that Rei1 facilitates Arx1 release by establishing a network of interactions near the tunnel exit that allows the recruitment of additional release factors such as Jjj1 and Ssa (see below), rather than by simple destabilization of the 60S-Arx1 interaction. The high-resolution structure of the 60S-Arx1-Alb1-Rei1 complex also shows that Rei1 inserts its C terminus deeply into the polypeptide exit tunnel (Fig. 7C,D) and forms extensive interactions with the tunnel walls. Rei1 reaches beyond the constriction site of the tunnel formed by proteins uL4 and uL22 (Nissen et al. 2000), and to within 15 Å of the PTC, probing the tunnel along almost its entire length (Fig. 7D; Greber et al. 2016). The insertion of Rei1 into the tunnel is a prerequisite for downstream maturation of the pre-60S particle because preventing tunnel-insertion by fusion of bulky domains to the Rei1 C terminus leads to 60S assembly defects and retention of maturation factors on cytoplasmic 60S biogenesis intermediates (Greber et al. 2016). This indicates that Rei1 may check the integrity of the tunnel and delay maturation in case of defects in tunnel assembly (see section “Recent insight into biogenesis factors involved in the assembly and probing of the polypeptide tunnel”).
In addition to Rei1, the Hsp70-type ATPase Ssa and its J-protein co-chaperone Jjj1 have been shown to participate in the release of Arx1 from the pre-60S particle (Demoinet et al. 2007; Meyer et al. 2007, 2010). Remarkably, the recent biochemical analysis of mutants of the human Jjj1 homolog DNAJC21 indicates that the components of this pathway and their functions in recycling of the Ebp1, the homolog of Arx1, are conserved in humans (Tummala et al. 2016). The low-resolution reconstruction of a 60S-Arx1-Rei1-Jjj1 complex shows additional density for Jjj1 near the contact site between Rei1 and Arx1 and close to ribosomal protein eL31 in proximity to the tunnel exit (Fig. 7E; Greber et al. 2012). This suggests that Jjj1 may recruit Ssa to this area to enable remodeling of the pre-60S particle and release of Arx1 (Fig. 7F). Further biochemical and structural studies are required to elucidate whether Ssa releases Arx1 by breaking the interaction between Arx1 and Rei1, or if Arx1 and Rei1 are released simultaneously, possibly after extraction of the Rei1 C terminus from the tunnel by Ssa. It is interesting to note that the J-protein component Zuo1 of the ribosome-associated complex (RAC) (Yan et al. 1998; Gautschi et al. 2001), which recruits the Hsp70-type chaperone Ssb to nascent chains emanating from the tunnel exit (Gautschi et al. 2002), also binds to eL31 (Peisker et al. 2008; Leidig et al. 2013; Zhang et al. 2014), suggesting that the two ribosome-bound J-domain proteins Jjj1 and Zuo1 might be architecturally related (Fig. 7E; Kaschner et al. 2015). Both Jjj1 and RAC have also been implicated in early, nuclear steps of 60S subunit biogenesis, where they may play a role in pre-rRNA processing, unrelated to Arx1 recycling or nascent chain folding (Albanèse et al. 2010). Even though further functional data are required to precisely define the nuclear roles of Jjj1-Ssa and RAC-Ssb, the available data suggest both architectural and functional overlap between these two ribosome-bound chaperone systems.
The release of eIF6 and its connection to ribosomopathies
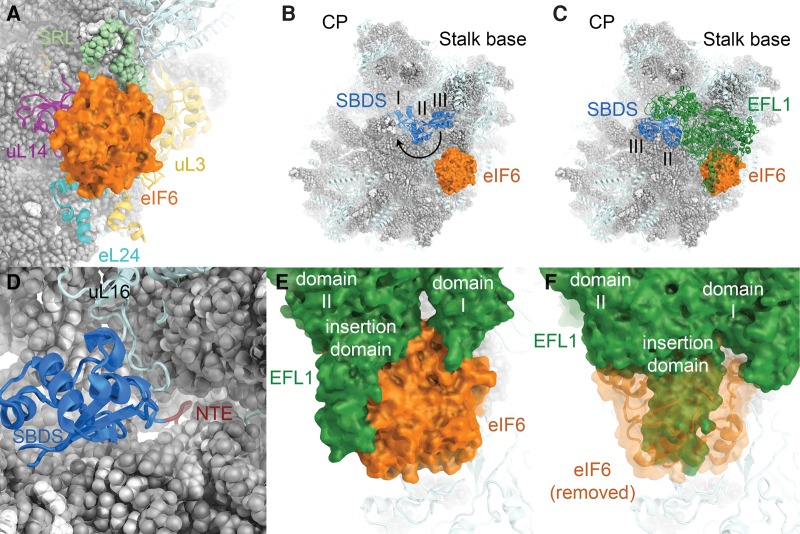
The release of Arx1 from the tunnel exit region and the assembly of the P-stalk license the final maturation of the subunit interface, including the release of eIF6 and Nmd3 (Lo et al. 2010). eIF6 is a multifunctional protein that has a conserved role in large subunit biogenesis (Sanvito et al. 1999; Si and Maitra 1999; Basu et al. 2001), acts to increase the efficiency of translation initiation in mammalian cells, possibly by providing a free pool of 60S subunits (Gandin et al. 2008; Miluzio et al. 2009), and has been linked to cancer cell transformation and tumor progression (Gandin et al. 2008; Brina et al. 2015). Structures of eIF6 bound to the 60S ribosomal subunit show that it binds to ribosomal proteins uL3, uL14, and eL24, as well as to the SRL of the 25S rRNA (Fig. 8A; Gartmann et al. 2010; Klinge et al. 2011; Weis et al. 2015). On the molecular level, eIF6 acts as an anti-association factor that prevents the joining of the (pre-)60S subunit with 40S subunits (Russell and Spremulli 1979; Valenzuela et al. 1982) by inhibiting intersubunit bridge formation near 60S ribosomal protein uL14 (Gartmann et al. 2010). The release of eIF6 from the pre-60S particle occurs during late cytoplasmic maturation and requires the action of the elongation factor-like GTPase EFL1 (Efl1/Ria1 in yeast) (Bécam et al. 2001; Senger et al. 2001) and the SBDS protein (Sdo1 in yeast) (Menne et al. 2007).
FIGURE 8.
Insights into the binding of eIF6 to the 60S subunit and its release by SBDS and EFL1. (A) Structure of eIF6 bound to the 60S subunit (PDB ID 4V8P). (B) Structure of the 60S-eIF6-SBDS complex (PDB ID 5AN9). SBDS domains are numbered. 60S subunit visualization based on the yeast 60S subunit (PDB ID 4V88). SBDS closed-to-open transition indicated by an arrow. (C) Visualization of the 60S-eIF6-SBDS-EFL1 complex (PDB ID 5ANB). (D) The SBDS N-terminal extension (NTE, dark red) enters the polypeptide tunnel near the PTC. For clarity, only the N-terminal domain I of SBDS is shown. (E) EFL1-eIF6 interactions in the 60S-eIF6-SBDS-EFL1 complex. EFL1 domains I and II as well as an insertion domain in domain II are indicated. (F) In the post-eIF6 release conformation, the position of EFL1 is incompatible with eIF6 binding due to extensive overlap (PDB ID 5ANC).
