Abstract
It is common wisdom that codon usage bias has evolved in the selection for efficient translation, in which highly expressed genes are encoded predominantly by optimal codons. However, a growing body of evidence suggests regulatory roles for non-optimal codons in translation dynamics. Here we report that in mammalian cells, non-optimal codons play a critical role in promoting selective mRNA translation during amino acid starvation. During starvation, in contrast to genes encoding ribosomal proteins whose translation is highly sensitive to amino acid deprivation, translation of genes involved in the cellular protein degradation pathways remains unaffected. We found that these two gene groups bear different codon composition, with non-optimal codons being highly enriched in genes encoding the ubiquitin–proteasome system. Supporting the selective tRNA charging model originally proposed in Escherichia coli, we demonstrated that tRNA isoacceptors decoding rare codons are maintained in translating ribosomes under amino acid starvation. Finally, using luciferase reporters fused with endogenous gene-derived sequences, we show that codon optimality contributes to differential mRNA translation in response to amino acid starvation. These results highlight the physiological significance of codon usage bias in cellular adaptation to stress.
Keywords: translation, genetic code, codon bias, tRNA, starvation, adaptation
INTRODUCTION
In a post-genomic era, the importance of translational control in gene expression has been increasingly appreciated (Schwanhausser et al. 2011). Under adverse conditions, many stress-signaling pathways converge on key translational factors, thereby attenuating global protein synthesis (Sonenberg and Hinnebusch 2009; Spriggs et al. 2010). However, subsets of mRNAs undergo selective translation to produce proteins that are vital for cell survival and stress recovery (Liu and Qian 2014). This stress adaptation is particularly important under nutrient deprivation. In response to scarcity of essential nutrients like amino acids, cells undergo reprogramming to up-regulate cellular degradation pathways including the ubiquitin–proteasome pathway (UPP) and autophagy (He and Klionsky 2009). As a result, efficient degradation of cellular proteins ensures recycling of amino acids when the external supply is lost. Amino acids are building blocks for protein synthesis. It is not surprising that mRNA translation is tightly coupled to nutrient sensing systems such as the mammalian target of rapamycin complex I (mTORC1) and the general control nonderepressible 2 (GCN2) kinases (Wek et al. 2006; Ma and Blenis 2009). mTORC1 phosphorylates multiple targets that concertedly regulate protein metabolism. For instance, through phosphorylation of 4E-binding protein (4E-BP), mTORC1 increases cap-dependent translation (Ma and Blenis 2009; Zoncu et al. 2011). Conversely, through inhibition of Unc51-like kinases 1/2 (Ulk1/2), mTORC1 suppresses autophagy (Mizushima 2010). Upon nutrient deprivation, suppressed mTORC1 signaling not only reduces global protein synthesis but also triggers activation of the autophagy pathway (He and Klionsky 2009). Additionally, phosphorylation of the serine amino acid residue 51 (Ser-51) in the eukaryotic initiation factor 2α by GCN2 mediates translation control during nutrient deprivation (Dever et al. 1992; Berlanga et al. 1999; Sood et al. 2000). Despite the progress in our understanding of these signaling pathways, it remains puzzling how cells maintain continuous synthesis of proteins essential for the degradation pathway under the limited supply of amino acids. Neither the specific translation-promoting features of these messages nor the regulatory mechanism has been clearly defined.
During translation, ribosomes rely on aminoacylated (charged) tRNAs to read codons in mRNAs. Since there are 61 sense codons for only 20 amino acids, each amino acid is often encoded by multiple synonymous codons. Notably, synonymous codons are not present in equal frequencies and such codon usage bias is pervasive across species, genomes, as well as individual genes (Plotkin and Kudla 2011). Similarly, tRNA isoacceptors carrying the same amino acid are not uniformly expressed in cells (Bulmer 1987). Recent studies reported differential tRNA expression levels between cellular proliferation and differentiation (Gingold et al. 2014), highlighting the critical role of tRNA concentrations in fundamental biological processes. The concept of codon optimality reflects the balance between the supply of charged tRNAs in the cytosolic pool and the demand of tRNA usage by translating ribosomes. It is generally believed that optimal codons are decoded faster and more accurately by the ribosome than non-optimal codons (Drummond and Wilke 2009). Consistent with this notion, there is a predominant use of optimal codons in highly expressed genes (Sharp and Li 1986; Hershberg and Petrov 2008; Tuller et al. 2010). In contrast, non-optimal codons are postulated to slow down translation elongation. By tuning the elongation rate, codon usage bias has been reported to influence the behavior of nascent chains, including cotranslational folding as well as interaction with the signal recognition particle (SRP) (Pechmann et al. 2014; Yu et al. 2015).
