Abstract
RNA nanotechnology is rapidly emerging. Due to advantageous pharmacokinetics and favorable in vivo biodistribution, RNA nanoparticles have shown promise in targeted delivery of therapeutics. RNA nanotechnology applies bottom-up assembly, thus elucidation of the mechanism of interaction between multiple components is of fundamental importance. The tendency of diminishing concern about RNA instability has accelerated by the finding of the novel thermostable three-way junction (3WJ) motif of the phi29 DNA-packaging motor. The kinetics of these three components, each averaging 18 nucleotides (nt), was investigated to elucidate the mechanism for producing the stable 3WJ. The three fragments coassembled into the 3WJ with extraordinary speed and affinity via a two-step reaction mechanism, 3WJb + 3WJc ↔ 3WJbc + 3WJa ↔ 3WJabc. The first step of reaction between 3WJb and 3WJc is highly dynamic since these two fragments only contain 8 nt for complementation. In the second step, the 3WJa, which contains 17 nt complementary to the 3WJbc complex, locks the unstable 3WJbc complex into a highly stable 3WJ. The resulting pRNA-3WJ is more stable than any of the dimer species as shown in the much more rapid association rates and slowest dissociation rate constant. The second step occurs at a very high association rate that is difficult to quantify, resulting in a rapid formation of a stable 3WJ. Elucidation of the mechanism of three-component collision in producing the ultrastable 3WJ proves a promising platform for bottom-up assembly of RNA nanoparticles as a new class of anion polymers for material science, electronic elements, or therapeutic reagents.
Keywords: RNA nanotechnology, nanobiotechnology, bacteriophage, phi29, RNA nanoparticle, viral DNA packaging motor
INTRODUCTION
The field of RNA nanotechnology has recently undergone rapid expansion mainly due to the fact that RNA has the simplicity characteristic of DNA yet the versatile functionality of proteins (Masquida et al. 1997; Guo et al. 1998; Lilley 2000; Leontis and Westhof 2003; Lescoute and Westhof 2006; Guo 2010; Bindewald et al. 2011; Ye et al. 2012). With the combination of the advantage of these two materials, RNA can easily be designed and constructed through bottom-up assembly and plays diverse roles (Felden et al. 1996; Chworos et al. 2004; Shu et al. 2004; Khaled et al. 2005; Jaeger and Chworos 2006; Afonin et al. 2008). The wide-ranging functionalities of RNA make it a prime candidate for applications in sensing operates, computer parts (Rinaudo et al. 2007; Breaker 2008; Win and Smolke 2008; Benenson 2009; Xie et al. 2010, 2011; Qiu et al. 2013; Shu et al. 2014a), imaging reagents (Calzada et al. 2012; Chen et al. 2012; Paige et al. 2012; Kellenberger et al. 2013), NEM devices (Noy 2011), and therapeutics including the delivery, specific targeting, and treatment of cancer and viral infections (Guo et al. 1998; Shu et al. 2003, 2004, 2011a, 2013; Yingling and Shapiro 2007; Severcan et al. 2009; Afonin et al. 2011; Chang et al. 2012; Delebecque et al. 2012; Haque et al. 2012; Lee et al. 2012, 2015b; Tabernero et al. 2013). The use of RNA as a delivery vehicle provides several benefits over other systems and technologies. Firstly, RNA nanoparticles can be produced with known stoichiometry and high reproducibility (Guo et al. 1998; Lilley 2000; Shu et al. 2004, 2011a; Afonin et al. 2010; Jasinski et al. 2014; Khisamutdinov et al. 2014a,b), as RNA is composed of simple building blocks with predictable secondary structure. RNA aptamers can be used for specific targeting of cell groups through binding to cell surface receptors much like a protein ligand or antibody (Cerchia et al. 2009; Abdelmawla et al. 2011; Shu et al. 2011b). Furthermore, the replacement of protein-targeting reagents with nucleic acids prevents the induction of antibodies (Abdelmawla et al. 2011; Guo et al. 2012; Shu et al. 2014b), allowing for repeated administration. RNA nanoparticles are typically between 10 and 50 nanometers in size, meaning they are retained in the human body yet are small enough to pass through cell membranes through endocytosis (Maeda 2001; Gao et al. 2005; Jain 2005; Li and Szoka 2007; Maeda et al. 2013; Jasinski et al. 2014; Khisamutdinov et al. 2014b). In in vivo testing, RNA nanoparticles have shown favorable pharmacokinetic and biodistribution properties, as they are able to avoid accumulation in healthy organs and tissues with efficient retention in tumors (Shu et al. 2011a; Haque et al. 2012). Additionally, RNA possess unique in vivo attributes: (i) transcription, termination, splicing, and processing allow for in vivo RNA production; (ii) riboswitches (Mulhbacher et al. 2010) and ribozymes (Sarver et al. 1990; Hoeprich et al. 2003; Lilley 2003; Winkler et al. 2004; Liu et al. 2007) result in in vivo processing and possible assembly into nanoparticles in cell for special functions such as intracellular computation (Rinaudo et al. 2007; Breaker 2008; Win and Smolke 2008; Benenson 2009; Xie et al. 2010, 2011; Qiu et al. 2013; Shu et al. 2014a); (iii) RNA can self-assemble without external forces (Shu et al. 2014a); and (iv) RNA is stable in acidic environments allowing for survival in endosome (Guo et al. 2012). Even with these advantages, RNA nanotechnology still has many hurdles that it must overcome in order to be used in therapeutics. Most notably, RNA is known to have stability issues and is prone to degradation by RNase that has since been solved through backbone modifications, such as 2′-fluoro (Pieken et al. 1991; Liu et al. 2011), 2′-O-methyl (Czauderna et al. 2003), or locked nucleotides (Kaur et al. 2006). The dissociation of the self-assembled RNA nanoparticles at low concentrations and elevated temperatures in in vivo environments makes it obligatory to address the dynamics and kinetics of RNA nanoparticle assembly (Privalov and Filiminov 1978; Freier et al. 1986; Jaeger et al. 1993; Kawasaki et al. 1993; Lesnik and Freier 1995; Sugimoto et al. 1995; Gyi et al. 1996; Diamond et al. 2001; Brunel et al. 2002).
