Abstract
The replication-dependent histone mRNAs end in a stem–loop instead of the poly(A) tail present at the 3′ end of all other cellular mRNAs. Following processing, the 3′ end of histone mRNAs is trimmed to 3 nucleotides (nt) after the stem–loop, and this length is maintained by addition of nontemplated uridines if the mRNA is further trimmed by 3′hExo. These mRNAs are tightly cell-cycle regulated, and a critical regulatory step is rapid degradation of the histone mRNAs when DNA replication is inhibited. An initial step in histone mRNA degradation is digestion 2–4 nt into the stem by 3′hExo and uridylation of this intermediate. The mRNA is then subsequently degraded by the exosome, with stalled intermediates being uridylated. The enzyme(s) responsible for oligouridylation of histone mRNAs have not been definitively identified. Using high-throughput sequencing of histone mRNAs and degradation intermediates, we find that knockdown of TUT7 reduces both the uridylation at the 3′ end as well as uridylation of the major degradation intermediate in the stem. In contrast, knockdown of TUT4 did not alter the uridylation pattern at the 3′ end and had a small effect on uridylation in the stem–loop during histone mRNA degradation. Knockdown of 3′hExo also altered the uridylation of histone mRNAs, suggesting that TUT7 and 3′hExo function together in trimming and uridylating histone mRNAs.
Keywords: histone mRNA, mRNA degradation, uridylation
INTRODUCTION
Replication-dependent histone mRNAs are the only known eukaryotic mRNAs that are not polyadenylated. Instead, they end in a highly conserved 3′ stem–loop (SL). These histone mRNAs are tightly cell-cycle regulated, and the stem–loop is the major cis-element that controls both the increased expression at the beginning of S-phase and the rapid degradation at the end of S-phase (Harris et al. 1991). Since the replication-dependent histone genes do not contain introns, the only RNA processing step in biosynthesis of histone mRNA is 3′ end processing, endonucleolytic cleavage 5 nt after the stem–loop, which requires both the stem–loop binding protein (SLBP) (Wang et al. 1996) and the U7 snRNP (Mowry and Steitz 1987).
The stem–loop fulfills most of the functions that the poly(A) tail does for other mRNAs. It is required for translation of histone mRNA (Sanchez and Marzluff 2002) and is the cis-element that is required for rapid degradation of histone mRNA when DNA replication is inhibited (Pandey and Marzluff 1987). Translation is also required for degradation, and it is critical that the stem–loop be located close to the stop codon (Graves et al. 1987; Kaygun and Marzluff 2005b).
Deadenylation is the first step in the degradation of most polyadenylated mRNAs. This process does not remove the poly(A) tail entirely, but leaves an oligo(A) tail that can associate with Lsm1-7. Lsm1-7 can stimulate decapping, while the deadenylation also primes the message for degradation by the exosome. Thus, the message can be bidirectionally degraded, from 5′–3′ after decapping and 3′–5′ by the exosome (Parker and Song 2004; Garneau et al. 2007). Since histone mRNAs do not end in a poly(A) tail, degradation must be initiated differently. We previously found that oligo(U) tails are also added to the 3′ end of histone mRNA to initiate degradation (Mullen and Marzluff 2008) and are added at multiple sites in the mRNA as it is being degraded 3′–5′ (Slevin et al. 2014). Lsm1-7 is required for histone mRNA degradation and can bind the oligo(U) tail. The C-terminal tail of Lsm4 in the Lsm1-7 ring binds directly to SLBP and 3′hExo. This interaction is required for efficient degradation of histone mRNAs, suggesting that SLBP also plays a direct role in histone mRNA degradation (Lyons et al. 2014).
A second protein, 3′hExo (Eri1), also binds the 3′ end of histone mRNA and forms a ternary complex on the 3′ end of histone mRNA with SLBP. In vitro 3′hExo trims the last 2–3 nt off the 3′ end of histone mRNA after processing (Dominski et al. 2003; Yang et al. 2006). This trimmed histone mRNA is the predominant version of histone mRNA present in the cytoplasm (Mullen and Marzluff 2008). Knockout of 3′hExo prevents the trimming of processed histone mRNA, as well as the rapid degradation of histone mRNA after inhibition of DNA replication (Hoefig et al. 2013). 3′hExo is likely responsible for the initial step in degradation of the 3′ end of histone mRNA, forming an intermediate which has been degraded into the 3′ side of the stem and which is uridylated (Hoefig et al. 2013; Slevin et al. 2014).
The enzyme(s) that add the oligouridine tails to histone mRNAs have not been definitively identified. Seven noncanonical poly(A) polymerases (also known as terminal uridylyl transferases or TUTases) are present in metazoans, which are capable of adding either adenosines or uridines and sometimes either. Oligouridylation by some of these proteins was initially identified in Schizosaccharomyces pombe (Rissland et al. 2007) and in biochemical studies using Xenopus oocytes (Kwak and Wickens 2007). These seven enzymes have a wide variety of functions in the life cycles of snRNAs, miRNAs, and snoRNAs. They participate in cytoplasmic polyadenylation (Rouhana et al. 2005), RNA degradation through the TRAMP complex (Hamill et al. 2010), addition of uridines to the 3′ end of U6 snRNA (Trippe et al. 2006) or adenosines to the 3′ end of snoRNA precursors (Berndt et al. 2012), and uridylation or adenylation of miRNAs and pre-miRNAs (Heo et al. 2008, 2012; D'Ambrogio et al. 2012).
Recently, it has been shown that TUT4 (ZCCHC11) and TUT7 (ZCCHC6) add short oligo U-tails to the oligo(A) tail remaining on mRNAs after deadenylation in mammalian cells, potentially marking them for degradation (Chang et al. 2014; Lim et al. 2014). Several different TUTases have been implicated in histone mRNA degradation by RNA interference experiments (Mullen and Marzluff 2008; Schmidt et al. 2011), although these studies were not definitive.
To better understand the nature of the oligouridylation of the 3′ end of histone mRNA, our laboratory developed a high-throughput, next-generation sequencing strategy to sequence histone mRNA degradation intermediates (EnD-seq) and a computational algorithm (AppEnD) to identify untemplated 3′ additions to RNAs (Welch et al. 2015). Our initial experiments with this method identified a broad range of uridylated degradation intermediates (Slevin et al. 2014) and showed that there is extensive mono- and diuridylation at the 3′ end of histone mRNA in S-phase to maintain the proper length of histone mRNA (Welch et al. 2015).
The goal of this study is to determine which TUTase or TUTases participate in uridylation of the 3′ end of histone mRNAs. Using RNAi to knock down different TUTases, combined with high-throughput sequencing to examine changes in degradation intermediates, we have identified TUT7 as the primary enzyme that adds uridines to both mature histone mRNA to maintain the proper length of the 3′ end and the initial degradation intermediates formed by 3′hExo. We also show evidence that suggests that TUT7 functions together with 3′hExo to coordinate the initial steps of degradation and urididylation. In contrast, knockdown of TUT4 had little effect on the uridylation at the 3′ end and only a small effect on uridylation further into the stem–loop.
RESULTS
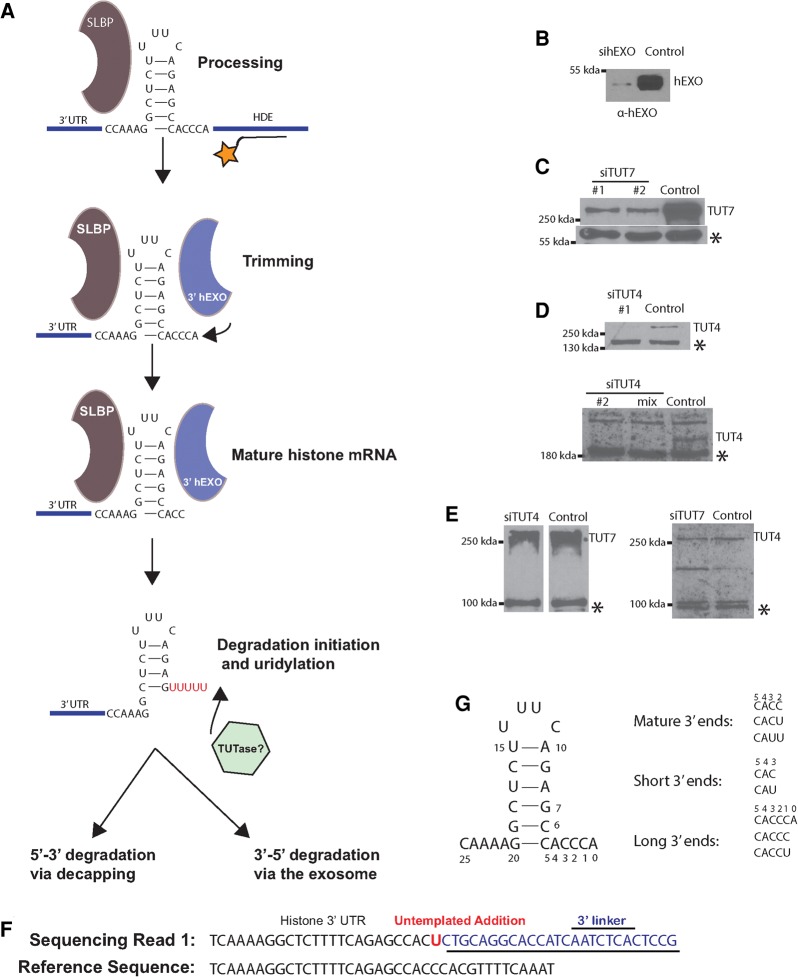
The metabolism of the 3′ end of histone mRNA is outlined in Figure 1A. Following cleavage 5 nt after the stem–loop in the nucleus, 2–3 nt are removed from the 3′ end by 3′hExo, resulting in the cytoplasmic histone mRNA. The length of the 3′ end of the mRNA is maintained by uridylation of any RNAs that are further trimmed. When DNA replication is inhibited, histone mRNA is rapidly degraded. The initial step in degradation is addition of an oligo(U) tail and degradation by the histone mRNA 3–4 nt into the stem. This degradation intermediate accumulates and is uridylated, resulting in rapid subsequent degradation of the histone mRNA by the exosome (Slevin et al. 2014).