Despite several reported effects, it is not clearly established whether synonymous codon choices imbue certain messages with different translational potential, and whether this can influence their expression during stress conditions, especially during starvation. Intriguingly, amino acid starvation induces selective charging of rare tRNA isoacceptors in Escherichia coli, as initially predicted by a theoretical model (Elf et al. 2003) and later confirmed by experimental evidence (Dittmar et al. 2005). Hence it seems likely that the codon composition in eukaryotic genomes could contribute to selective mRNA translation in response to amino acid deprivation. In this study, we have addressed this important question in the context of messages encoding the UPP system. We demonstrate that codon optimality has a broad and powerful contribution to differential translation of these genes in response to amino acid starvation in mammalian cells. To gain mechanistic insights into starvation adaptation in mammalian cells, we took advantage of genome-wide ribosome profiling, global codon usage analysis, quantitative tRNA arrays, and luciferase reporter assays. Our data reveal a previously unappreciated mechanism underlying cellular adaptation to amino acid shortage at the level of mRNA translation.
RESULTS
Differential mRNA translation during amino acid starvation
To determine the global effect of amino acid starvation on mRNA translation, we performed ribosome profiling (Ribo-seq) on HEK293 cells grown either in complete media or in amino acid-depleted media for 1 h (Gao et al. 2015). Ribo-seq provides a snapshot of ribosome positions on the mRNA at a given time. We computed the relative translation efficiency (TE) of individual transcripts by normalizing Ribo-seq data to mRNA levels measured by RNA-seq. While a large group of genes underwent translational attenuation upon starvation (486 genes with a decrease of more than threefold), a considerable number of genes exhibited translational up-regulation (735 genes with an increase of more than threefold) (Fig. 1A). In line with previous studies (Ingolia et al. 2009; Gao et al. 2015), genes with substantial translational suppression are enriched with ribosome biogenesis and translation as revealed by gene ontology (GO) analysis (Supplemental Fig. S1A). Wilcoxon test showed that the fold change of translation genes is significantly lower than the genome average (0.33 of translation vs. 1.42 of genome, P < 2.2 × 10−16). Genes showing resistance to translational attenuation demonstrated a rather broad category and none of the GO terms were significantly enriched (Supplemental Fig. S1A). However, despite the lack of a significant difference with respect to the genome (Wilcoxon test value 1.40 of UPP genes vs. 1.42 of genome, P = 0.85), nearly all the genes encoding UPP and autophagy pathways maintained or up-regulated their translation efficiency during amino acid deprivation (Fig. 1B).
FIGURE 1.
Differential mRNA translation during amino acid starvation. (A) HEK293 cells with or without amino acid starvation (1 h) were subjected to Ribo-seq and RNA-seq followed by computation of relative translation efficiency (TE). Distribution of TE fold change upon starvation is presented with up-regulated transcripts (n = 735) shown in red and down-regulated transcripts (n = 486) shown in green. The TE fold change threshold is set at threefold, which is shown in log2 scale. (B) TE fold change of transcripts encoding translation (green) and the ubiquitin-proteasome system (red). The genes involved in the ubiquitin-proteasome system and translation pathways were identified based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Supplemental Tables S2, S3). (C) Polysome profile for HEK293 cells with or without amino acid starvation was subjected to sucrose gradient sedimentation. Equal volumes of fractions were analyzed for the distribution of various mRNAs using RT-qPCR. The ratio of polyribosome-associated mRNA relative to the monosome-associated is presented for each gene. Data shown here are representative of three biological replicates. Error bar, ±SD; (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, n = 3.