Recently, a novel ultrastable RNA motif was found in the packaging RNA (pRNA) of the phi29 dsDNA packaging motor (Fig. 1) that has since diminished the concerns of RNA nanoparticles dissociating at low concentrations found in vivo because of its unusually high thermostability (Shu et al. 2011a). This motif is a three-way junction (3WJ) that serves as the central core structure of the pRNA and connects the helical domain (Zhang et al. 1994) with the two interlocking looped regions of the pRNA (Reid et al. 1994). It has been determined that the 3WJ serves as the central folding domain and provides the overall high stability seen in pRNA. The motif can be formed from three individual RNA oligos at a high yield even in the absence of metal ions (Shu et al. 2011a). The novelty behind this RNA structure is in the ease of formation, a remarkably low Gibbs free energy (ΔG°) of −28 kcal/mol, and a high melting temperature (Tm) of 59.3°C (Binzel et al. 2014). Furthermore, it has been proven that the pRNA-3WJ allows the inclusion of RNA functional moieties such as receptor-binding aptamer, siRNA, ribozyme, miRNA, or riboswitch, resulting in the production of functional nanoparticles (Shu et al. 2011a; Lee et al. 2015a; Rychahou et al. 2015). The addition of functional groups did not interfere with the proper folding of the 3WJ or the added functional moieties. Previously, attempts have been made to elucidate the mechanism for the kinetic stability of the pRNA-3WJ, yet the 3WJ was found to be too stable to detect its dissociation by the current technology of radioactive chasing (Shu et al. 2011a). However, thermodynamic studies were previously completed on the pRNA-3WJ (Binzel et al. 2014). From the association and melting profiles in the thermodynamic studies, a single transition step was seen, hinting to the pRNA-3WJ folding in a single step of all three strands. Furthermore, there was no evidence of a dimer intermediate in the 3WJ formation.
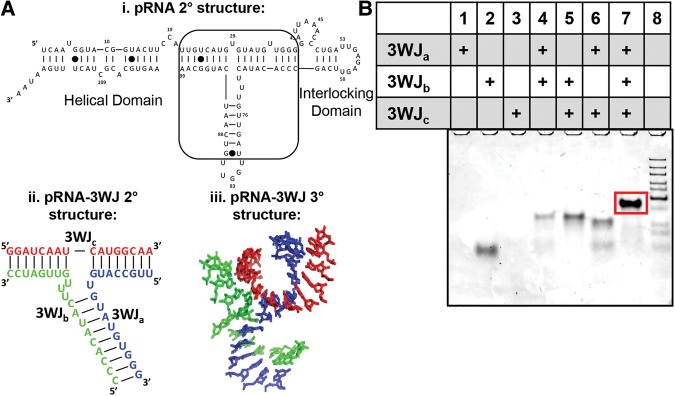
FIGURE 1.
Overview of the Phi29 pRNA and the three-way junction (3WJ). (A) (i) Secondary structure of the phi29 pRNA monomer with the pRNA-3WJ outlined by the box, which connects the helical domain to the interlocking procapsid binding domains. (ii) Secondary structure and sequence of the pRNA-3WJ and (iii) the crystal structure of the pRNA-3WJ. (B) Assembly gel of the pRNA-3WJ from the three short RNA oligo strands on 12% native PAGE. The 3WJ (lane 7) shows efficiency of folding and particle homogeneity before purification.
The understanding of the mechanism of assembly of the pRNA-3WJ and the interaction of the RNA strands is of fundamental importance, but the study of three-component collisions has not been well-studied in chemistry, biology, or material sciences. Here, we investigated the kinetic properties of the pRNA-3WJ using surface plasmon resonance (SPR) to study its association and disassociation using bottom-up RNA construction nanotechnics. We provided real-time examination of the three-component collision of the pRNA-3WJ. Through these studies, the pRNA-3WJ proves to be ultrastable and only disassociates at extremely low concentration, which is atypical to other well-studied double-stranded RNA or dimeric macromolecules. Furthermore, the pRNA central motif relies heavily on each of the three strands for rapid folding and slow dissociation rates.
RESULTS AND DISCUSSION
Determination of kinetic parameters of the pRNA-3WJ
The Bio-Rad ProteOn XPR36 allows for real-time monitoring and analysis of molecule association and dissociation. SPR allows for examination of interactions between molecules on a prism surface. In this case, the ligand RNA strand is bound through a biotin label to the neutravidin-labeled chip surface. As mass accrues or decreases on the chip surface either by ligand labeling or interactions between the ligand and analyte, the angle of light refraction through the chip prism and resonance is changed, thus creating a response that then can be fit with kinetic models. The Bio-Rad ProteOn XPR36 system allows for the observation of 36 interactions at the same time while including controls of ligand and analyte free spots, thus providing a platform for elaborate studies of the association and dissociation of the pRNA-3WJ. Through real-time analysis, we are able to calculate the dissociation constant (KD) by measuring the rates of reactions moving toward 3WJ complex formation and dissociation to single RNA strands.
Through classic definitions of kinetics, all chemical reactions are viewed to reach equilibrium where there is a balance in the production of products and reactants. In the case of the pRNA three-way junction, a single-step chemical reaction is described by Equation 1 below:
| (1) |
and the rate of forming pRNA-3WJ can be written as
| (2) |
where ka is the rate constant of the pRNA-3WJ formation, and kd is the rate constant of dissociation into individual RNA strands in units of M−2 s−1 and s−1, respectively. At equilibrium, the rates of forward and backward reactions are equal to each other, creating a balance between formation of the pRNA-3WJ and single-stranded RNA. Together, the two rate constants can be combined to define the disassociation and association constants.