The human SBDS protein is mutated in Shwachman-Bodian-Diamond syndrome (Boocock et al. 2003), an autosomal recessive congenital disorder characterized by skeletal abnormalities, exocrine pancreatic insufficiency, bone marrow failure, and a predisposition to leukemia (Dror 2005). It has been shown that disease mutations in SBDS cause ribosomal subunit joining defects and uncoupling of the GTPase activity of EFL1 from eIF6 release, indicating that the persistence of eIF6 on nascent 60S subunits is the cause of the disease (Finch et al. 2011; Johnson and Ellis 2011; Wong et al. 2011; Burwick et al. 2012). Shwachman-Bodian-Diamond syndrome has therefore been classified as a ribosomopathy, a class of diseases that also includes Diamond-Blackfan anemia and that is characterized by defective ribosome biosynthesis, assembly, or function (Narla and Ebert 2010; Ball 2011; Finch et al. 2011). Nucleolar stress, p53 activation due to failed ribosome biogenesis, bone marrow failure, and a predisposition to cancers are commonly observed in ribosomopathies (Liu and Ellis 2006; Narla and Ebert 2010; Danilova and Gazda 2015). The latest addition to the list of ribosome assembly factors involved in ribosomopathies is DNAJC21, the human homolog of the yeast 60S maturation factor Jjj1, mutations of which have been found to be associated with an inherited bone marrow failure syndrome (Tummala et al. 2016).
The paradoxical observation that ribosome assembly deficits in humans often lead not only to hypoproliferative diseases, such as anemia, but also to hyperproliferative disorders, such as leukemias and cancers, has recently been addressed using a yeast model of the ribosomal protein uL16 R98S mutation that leads to T-cell acute lymphoblastic leukemias (T-ALL) in humans (De Keersmaecker et al. 2013; Sulima et al. 2014b). It was found that this mutation causes a 60S subunit biogenesis deficit that can be suppressed by mutations in ribosome assembly factors (De Keersmaecker et al. 2013; Sulima et al. 2014b), leading to the release of functionally compromised subunits into the pool of translating ribosomes (Sulima et al. 2014b). It has been shown that the absence of a single protein from the eukaryotic ribosome can perturb the expression of specific sets of genes, even if global translation is unchanged (Kondrashov et al. 2011). Therefore, the selective pressure exerted by ribosome synthesis-impairing mutations may lead to the selection of suppressor mutations, which may themselves be harmful or may lead to aberrant mRNA translation due to the presence of faulty ribosomal subunits in the cell, ultimately causing the hyperproliferative phenotype (Sulima et al. 2014b; De Keersmaecker et al. 2015).
The mechanism of eIF6 release
An initial low-resolution cryo-EM reconstruction of 60S-bound eIF6 (Gartmann et al. 2010) and its comparison with ribosome-bound eEF-2 suggested that EFL1 may induce the release of eIF6 by steric exclusion (Gartmann et al. 2010). Detailed mechanistic insight into the molecular rearrangements culminating in the release of eIF6 from the pre-60S subunit came from the structural analysis of a heterologous system assembled from natively purified D. discoideum 60S-eIF6 complexes and human SBDS and EFL1 (Fig. 8B,C; Weis et al. 2015). These structures suggest that eIF6 is released by EFL1 according to a cofactor-dependent conformational switching mechanism (Hauryliuk et al. 2008; Weis et al. 2015). The SBDS protein is comprised of three domains that show considerable flexibility relative to each other (Shammas et al. 2005; de Oliveira et al. 2010; Finch et al. 2011). In the absence of EFL1, SBDS binds to the pre-60S particle in a closed conformation. Its N-terminal domain (domain I) is located in the P-site, where six N-terminal residues stretch across the PTC into the polypeptide tunnel (Fig. 8D). Domains II and III extend toward the GTPase activating region of the 60S subunit, where the C-terminal domain III of SBDS occupies a binding site near uL11 at the P-stalk base (Fig. 8B; Weis et al. 2015). EFL1 binding induces an open conformation of SBDS, in which SBDS domain III is displaced from the P-stalk base and rotated toward the subunit interface side (Fig. 8B,C). EFL1 itself is bound similarly to the homologous translation GTPases eEF-2 (Spahn et al. 2004) and EF-G (Gao et al. 2009), forming extensive interactions with the 60S subunit, SBDS, and eIF6 (Fig. 8C,E; Weis et al. 2015). The closed-to-open transition of SBDS allows EFL1 domain V to associate with the vacated binding site at the P-stalk base and has been proposed to drive EFL1 toward a conformation that is incompatible with the presence of eIF6, which is thereby evicted from its binding site (Fig. 8F). eIF6 release is followed by GTP hydrolysis and EFL1 dissociation (Weis et al. 2015).
In addition to clarifying the mechanism of eIF6 release from the pre-60S particle, these structures also suggest that disease-associated mutations in SBDS and uL16 causing Shwachman-Bodian-Diamond syndrome and T-ALL, respectively, exert their deleterious effects by perturbing the binding of SBDS to the pre-60S subunit or the conformational dynamics of SBDS that are required to allow binding of EFL1 to the P-stalk base (Weis et al. 2015).
The findings from the high-resolution reconstructions of the 60S-eIF6-SBDS-EFL1 complexes, which were obtained using a heterologous system, are in agreement with the low-resolution reconstruction of a homologous yeast 60S-Sdo1 complex, which confirmed the binding of Sdo1 domain I to the P-site and the structural dynamics of domains II and III (Ma et al. 2016). However, this study additionally suggested that Sdo1 might induce dimerization of 60S subunits and raises the possibility that these 60S dimers might represent a storage form of the 60S subunit induced by stress conditions (Ma et al. 2016). In the context of the observed binding of other maturation factors, such as Nmd3, Arx1, Alb1, Rei1, and eIF6, to mature 60S particles in vivo (Ho et al. 2000a; Merl et al. 2010), this is an intriguing hypothesis. However, further experimental evidence is required to establish the physiological significance of this observation.
Probing and proofreading of functional centers of the nascent 60S subunit
Recent biochemical, genetics, and structural data have led to the emerging view that all functional centers of the 60S subunit are first blocked during ribosome assembly to prevent premature translation or other unproductive interactions, and then proofread and functionally tested before the subunit is licensed to participate in translation (Johnson et al. 2002; Lo et al. 2010; Bradatsch et al. 2012; Bussiere et al. 2012; Greber et al. 2012, 2016; Karbstein 2013). This concept also applies to the small ribosomal subunit, where biogenesis factors block the access to the tRNA- and mRNA-binding sites in the pre-40S particle (Strunk et al. 2011), and where a “test-drive” involving mature 60S subunits is required for final activation of the 40S subunit (Lebaron et al. 2012; Strunk et al. 2012; García-Gómez et al. 2014).
The structural data discussed in this review reveal many instances of structural probing of certain assembly steps before downstream processes are initiated, such as the monitoring of the conformation of the CP that controls the Rea1-dependent remodeling of the nuclear pre-60S particle (Barrio-Garcia et al. 2016) or the ordered progression of cytoplasmic maturation (Lo et al. 2010). In addition to these continuous checks of 60S maturation, the available data suggest that there are at least two major events where concerted structural probing and functional proofreading of the functional centers of the pre-60S subunit play a central role: nuclear export and final cytoplasmic maturation.