FIGURE 1.
Histone mRNA metabolism. (A) A diagram of histone mRNA metabolism showing the steps in histone mRNA biosynthesis that result in a cytoplasmic histone mRNA ending 3 nt after the stem–loop, as a result of trimming the nuclear mRNA after processing by 3′hExo. The length of this 3′ end is maintained by addition of uridines to the 3′ ends of molecules that have been trimmed further. Degradation initiated by uridylation of the 3′ end of histone mRNA and exonucleolytic degradation into the stem by 3′hExo result in the accumulation of oligouridylated intermediates that are degraded 3′–5′ by the exosome or 5′–3′ by decapping. (B–E) Cells were treated with the indicated siRNAs or control siRNA for 2 (TUT7, 3′hExo) or 3 d (TUT4) and total cell protein analyzed by Western blotting. (B) 3′hExo, (C) TUT7, (D) TUT4 (top, siRNA 1; bottom, siRNA 2 and mix of siRNAs 1 and 2). (E, left) TUT4 depletion analyzed for TUT7. (Right) TUT7 depletion analyzed for TUT4. (*) Indicates cross-reacting bands that serve as a loading control. (F) Example of a sequencing read that allows unambiguous identification of untemplated nts at the 3′ end of a histone mRNA molecule. A primer is ligated onto the 3′ end of the RNA, and the sequences obtained contain the primer/RNA junction as previously described. Untemplated nts (in this case a single U) are nts between the last nt of genomic DNA and the first nt of the primer (Welch et al. 2015). (G) Structure of the most abundant 3′ ends on cytoplasmic histone mRNA with the numbering system for the 3′ SL used in this paper.
We examined two points in the histone mRNA life cycle: (i) the exonucleolytic trimming and uridylation of histone mRNA in the cytoplasm, and (ii) uridylation of degradation intermediates during 3′–5′ degradation of the histone mRNA. We used siRNAs to knock down 3′hExo (Fig. 1B), TUT7 (Fig. 1C), and TUT4 (Fig. 1D,E). We also determined that knockdown of TUT4 or TUT7 did not affect the level of the other TUTase (Fig. 1E). We used our high-throughput sequencing strategy (EnD-seq) and a bioinformatics analysis pipeline (AppEnD) to identify untemplated 3′ additions to histone mRNAs (Fig. 1F; Slevin et al. 2014; Welch et al. 2015) and to analyze changes in the 3′ ends of histone mRNAs that resulted from knockdown of different TUTases and 3′hExo. From the sequencing data, we determined the position of the 3′ end of RNAs and whether there were any untemplated nts. We plotted the data showing the position of the last templated nucleotide. The last nt of the processed histone mRNA (ending in ACCCA) is nt 0.
3′hExo and TUT7 create the short U-tails at the 3′ end of histone mRNA
We recently reported that one to two uridines are added to the 3′ end of histone mRNAs if it is trimmed to 1 or 2 nt from the end of the stem to maintain the overall length of histone mRNA (Welch et al. 2015). To determine how the short U tails are formed on the 3′ end of mature histone mRNA, we knocked down 3′hExo, TUT4, and TUT7 using siRNAs (Fig. 1B–E) and then sequenced and mapped the 3′ end of histone mRNAs using EnD-seq and AppEnD.
An example of a sequencing read that allowed unambiguous identification of a single nt tail is shown in Figure 1F. The 5′ end of the RNA sequence is aligned to the genomic DNA and the 3′ end to the linker ligated to the 3′ end, allowing identification of any untemplated nts. The various types of 3′ ends we observed on histone mRNA in exponentially growing cells are shown in Figure 1G. The majority of 3′ ends extend 3 nt beyond the stem, ending in ACC, ACU, or AUU. The ACU and AUU result from nontemplated additions of uridines after premature shortening of the histone mRNA.
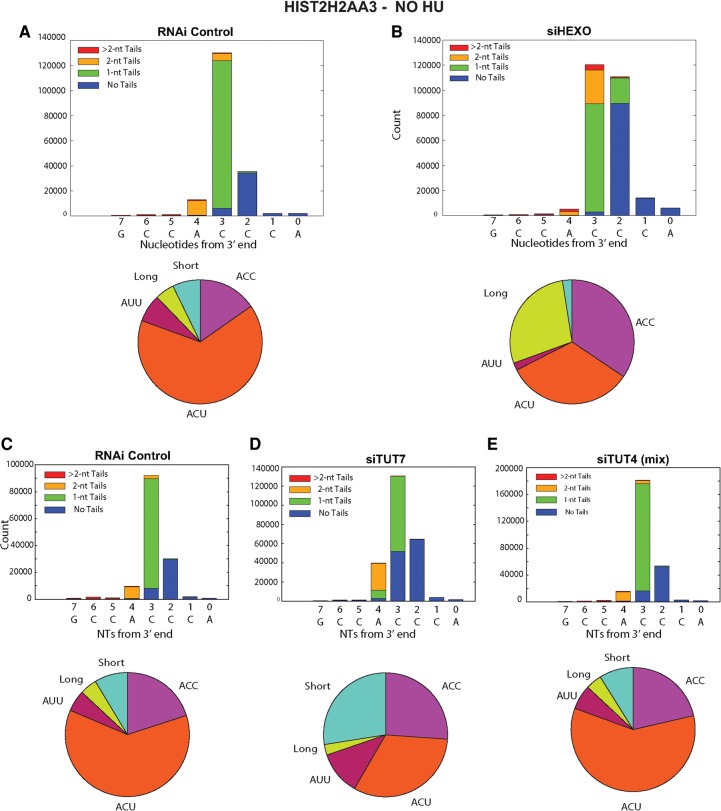
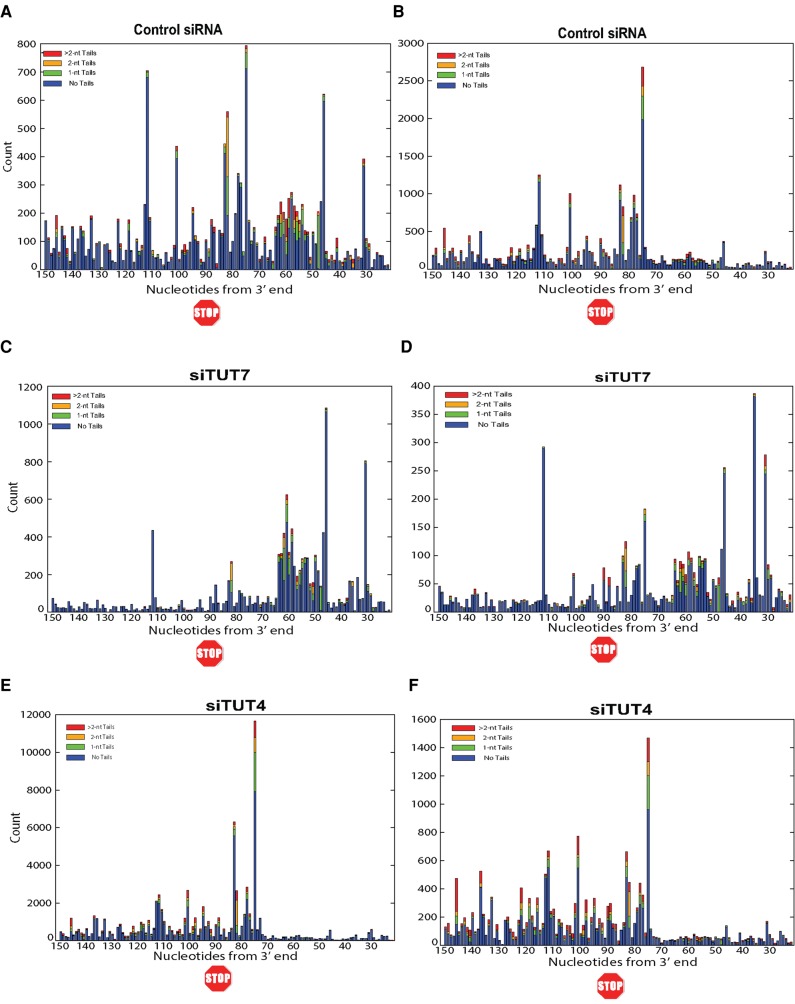
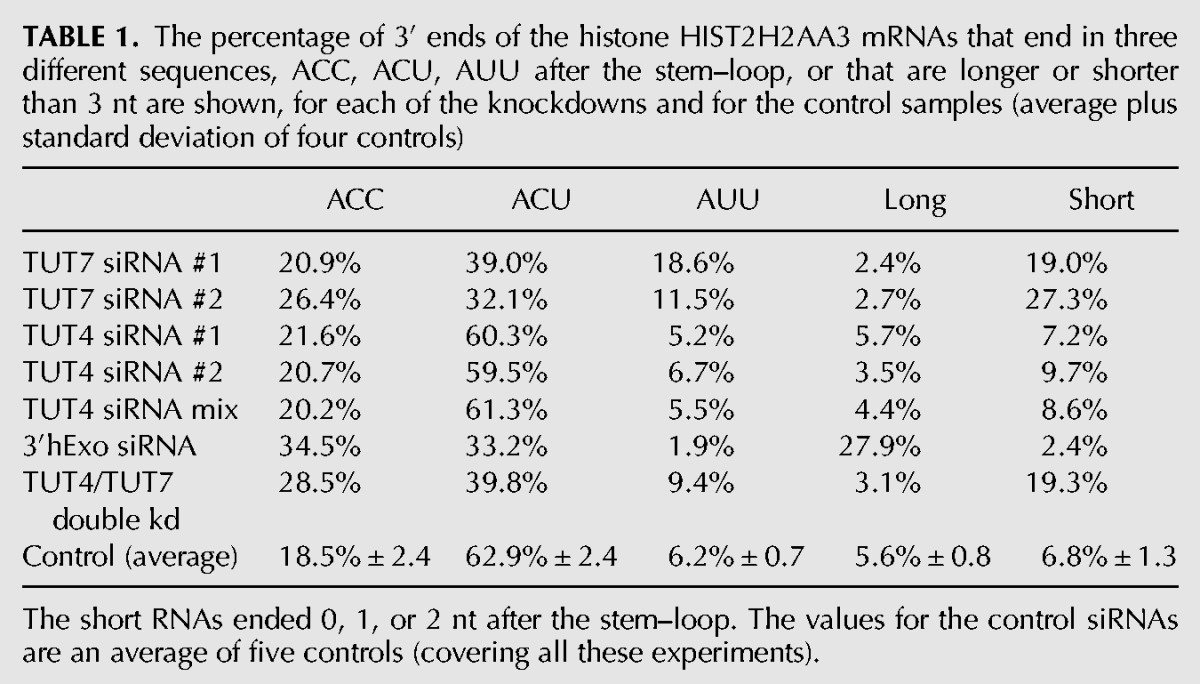
We made libraries from cells treated with a control siRNA and from knockdowns of TUT7, 3′hExo, and TUT4. The sequencing results for the HIST2H2AA3 mRNA are displayed in stacked bar plots of the 3′ end of the stem–loop (Fig. 2), showing the positions of the last templated nt, with the different colors representing untailed RNAs, RNAs with a single nontemplated U, two nontemplated U's, or more than two nontemplated U's (Fig. 2). Below each bar graph is a pie chart showing the distribution of the 3′ ends on the mRNA, and the results of the pie charts are summarized in Table 1 for both Figure 2 and Supplemental Figure 1. In Figure 2A, the RNA from control cells has primarily 3 nt after the stem–loop, ending primarily in ACC, ACU, or AUU, with the most frequent being ACU.