| (3) |
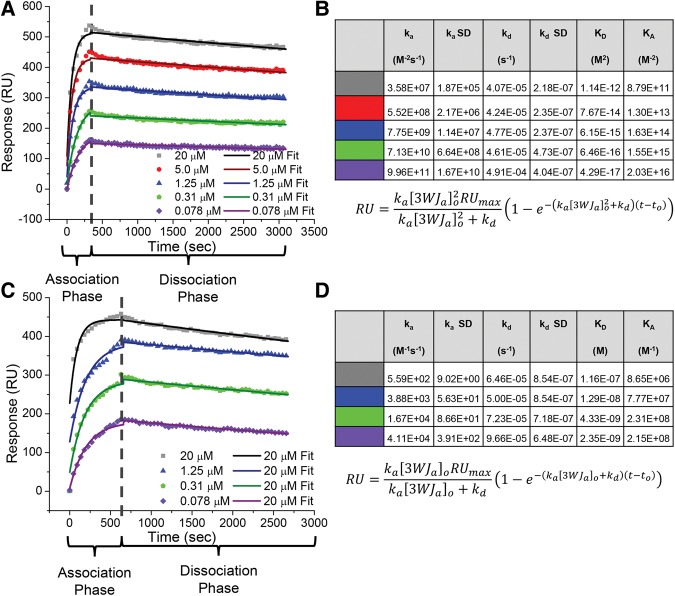
Based on previous experiments (Binzel et al. 2014), it was believed that the pRNA-3WJ was undergoing a single association and dissociation step following what is seen in Equations 1 and 5 (Shu et al. 2011a; Binzel et al. 2014). Using SPR, association and disassociation of the pRNA-3WJ was examined using the 3WJa strand immobilized onto the chip surface, hence acting as the ligand, followed by co-injecting 3WJb and 3WJc strands, acting as analytes. A single transition is seen in the association over a range of concentrations from 20 μM to 78 nM. Based on the previous idea that the pRNA-3WJ formed in a single step from the three components, the data were fit to a Langmuir three-component pseudo-first-order model using Equation 5, solving for the reaction rate constants (Fig. 2A,B). In these experiments, a relatively constant dissociation averaged 4.52 × 10−5 s−1; this value is within expected range for biological samples. However, the association rate constant of the pRNA-3WJ was seen changing on order of magnitudes, meaning a constant association rate across tested concentrations of 20–0.078 μM. This clearly negates first-order chemical reactions and indicates error in the assembly kinetic parameters, as one would expect the reaction rate to change with varying concentration leading to a constant ka.
FIGURE 2.
SPR of the pRNA-3WJ. (A) Assembly of the 3WJ of three-component strands. 5′-Biotin-labeled 3WJa was first immobilized to the SPR chip surface. 3WJb and 3WJc strands were mixed at equal, varying concentrations, ranging from 20 μM to 78 nM, and injected across the chip surface for 330 sec (association phase). Following the interaction of the 3WJ strands, blank TMS buffer was injected across the chip surface for 2700 sec (dissociation phase). The association and dissociation curves were fit using a three-component pseudo-first-order model. (B) Calculated kinetic parameters from SPR curve following a three-component pseudo-first-order kinetic model. (C) SPR assay for pRNA-3WJ under the assumption of previous dimer formation data. 5′-Biotin-labeled 3WJa was first bound to the SPR chip surface. 3WJb and 3WJc strands were mixed at equal, varying concentrations, ranging from 20 μM to 78 nM, and were injected across the chip surface for 660 sec followed by blank TMS buffer for 2700 sec. Association and dissociation curves were fit using a two-component pseudo-first-order model. (D) Kinetic parameters from SPR curves following a two-component pseudo-first-order kinetic model.
KD calculation of pRNA-3WJ dimers
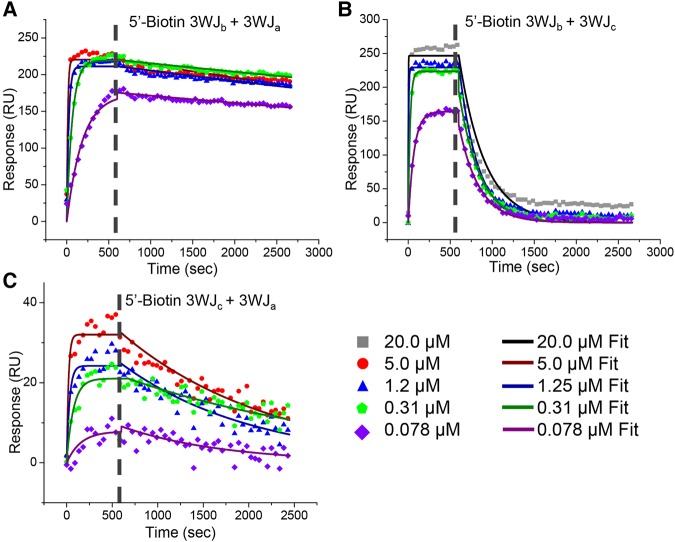
In order to confirm RNA/RNA interactions on the SPR chip were in fact the formation of the 3WJ, dimers of the pRNA-3WJ were tested and assembled (Table 1; Fig. 3). In each of the dimer interactions (3WJab, 3WJac, and 3WJbc), association and dissociation periods were produced. Furthermore, once the data were fit with a pseudo-first-order Langmuir model due to the expected 1:1 interaction between ligand and analyte, it was seen that each of the calculated kas and kds produced were constant and concentration independent as expected by kinetics. This means the pRNA-3WJ dimers were forming properly on the chip surface. Interestingly, in comparing the association rate constants of each of the dimer species, the 3WJbc dimer produced the most rapid “on” rate with a ka of 1.37 × 105 M−1 s−1; this is nearly an order of magnitude higher than either the 3WJab or the 3WJac dimers. Here the association rate constant changes units to M−1 s−1 as only two components are interacted, whereas before three components were fused, resulting in the M−2 s−1 unit. This increase in association rate may be due to less internal structuring of the 3WJc strand, as we see no ethidium bromide staining of the single strand, thus giving a better diffusion to the 3WJb compared to the 3WJa forming the 3WJab dimer (Fig. 1A). In comparing the dissociation rate constants (kd) of the dimers, the 3WJab dimer is seen to be the most stable (6.78 × 10−5 s−1), followed by 3WJac with a kd of 6.34 × 10−4 s−1, and finally the weakest dimer being the 3WJbc (3.95 × 10−3 s−1). This indicates that the 3WJbc dimer not only would form the quickest but also dissociates the most rapidly. Additionally, out of concern that the biotin end label on the ligand RNA interfered or altered the binding of RNA complexes on the SPR chip surface, duplicate studies of dimer interactions were completed by switching the ligand strand to analyte and analyte to ligand through changing the biotin label to the second strand. Through these interactions no obvious change in association profiles or ka values were seen indicating the biotin label allows for the binding of the ligand strand to the chip surface, showing no significant effect on RNA folding.
TABLE 1.
Kinetic parameters of pRNA-3WJ dimer species

FIGURE 3.
SPR assay for pRNA-3WJ dimers. Dimer species that make up the pRNA-3WJ were examined through SPR at varying concentrations ranging from 20 μM to 78 nM. The injected RNA strand was injected for 660 sec (association phase) followed by blank TMS buffer for 2700 sec (dissociation phase). The histographs were then fit using a two-component pseudo-first-order Langmuir model for bound 5′-biotin 3WJb and 3WJa (A), bound 5′-biotin 3WJb and 3WJc (B), bound 5′-biotin 3WJc and 3WJa (C).