Nuclear export is a key irreversible step in ribosome biogenesis, which in yeast requires the recruitment of at least five export factors or export factor complexes to the maturing 60S subunit (see section “Overview of nuclear export and cytoplasmic maturation of the pre-60S particle”). The requirement for the recruitment of multiple export factors may enable an extensive structural probing of the correct assembly of the pre-60S particle. Arx1 bound near the tunnel exit may sense the presence of a properly assembled binding platform for the Sec translocon and SRP (Bradatsch et al. 2012; Greber et al. 2012, 2016), while binding of Nmd3 to the subunit interface (Sengupta et al. 2010) and interactions of Mex67-Mtr2 with the 5S rRNA at the CP (Yao et al. 2007) likely only occur if these regions of the pre-60S particle show conformations corresponding to the appropriate maturation stage. Additionally, Nmd3 binding to the subunit interface is controlled by a Nog2-dependent maturation checkpoint that also shows some cross talk with the Rea1 remodeling machinery at the CP (Matsuo et al. 2014; Barrio-Garcia et al. 2016). The concerted action of these multiple instances of structural probing may enable a comprehensive check of the assembly of all major domains of the pre-60S subunit to prevent the nuclear export of faulty or immature pre-60S particles (Johnson et al. 2002), segregating them away from the cytoplasmic assembly factors acting on the pool of pre-ribosomal particles undergoing final maturation. After nuclear export, this arrangement of assembly and export factors, combined with bound Mrt4 and eIF6 that block the P-stalk base and impede access to the SRL (see above), ensures that the freshly exported pre-60S particles cannot interact with 40S subunits or translation factors, which they will encounter in high concentrations in the cytoplasm (Fig. 9).
FIGURE 9.
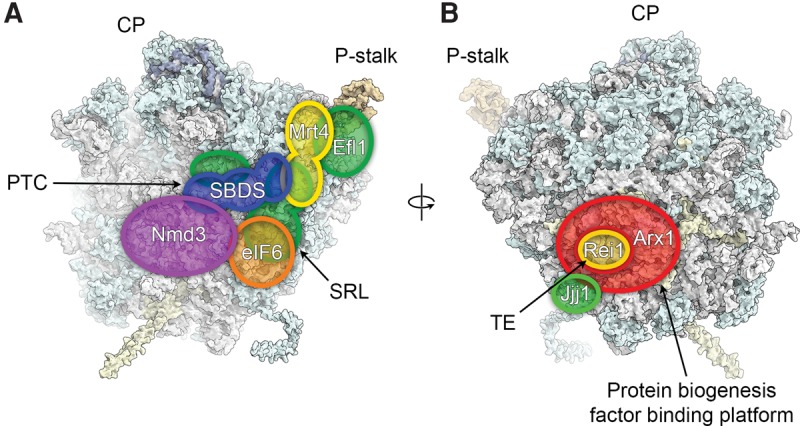
Blocking, probing, and proofreading of functional centers during cytoplasmic 60S maturation. Schematic footprints of assembly and maturation factors that are released or act during cytoplasmic maturation of the 60S subunit are indicated. (A) Nmd3 and eIF6 block access to the subunit interface and the SRL while Mrt4 functions as a placeholder for the P-stalk protein P0. SBDS and EFL1 proofread the PTC, the SRL, and the P-stalk during eIF6 release. (B) Rei1 and Jjj1 probe the exit tunnel and the RAC binding site during release of Arx1, which likely functions mostly in shielding access to the tunnel at these late stages of maturation.
The final activation of the functional centers of the 60S subunit as well as the functional proofreading of their activity occurs during cytoplasmic maturation. Both SBDS and EFL1 are likely to play a key role during this final phase of 60S subunit maturation: SBDS interacts with the P-stalk base, the P-site, the PTC, and the entrance of the exit tunnel, while EFL1 probably requires a functional P-stalk for its recruitment and the SRL for efficient activation of GTP hydrolysis after removal of eIF6 (Fig. 9; Bussiere et al. 2012; Weis et al. 2015). Therefore, eIF6 release likely only occurs in the context of a fully functional 60S subunit with a properly assembled PTC and the capability of translation GTPase recruitment. Similarly, the release of Arx1 may require a functional chaperone-docking site near the tunnel exit to recruit Jjj1-Ssa during the release reaction, thereby ensuring that the RAC-Ssb chaperone system can properly access its substrates during protein translation. Additionally, free passage through the polypeptide tunnel may be monitored by Rei1, possibly supported by SBDS at the other end of the tunnel (see next section).
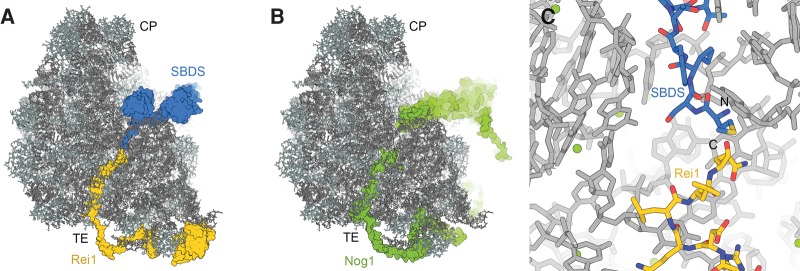
Recent insight into biogenesis factors involved in the assembly and probing of the polypeptide tunnel
The structure of polypeptides in the ribosomal tunnel, particularly in the case of regulatory nascent chains that are able to induce translational stalling, has been extensively studied, revealing that the tunnel is not merely a passive conduit, but allows secondary structure formation and provides interaction sites for some nascent chains (Bhushan et al. 2010; Matheisl et al. 2015; Wilson et al. 2016). It is therefore appropriate to think of the tunnel as an important functional center of the large ribosomal subunit with a well-defined structure that may require monitoring and proofreading during ribosome assembly, a process that may involve Rei1, SBDS, and Nog1. The recent cryo-EM studies of 60S assembly have revealed that Rei1 and Nog1 enter the tunnel from the exit side, while SBDS enters from the side of the PTC (Fig. 10A,B; Weis et al. 2015; Greber et al. 2016; Wu et al. 2016). Notably, all factors assume an orientation that places the N terminus of the inserted sequences closer to the tunnel exit and the C terminus closer to the PTC, mimicking the orientation of a nascent polypeptide chain. Therefore, the Nog1, Rei1, and SBDS segments in the tunnel are probably able to sample a similar conformational space as emerging nascent chains, enabling them to check the ability of the tunnel to act as a conduit for newly synthesized proteins. Nog1 binds to the pre-60S particle already in the nucleus, where it may ensure correct tunnel assembly, while Rei1 and SBDS may perform a final check during cytoplasmic maturation. The binding of Nog1 and Rei1 is likely mutually exclusive due to extensive steric overlap in the tunnel exit region (Greber et al. 2016; Wu et al. 2016). The removal of Nog1 has been suggested to be coupled to the release of Rlp24 (Wu et al. 2016), which may explain why the Drg1-dependent removal of Rlp24 is a prerequisite for Rei1 binding to the pre-60S particle.
FIGURE 10.
Binding of Rei1, SBDS, and Nog1 in the polypeptide exit tunnel. (A) The superposition of structures of 60S-bound yeast Rei1 (PDB ID 5APN) and human SBDS (PDB ID 5ANB) reveals that the two proteins could fill the entire exit tunnel. (B) Nog1 approaches the PTC with its N-terminal helical bundle domain and enters the tunnel exit with its C-terminal extension (PDB 3JCT). (C) Rei1 and SBDS could occupy the tunnel without steric overlap (see text for details).
A superposition of the human SBDS and yeast Rei1 structures in the tunnel (Weis et al. 2015; Greber et al. 2016) shows that the SBDS N terminus and the Rei1 C terminus would be in close proximity, although without overlap, if the two biogenesis factors were to occupy the same pre-60S particles (Fig. 10C). Because the C terminus of Rei1 and the N terminus of SBDS are highly conserved between yeast and humans, suggesting that they are similarly positioned in the polypeptide tunnel across species, this observation probably also applies to the homologous complexes formed by human SBDS and Znf622 or yeast Sdo1 and Rei1, respectively. This arrangement of these two maturation factors may enable them to ensure that the entire passage through the ribosomal tunnel is free of obstructions, which might arise from misfolded rRNA or misincorporated ribosomal proteins (Weis et al. 2015; Greber et al. 2016). The existence of a checkpoint that detects errors in the incorporation of uL4 into the pre-60S particle, where it forms part of the constriction site inside the tunnel, has been suggested recently (Stelter et al. 2015). It remains to be established whether Rei1 and SBDS are involved in this putative checkpoint and whether the two proteins act on the same pre-ribosomal particles or not.