FIGURE 2.
Effect of knockdown of 3′hExo, TUT4, and TUT7 on the 3′ ends of HIST2H2AA3 mRNA. (A–E) The results of high-throughput sequencing showing the last 8 nt (with nt 0 the last A of the ACCCA) of the HIST2H2AA3 mRNA from control (A,C), 3′hExo knockdown (B), TUT7 knockdown siRNA 2 (D), and TUT4 knockdown with siRNA mix (E). The y-axis gives the number of sequencing reads at each position. The lengths of nontemplated tails (all uridines) at each position are indicated by the different shades in the inset. The RNAs shown in A and B were from cells cultured in parallel with the indicated siRNAs, and the RNAs shown in C–E were cultured in parallel. Pie charts showing the proportion of three different sequences, ACC, ACU, and AUU, of mRNAs ending 3 nt after the stem, and all longer or shorter mRNAs are under each of the bar graphs. The results are quantified in Table 1.
TABLE 1.
The percentage of 3′ ends of the histone HIST2H2AA3 mRNAs that end in three different sequences, ACC, ACU, AUU after the stem–loop, or that are longer or shorter than 3 nt are shown, for each of the knockdowns and for the control samples (average plus standard deviation of four controls)
Because 3′hExo has been implicated in trimming the 3′ end of processed histone mRNA, we anticipated that the distribution of 3′ ends on histone mRNAs in S-phase cells would be altered by knocking down 3′hExo. We knocked down human 3′hExo >90% by siRNA (Fig. 1B), then sequenced and mapped the 3′ end of histone mRNAs using EnD-seq and AppEnD. While this level of knockdown was shown in the past to not significantly affect the overall rate of histone mRNA degradation (Mullen and Marzluff 2008), a knockout of 3′hExo stabilizes histone mRNA and results in cytoplasmic histone mRNAs that end in 5 nt after the stem–loop (Hoefig et al. 2013).
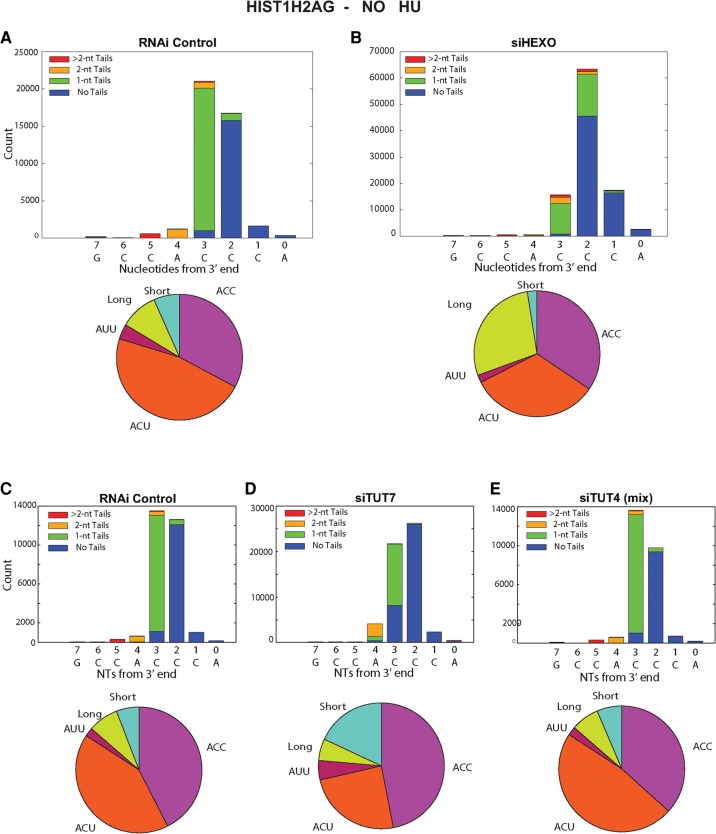
The knockdown resulted in several changes in the 3′ ends of histone mRNA compared with control siRNA, consistent with less 3′ trimming due to the decreased levels of 3′hExo (Fig. 2B). There is a small amount of histone mRNA that is untrimmed (ending in ACCCA), and a large increase in the histone message that is trimmed back by 1 nt to ACCC, and by 2 nt to ACC (Fig. 2B), resulting in an overall increase in long histone mRNAs. We quantified the changes in histone mRNA 3′ ends in the pie charts in Figure 2B and Table 1. In control cells only 18% of the HIST2H2AA3 message ends in ACC, compared to 35% of mRNA in the 3′hExo knockdown cells. A similar change occurred in the HIST1H2AG mRNA (Table 2; Fig. 3B) with an increase in message ending in ACC, from 33% to 45%. There are multiple nonallelic mRNAs for each core histone (Marzluff et al. 2002), and each mRNA showed a characteristic reproducible pattern of uridylation, which differed slightly for each mRNA, but the direction of the changes that occurred as a result of the TUT7 knockdown was similar for each histone mRNA (Table 3). All the mRNAs showed a decrease in the percentage of ends ending in ACU and an increase in the number of short histone mRNAs.
TABLE 2.
The percentage of 3′ ends of the histone HIST1H2AG mRNAs for the same samples analyzed in Table 1
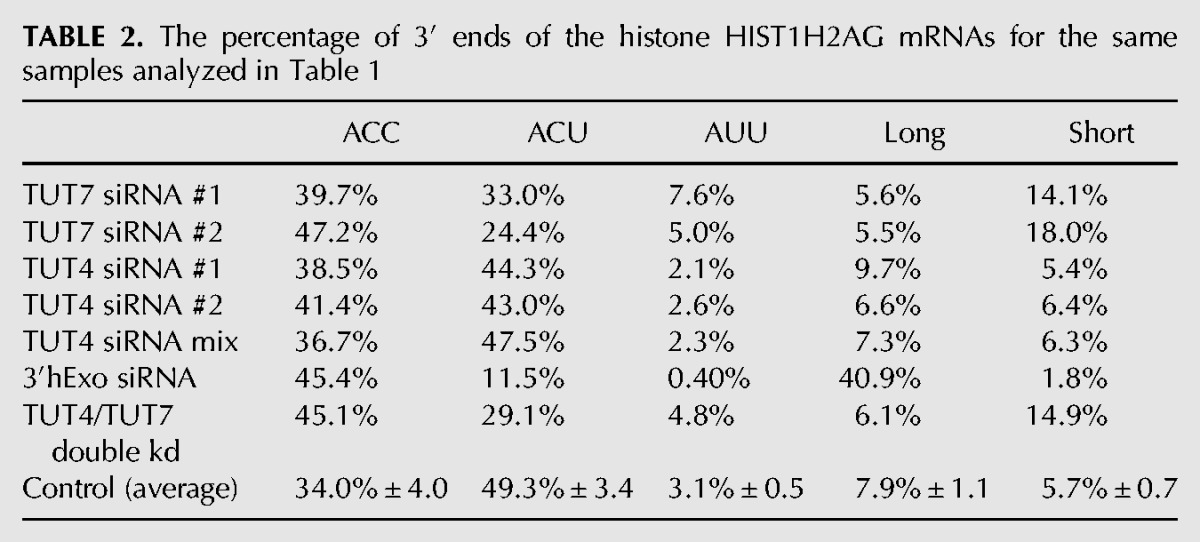
FIGURE 3.
Effect of knockdown of 3′hExo, TUT4, and TUT7 on the 3′ ends of HIST1H2AG mRNA. The results for the HIST1H2AG mRNA from the same sequencing experiments in Fig. 2 are shown from control (A,C), 3′hExo knockdown (B), TUT7 knockdown siRNA 2 (D), and TUT4 knockdown with siRNA mix (E) cells. The pie charts below the figure summarize the data. These are quantified in Table 2.
TABLE 3.