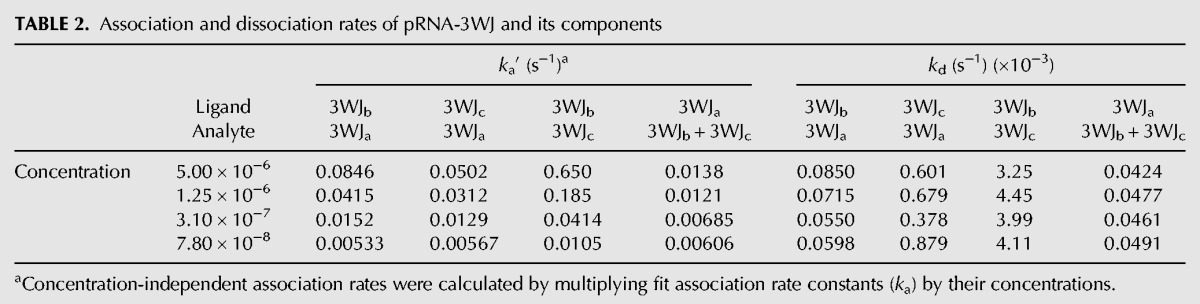
Next, in hopes of gaining insight into the assembly mechanism of the pRNA-3WJ, the association rates of the dimer species were compared with the association rate of the tested three-component one-step reaction of the pRNA-3WJ. This was completed by calculating concentration-independent association rates (ka′) by taking the fitted rate constants and multiplying by the concentration of the studied interaction resulting in an s−1 unit (Table 2). It is important to note that the pRNA-3WJ was multiplied by a concentration squared value while dimers only by concentration, due to the difference in association reaction orders. Surprisingly, it is seen that the pRNA-3WJ produces a rate of reaction on average an order of magnitude below the 3WJbc dimer. This indicates that the formation of the dimer species is occurring before the formation of the pRNA-3WJ, thus creating an intermediate step that was previously not seen in thermodynamic studies (Binzel et al. 2014).
TABLE 2.
Association and dissociation rates of pRNA-3WJ and its components
Determination of time constants of each pRNA-3WJ strand
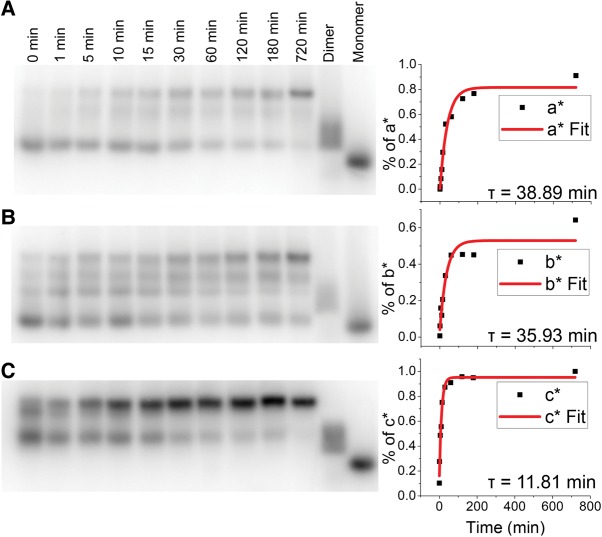
To further gain insight into the formation of a dimer species, time-based strand displacement was completed on each of the individual pRNA-3WJ strands. Using a time-based strand replacement electrophoretic mobility shift assay (EMSA), time constants were calculated for each of the RNA strands in order to examine which, if any, of the strands is responsible for the fast folding that has been seen in the pRNA-3WJ and the stability at high and low concentrations. The results show that the 3WJa and 3WJb strands produce a similar τ of 38.89 min and 35.93 min, respectively, while the 3WJc showed a τ of only 11.81 min (Fig. 4). The longer times for the 50% strand replacement of the 3WJa and 3WJb strands suggest that the structure and property of the 3WJc strand is special and unique. Using ethidium bromide or SYBR-green staining within gels, the 3WJc is undetectable, whereas both 3WJa and 3WJb are detected, indicating a lack in internal structuring of the 3WJc strand (Shu et al. 2011b). From these studies, it was determined that 3WJa and 3WJb strands are essential for the strong stability of the pRNA-3WJ motif; however, the high reactivity of the 3WJc strand allows for the rapid association seen in SPR studies and previous thermodynamic studies (Binzel et al. 2014). These results further confirm and support the SPR dimer studies showing a rapid rate of reaction from the 3WJc-containing dimers with a strong stability or dissociation of the 3WJab dimer species.
FIGURE 4.
Time constants of 3WJ strands by electrophoretic mobility shift assays (EMSA). The half-life of each 3WJ strand in the pRNA-3WJ was calculated by EMSA using 32P-labeled free strands, replacing unlabeled strands on assembled pRNA-3WJ. The transition of monomer to 3WJ was then plotted, and the data were fitted to calculate τ. (A) 3WJa, (B) 3WJb, and (C) 3WJc.
Association mechanism of the pRNA-3WJ
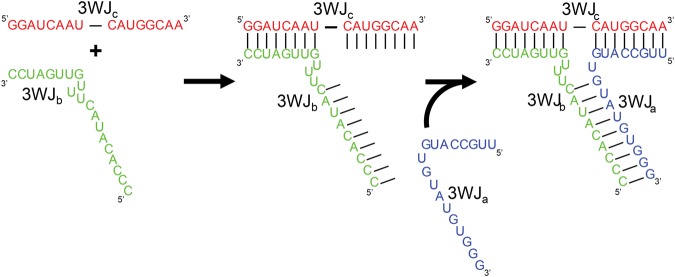
The previous thermodynamic studies on the pRNA-3WJ showed that the three RNA fragments associated very rapidly and the formation of a dimer intermediate was undetected. While a one-step association between the three strands seemed unlikely, it was believed that the pRNA-3WJ formed through the initiation of a pseudo-dimer with a very rapid addition of the third RNA strand. Here we have found that in fact the 3WJb and 3WJc strands form a dimer species at a rapid rate and faster than the modeled association as three strands together. Thus, the two analytes (3WJb and 3WJc) were forming into a dimer complex before reaching the chip surface and being able to interact with the 3WJa ligand strand on the chip surface. With this idea the pRNA-3WJ data were reanalyzed to follow a pseudo-first-order reaction that would result in an assembly mechanism of 3WJb + 3WJc ↔ 3WJbc + 3WJa ↔ 3WJabc (Fig. 2C,D). Here the same dissociation rates were observed as in the three-component, one-step system. Additionally, the rates of association were independent of concentrations tested, leading to varying rate constants. This again negates first-order reaction laws; however, the varying rate of association constants can be attributed to using a range of working concentrations and thus was observed kas several orders of magnitude above the real association rate constant. This means that the rate of the reaction for the 3WJbc + 3WJa ↔ 3WJabc is occurring much quicker than the observable level on SPR and the required concentrations needed to slow the reaction to an observable level would be too low to be sensed on the SPR chip surface. This situation differs from the previous changing association rate constants seen in the one-step association fitting due to the fact that here we know from the timing scale and the dimer studies that the 3WJbc dimer is forming before the two strands are able to interact with the 3WJa ligand bound to the chip. This leads to a simple 1:1 interaction of the 3WJa ligand with the 3WJbc dimer flowing across the chip. Additionally, through electrophoresis studies, a 1:1:1 interaction is always seen further supporting the Langmuir model used in the SPR studies.