Future perspectives
The functional and structural data reviewed here have uncovered fascinating details of the assembly and maturation of pre-ribosomal particles. Nevertheless, obtaining in-depth mechanistic insights into ribosome biogenesis will remain a formidable challenge, particularly for nucleolar and early nuclear assembly intermediates, due to the enormous complexity and highly dynamic nature of rapidly maturing pre-ribosomal particles. The recent cryo-EM maps of ribosome biogenesis complexes at near-atomic resolution (Weis et al. 2015; Greber et al. 2016; Wu et al. 2016) allowed the building of complete atomic models of assembly factors for which no previous high-resolution X-ray or NMR structures were available, providing detailed insight into the molecular interactions and mechanisms that govern pre-ribosome maturation. With the rise of cryo-EM as the method of choice for determination of high-resolution structures of large assemblies (Bai et al. 2015a; Nogales and Scheres 2015) and the continued technical advances of this method (Merk et al. 2016), including the development of better electron detectors (McMullan et al. 2014) and more powerful particle sorting algorithms (Bai et al. 2015b), the determination of structures of more dynamic or more transient ribosome biogenesis intermediates at near-atomic resolution will become feasible in the future. Applied to cases where the resolution is not fully sufficient for de novo tracing of protein chains and building of complete atomic models, the combination of cryo-EM and CX-MS (Walzthoeni et al. 2013) holds great promise for the detailed analysis of even highly dynamic molecular assemblies, including biogenesis intermediates that are located far upstream in the pathway. In addition to state-of-the-art structural biology methods, innovative techniques may need to be developed and applied to trap and isolate these early intermediates (Chaker-Margot et al. 2015).
The recent addition of high-resolution cryo-EM to the toolbox used to study ribosome assembly will greatly facilitate progress toward a detailed understanding of the compositional and structural transitions of pre-ribosomal particles on their journey from the nucleolus to their functional activation in the cytoplasm. Such insight will greatly benefit our understanding of cellular physiology and growth control, and may also serve as a conceptual framework for the study of the self-assembly of highly complex molecular systems. Additionally, this knowledge may aid in conceiving strategies or developing compounds for treatment of diseases that are associated with failed or up-regulated ribosome synthesis, such as ribosomopathies and cancer.
ACKNOWLEDGMENTS
I thank Nenad Ban for support and advice, Vikram G. Panse for discussions, Avinash B. Patel for critical reading of the manuscript, and Roland Beckmann and Clara Barrio-Garcia for providing PDB coordinate files. I apologize to all authors whose contributions to the field could not be discussed within the scope of this review. Molecular graphics were rendered using UCSF CHIMERA (Pettersen et al. 2004) or PYMOL (The PyMol Molecular Graphics System, version 1.7.4., Schrödinger LLC). I was supported by an Advanced Postdoc Mobility Fellowship from the Swiss National Science Foundation (project P300PA_160983).
Footnotes
Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.057927.116.
REFERENCES
- Albanèse V, Reissmann S, Frydman J. 2010. A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. J Cell Biol 189: 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altvater M, Chang Y, Melnik A, Occhipinti L, Schütz S, Rothenbusch U, Picotti P, Panse VG. 2012. Targeted proteomics reveals compositional dynamics of 60S pre-ribosomes after nuclear export. Mol Syst Biol 8: 628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano N, Kato K, Nakamura A, Komoda K, Tanaka I, Yao M. 2015. Structural and functional analysis of the Rpf2-Rrs1 complex in ribosome biogenesis. Nucleic Acids Res 43: 4746–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiano R, de la Cruz J. 2010. Ribosomal protein L35 is required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res 38: 5177–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiano R, Gamalinda M, Woolford JL, de la Cruz J. 2012. Saccharomyces cerevisiae ribosomal protein L26 is not essential for ribosome assembly and function. Mol Cell Biol 32: 3228–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiano R, Badis G, Saveanu C, Namane A, Doyen A, Diaz-Quintana A, Jacquier A, Fromont-Racine M, de la Cruz J. 2013. Yeast ribosomal protein L7 and its homologue Rlp7 are simultaneously present at distinct sites on pre-60S ribosomal particles. Nucleic Acids Res 41: 9461–9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X-C, McMullan G, Scheres SHW. 2015a. How cryo-EM is revolutionizing structural biology. Trends Biochem Sci 40: 49–57. [DOI] [PubMed] [Google Scholar]
- Bai X-C, Rajendra E, Yang G, Shi Y, Scheres S. 2015b. Sampling the conformational space of the catalytic subunit of human γ-secretase. eLife 4: e11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball S. 2011. Diamond Blackfan anemia. Hematology Am Soc Hematol Educ Program 2011: 487–491. [DOI] [PubMed] [Google Scholar]
- Ban N, Beckmann R, Cate JHD, Dinman JD, Dragon F, Ellis SR, Lafontaine DLJ, Lindahl L, Liljas A, Lipton JM, et al. 2014. A new system for naming ribosomal proteins. Curr Opin Struct Biol 24: 165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrio-Garcia C, Thoms M, Flemming D, Kater L, Berninghausen O, Bassler J, Beckmann R, Hurt E. 2016. Architecture of the Rix1-Rea1 checkpoint machinery during pre-60S-ribosome remodeling. Nat Struct Mol Biol 23: 37–44. [DOI] [PubMed] [Google Scholar]
- Bassler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol Cell 8: 517–529. [DOI] [PubMed] [Google Scholar]
- Bassler J, Kallas M, Pertschy B, Ulbrich C, Thoms M, Hurt E. 2010. The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol Cell 38: 712–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler J, Klein I, Schmidt C, Kallas M, Thomson E, Wagner MA, Bradatsch B, Rechberger G, Strohmaier H, Hurt E, et al. 2012. The conserved Bud20 zinc finger protein is a new component of the ribosomal 60S subunit export machinery. Mol Cell Biol 32: 4898–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler J, Paternoga H, Holdermann I, Thoms M, Granneman S, Barrio-Garcia C, Nyarko A, Stier G, Clark SA, Schraivogel D, et al. 2014. A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation. J Cell Biol 207: 481–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu U, Si K, Warner JR, Maitra U. 2001. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol Cell Biol 21: 1453–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bécam AM, Nasr F, Racki WJ, Zagulski M, Herbert CJ. 2001. Ria1p (Ynl163c), a protein similar to elongation factors 2, is involved in the biogenesis of the 60S subunit of the ribosome in Saccharomyces cerevisiae. Mol Genet Genomics 266: 454–462. [DOI] [PubMed] [Google Scholar]
- Becker T, Bhushan S, Jarasch A, Armache J-P, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, et al. 2009. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science 326: 1369–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann R, Spahn CM, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G. 2001. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107: 361–372. [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. 2011. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334: 1524–1529. [DOI] [PubMed] [Google Scholar]
- Bhushan S, Gartmann M, Halic M, Armache J-P, Jarasch A, Mielke T, Berninghausen O, Wilson DN, Beckmann R. 2010. α-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat Struct Mol Biol 17: 313–317. [DOI] [PubMed] [Google Scholar]
- Boocock GRB, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, Rommens JM. 2003. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet 33: 97–101. [DOI] [PubMed] [Google Scholar]
- Bradatsch B, Katahira J, Kowalinski E, Bange G, Yao W, Sekimoto T, Baumgärtel V, Boese G, Bassler J, Wild K, et al. 2007. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol Cell 27: 767–779. [DOI] [PubMed] [Google Scholar]