Changes in 3′ ends of H2a and H2b mRNAs as a result of knockdown of TUT7
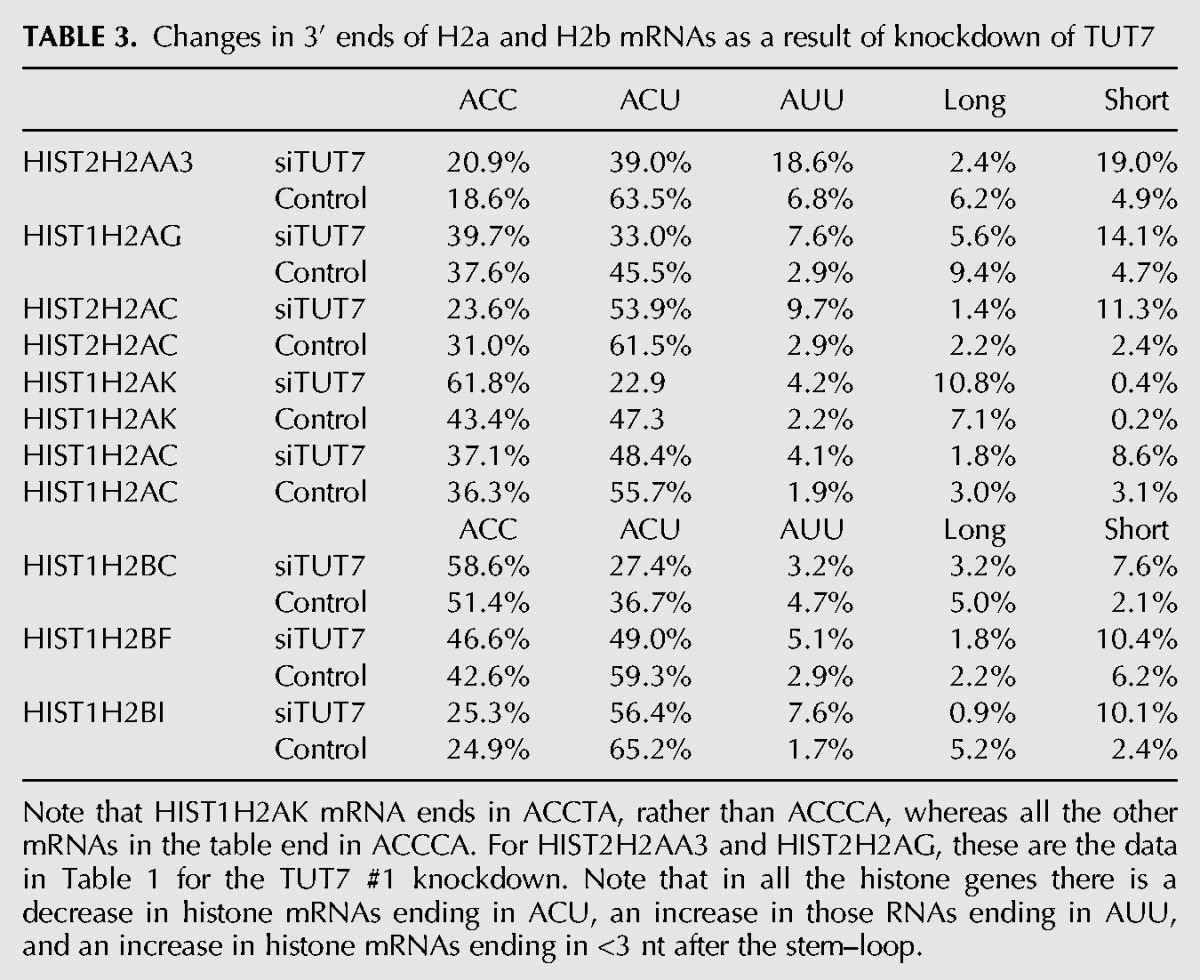
The 3′hExo knockdown also affected the number and position of the 1 and 2 nt U-tails added to the 3′ end of histone mRNA. In control cells, the majority of the 3′ ends have a short U-tail replacement of the cytosine residues, ending in ACU or, less commonly, AUU, resulting in the majority of the mRNAs (86%) extending 3 nt after the stem–loop. A similar distribution is seen in the HIST1H2AG gene, with ACC being more prevalent than in the HIST2H2AA3 mRNA, but with ACU still the major 3′ end (Fig. 3). In the 3′hExo knockdown, there was a four- to fivefold increase in the number of mRNAs that ended >3 nt after the stem–loop in both HIST2H2AA3 (28% compared with 6%) and HIST1H2AG (41% compared to 8%) (Fig. 3B). This increase resulted in fewer “mature” histone mRNAs that end 3 nt after the stem (70%, down from 88%) and fewer “short” messages ending 2 or fewer nts after the stem (2.4%, down from 6.8%), as seen in Table 1. Again, most (72%) of the HIST2H2AA3 molecules have been uridylated, with the uridylation often resulting in a 4 nt extension beyond the stem–loop comprised of both templated and untemplated nts.
The presence of RNAs with longer U tails in the 3′hExo knockdown suggests that the 1 and 2 nt tails that make up a large number of the reads at the 3′ end in the control result from a balance of uridylation and exonucleolytic trimming and not from addition of a specific number of nts by the TUTase.
TUT7 knockdown alters the pattern of U-tails at the 3′ end of histone mRNAs
We knocked down TUT7 >80% by siRNA (Fig. 1C), a knockdown with efficiency similar to that of the 3′hExo knockdown, although there was still residual enzyme present in the cells. The knockdown had two major effects on the 3′ end of histone mRNA in S-phase cells. There was a threefold increase in the proportion of H2AA3 mRNAs in the TUT7 knockdowns that were shorter than the ACC/ACU/AUU 3′ ends (from 7% in the control to >20% in the knockdown) (Table 1; Fig. 2D), suggesting that 3′hExo shortened the 3′ end but the depletion of TUT7 left the cell with a reduced ability to uridylate the 3′ end and properly restore the length of histone mRNA. This treatment resulted in reduction of both the number of mature and “long” histone mRNAs (Table 1; Fig. 2D). In the control siRNA cells, ∼75% of the HIST2H2AA3 mRNAs had nontemplated uridines at the 3′ end (Table 1; Fig. 2C). When TUT7 was depleted, that number dropped to 61%. There was a similar effect in H2AG mRNAs (Table 2; Fig. 3D).
Note that there was an increase in the percentage of histone mRNAs ending in AUU on all the histone mRNAs after the TUT7 knockdown (Tables 1–3). This paradoxical finding could result from an increased likelihood of degradation by 3′hExo further into the tail, followed by addition of 2 U's to restore the proper length of the 3′ end.
Knockdown of TUT4 had minimal effects on uridylation of histone mRNAs at the 3′ end
TUT4 is a very similar protein to TUT7, both structurally and functionally. It has been previously suggested that TUT4 and TUT7 can compensate for each other in uridylation of miRNAs and pre-miRNAs (Thornton et al. 2012). When we knocked down TUT4, there was little effect on the 3′ end of H2AA3 mRNAs when compared to the siRNA control. Neither the length of the mRNAs nor the pattern of uridylation was changed on HIST2H2AA3 mRNA or HIST1H2AG mRNA (Tables 1, 2; Figs. 2E, 3E).
Inhibition of DNA replication does not alter uridylation at the 3′ end of histone mRNA
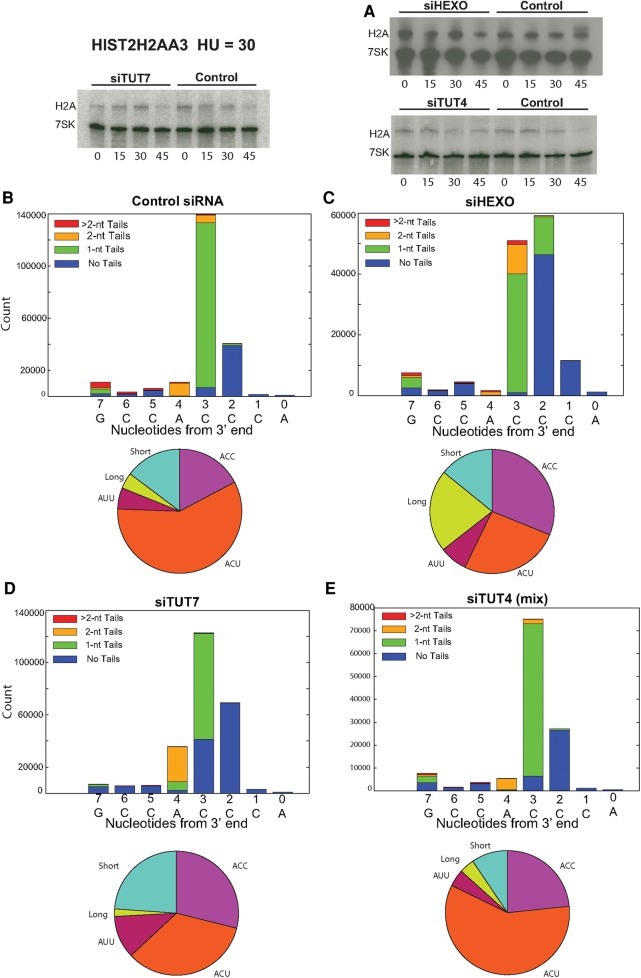
We treated cells with hydroxyurea (HU) to inhibit DNA replication and initiate histone mRNA degradation, which results in extensive uridylation of histone mRNA degradation intermediates (Slevin et al. 2014). Knockdowns of TUT7, TUT4, or 3′hExo each had minimal effects on the overall kinetics of histone mRNA degradation when DNA replication was inhibited (Fig. 4A). Previously, we had analyzed knockdown of 3′hExo, TUT4, and TUT7, and found no significant effect on the overall rate of histone mRNA degradation (Mullen and Marzluff 2008), although a subsequent knockout of 3′hExo in mice demonstrated that it is essential for histone mRNA degradation (Hoefig et al. 2013). We reasoned that although the small amount of residual enzyme was sufficient to promote degradation, that reduction in enzyme levels might alter the pattern of degradation intermediates. To determine the effect of knockdown of 3′hExo, TUT4, or TUT7 on the pathway of histone mRNA degradation, we analyzed the spectrum of degradation intermediates found during histone mRNA degradation by high-throughput sequencing. The samples analyzed had <50% of the histone mRNA degraded as analyzed by Northern blotting. In addition to uridylation of the 3′ ends, there is extensive uridylation of degradation intermediates in the 3′ side of the stem.