Therefore, it is proposed that the pRNA-3WJ undergoes a two-step reaction, in which the second reaction takes place immediately following the formation of the dimer intermediate at a rate that is too fast to currently quantify on SPR (Fig. 5). This high reaction rate is due to the fact that the initiation of 3WJbc formation increases the number of bases for binding from eight or nine bases from the single strand to 17 bases on the duplex for interaction with the 3WJa strand (Fig. 5, middle panel). This increase in bases results in a higher probability of reaction per collision between the RNA dimer and the third strand, giving a significantly increased rate. Furthermore, this increase in base-pairing from 8 to 17 nucleotides (nt) significantly lowers the Gibbs free energy of formation (ΔG°), indicating the reaction will occur more spontaneously. With the increase of nucleotides upon formation of the 3WJbc, the 3WJa is recruited to form the completed 3WJ with a much higher affinity. Thus, this second high-speed reaction relies on the formation of the 3WJbc dimer that is the rate-limiting factor for 3WJ assembly. Additionally, due to the rapid reaction of the second step, only a low concentration of the dimer intermediate is needed to drive the 3WJ assembly. Thus, to the naked eye the assembly of the pRNA-3WJ is seemingly a singular step reaction without the evidence of an intermediate step.
FIGURE 5.
Assembly mechanism of pRNA-3WJ. The pRNA-3WJ assembles through a two-step association mechanism in which the 3WJbc dimer first forms, as shown in the first panel. This association step greatly increases the attraction of the 3WJa strand through the doubling of nucleotides presented for binding resulting in a dropped ΔG°. This second reaction occurs at an unobservable rate and results in forming a stable nanostructure, in which each of the three strands is locked into the structure by two areas of base-pairing.
Furthermore, each strand and helical region of the pRNA-3WJ plays an important role in rapid formation and high stability. As discussed earlier, from the τ values we know that the 3WJc strand is highly reactive; this results in the highest ka of the 3WJbc dimer. Although the dimer between the 3WJb and the 3WJc strands forms the quickest, it also displays the shortest dissociation rate, showing that the pRNA-3WJ stability is provided through the other helical domains. The 3WJa and 3WJc strands provide the high stability of the 3WJ, as shown in their long half-lives and the slow kd seen in SPR studies of the 3WJab. However, although the pRNA-3WJ relies heavily on the 3WJab helix region for its stability, the pRNA-3WJ itself is the most stable motif, as shown in having the slowest “off” rate (kd) (Table 2). This can be attributed to the fact that if one helical region of the pRNA-3WJ breaks, the local concentration of the strand is very high compared to what would be seen in solution, thus initiating an immediate refolding of that helical region as the second half of the strand is still attached to the pRNA-3WJ. As a result of these kinetic studies, the high association rate and slow dissociation rate of the pRNA-3WJ result from contributions of each of the RNA strands, leading to a three-component collision.
Conclusions
The pRNA-3WJ formation follows a two-step assembly mechanism of 3WJb + 3WJc ↔ 3WJbc + 3WJa ↔ 3WJabc. The 3WJbc dimer formation is the first step, occurring at a slower and less dynamic rate, while the formation of the pRNA-3WJ occurs at a much faster rate since each of the two-component pairs only contain 8 nt for complementation. While assembly of each dimer resulted in an uninterrupted 17 nt sequence, the third complementary component locks the unstable 3WJbc complex into a highly stable 3WJabc. It is suggestive that dimeric intermediates might produce the 3WJabc at a fast rate, and formation of any pair of dimers can promote a fast assembly with the third components to produce the 3WJabc when all three strands are incubated in the solution. Additionally, this mechanism relies on having a balanced concentration of the pRNA-3WJ strands. Thus the pRNA-3WJ shows extraordinary stability and ease of formation, resulting in a branched motif that can be used for the construction of stable RNA nanoparticles.
MATERIALS AND METHODS
Oligonucleotides and assembly of 3WJs
The pRNA-3WJ was constructed from three RNA oligo fragments. RNA strands were produced by Integrated DNA Technologies (IDT). Assembly of the pRNA-3WJ was completed by mixing each of the three RNA strands (3WJa, 3WJb, 3WJc) at equal molar concentrations at room temperature in TMS buffer (50 mM Tris pH 8.0, 100 mM NaCl, 10 mM MgCl2). Samples were then run on a 12% native polyacrylamide gel at 100 V for 2 h at 4°C in TBM running buffer (89 mM Tris, 200 mM borate acid, and 5 mM MgCl2) to ensure pRNA-3WJ size and folding.
SPR studies of the pRNA-3WJ
The Bio-Rad ProteOn XPR36 was used to complete real-time interaction studies of the pRNA-3WJ motif. A neutravidin-coated HLC chip (Bio-Rad) was used to immobilize one of the three 3WJ strands with a 5′ biotin label. Several studies were completed by immobilizing the 3WJa and 3WJc strands to the chip surface, followed by injection and interaction of the remaining two 3WJ strands. All studies were conducted at room temperature and in TMS buffer (50 mM Tris, pH 8.0, 100 mM NaCl, 10 mM MgCl2) with added 0.1% Tween. RNA strands were injected at a concentration ranging from 20.0 μM to 78.0 nM at a constant flow rate of 25 μL/min for a total of 660 sec. This assures the analyte RNAs are in high excess in the association phase followed by injection of blank TMS buffer for 2700 sec at 25 μL/min in the dissociation phase. Real-time association and dissociation was observed and displayed in a histograph of total response (relationship on mass on chip surface) and time.