- Bradatsch B, Leidig C, Granneman S, Gnädig M, Tollervey D, Böttcher B, Beckmann R, Hurt E. 2012. Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat Struct Mol Biol 19: 1234–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brina D, Miluzio A, Ricciardi S, Biffo S. 2015. eIF6 anti-association activity is required for ribosome biogenesis, translational control and tumor progression. Biochim Biophys Acta 1849: 830–835. [DOI] [PubMed] [Google Scholar]
- Burwick N, Coats SA, Nakamura T, Shimamura A. 2012. Impaired ribosomal subunit association in Shwachman-Diamond syndrome. Blood 120: 5143–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussiere C, Hashem Y, Arora S, Frank J, Johnson AW. 2012. Integrity of the P-site is probed during maturation of the 60S ribosomal subunit. J Cell Biol 197: 747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calviño FR, Kharde S, Ori A, Hendricks A, Wild K, Kressler D, Bange G, Hurt E, Beck M, Sinning I. 2015. Symportin 1 chaperones 5S RNP assembly during ribosome biogenesis by occupying an essential rRNA-binding site. Nat Commun 6: 6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaker-Margot M, Hunziker M, Barandun J, Dill BD, Klinge S. 2015. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat Struct Mol Biol 22: 920–923. [DOI] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E. 2007. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem 76: 647–671. [DOI] [PubMed] [Google Scholar]
- Danilova N, Gazda HT. 2015. Ribosomopathies: how a common root can cause a tree of pathologies. Dis Model Mech 8: 1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keersmaecker K, Atak ZK, Li N, Vicente C, Patchett S, Girardi T, Gianfelici V, Geerdens E, Clappier E, Porcu M, et al. 2013. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet 45: 186–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keersmaecker K, Sulima SO, Dinman JD. 2015. Ribosomopathies and the paradox of cellular hypo- to hyperproliferation. Blood 125: 1377–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Karbstein K, Woolford J, John L. 2015. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu Rev Biochem 84: 93–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira JF, Sforça ML, Blumenschein TMA, Goldfeder MB, Guimarães BG, Oliveira CC, Zanchin NIT, Zeri AC. 2010. Structure, dynamics, and RNA interaction analysis of the human SBDS protein. J Mol Biol 396: 1053–1069. [DOI] [PubMed] [Google Scholar]
- Dembowski JA, Ramesh M, McManus CJ, Woolford JL. 2013. Identification of the binding site of Rlp7 on assembling 60S ribosomal subunits in Saccharomyces cerevisiae. RNA 19: 1639–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demoinet E, Jacquier A, Lutfalla G, Fromont-Racine M. 2007. The Hsp40 chaperone Jjj1 is required for the nucleo-cytoplasmic recycling of preribosomal factors in Saccharomyces cerevisiae. RNA 13: 1570–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C, Houseley J, Tollervey D. 2006. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J 25: 1534–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaconu M, Kothe U, Schlünzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. 2005. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell 121: 991–1004. [DOI] [PubMed] [Google Scholar]
- Dragon F, Gallagher JEG, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417: 967–970. [DOI] [PubMed] [Google Scholar]
- Dror Y. 2005. Shwachman-Diamond syndrome. Pediatr Blood Cancer 45: 892–901. [DOI] [PubMed] [Google Scholar]
- Fatica A, Cronshaw AD, Dlakić M, Tollervey D. 2002. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol Cell 9: 341–351. [DOI] [PubMed] [Google Scholar]
- Fernandez-Pevida A, Kressler D, de la Cruz J. 2015. Processing of preribosomal RNA in Saccharomyces cerevisiae. Wiley Interdiscip Rev RNA 6: 191–209. [DOI] [PubMed] [Google Scholar]
- Finch AJ, Hilcenko C, Basse N, Drynan LF, Goyenechea B, Menne TF, González Fernández A, Simpson P, D'Santos CS, Arends MJ, et al. 2011. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev 25: 917–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E. 2001. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol 21: 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani K, Nissan TA, Petfalski E, Tollervey D, Hurt E. 2004. Rea1, a dynein-related nuclear AAA-ATPase, is involved in late rRNA processing and nuclear export of 60 S subunits. J Biol Chem 279: 55411–55418. [DOI] [PubMed] [Google Scholar]
- Gamalinda M, Jakovljevic J, Babiano R, Talkish J, de la Cruz J, Woolford JL. 2013. Yeast polypeptide exit tunnel ribosomal proteins L17, L35 and L37 are necessary to recruit late-assembling factors required for 27SB pre-rRNA processing. Nucleic Acids Res 41: 1965–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalinda M, Ohmayer U, Jakovljevic J, Kumcuoglu B, Woolford J, Mbom B, Lin L, Woolford JL. 2014. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev 28: 198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Miluzio A, Barbieri AM, Beugnet A, Kiyokawa H, Marchisio PC, Biffo S. 2008. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature 455: 684–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y-G, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. 2009. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326: 694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbarino JE, Gibbons IR. 2002. Expression and genomic analysis of midasin, a novel and highly conserved AAA protein distantly related to dynein. BMC Genomics 3: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Gómez JJ, Fernandez-Pevida A, Lebaron S, Rosado IV, Tollervey D, Kressler D, de la Cruz J. 2014. Final pre-40S maturation depends on the functional integrity of the 60S subunit ribosomal protein L3. PLoS Genet 10: e1004205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartmann M, Blau M, Armache J-P, Mielke T, Topf M, Beckmann R. 2010. Mechanism of eIF6-mediated inhibition of ribosomal subunit joining. J Biol Chem 285: 14848–14851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasse L, Flemming D, Hurt E. 2015. Coordinated ribosomal ITS2 RNA processing by the Las1 complex integrating endonuclease, polynucleotide kinase, and exonuclease activities. Mol Cell 60: 808–815. [DOI] [PubMed] [Google Scholar]
- Gautschi M, Lilie H, Fünfschilling U, Mun A, Ross S, Lithgow T, Rücknagel P, Rospert S. 2001. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc Natl Acad Sci 98: 3762–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi M, Mun A, Ross S, Rospert S. 2002. A functional chaperone triad on the yeast ribosome. Proc Natl Acad Sci 99: 4209–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerlings TH, Vos JC, Raué HA. 2000. The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5′–3′ exonucleases. RNA 6: 1698–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardy S, Menet AM, Peña C, Petkowski JJ, Panse VG. 2014. Assembly and nuclear export of pre-ribosomal particles in budding yeast. Chromosoma 123: 327–344. [DOI] [PubMed] [Google Scholar]
- Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schäfer T, Kuster B, Tschochner H, Tollervey D, et al. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol Cell 10: 105–115. [DOI] [PubMed] [Google Scholar]
- Granneman S, Petfalski E, Tollervey D. 2011. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J 30: 4006–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Montellese C, Ban N. 2012. Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat Struct Mol Biol 19: 1228–1233. [DOI] [PubMed] [Google Scholar]
- Greber BJ, Gerhardy S, Leitner A, Leibundgut M, Salem M, Boehringer D, Leulliot N, Aebersold R, Panse VG, Ban N. 2016. Insertion of the biogenesis factor Rei1 probes the ribosomal tunnel during 60S maturation. Cell 164: 91–102. [DOI] [PubMed] [Google Scholar]
- Grossman E, Medalia O, Zwerger M. 2012. Functional architecture of the nuclear pore complex. Annu Rev Biophys 41: 557–584. [DOI] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CMT, Grassucci RA, Frank J, Beckmann R. 2004. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427: 808–814. [DOI] [PubMed] [Google Scholar]
- Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, Meagher D, Imai B, Guo Y, Brame CJ, et al. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol Cell 8: 505–515. [DOI] [PubMed] [Google Scholar]
- Hauryliuk V, Hansson S, Ehrenberg M. 2008. Cofactor dependent conformational switching of GTPases. Biophys J 95: 1704–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges J, West M, Johnson AW. 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J 24: 567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gérus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65: 2334–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Plisson-Chastang C, O'Donohue M-F, Chakraborty A, Gleizes P-E. 2014. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 6: 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JH, Kallstrom G, Johnson AW. 2000a. Nascent 60S ribosomal subunits enter the free pool bound by Nmd3p. RNA 6: 1625–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JH, Kallstrom G, Johnson AW. 2000b. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol 151: 1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JM, Ares M. 1991. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J 10: 4231–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung N-J, Johnson AW. 2006. Nuclear recycling of the pre-60S ribosomal subunit-associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae. Mol Cell Biol 26: 3718–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung N-J, Lo K-Y, Patel SS, Helmke K, Johnson AW. 2008. Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast. Mol Biol Cell 19: 735–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E, Hannus S, Schmelzl B, Lau D, Tollervey D, Simos G. 1999. A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants. J Cell Biol 144: 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovljevic J, Ohmayer U, Gamalinda M, Talkish J, Alexander L, Linnemann J, Milkereit P, Woolford JL. 2012. Ribosomal proteins L7 and L8 function in concert with six A3 assembly factors to propagate assembly of domains I and II of 25S rRNA in yeast 60S ribosomal subunits. RNA 18: 1805–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeeninga RE, Van Delft Y, de Graaff-Vincent M, Dirks-Mulder A, Venema J, Raué HA. 1997. Variable regions V13 and V3 of Saccharomyces cerevisiae contain structural features essential for normal biogenesis and stability of 5.8S and 25S rRNA. RNA 3: 476–488. [PMC free article] [PubMed] [Google Scholar]
- Jensen BC, Wang Q, Kifer CT, Parsons M. 2003. The NOG1 GTP-binding protein is required for biogenesis of the 60 S ribosomal subunit. J Biol Chem 278: 32204–32211. [DOI] [PubMed] [Google Scholar]
- Johnson AW, Ellis SR. 2011. Of blood, bones, and ribosomes: is Swachman-Diamond syndrome a ribosomopathy? Genes Dev 25: 898–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AW, Lund E, Dahlberg J. 2002. Nuclear export of ribosomal subunits. Trends Biochem Sci 27: 580–585. [DOI] [PubMed] [Google Scholar]
- Joseph N, Krauskopf E, Vera MI, Michot B. 1999. Ribosomal internal transcribed spacer 2 (ITS2) exhibits a common core of secondary structure in vertebrates and yeast. Nucleic Acids Res 27: 4533–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappel L, Loibl M, Zisser G, Klein I, Fruhmann G, Gruber C, Unterweger S, Rechberger G, Pertschy B, Bergler H. 2012. Rlp24 activates the AAA-ATPase Drg1 to initiate cytoplasmic pre-60S maturation. J Cell Biol 199: 771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein K. 2013. Quality control mechanisms during ribosome maturation. Trends Cell Biol 23: 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschner LA, Sharma R, Shrestha OK, Meyer AE, Craig EA. 2015. A conserved domain important for association of eukaryotic J-protein co-chaperones Jjj1 and Zuo1 with the ribosome. Biochim Biophys Acta 1853: 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmler S, Occhipinti L, Veisu M, Panse VG. 2009. Yvh1 is required for a late maturation step in the 60S biogenesis pathway. J Cell Biol 186: 863–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharde S, Calviño FR, Gumiero A, Wild K, Sinning I. 2015. The structure of Rpf2-Rrs1 explains its role in ribosome biogenesis. Nucleic Acids Res 43: 7083–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatter H, Myasnikov AG, Natchiar SK, Klaholz BP. 2015. Structure of the human 80S ribosome. Nature 520: 640–645. [DOI] [PubMed] [Google Scholar]
- Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. 2011. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334: 941–948. [DOI] [PubMed] [Google Scholar]
- Klinge S, Voigts-Hoffmann F, Leibundgut M, Ban N. 2012. Atomic structures of the eukaryotic ribosome. Trends Biochem Sci 37: 189–198. [DOI] [PubMed] [Google Scholar]
- Knockenhauer KE, Schwartz TU. 2016. The nuclear pore complex as a flexible and dynamic gate. Cell 164: 1162–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. 2011. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145: 383–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornprobst M, Turk M, Kellner N, Cheng J, Flemming D, Koš-Braun I, Kos M, Thoms M, Berninghausen O, Beckmann R, et al. 2016. Architecture of the 90S pre-ribosome: a structural view on the birth of the eukaryotic ribosome. Cell 166: 380–393. [DOI] [PubMed] [Google Scholar]
- Kos M, Tollervey D. 2010. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell 37: 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E, Bange G, Bradatsch B, Hurt E, Wild K, Sinning I. 2007. The crystal structure of Ebp1 reveals a methionine aminopeptidase fold as binding platform for multiple interactions. FEBS Lett 581: 4450–4454. [DOI] [PubMed] [Google Scholar]
- Kramer G, Boehringer D, Ban N, Bukau B. 2009. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol 16: 589–597. [DOI] [PubMed] [Google Scholar]
- Kressler D, Bange G, Ogawa Y, Stjepanovic G, Bradatsch B, Pratte D, Amlacher S, Strauss D, Yoneda Y, Katahira J, et al. 2012a. Synchronizing nuclear import of ribosomal proteins with ribosome assembly. Science 338: 666–671. [DOI] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Bergler H, Bassler J. 2012b. The power of AAA-ATPases on the road of pre-60S ribosome maturation—molecular machines that strip pre-ribosomal particles. Biochim Biophys Acta 1823: 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Peng W-T, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell 13: 225–239. [DOI] [PubMed] [Google Scholar]
- Lebaron S, Schneider C, van Nues RW, Swiatkowska A, Walsh D, Böttcher B, Granneman S, Watkins NJ, Tollervey D. 2012. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat Struct Mol Biol 19: 744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton A, Saveanu C, Decourty L, Jacquier A, Fromont-Racine M. 2006a. Nsa2 is an unstable, conserved factor required for the maturation of 27 SB pre-rRNAs. J Biol Chem 281: 27099–27108. [DOI] [PubMed] [Google Scholar]
- Lebreton A, Saveanu C, Decourty L, Rain J-C, Jacquier A, Fromont-Racine M. 2006b. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J Cell Biol 173: 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidig C, Bange G, Kopp J, Amlacher S, Aravind A, Wickles S, Witte G, Hurt E, Beckmann R, Sinning I. 2013. Structural characterization of a eukaryotic chaperone—the ribosome-associated complex. Nat Struct Mol Biol 20: 23–28. [DOI] [PubMed] [Google Scholar]
- Leidig C, Thoms M, Holdermann I, Bradatsch B, Berninghausen O, Bange G, Sinning I, Hurt E, Beckmann R. 2014. 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle. Nat Commun 5: 3491. [DOI] [PubMed] [Google Scholar]
- Liu JM, Ellis SR. 2006. Ribosomes and marrow failure: coincidental association or molecular paradigm? Blood 107: 4583–4588. [DOI] [PubMed] [Google Scholar]