FIGURE 4.
The 3′ ends of the full-length histone mRNAs are not affected by inhibiting DNA replication. Exponentially growing cells treated with a control siRNA or with siRNAs against 3′hExo, TUT4, or TUT7 were treated with HU and cells harvested before treatment and 15, 30, or 45 min after HU treatment. (A) Histone mRNA levels were determined by Northern blotting and the blot probed by hybridization with probes for histone H2a mRNA and 7SK RNA as a loading control. (B—E) Libraries were prepared from RNA isolated 30 min after HU treatment from control (B), 3′hExo knockdown (C), TUT7 knockdown (D), and TUT4 knockdown (E) cultures. The reads from the 3′ end of the mRNAs (nts 1–8) were plotted as in Figure 2. The TUT4 siRNA and TUT7 siRNAs were the siRNAs used in Figure 2.
After HU treatment, there was no change in the distribution of 3′ ends (nts 1–4 after the loop) compared to the same cells analyzed before HU treatment (cf. Fig. 4B–E and Fig. 2A–D; cf. Table 1 and Table 4). This was true for the control cells, as well as the TUT7, 3′hExo, and TUT4 knockdown cells. Note that the increase in molecules seen at positions 5–7 in Figure 4 is due to degradation intermediates (see Fig. 5), which are not included in Table 4. This result suggests that there is not a preference for initiating degradation among the histone mRNAs ending in AUU, ACU, or ACC. Thus the mono- and diuridylations that maintain the histone mRNA at the same length as the “normal,” trimmed ACC message are not degradation intermediates, but rather mature histone mRNAs with slightly different 3′ ends, which are formed by the activity of 3′hExo and TUT7.
TABLE 4.
The percentage of 3′ ends of the histone HIST2H2AA3 mRNAs after treatment of the cells in Table 1 with HU for 30 min
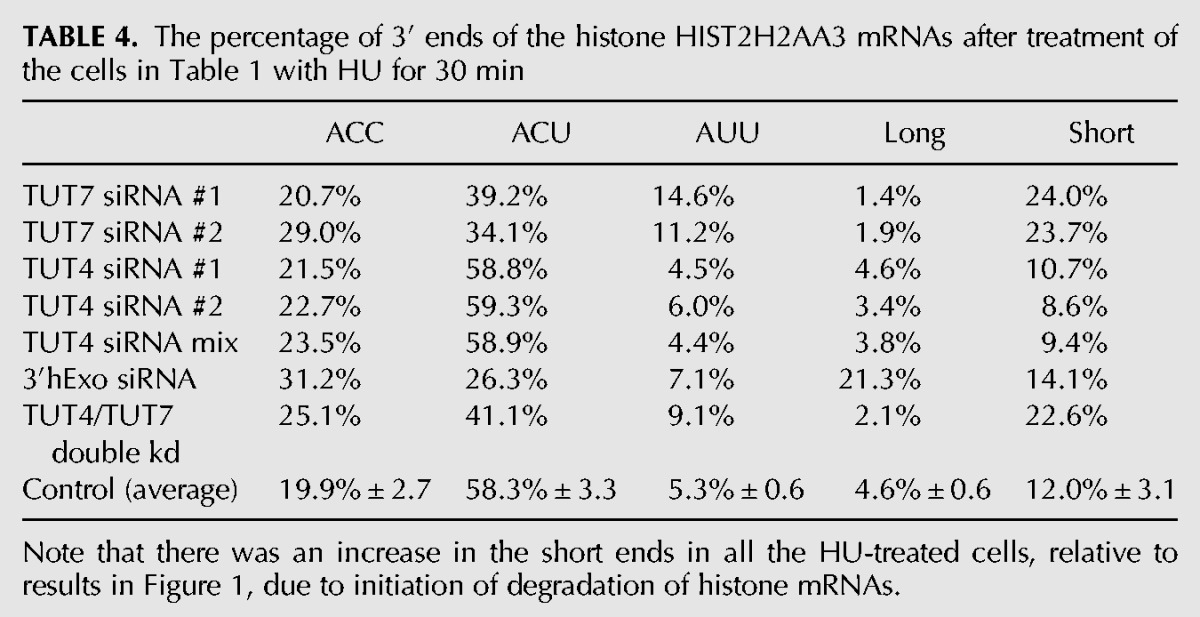
FIGURE 5.
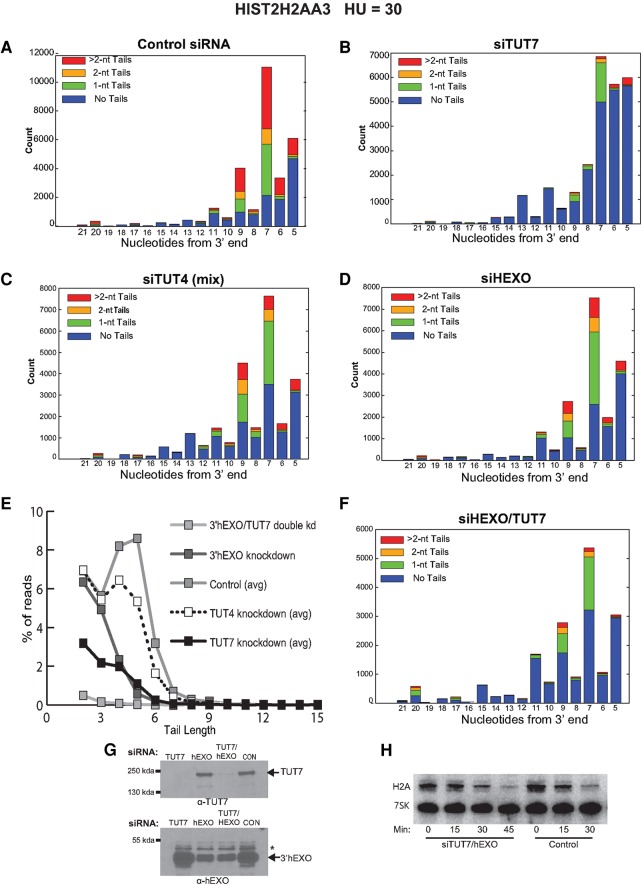
Knockdown of TUT7 and 3′hExo changes the distribution of HIST2H2AA3 degradation intermediates in the stem–loop. (A–D) The distribution of total reads for the HIST2H2AA3 gene in each experiment in Figure 4B–E between nts 5 and 21 for the experiments shown in Figure 4 is shown. Nucleotide 5 is the C at the 3′ side of the base of the stem; nt 20 is the G at the 5′ side of the base of the stem (Fig. 1B). Control sample (A), knockdown of TUT7 (B), TUT4 (C), and 3′hExo (D). The red portion of the bar graphs has nontemplated U-tails >2 nt. (E) The percentage of total reads with the indicated tail lengths is plotted for the H2AA3 gene for five controls (average) and the experiments in A–D and F. (F) TUT7 and 3′hExo were simultaneously knocked down, the cells treated with HU, and the sequences of the 3′ ends of the histone HIST2H2AA3 mRNAs between nts 5 and 21 plotted. (G, left panel) Protein extracts from TUT7, 3′hExo, and the TUT7/3′hExo double knockdown were analyzed by Western blotting after resolution on a 6% SDS-polyacrylamide gel. The filter was cut and incubated with either the TUT7 (top) or the 3′hExo (bottom) antibody. (H) Northern blot analysis after the RNA from control siRNA-treated cells and the TUT7/3′hExo double knockdown cells is shown, 15, 30, and 45 min after treatment with HU.
Knockdown of TUT7 affects the initial step in degradation of histone mRNA
The initial intermediate in histone mRNA that accumulates results from degradation into the 3′ side of the stem by 3′hExo (Fig. 5A; Hoefig et al. 2013; Slevin et al. 2014). In Figure 5A–E, we show the degradation intermediates present at nts 5–20 in each of the siRNA knockdowns, removing the remaining mature histone mRNAs from the analysis. In control cells, the bulk of the intermediates are found at positions 5–9 from the 3′ end (Slevin et al. 2014), with the most abundant intermediate and the most tails found at position 7, the G 3 nt into the stem. These intermediates are extensively uridylated (Fig. 5A) and have a large fraction of longer tails (4–6 nt), with tails extending to >10 nt (Fig. 5F). In control cells, there were very few intermediates between nts 10 and 19, encompassing the loop and 5′ side of the stem. This suggests that once degradation of this intermediate is initiated, degradation proceeds rapidly through the rest of the stem–loop. Our previous study suggested that these uridylated intermediates are targets for degradation by the exosome (Slevin et al. 2014).
When TUT7 was knocked down, there was both a change in the distribution of the intermediates as well as many fewer uridylated RNAs. In addition, the average tail length of the uridylated RNAs was shorter (Fig. 5B,E). There was a substantial increase in intermediates that had only been degraded to nts 5–6 and an increase in intermediates that had been degraded >9 nt into the stem–loop. Surprisingly, knockdown of 3′hExo also affected uridylation of the degradation intermediates (Fig. 5D,E). The pattern of intermediates was similar to control cells, but both the number and length of the tails were reduced (Fig. 5D,E). A difference between the 3′hExo knockdown and the TUT7 knockdown was that in the 3′hExo knockdown there was no further degradation into the stem–loop—the degradation intermediates accumulated in the 7–9 nt region, as they did in the controls. When we knocked down both TUT7 and 3′hExo simultaneously (Fig. 5F), we saw a further decrease in the number of tails as well as fewer molecules degraded further into the stem–loop compared with the TUT7 knockdown. This result is consistent with degradation past nt 9 resulting from additional digestion by 3′hExo. Note that the knockdown of TUT7 did not affect the levels of 3′hExo and vice versa (Fig. 5G), and the double knockdown did not have an effect on the overall rate of histone mRNA degradation (Fig. 5H). Taken together, these results suggest that 3′hExo and TUT7 work together in degrading the histone mRNA into the stem–loop (see Discussion).