SPR studies of the pRNA-3WJ dimers
Utilizing a similar approach as the pRNA-3WJ, the formation and dissociation of dimers from the pRNA-3WJ were analyzed. In order to remain consistent with the studies, the same concentration gradients were used to examine the formation of two of the three strands coming together within the three-way junction.
Kinetic modeling of surface plasmon data
Data were extracted from the Bio-Rad ProteOn XPR36 and modeled using Igor Pro 6.37. The association periods of the SPR plots were modeled using a Langmuir pseudo-first-order model consisting of two and three components using Equations 4 and 5, respectively. These models were selected due to the nature of SPR studies, in that we are observing a 1:1 interaction (thus Langmuir) in which the interacting strand is in large excess to the bound strand, creating a pseudo-first-order interaction. The dissociation of all RNA motifs was modeled using a Langmuir zero-order reaction as shown in Equation 6.
| (4) |
| (5) |
| (6) |
where R is the SPR response in terms of t, time in seconds. These kinetic models are derived in the Supplemental Material. In fitting the data, the global fitting package was used in linking the association and dissociation of each sample through the kd and Rmax and Ro.
Calculation of time constant (τ) of pRNA-3WJa, 3WJb, and 3WJc
The time constant (τ) of the pRNA-3WJ core strand was determined through electrophoresis mobility shift assays (EMSA). A single strand of the pRNA-3WJ fragments was radiolabeled at the 5′-terminus using γ-32P ATP (PerkinElmer) and denoted with an asterisk, as previously described (Binzel et al. 2014). The radiolabeled oligo was held at a constant concentration of 10 nM and incubated over varying time points ranging from 0 to 720 min with completed pRNA-3WJ structure at 100 nM concentrations. Samples were snap frozen on dry ice and then an electrophoresis 12% native polyacrylamide gel at 100 V for 2 h at 4°C in TBM running buffer was run. The gel was then imaged by transferring the radio signal to a phosphor screen for 12 h at −80°C and visualized using a Typhoon 7000 (GE). Band quantification was completed using ImageJ. The ratio of single strand to 3WJ was then calculated and plotted against time in OriginPro 8.5. The curve was fit using Equation 7 below:
| (7) |
where t is the time in seconds, and τ is the time constant at which 50% of the labeled 3WJ strand was in single-strand state and 50% had replaced unlabeled strand in the 3WJ complex (Novikova et al. 2010). These studies were completed as a way to compare the strand displacement between each of the pRNA-3WJ strands.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by National Institutes of Health grants EB003730 and CA151648 to P.G., as well as National Institutes of Health grant R25CA153954 to Brad Anderson. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. Funding for Dr. Guo's Endowed Chair in Nanobiotechnology position is from the William Fairish Endowment Fund, and Dr. Guo's Sylvan G. Frank Endowed Chair position is funded by the C.M. Chen Foundation. Dr. Guo is a consultant of Matt-Holding Ltd., Nanobio Delivery Pharmaceutics Co. Ltd., and Oxford Nanopore Technology Ltd.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.057646.116.
REFERENCES
- Abdelmawla S, Guo S, Zhang L, Pulukuri S, Patankar P, Conley P, Trebley J, Guo P, Li QX. 2011. Pharmacological characterization of chemically synthesized monomeric pRNA nanoparticles for systemic delivery. Mol Ther 19: 1312–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin KA, Danilov EO, Novikova IV, Leontis NB. 2008. TokenRNA: a new type of sequence-specific, label-free fluorescent biosensor for folded RNA molecules. Chembiochem 9: 1902–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, Shapiro BA, Jaeger L. 2010. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat Nanotechnol 5: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin KA, Grabow WW, Walker FM, Bindewald E, Dobrovolskaia MA, Shapiro BA, Jaeger L. 2011. Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat Protoc 6: 2022–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benenson Y. 2009. RNA-based computation in live cells. Curr Opin Biotechnol 20: 471–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindewald E, Afonin K, Jaeger L, Shapiro BA. 2011. Multistrand RNA secondary structure prediction and nanostructure design including pseudoknots. ACS Nano 5: 9542–9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binzel DW, Khisamutdinov EF, Guo P. 2014. Entropy-driven one-step formation of Phi29 pRNA 3WJ from three RNA fragments. Biochemistry 53: 2221–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. 2008. Complex riboswitches. Science 319: 1795–1797. [DOI] [PubMed] [Google Scholar]
- Brunel C, Marquet R, Romby P, Ehresmann C. 2002. RNA loop-loop interactions as dynamic functional motifs. Biochimie 84: 925–944. [DOI] [PubMed] [Google Scholar]
- Calzada V, Zhang X, Fernandez M, az-Miqueli A, Iznaga-Escobar N, Deutscher SL, Balter H, Quinn TP, Cabral P. 2012. A potencial theranostic agent for EGF-R expression tumors: (177)Lu-DOTA-nimotuzumab. Curr Radiopharm 5: 318–324. [DOI] [PubMed] [Google Scholar]
- Cerchia L, Giangrande PH, McNamara JO, de Franciscis V. 2009. Cell-specific aptamers for targeted therapies. Methods Mol Biol 535: 59–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CI, Lee TY, Kim S, Sun X, Hong SW, Yoo JW, Dua P, Kang HS, Kim S, Li CJ, et al. 2012. Enhanced intracellular delivery and multi-target gene silencing triggered by tripodal RNA structures. J Gene Med 14: 138–146. [DOI] [PubMed] [Google Scholar]
- Chen Z, Penet MF, Nimmagadda S, Li C, Banerjee SR, Winnard PT Jr, Artemov D, Glunde K, Pomper MG, Bhujwalla ZM. 2012. PSMA-targeted theranostic nanoplex for prostate cancer therapy. ACS Nano 6: 7752–7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. 2004. Building programmable jigsaw puzzles with RNA. Science 306: 2068–2072. [DOI] [PubMed] [Google Scholar]