- Lo K-Y, Li Z, Wang F, Marcotte EM, Johnson AW. 2009. Ribosome stalk assembly requires the dual-specificity phosphatase Yvh1 for the exchange of Mrt4 with P0. J Cell Biol 186: 849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo K-Y, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW. 2010. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell 39: 196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Yan K, Tan D, Li N, Zhang Y, Yuan Y, Li Z, Dong M-Q, Lei J, Gao N. 2016. Structural dynamics of the yeast Shwachman-Diamond syndrome protein (Sdo1) on the ribosome and its implication in the 60S subunit maturation. Protein Cell 7: 187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madru C, Lebaron S, Blaud M, Delbos L, Pipoli J, Pasmant E, Réty S, Leulliot N. 2015. Chaperoning 5S RNA assembly. Genes Dev 29: 1432–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheisl S, Berninghausen O, Becker T, Beckmann R. 2015. Structure of a human translation termination complex. Nucleic Acids Res 43: 8615–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, Granneman S, Thoms M, Manikas R-G, Tollervey D, Hurt E. 2014. Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature 505: 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullan G, Faruqi AR, Clare D, Henderson R. 2014. Comparison of optimal performance at 300 keV of three direct electron detectors for use in low dose electron microscopy. Ultramicroscopy 147: 156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M. 2012. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol 19: 560–567. [DOI] [PubMed] [Google Scholar]
- Menne TF, Goyenechea B, Sánchez-Puig N, Wong CC, Tonkin LM, Ancliff PJ, Brost RL, Costanzo M, Boone C, Warren AJ. 2007. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat Genet 39: 486–495. [DOI] [PubMed] [Google Scholar]
- Merk A, Bartesaghi A, Banerjee S, Falconieri V, Rao P, Davis MI, Pragani R, Boxer MB, Earl LA, Milne JLS, et al. 2016. Breaking cryo-EM resolution barriers to facilitate drug discovery. Cell 165: 1698–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merl J, Jakob S, Ridinger K, Hierlmeier T, Deutzmann R, Milkereit P, Tschochner H. 2010. Analysis of ribosome biogenesis factor-modules in yeast cells depleted from pre-ribosomes. Nucleic Acids Res 38: 3068–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AE, Hung N-J, Yang P, Johnson AW, Craig EA. 2007. The specialized cytosolic J-protein, Jjj1, functions in 60S ribosomal subunit biogenesis. Proc Natl Acad Sci 104: 1558–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AE, Hoover LA, Craig EA. 2010. The cytosolic J-protein, Jjj1, and Rei1 function in the removal of the pre-60 S subunit factor Arx1. J Biol Chem 285: 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles TD, Jakovljevic J, Horsey EW, Harnpicharnchai P, Tang L, Woolford JL. 2005. Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol Cell Biol 25: 10419–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miluzio A, Beugnet A, Volta V, Biffo S. 2009. Eukaryotic initiation factor 6 mediates a continuum between 60S ribosome biogenesis and translation. EMBO Rep 10: 459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Tollervey D. 1996. The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev 10: 502–513. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. 1997. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-5′ exoribonucleases. Cell 91: 457–466. [DOI] [PubMed] [Google Scholar]
- Morita D, Miyoshi K, Matsui Y, Toh-e A, Shinkawa H, Miyakawa T, Mizuta K. 2002. Rpf2p, an evolutionarily conserved protein, interacts with ribosomal protein L11 and is essential for the processing of 27 SB Pre-rRNA to 25 S rRNA and the 60 S ribosomal subunit assembly in Saccharomyces cerevisiae. J Biol Chem 277: 28780–28786. [DOI] [PubMed] [Google Scholar]
- Narla A, Ebert BL. 2010. Ribosomopathies: human disorders of ribosome dysfunction. Blood 115: 3196–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J 21: 5539–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan TA, Galani K, Maco B, Tollervey D, Aebi U, Hurt E. 2004. A pre-ribosome with a tadpole-like structure functions in ATP-dependent maturation of 60S subunits. Mol Cell 15: 295–301. [DOI] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930. [DOI] [PubMed] [Google Scholar]
- Nogales E, Scheres SHW. 2015. Cryo-EM: a unique tool for the visualization of macromolecular complexity. Mol Cell 58: 677–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Occhipinti L, Chang Y, Altvater M, Menet AM, Kemmler S, Panse VG. 2013. Non-FG mediated transport of the large pre-ribosomal subunit through the nuclear pore complex by the mRNA export factor Gle2. Nucleic Acids Res 41: 8266–8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Dlakić M, Tollervey D. 2004. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes Dev 18: 196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. 2004. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell 16: 943–954. [DOI] [PubMed] [Google Scholar]
- Panse VG, Johnson AW. 2010. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem Sci 35: 260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pausch P, Singh U, Ahmed YL, Pillet B, Murat G, Altegoer F, Stier G, Thoms M, Hurt E, Sinning I, et al. 2015. Co-translational capturing of nascent ribosomal proteins by their dedicated chaperones. Nat Commun 6: 7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisker K, Braun D, Wölfle T, Hentschel J, Fünfschilling U, Fischer G, Sickmann A, Rospert S. 2008. Ribosome-associated complex binds to ribosomes in close proximity of Rpl31 at the exit of the polypeptide tunnel in yeast. Mol Biol Cell 19: 5279–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertschy B, Saveanu C, Zisser G, Lebreton A, Tengg M, Jacquier A, Liebminger E, Nobis B, Kappel L, van der Klei I, et al. 2007. Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol Cell Biol 27: 6581–6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- Phipps KR, Charette JM, Baserga SJ. 2011. The small subunit processome in ribosome biogenesis—progress and prospects. Wiley Interdiscip Rev RNA 2: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöll G, Braun T, Jakovljevic J, Neueder A, Jakob S, Woolford JL, Tschochner H, Milkereit P. 2009. rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PLoS One 4: e8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Séraphin B. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24: 218–229. [DOI] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. 2011. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331: 730–736. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Mateos M, Abia D, García-Gómez JJ, Morreale A, de la Cruz J, Santos C, Remacha M, Ballesta JPG. 2009. The amino terminal domain from Mrt4 protein can functionally replace the RNA binding domain of the ribosomal P0 protein. Nucleic Acids Res 37: 3514–3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DW, Spremulli LL. 1979. Purification and characterization of a ribosome dissociation factor (eukaryotic initiation factor 6) from wheat germ. J Biol Chem 254: 8796–8800. [PubMed] [Google Scholar]
- Sahasranaman A, Dembowski J, Strahler J, Andrews P, Maddock J, Woolford JL. 2011. Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: role of factors required for 27S pre-rRNA processing. EMBO J 30: 4020–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanvito F, Piatti S, Villa A, Bossi M, Lucchini G, Marchisio PC, Biffo S. 1999. The β4 integrin interactor p27BBP/eIF6 is an essential nuclear matrix protein involved in 60S ribosomal subunit assembly. J Cell Biol 144: 823–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu C, Bienvenu D, Namane A, Gleizes PE, Gas N, Jacquier A, Fromont-Racine M. 2001. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J 20: 6475–6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu C, Namane A, Gleizes P-E, Lebreton A, Rousselle J-C, Noaillac-Depeyre J, Gas N, Jacquier A, Fromont-Racine M. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol Cell Biol 23: 4449–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Ramakrishnan V. 2009. What recent ribosome structures have revealed about the mechanism of translation. Nature 461: 1234–1242. [DOI] [PubMed] [Google Scholar]
- Senger B, Lafontaine DL, Graindorge JS, Gadal O, Camasses A, Sanni A, Garnier JM, Breitenbach M, Hurt E, Fasiolo F. 2001. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol Cell 8: 1363–1373. [DOI] [PubMed] [Google Scholar]