In contrast to the TUT7 knockdown, knockdown of TUT4 did not change the distribution of degradation intermediates between nts 5 and 9, although it did have an effect on the overall level of uridylation in that region (Fig. 5C,E). There was also an increase in the intermediates past nt 11, although not as large as the increase in the TUT7 knockdown. These results suggest that TUT4 contributes to the uridylation of histone mRNA but the effect is minor compared to the effect of TUT7. In particular, TUT7 has a much larger effect on the distribution of degradation intermediates (accumulating more intermediates ending at nts 5–7), while the distribution of intermediates in the TUT4 knockdown was similar to the control. These results are consistent with TUT7 being the primary enzyme that adds the U-tails to the stem–loop during degradation of histone mRNA.
TUT7 depletion does not result in accumulation of intermediates in the coding region
Degradation of histone mRNA 3′–5′ can be divided into two stages: (i) degradation into the stem–loop by 3′hExo with concomitant uridylation of the degradation intermediates, and (ii) rapid degradation to just 3′ of the ribosome by the exosome, followed by pausing and uridylation of the degradation intermediates, which continues as the exosome goes through the coding region (Slevin et al. 2014). In control cells, there are fewer degradation intermediates detected between the 3′ side of the stem–loop and a position in the 3′ UTR ∼15 nt from the termination codon (Fig. 6A,B) than in positions further into the 3′ UTR, which we have previously argued is likely due to arrest of degradation by the terminating ribosome (Slevin et al. 2014). The relative proportion of these two degradation intermediates reflects the flux of intermediates through the pathway. A change in proportion of the two populations of intermediates will occur when there is inhibition or acceleration of one of the steps. For example, when the exosome component Pml/Sc100 was knocked down, there was an increased accumulation of uridylated intermediates in the stem–loop (Slevin et al. 2014), consistent with degradation through the 3′ UTR and coding region being catalyzed by the exosome.
FIGURE 6.
Effect of TUT7 and TUT4 knockdown on the distribution of histone mRNA degradation intermediates in the 3′ UTR and open reading frame. RNA was prepared from cells treated with control siRNA (A,B), TUT7 knockdown cells (C,D), and TUT4 knockdown cells (E,F). Cells were treated with HU and RNA prepared from cells 30 min after HU treatment; panels A,C,E are the total reads from the HIST2H2AA3 mRNA from nts 21 to 150 for the samples shown in Figure 4; and panels B,D,F are from a parallel set of experiments.
In the TUT7 knockdown, there were many more intermediates before the ribosome stalling site, with a prominent number of reads between nts 50 and 65 (Fig. 6C,D) and with many fewer reads after nt 75 than in the control cells. This is consistent with TUT7 playing a critical role in the accumulation and ultimate degradation of the intermediates in the stem–loop, as a result of uridylation of the intermediates. Thus, the generation of intermediates in the 3′ UTR and coding region is slowed, and they may be removed by the exosome in a TUT7-independent manner.
A different effect was seen in the TUT4 knockdown. The TUT4 knockdown shows a similar pattern to controls, with a gap between the stem–loop and a prominent peak 15 nt 3′ of the terminating ribosome, and intermediates through the coding region (Fig. 6E,F). There was not a significant effect of TUT4 or TUT7 knockdown on the proportion of intermediates that were uridylated or the length of the U-tails in this region. Whether TUT4 plays a role in degradation of these intermediates (and whether uridylation of these intermediates is required for their degradation) could not be determined.
We conclude that TUT7 plays a major role in histone mRNA metabolism. It is responsible for both the uridylation of the 3′ end of the histone mRNA to maintain the proper length of histone mRNA when the mRNA is relatively stable, and for the uridylation of degradation intermediates during the initial steps of degradation by 3′hExo into the stem. In both these steps, TUT7 and 3′hExo likely work together. Knockdown of TUT4 has no effect on the uridylation of the 3′ end of histone mRNA in growing cells, and only a small effect on uridylation of histone mRNA degradation intermediates, and pattern of degradation intermediates.
DISCUSSION
A major regulatory step in histone mRNA metabolism is regulation of histone mRNA half-life to maintain the coupling of histone mRNA levels with the rate of DNA replication. There is coordinated degradation of the family of replication-dependent histone mRNAs, which is mediated through the stem–loop at the 3′ end of the mRNA, and the proteins bound to the mRNA, SLBP, and 3′hExo. This degradation of histone mRNA requires that the mRNA be actively translated (Graves et al. 1987; Kaygun and Marzluff 2005b). Histone mRNA degradation also requires many of the same factors required for degradation of polyadenylated mRNAs, including Lsm1-7 and the exosome. Recruitment of Upf1, a key factor in nonsense-mediated decay, is an early step in triggering degradation (Kaygun and Marzluff 2005a). Individual knockdowns of Upf1, Lsm1-7, and the exosome all have a greater effect on degradation than knocking down components of the 5′–3′ decay pathway, consistent with a major pathway of histone mRNA degradation proceeding 3′–5′ (Mullen and Marzluff 2008). In addition, the early steps of histone mRNA degradation occur while the mRNA is capped and associated with polyribosomes (Slevin et al. 2014).
Uridylation of the 3′ end of histone mRNA plays a prominent role in histone mRNA metabolism. Following 3′ end formation in the nucleus, resulting in histone mRNAs ending in ACCCA, SLBP and 3′hExo form a stable complex on the 3′ end of histone mRNA (Yang et al. 2006; Tan et al. 2013), and 3′hExo trims 2 nt off histone mRNA, resulting in the major cytoplasmic histone mRNA that ends 3 nt after the stem–loop (Mullen and Marzluff 2008; Hoefig et al. 2013). It is not clear whether export precedes trimming, since 3′hExo is found in both the nucleus and the cytoplasm. Our data shows that the length of cytoplasmic histone mRNA is maintained by uridylation of the mRNA by TUT7. If the mRNA is trimmed to shorter than 3′ nt after the stem–loop, uridines are added to restore the length. In S-phase, the most abundant 3′ ends for many histone mRNAs have a nontemplated uridine on the 3′ end. Knockdown of TUT7 results in a larger number of histone mRNAs with fewer than 3 nt after the stem–loop. TUT4 knockdown had no effect on the uridylation or length of the cytoplasmic histone mRNAs. If >3 nt are removed at this step, uridylation by TUT7 restores the length of the mRNA to 3 nt beyond the stem–loop (Fig. 7). This process may be reiterated during the life span of a histone mRNA.
FIGURE 7.
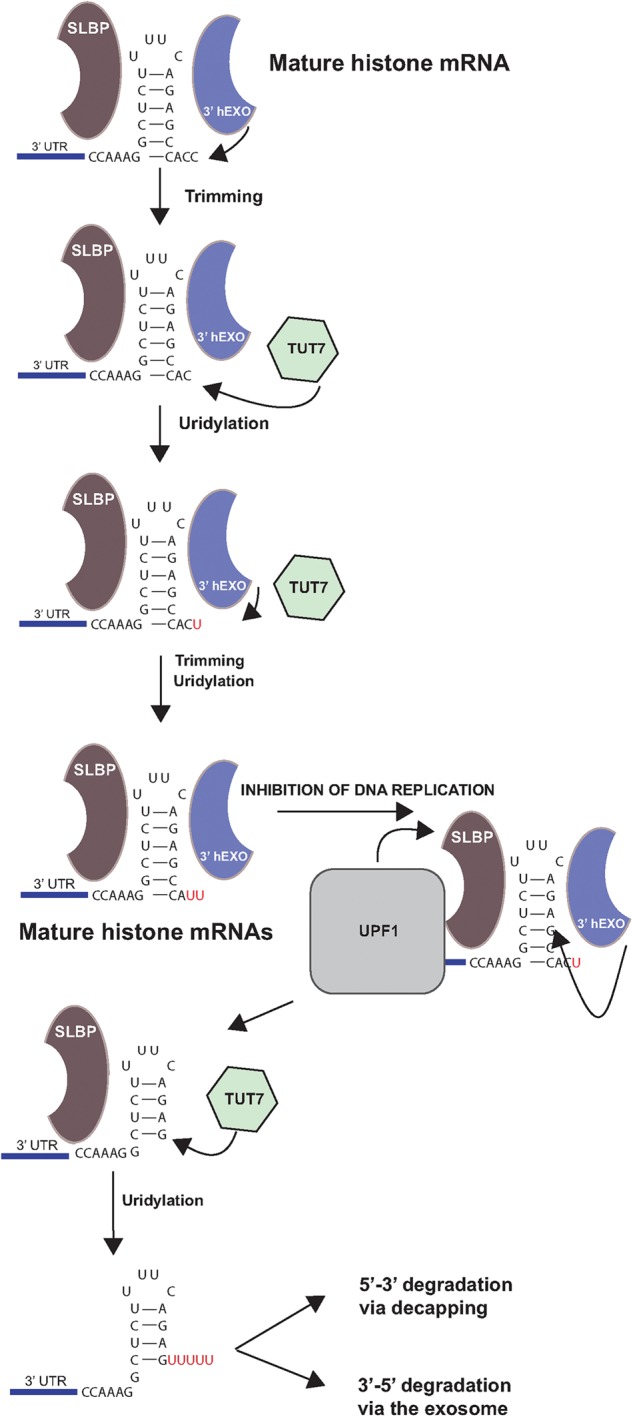
Model for uridylation of the 3′ end of histone mRNA. The 3′ end of histone mRNA is formed by trimming by 3′hExo. If 3′hExo continues trimming, the length of the 3′ end is restored by the action of TUT7 to add uridines to return them to the proper length. Several cycles of this may occur. When DNA replication is inhibited, Upf1 is recruited to the 3′ end of histone mRNA, the 3′ end of the mRNA is uridylated and then 3′hExo degrades into the stem. This intermediate is uridylated by TUT7 and then subsequently degraded either 5′–3′ or 3′–5′.