- Czauderna F, Fechtner M, Dames S, Aygun H, Klippel A, Pronk GJ, Giese K, Kaufmann J. 2003. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res 31: 2705–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delebecque CJ, Silver PA, Lindner AB. 2012. Designing and using RNA scaffolds to assemble proteins in vivo. Nat Protoc 7: 1797–1807. [DOI] [PubMed] [Google Scholar]
- Diamond JM, Turner DH, Mathews DH. 2001. Thermodynamics of three-way multibranch loops in RNA. Biochemistry 40: 6971–6981. [DOI] [PubMed] [Google Scholar]
- Felden B, Florentz C, Giege R, Westhof E. 1996. A central pseudoknotted three-way junction imposes tRNA-like mimicry and the orientation of three 5′ upstream pseudoknots in the 3′ terminus of tobacco mosaic virus RNA. RNA 2: 201–212. [PMC free article] [PubMed] [Google Scholar]
- Freier SM, Kierzek R, Jaeger JA, Sugimoto N, Caruthers MH, Neilson T, Turner DH. 1986. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci 83: 9373–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Shi W, Freund LB. 2005. Mechanics of receptor-mediated endocytosis. Proc Natl Acad Sci 102: 9469–9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P. 2010. The emerging field of RNA nanotechnology. Nat Nanotechnol 5: 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Zhang C, Chen C, Trottier M, Garver K. 1998. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell 2: 149–155. [DOI] [PubMed] [Google Scholar]
- Guo P, Haque F, Hallahan B, Reif R, Li H. 2012. Uniqueness, advantages, challenges, solutions, and perspectives in therapeutics applying RNA nanotechnology. Nucleic Acid Ther 22: 226–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyi JI, Conn GL, Lane AN, Brown T. 1996. Comparison of the thermodynamic stabilities and solution conformations of DNA center dot RNA hybrids containing purine-rich and pyrimidine-rich strands with DNA and RNA duplexes. Biochemistry 35: 12538–12548. [DOI] [PubMed] [Google Scholar]
- Haque F, Shu D, Shu Y, Shlyakhtenko L, Rychahou P, Evers M, Guo P. 2012. Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today 7: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeprich S, ZHou Q, Guo S, Qi G, Wang Y, Guo P. 2003. Bacterial virus phi29 pRNA as a hammerhead ribozyme escort to destroy hepatitis B virus. Gene Ther 10: 1258–1267. [DOI] [PubMed] [Google Scholar]
- Jaeger L, Chworos A. 2006. The architectonics of programmable RNA and DNA nanostructures. Curr Opin Struct Biol 16: 531–543. [DOI] [PubMed] [Google Scholar]
- Jaeger JA, SantaLucia JJ, Tinoco IJ. 1993. Determination of RNA structure and thermodynamics. Annu Rev Biochem 62: 255–285. [DOI] [PubMed] [Google Scholar]
- Jain KK. 2005. The role of nanobiotechnology in drug discovery. Drug Discov Today 10: 1435–1442. [DOI] [PubMed] [Google Scholar]
- Jasinski D, Khisamutdinov EF, Lyubchenko YL, Guo P. 2014. Physicochemically tunable poly-functionalized RNA square architecture with fluorogenic and ribozymatic properties. ACS Nano 8: 7620–7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, Arora A, Wengel J, Maiti S. 2006. Thermodynamic, counterion, and hydration effects for the incorporation of locked nucleic acid nucleotides into DNA duplexes. Biochemistry 45: 7347–7355. [DOI] [PubMed] [Google Scholar]
- Kawasaki AM, Casper MD, Freier SM, Lesnik EA, Zounes MC, Cummins LL, Gonzalez C, Cook PD. 1993. Uniformly modified 2′-deoxy-2′-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J Med Chem 36: 831–841. [DOI] [PubMed] [Google Scholar]
- Kellenberger CA, Wilson SC, Sales-Lee J, Hammond MC. 2013. RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP. J Am Chem Soc 135: 4906–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaled A, Guo S, Li F, Guo P. 2005. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano Lett 5: 1797–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khisamutdinov EF, Jasinski DL, Guo P. 2014a. RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry. ACS Nano 8: 4771–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khisamutdinov E, Li H, Jasinski D, Chen J, Fu J, Guo P. 2014b. Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square, and pentagon nanovehicles. Nucleic Acids Res 42: 9996–10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JB, Hong J, Bonner DK, Poon Z, Hammond PT. 2012. Self-assembled RNA interference microsponges for efficient siRNA delivery. Nat Mater 11: 316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TJ, Haque F, Shu D, Yoo JY, Li H, Yokel RA, Horbinski C, Kim TH, Kim SH, Nakano I, et al. 2015a. RNA nanoparticles as a vector for targeted siRNA delivery into glioblastoma mouse model. Oncotarget 6: 14766–14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Yagati AK, Pi F, Sharma A, Choi JW, Guo P. 2015b. Construction of RNA-quantum dot chimera for nanoscale resistive biomemory application. ACS Nano 9: 6675–6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. 2003. Analysis of RNA motifs. Curr Opin Struct Biol 13: 300–308. [DOI] [PubMed] [Google Scholar]
- Lescoute A, Westhof E. 2006. Topology of three-way junctions in folded RNAs. RNA 12: 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesnik EA, Freier SM. 1995. Relative thermodynamic stability of DNA, RNA, and DNA:RNA hybrid duplexes: relationship with base composition and structure. Biochemistry 34: 10807–10815. [DOI] [PubMed] [Google Scholar]
- Li W, Szoka F. 2007. Lipid-based nanoparticles for nucleic acid delivery. Pharm Res 24: 438–449. [DOI] [PubMed] [Google Scholar]
- Lilley DM. 2000. Structures of helical junctions in nucleic acids. Q Rev Biophys 33: 109–159. [DOI] [PubMed] [Google Scholar]
- Lilley DM. 2003. The origins of RNA catalysis in ribozymes. Trends Biochem Sci 28: 495–501. [DOI] [PubMed] [Google Scholar]
- Liu H, Guo S, Roll R, Li J, Diao Z, Shao N, Riley MR, Cole AM, Robinson JP, Snead NM, et al. 2007. Phi29 pRNA vector for efficient escort of hammerhead ribozyme targeting survivin in multiple cancer cells. Cancer Biol Ther 6: 697–704. [DOI] [PubMed] [Google Scholar]