- Sengupta J, Bussiere C, Pallesen J, West M, Johnson AW, Frank J. 2010. Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J Cell Biol 189: 1079–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shammas C, Menne TF, Hilcenko C, Michell SR, Goyenechea B, Boocock GRB, Durie PR, Rommens JM, Warren AJ. 2005. Structural and mutational analysis of the SBDS protein family. Insight into the leukemia-associated Shwachman-Diamond Syndrome. J Biol Chem 280: 19221–19229. [DOI] [PubMed] [Google Scholar]
- Si K, Maitra U. 1999. The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor. Mol Cell Biol 19: 1416–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CMT, Gomez-Lorenzo MG, Grassucci RA, Jørgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JPG, Frank J. 2004. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J 23: 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P, Huber FM, Kunze R, Flemming D, Hoelz A, Hurt E. 2015. Coordinated ribosomal L4 protein assembly into the pre-ribosome is regulated by its eukaryote-specific extension. Mol Cell 58: 854–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL, Karbstein K, Skiniotis G. 2011. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science 333: 1449–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Novak MN, Young CL, Karbstein K. 2012. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell 150: 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulima SO, Gülay SP, Anjos M, Patchett S, Meskauskas A, Johnson AW, Dinman JD. 2014a. Eukaryotic rpL10 drives ribosomal rotation. Nucleic Acids Res 42: 2049–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulima SO, Patchett S, Advani VM, De Keersmaecker K, Johnson AW, Dinman JD. 2014b. Bypass of the pre-60S ribosomal quality control as a pathway to oncogenesis. Proc Natl Acad Sci 111: 5640–5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney R, Chen L, Yao MC. 1994. An rRNA variable region has an evolutionarily conserved essential role despite sequence divergence. Mol Cell Biol 14: 4203–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkish J, Zhang J, Jakovljevic J, Horsey EW, Woolford JL. 2012. Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res 40: 8646–8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Sahasranaman A, Jakovljevic J, Schleifman E, Woolford JL. 2008. Interactions among Ytm1, Erb1, and Nop7 required for assembly of the Nop7-subcomplex in yeast preribosomes. Mol Biol Cell 19: 2844–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng T, Thomas G, Mercer CA. 2013. Growth control and ribosomopathies. Curr Opin Genet Dev 23: 63–71. [DOI] [PubMed] [Google Scholar]
- Thomas F, Kutay U. 2003. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J Cell Sci 116: 2409–2419. [DOI] [PubMed] [Google Scholar]
- Thoms M, Thomson E, Bassler J, Gnädig M, Griesel S, Hurt E. 2015. The exosome is recruited to RNA substrates through specific adaptor proteins. Cell 162: 1029–1038. [DOI] [PubMed] [Google Scholar]
- Thoms M, Ahmed YL, Maddi K, Hurt E, Sinning I. 2016. Concerted removal of the Erb1-Ytm1 complex in ribosome biogenesis relies on an elaborate interface. Nucleic Acids Res 44: 926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapman J, Retèl J, Planta RJ. 1975. Ribosomal precursor particles from yeast. Exp Cell Res 90: 95–104. [DOI] [PubMed] [Google Scholar]
- Tummala H, Walne AJ, Williams M, Bockett N, Collopy L, Cardoso S, Ellison A, Wynn R, Leblanc T, Fitzgibbon J, et al. 2016. DNAJC21 mutations link a cancer-prone bone marrow failure syndrome to corruption in 60S ribosome subunit maturation. Am J Hum Genet 99: 115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udem SA, Warner JR. 1972. Ribosomal RNA synthesis in Saccharomyces cerevisiae. J Mol Biol 65: 227–242. [DOI] [PubMed] [Google Scholar]
- Ulbrich C, Diepholz M, Bassler J, Kressler D, Pertschy B, Galani K, Böttcher B, Hurt E. 2009. Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell 138: 911–922. [DOI] [PubMed] [Google Scholar]
- Valenzuela DM, Chaudhuri A, Maitra U. 1982. Eukaryotic ribosomal subunit anti-association activity of calf liver is contained in a single polypeptide chain protein of Mr = 25,500 (eukaryotic initiation factor 6). J Biol Chem 257: 7712–7719. [PubMed] [Google Scholar]
- Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. 2010. The mechanism for activation of GTP hydrolysis on the ribosome. Science 330: 835–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Fernandez IS, Scheres SHW, Hegde RS. 2014. Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution. Cell 157: 1632–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss NR, Gerstein M, Steitz TA, Moore PB. 2006. The geometry of the ribosomal polypeptide exit tunnel. J Mol Biol 360: 893–906. [DOI] [PubMed] [Google Scholar]
- Walzthoeni T, Leitner A, Stengel F, Aebersold R. 2013. Mass spectrometry supported determination of protein complex structure. Curr Opin Struct Biol 23: 252–260. [DOI] [PubMed] [Google Scholar]
- Warner JR. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440. [DOI] [PubMed] [Google Scholar]
- Wegrecki M, Rodriguez-Galan O, de la Cruz J, Bravo J. 2015. The structure of Erb1-Ytm1 complex reveals the functional importance of a high-affinity binding between two β-propellers during the assembly of large ribosomal subunits in eukaryotes. Nucleic Acids Res 43: 11017–11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis F, Giudice E, Churcher M, Jin L, Hilcenko C, Wong CC, Traynor D, Kay RR, Warren AJ. 2015. Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat Struct Mol Biol 22: 914–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells GR, Weichmann F, Colvin D, Sloan KE, Kudla G, Tollervey D, Watkins NJ, Schneider C. 2016. The PIN domain endonuclease Utp24 cleaves pre-ribosomal RNA at two coupled sites in yeast and humans. Nucleic Acids Res 44: 5399–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M, Hedges JB, Chen A, Johnson AW. 2005. Defining the order in which Nmd3p and Rpl10p load onto nascent 60S ribosomal subunits. Mol Cell Biol 25: 3802–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN, Arenz S, Beckmann R. 2016. Translation regulation via nascent polypeptide-mediated ribosome stalling. Curr Opin Struct Biol 37: 123–133. [DOI] [PubMed] [Google Scholar]
- Wong CC, Traynor D, Basse N, Kay RR, Warren AJ. 2011. Defective ribosome assembly in Shwachman-Diamond syndrome. Blood 118: 4305–4312. [DOI] [PubMed] [Google Scholar]
- Woolford JL, Baserga SJ. 2013. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195: 643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Tutuncuoglu B, Yan K, Brown H, Zhang Y, Tan D, Gamalinda M, Yuan Y, Li Z, Jakovljevic J, et al. 2016. Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature 534: 133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Schilke B, Pfund C, Walter W, Kim S, Craig EA. 1998. Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J 17: 4809–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W, Roser D, Köhler A, Bradatsch B, Bassler J, Hurt E. 2007. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol Cell 26: 51–62. [DOI] [PubMed] [Google Scholar]
- Yao Y, Demoinet E, Saveanu C, Lenormand P, Jacquier A, Fromont-Racine M. 2010. Ecm1 is a new pre-ribosomal factor involved in pre-60S particle export. RNA 16: 1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp I, Kutay U. 2007. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett 581: 2783–2793. [DOI] [PubMed] [Google Scholar]
- Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, Rout MP, Hiley SL, Hughes T, Woolford JL. 2007. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev 21: 2580–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ma C, Yuan Y, Zhu J, Li N, Chen C, Wu S, Yu L, Lei J, Gao N. 2014. Structural basis for interaction of a cotranslational chaperone with the eukaryotic ribosome. Nat Struct Mol Biol 21: 1042–1046. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wu C, Cai G, Chen S, Ye K. 2016. Stepwise and dynamic assembly of the earliest precursors of small ribosomal subunits in yeast. Genes Dev 30: 718–732. [DOI] [PMC free article] [PubMed] [Google Scholar]