Thirty years ago Jeff Ross and coworkers provided substantial evidence that histone mRNA degradation is initiated by exonucleolytic degradation of the 3′ end of histone mRNA (Ross and Kobs 1986; Ross et al. 1986) and partially purified a polyribosome-associated exonuclease, which most likely was 3′hExo (Caruccio and Ross 1994). Initiation of histone mRNA degradation requires oligouridylation of histone mRNA, and oligouridylated full-length (5 nt after the stem–loop) histone mRNA accumulates in the 3′hExo knockout (Hoefig et al. 2013), suggesting that uridylation of the 3′ end is a critical step in initiating degradation (Su et al. 2013). The initial intermediate that accumulates during degradation results from partial degradation into the stem–loop (Ross et al. 1986; Hoefig et al. 2013; Slevin et al. 2014) by 3′hExo. This intermediate is extensively uridylated (Hoefig et al. 2013; Slevin et al. 2014). Since cytoplasmic histone mRNAs end in a stem–loop with only 2–3 nt after the stem, they cannot bind Lsm1-7 without the addition of >5 nt to the cytoplasmic mRNA (Lyons et al. 2014). The requirement for Lsm1-7 in degradation of histone mRNAs results from the addition of an oligo(U) tail to the 3′ end and/or to the initial degradation intermediate in the stem, providing a binding site for Lsm1-7. The tails that accumulate on this intermediate are close to the size needed to bind Lsm1-7, suggesting that once they get long enough to bind Lsm1-7, further degradation can take place.
Previous studies using RNAi knockdown of the enzymes potentially involved in histone mRNA degradation (3′hExo, different TUTases) showed at best modest effects on the overall rate of histone mRNA degradation (Mullen and Marzluff 2008; Schmidt et al. 2011). More significant effects are observed with knockdown of factors also required for degradation of many other mRNAs, such as Upf1, Lsm1, and the exosome components (Kaygun and Marzluff 2005a; Mullen and Marzluff 2008; Slevin et al. 2014). The results we obtained with knockdown of 3′hExo illustrate this most clearly since very effective knockdown of this enzyme (>90%) had no significant effect on the rate of histone mRNA degradation, as detected by Northern blotting (Dominski et al. 2003; Mullen and Marzluff 2008). Only when 3′hExo was knocked out was histone mRNA degradation severely affected (Hoefig et al. 2013). The knockout cells contained histone mRNA that was not trimmed (had 5 nt after the stem) and was uridylated at the 3′ end (but not degraded) demonstrating a critical role for 3′hExo in both forming the 3′ end of cytoplasmic histone mRNA and initiating histone mRNA degradation (Hoefig et al. 2013).
We and others have obtained similar inconclusive results in RNAi knockdowns of the putative TUTases, with at best modest effects on histone mRNA degradation (Mullen and Marzluff 2008; Schmidt et al. 2011). The development of methods to analyze degradation intermediates by high-throughput sequencing has allowed us to look for changes in the distribution of intermediates and uridylation in mRNA degradation as a result of knockdown of different factors. Since we have identified discrete intermediates in histone mRNA degradation, we were able to assess changes in the relative amounts of different classes of intermediates, which are reflected in the relative proportion of different intermediates. For example, we previously showed that when the exosome subunit Pml-Scl100 is knocked down, the relative proportion of uridylated intermediates in the stem increased (Slevin et al. 2014), consistent with their being the initial targets of the exosome.
The experiments reported here show that knockdown of TUT7 and 3′hExo result in significant changes in the uridylation of the 3′ end of histone mRNA to maintain the normal length of mature histone mRNA and in the distribution of mRNA degradation intermediates. These results strongly suggest that TUT7, and not TUT4, plays the major role in uridylation of histone mRNA, both to maintain the normal length of the 3′ end in S-phase cells and to uridylate prominent degradation intermediates. Our data do suggest a minor role for TUT4 in the uridylation of histone mRNA degradation intermediates, but the results indicate that it is a secondary or complementary role to the major role played by TUT7.
TUT4 and TUT7 are similar proteins, which have been reported to uridylate both pre-miRNAs and mature miRNAs (Heo et al. 2009; Thornton et al. 2012; Kim et al. 2015). The relative amounts of TUT4 and TUT7 proteins in cells are not known. Inspection of sequencing data for several human cells and tissues shows that TUT7 mRNA is present in about 1.3 times higher levels than TUT4 mRNA, consistent with the two proteins present in similar amounts in most cells. In a recent study, Narry Kim and coworkers showed that both TUT4 and TUT7 add the U-tails present on some oligoadenylated mRNAs after deadenylation (Chang et al. 2014; Lim et al. 2014), marking them for degradation. These enzymes seem to be largely interchangeable for these uridylations, as shown by RNAi and in vitro uridylation experiments. The enzymes have also been reported to have specific functions as well. TUT7 has been shown to add a single uridine to specific miRNA precursors by Kim and coworkers (Heo et al. 2012). They also reported that TUT4 specifically uridylates let-7 pre-miRNA through an interaction with Lin-28 (Heo et al. 2009), although it has also been reported that TUT7 can compensate for this function of TUT4 (Thornton et al. 2012).
Knockdown of TUT7 dramatically changed the uridylation pattern of histone mRNAs, both in the mature mRNAs and in the initial intermediates formed after inhibition of DNA replication, while knockdown of TUT4 had no effect on uridylation in growing cells and only a small effect after the inhibition of replication. The selectivity for TUT7 in these reactions suggests that TUT7 may interact directly with components involved specifically in cytoplasmic histone mRNA metabolism, SLBP, and 3′hExo. The small effect observed on the degradation intermediates with TUT4 knockdown suggests that it may play a minor role in this step. TUT4 knockdown resulted in slightly shorter U tails and an increase in intermediates farther into the stem–loop. The accumulation of the intermediates in the stem–loop is likely due to the continued presence of SLBP and 3′hExo on the 3′ end of the mRNA. Once these proteins are removed (possibly by the action of Upf1), there may no longer be any selectivity for TUT7. Thus, we conclude that TUT7 is the major enzyme in the cell that uridylates histone mRNA, both on the stem–loop to maintain the proper length of the 3′ end and in the stem after initial degradation by 3′hExo to trigger further degradation of histone mRNA.
The role of uridylation in degradation of the intermediates after the stem–loop is not clear. In the TUT7 knockdown, there is a greatly reduced number of intermediates just after the stop codon and in the coding region, consistent with slow degradation through the stem–loop, resulting in a slow rate of formation of these intermediates, allowing them to be efficiently removed. In the TUT4 knockdown these intermediates accumulate, but there is no effect on their uridylation (either in the proportion of molecules uridylated or in the length of the U-tails). It is possible that either TUT7 or TUT4 could uridylate these intermediates because they would look identical to any other capped RNAs that had been cleaved or deadenylated. In 3′–5′ degradation of the heat-shock mRNAs in Drosophila, many of the intermediates have short A tails added (Harnisch et al. 2016), and addition of nontemplated nts may be a common feature of RNAs undergoing 3′–5′ degradation.
TUT7 and 3′hExo may collaborate in the uridylation of histone mRNA
In addition to clearly defining roles for TUT7 and hExo in the uridylation and degradation of histone mRNA, several aspects of our data suggest that the two enzymes may work together during degradation. Knockdown of 3′hExo unexpectedly altered the pattern of uridylation on the degradation intermediates in the stem–loop, reducing both the number and length of the tails. This change in number and length of tails was similar to the TUT7 knockdown, even though the level of TUT7 was unchanged in these cells. Knockdown of 3′hExo also altered the uridylation of the 3′ end of histone mRNA in growing cells, with an increase in histone mRNAs ending in ACCU and ACUU, suggesting that TUT7 may add a nonspecific number of uridines to the histone mRNAs that are then trimmed back by 3′hExo. These results are consistent with the possibility that 3′hExo and TUT7 function together both to maintain the normal length of the 3′ end in S-phase cells and to uridylate the degradation intermediates in the stem–loop as part of the pathway degradation.
Figure 7 shows our current model for histone mRNA uridylation and initiation of degradation. In S-phase cells, TUT7 uridylates the 3′ end of histone mRNA to compensate for shortening of the 3′ end by 3′hExo. When DNA replication is inhibited, there is a change in the histone mRNP, due to Upf1 activity and/or modification of SLBP, allowing further digestion by 3′hExo. Once 3′hExo digests into the stem–loop and stalls, TUT7 adds a longer oligo(U) tail on the partially degraded stem–loop, which is able to bind Lsm1-7, marking the message for degradation.
An early step in histone mRNA degradation is likely the recruitment of Upf1 to the histone mRNP as a result of inefficient translation termination (Kaygun and Marzluff 2005b). Upf1 binds to the histone mRNP by binding to the 3′ UTR (Zund et al. 2013; Brooks et al. 2015) and to SLBP (Kaygun and Marzluff 2005a) after DNA synthesis is inhibited. How the initial degradation into the stem by 3′hExo is activated or how the TUTase is recruited is not known, but it requires the helicase activity of Upf1 which may perturb the binding of SLBP to the stem–loop, allowing initial degradation by 3′hExo into the stem. Since 3′hExo and SLBP both interact with the C-terminal tail of Lsm4 in Lsm1-7 (Lyons et al. 2014), TUT7 and 3′hExo together may also interact with the Lsm1-7 complex to promote recruitment of this complex to the 3′ end of the degradation intermediates in the stem–loop, which may then result in subsequent degradation by the exosome.
The initial step in degradation of polyadenylated mRNAs is deadenylation, leaving an oligoA-tail which can bind Lsm1-7. Recent studies found that a fraction of the shortened A tails have one or two uridines added to the 3′ end, both in fission yeast (Rissland and Norbury 2009) and in mammalian cells (Chang et al. 2014; Lim et al. 2014), suggesting that oligouridylation may participate in degradation of polyadenylated mRNAs as well, although as yet there are no examples of specificity for uridylation of specific subsets of mRNAs.