- Liu J, Guo S, Cinier M, Shlyakhtenko LS, Shu Y, Chen C, Shen G, Guo P. 2011. Fabrication of stable and RNase-resistant RNA nanoparticles active in gearing the nanomotors for viral DNA packaging. ACS Nano 5: 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H. 2001. The enhanced permeability and retention (EPR) effect in tumor vasculature: the key role of tumor-selective macromolecular drug targeting. Adv Enzyme Regul 41: 189–207. [DOI] [PubMed] [Google Scholar]
- Maeda H, Nakamura H, Fang J. 2013. The EPR effect for macromolecular drug delivery to solid tumors: improvement of tumor uptake, lowering of systemic toxicity, and distinct tumor imaging in vivo. Adv Drug Deliv Rev 65: 71–79. [DOI] [PubMed] [Google Scholar]
- Masquida B, Felden B, Westhof E. 1997. Context dependent RNA-RNA recognition in a three-dimensional model of the 16S rRNA core. Bioorg Med Chem 5: 1021–1035. [DOI] [PubMed] [Google Scholar]
- Mulhbacher J, St-Pierre P, Lafontaine DA. 2010. Therapeutic applications of ribozymes and riboswitches. Curr Opin Pharmacol 10: 551–556. [DOI] [PubMed] [Google Scholar]
- Novikova IV, Hassan BH, Mirzoyan MG, Leontis NB. 2010. Engineering cooperative tecto-RNA complexes having programmable stoichiometries. Nucleic Acids Res 39: 2903–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy A. 2011. Bionanoelectronics. Adv Mater 23: 807–820. [DOI] [PubMed] [Google Scholar]
- Paige JS, Nguyen-Duc T, Song W, Jaffrey SR. 2012. Fluorescence imaging of cellular metabolites with RNA. Science 335: 1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieken WA, Olsen DB, Benseler F, Aurup H, Eckstein F. 1991. Kinetic characterization of ribonuclease-resistant 2′-modified hammerhead ribozymes. Science 253: 314–317. [DOI] [PubMed] [Google Scholar]
- Privalov PL, Filiminov VV. 1978. Thermodynamic analysis of transfer RNA unfolding. J Mol Biol 122: 447–464. [DOI] [PubMed] [Google Scholar]
- Qiu M, Khisamutdinov E, Zhao Z, Pan C, Choi J, Leontis N, Guo P. 2013. RNA nanotechnology for computer design and in vivo computation. Phil Trans R Soc A 371: 20120310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid RJD, Bodley JW, Anderson D. 1994. Characterization of the prohead-pRNA interaction of bacteriophage ϕ29. J Biol Chem 269: 5157–5162. [PubMed] [Google Scholar]
- Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R, Benenson Y. 2007. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat Biotechnol 25: 795–801. [DOI] [PubMed] [Google Scholar]
- Rychahou P, Haque F, Shu Y, Zaytseva Y, Weiss HL, Lee EY, Mustain W, Valentino J, Guo P, Evers BM. 2015. Delivery of RNA nanoparticles into colorectal cancer metastases following systemic administration. ACS Nano 9: 1108–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarver NA, Cantin EM, Chang PS, Zaia JA, Ladne PA, Stephens DA, Rossi JJ. 1990. Ribozymes as potential anti-HIV-1 therapeutic agents. Science 24: 1222–1225. [DOI] [PubMed] [Google Scholar]
- Severcan I, Geary C, Jaeger L, Bindewald E, Kasprzak W, Shapiro BA. 2009. Computational and experimental RNA nanoparticle design. In Automation in genomics and proteomics: an engineering case-based approach (ed. Alterovitz G, Ramoni M), pp. 193–220. Wiley, Boston, MA. [Google Scholar]
- Shu D, Huang L, Hoeprich S, Guo P. 2003. Construction of phi29 DNA-packaging RNA (pRNA) monomers, dimers and trimers with variable sizes and shapes as potential parts for nano-devices. J Nanosci Nanotechnol 3: 295–302. [DOI] [PubMed] [Google Scholar]
- Shu D, Moll WD, Deng Z, Mao C, Guo P. 2004. Bottom-up assembly of RNA arrays and superstructures as potential parts in nanotechnology. Nano Lett 4: 1717–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. 2011a. Thermodynamically stable RNA three-way junctions for constructing multi-functional nanoparticles for delivery of therapeutics. Nat Nanotechnol 6: 658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Cinier M, Shu D, Guo P. 2011b. Assembly of multifunctional phi29 pRNA nanoparticles for specific delivery of siRNA and other therapeutics to targeted cells. Methods 54: 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Haque F, Shu D, Li W, Zhu Z, Kotb M, Lyubchenko Y, Guo P. 2013. Fabrication of 14 different RNA nanoparticles for specific tumor targeting without accumulation in normal organs. RNA 19: 766–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Khisamutdinov E, Zhang L, Guo P. 2014a. Programmable folding of fusion RNA in vivo and in vitro driven by pRNA 3WJ motif of phi29 DNA packaging motor. Nucleic Acids Res 42: e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Pi F, Sharma A, Rajabi M, Haque F, Shu D, Leggas M, Evers BM, Guo P. 2014b. Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Adv Drug Deliv Rev 66C: 74–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. 1995. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 34: 11211–11216. [DOI] [PubMed] [Google Scholar]
- Tabernero J, Shapiro GI, Lorusso PM, Cervantes A, Schwartz GK, Weiss GJ, Paz-Ares L, Cho DC, Infante JR, Alsina M, et al. 2013. First-in-man trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov 3: 406–417. [DOI] [PubMed] [Google Scholar]
- Win MN, Smolke CD. 2008. Higher-order cellular information processing with synthetic RNA devices. Science 322: 456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. 2004. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428: 281–286. [DOI] [PubMed] [Google Scholar]
- Xie Z, Liu SJ, Bleris L, Benenson Y. 2010. Logic integration of mRNA signals by an RNAi-based molecular computer. Nucleic Acids Res 38: 2692–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. 2011. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science 333: 1307–1311. [DOI] [PubMed] [Google Scholar]
- Ye X, Hemida M, Zhang HM, Hanson P, Ye Q, Yang D. 2012. Current advances in Phi29 pRNA biology and its application in drug delivery. Wiley Interdiscip Rev RNA 3: 469–481. [DOI] [PubMed] [Google Scholar]
- Yingling YG, Shapiro BA. 2007. Computational design of an RNA hexagonal nanoring and an RNA nanotube. Nano Lett 7: 2328–2334. [DOI] [PubMed] [Google Scholar]
- Zhang CL, Lee C-S, Guo P. 1994. The proximate 5′ and 3′ ends of the 120-base viral RNA (pRNA) are crucial for the packaging of bacteriophage φ29 DNA. Virology 201: 77–85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.