MATERIALS AND METHODS
Cell culture and RNA interference
RNA interference experiments were done in HeLa cells using Invitrogen's Lipofectamine RNAiMAX for reverse transfections. The sequences of the siRNAs used are shown in Table 5. In a six-well dish, 5 µL of the Lipofectamine reagent and 100 pmol of siRNA were mixed in 500 µL of serum-free OPTI-MEM media for 20 min. After 20 min, 200,000–250,000 cells were plated directly onto the transfection mix, a concentration that resulted in them being ∼70% confluent the next day. The day after transfection, the cells were trypsinized and replated on 10 cm2 plates so that they would be below 50% confluency for the ensuing HU treatment. A single plate was used for each time point. Forty-eight hours later, the cells were treated with 5 mM HU and RNA was harvested from the cells. A parallel plate was harvested to make lysates for protein analysis by Western blotting. Proteins were resolved by electrophoresis on 8% SDS-polyacrylamide gels (3′hExo knockdowns) or 5% SDS-polyacrylamide gels for some of the TUTase knockdowns. The antibody previously described (Dominski et al. 2003) that we raised against 3′hExo, an antibody to PTB (gift of Mariano Garcia-Blanco), or antibodies to TUT4 (Bethyl labs A302-636) or TUT7 (Proteintech 25196-1-AP) were used, with PTB serving as the loading control in most experiments.
TABLE 5.
siRNA sequences used in siRNA knockdown experiments

Preparation of samples for high-throughput sequencing
Forty-eight hours after siRNA transfection, the cells were treated with 7.5 mM hydroxyurea to stop DNA synthesis. RNA was harvested before HU addition and at 15, 30, and 45 min. The amount of degradation was analyzed by Northern blotting as previously described, mixing probes for histone H2a mRNA and 7SK RNA as an internal control (Mullen and Marzluff 2008) to determine the amount of degradation. Two time points from each experiment were chosen for high-throughput sequencing before HU treatment and usually at the 30 min time point, when about half of the histone mRNA had been degraded. Libraries specific for histone H2a and H2b mRNAs were prepared as previously described (Slevin et al. 2014; Welch et al. 2015) and analyzed on a MiSeq using 125 nt paired end sequencing.
Mapping EnD-seq data
We mapped EnD-seq data to hg19 to determine, for each sequencing read, the precise 3′ terminus of transcription and the presence, length, and sequence composition of any untemplated 3′ additions. The data were analyzed using AppEnD as previously described (Welch et al. 2015). Briefly, AppEnD aligns reads to the genome using bowtie2 in local mode and then identifies soft-clipped portions of the reads, corresponding to bases that do not match the genome. Untemplated 3′ additions (if any) and the precise 3′ terminus of transcription are then located using dynamic programming alignment of the 3′ linker sequence and the soft-clipped portion of the read.
DATA DEPOSITION
The data are deposited at GEO (GSE85361).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute of General Medical Sciences, National Institutes of Health grant GM29832 to W.F.M. J.D.W. was supported by National Science Foundation Graduate Research Fellowship DGE-1144081 and National Institutes of Health BD2K Fellowship T32 CA201159.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.058107.116.
REFERENCES
- Berndt H, Harnisch C, Rammelt C, Stohr N, Zirkel A, Dohm JC, Himmelbauer H, Tavanez JP, Huttelmaier S, Wahle E. 2012. Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. RNA 18: 958–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks L III, Lyons SM, Mahoney JM, Welch JD, Liu Z, Marzluff WF, Whitfield ML. 2015. A multiprotein occupancy map of the mRNP on the 3′ end of histone mRNAs. RNA 21: 1943–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruccio N, Ross J. 1994. Purification of a human polyribosome-associated 3′ to 5′ exonuclease. J Biol Chem 269: 31814–31821. [PubMed] [Google Scholar]
- Chang H, Lim J, Ha M, Kim VN. 2014. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol Cell 53: 1044–1052. [DOI] [PubMed] [Google Scholar]
- D'Ambrogio A, Gu W, Udagawa T, Mello CC, Richter JD. 2012. Specific miRNA stabilization by Gld2-catalyzed monoadenylation. Cell Rep. 2: 1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang X, Kaygun H, Marzluff WF. 2003. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol Cell 12: 295–305. [DOI] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. 2007. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8: 113–126. [DOI] [PubMed] [Google Scholar]
- Graves RA, Pandey NB, Chodchoy N, Marzluff WF. 1987. Translation is required for regulation of histone mRNA degradation. Cell 48: 615–626. [DOI] [PubMed] [Google Scholar]
- Hamill S, Wolin SL, Reinisch KM. 2010. Structure and function of the polymerase core of TRAMP, a RNA surveillance complex. Proc Natl Acad Sci 107: 15045–15050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnisch C, Cuzic-Feltens S, Dohm JC, Gotze M, Himmelbauer H, Wahle E. 2016. Oligoadenylation of 3′ decay intermediates promotes cytoplasmic mRNA degradation in Drosophila cells. RNA 22: 428–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ME, Böhni R, Schneiderman MH, Ramamurthy L, Schümperli D, Marzluff WF. 1991. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol Cell Biol 11: 2416–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho C, Ha M, Han J, Kim VN. 2008. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell 32: 276–284. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. 2009. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708. [DOI] [PubMed] [Google Scholar]
- Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. 2012. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell 151: 521–532. [DOI] [PubMed] [Google Scholar]
- Hoefig KP, Rath N, Heinz GA, Wolf C, Dameris J, Schepers A, Kremmer E, Ansel KM, Heissmeyer V. 2013. Eri1 degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nat Struct Mol Biol 20: 73–81. [DOI] [PubMed] [Google Scholar]
- Kaygun H, Marzluff WF. 2005a. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol 12: 794–800. [DOI] [PubMed] [Google Scholar]
- Kaygun H, Marzluff WF. 2005b. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol Cell Biol 25: 6879–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Ha M, Loeff L, Chang H, Simanshu DK, Li S, Fareh M, Patel DJ, Joo C, Kim VN. 2015. TUT7 controls the fate of precursor microRNAs by using three different uridylation mechanisms. EMBO J 34: 1801–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JE, Wickens M. 2007. A family of poly(U) polymerases. RNA 13: 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Ha M, Chang H, Kwon SC, Simanshu DK, Patel DJ, Kim VN. 2014. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 159: 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons SM, Ricciardi A, Guo AY, Kambach C, Marzluff WF. 2014. The C-terminal tail of Lsm4 interacts directly with the 3′ end of the histone mRNP and is required for efficient histone mRNA degradation. RNA 20: 88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Gongidi P, Woods KR, Jin JP, Maltais L. 2002. The human and mouse replication-dependent histone genes. Genomics 80: 487–498. [PubMed] [Google Scholar]
- Mowry KL, Steitz JA. 1987. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science 238: 1682–1687. [DOI] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF. 2008. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 22: 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey NB, Marzluff WF. 1987. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol Cell Biol 7: 4557–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Song H. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 11: 121–127. [DOI] [PubMed] [Google Scholar]
- Rissland OS, Norbury CJ. 2009. 3′ Uridylation precedes decapping in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol 16: 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Mikulaslova A, Norbury CJ. 2007. Efficient RNA polyuridylation by non-canonical poly(A) polymerases. Mol Cell Biol 27: 3612–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Kobs G. 1986. H4 histone messenger RNA decay in cell-free extracts initiates at or near the 3′ terminus and proceeds 3′ to 5′. J Mol Biol 188: 579–593. [DOI] [PubMed] [Google Scholar]
- Ross J, Peltz SW, Kobs G, Brewer G. 1986. Histone mRNA degradation in vivo: the first detectable step occurs at or near the 3′ terminus. Mol Cell Biol 6: 4362–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L, Wang L, Buter N, Kwak JE, Schiltz CA, Gonzalez T, Kelley AE, Landry CF, Wickens M. 2005. Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA 11: 1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Marzluff WF. 2002. The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol Cell Biol 22: 7093–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MJ, West S, Norbury CJ. 2011. The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA 17: 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slevin MK, Meaux S, Welch JD, Bigler R, de Marval PLM, Su W, Prins JF, Rhoads RE, Marzluff WF. 2014. Deep sequencing shows multiple oligouridylations are required for 3′ to 5′ degradation of histone mRNAs on polyribosomes. Mol Cell 53: 1020–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Slepenkov SV, Slevin MK, Lyons SM, Ziemniak M, Kowalska J, Darzynkiewicz E, Jemielity J, Marzluff WF, Rhoads RE. 2013. mRNAs containing the histone 3′ stem-loop are degraded primarily by decapping mediated by oligouridylation of the 3′ end. RNA 19: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D, Marzluff WF, Dominski Z, Tong L. 2013. Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3′hExo ternary complex. Science 339: 318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JE, Chang HM, Piskounova E, Gregory RI. 2012. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA 18: 1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ. 2006. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA 12: 1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-F, Whitfield ML, Ingledue TI, Dominski Z, Marzluff WF. 1996. The protein which binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev 10: 3028–3040. [DOI] [PubMed] [Google Scholar]
- Welch JD, Slevin MK, Tatomer DC, Duronio RJ, Prins JF, Marzluff WF. 2015. EnD-Seq and AppEnD: sequencing 3′ ends to identify nontemplated tails and degradation intermediates. RNA 21: 1375–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Purdy M, Marzluff WF, Dominski Z. 2006. Characterization of 3′hExo, a 3′ exonuclease specifically interacting with the 3′ end of histone mRNA. J Biol Chem 281: 30447–30454. [DOI] [PubMed] [Google Scholar]
- Zund D, Gruber AR, Zavolan M, Muhlemann O. 2013. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nat Struct Mol Biol 20: 936–943. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.