Eukaryotic 26S proteasome is an ATP-dependent protease that works in tandem with the ubiquitin system to break down proteins in the cell (Adams 2003; Murata et al. 2009). The proteasome is a multi-subunit protein, where three subunits have hydrolytic activity as threonine proteases for the cleavage of peptide bonds. These activities are caspase-like activity (or peptidylglutamyl-peptide hydrolyzing activity), trypsin-like activity, and chymotrypsin-like activity, respectively (Murata et al. 2009). Direct measurement of proteasome function showed a rapid increase of chymotrypsin activities in cells upon amino acid withdrawal (Supplemental Fig. S1B). Given the critical role of degradation pathways in recycling internal amino acids, it makes intuitive sense that the up-regulation of UPP and autophagy-related proteins is essential for cell survival during starvation.
To validate the ribosome profiling results, we directly measured the distribution of several UPP transcripts between monosome and polysome fractions in cells with or without starvation (Fig. 1C). A recent study by Heyer and Moore has shown that a certain subset of mRNAs in Saccharomyces cerevisiae can be translated by monosomes (Heyer and Moore 2016). However, the study did not show that UPP genes are translated by monosomes. Since it is generally believed that most actively translated mRNAs are enriched in polysomes, whereas inactive mRNAs are associated with monosomes (Arava et al. 2003), we used the polysome to monosome ratio to help us determine whether a particular mRNA is efficiently or poorly translated. As expected, during starvation genes encoding ubiquitin-activating enzymes (UBA1, UBA3), NEDD8-activating enzyme (NAE1), and autophagy-related gene (ATG7) showed little changes or even increased mRNA levels in the polysome (Fig. 1C; Supplemental Fig. S2). In contrast, genes encoding housekeeping proteins, and ribosomal proteins RPS5, RPS11, RPL6, and RPL28 showed evident reduction of their transcripts in polysome fractions upon amino acid starvation (Fig. 1C; Supplemental Fig. S2). Therefore, translation of UPP mRNA subsets is clearly resistant to amino acid starvation.
Codon optimality contributes to differential mRNA translation during amino acid starvation
Under adverse conditions like amino acid deprivation, translational response mainly occurs at the stage of initiation (Sonenberg and Hinnebusch 2009; Spriggs et al. 2010). However, for mRNAs undergoing selective translation, we lack a clear understanding of how elongation proceeds under the shortage of charged tRNAs. Having found the differential translation of mRNAs encoding the UPP genes, in contrast to ribosomal genes in response to amino acid deprivation, we considered whether the codon composition of these two gene groups might be different. We examined the relative synonymous codon usage (RSCU) profiles of UPP genes (up-regulated TE) and ribosomal genes (downregulated TE) (Fig. 2A). Using the codon usage frequency in the human genome as a reference, we separated rare codons from common codons. Although common codons exhibit no distinct patterns between the two gene groups, rare codons are more enriched in UPP genes. Some rare codons such as ATA (isoleucine), TTA, and CTA (leucine) are abundant in the majority of UPP genes, but absent in many genes encoding ribosomal proteins. We further confirmed this finding using the tRNA adaptation index (tAI) (dos Reis et al. 2004), an alternative measure of codon usage that is measured by taking into consideration the relative copy number of tRNA isoacceptors (Fig. 2B). The frequency of non-optimal codons is significantly higher in UPP genes than in ribosomal genes (Mann Whitney test, P < 0.0001). The frequency of non-optimal codons in UPP genes was also higher than the genome average (Fig. 2B).
FIGURE 2.

Codon composition contributes to differential mRNA translation during amino acid starvation. (A) The relative synonymous codon usage (RSCU) profile for UPP and translation genes is presented as a heatmap. Relative codon usage frequency for each codon in a gene was calculated by dividing the frequency of that particular codon by the total number of synonymous codons under the assumption of equal usage of the synonymous codons for an amino acid. The codons are divided into common and rare categories based on the human codon usage frequency. The absence of a codon is denoted by white. The color change from yellow to green denotes a lower to higher enrichment of a particular codon. (B) Boxplot showing the frequency of non-optimal codons found in the whole genome (whole human transcriptome), UPP genes, and translation genes. The codons are divided into optimal and non-optimal categories based on their tRNA adaptation index (tAI) values. tAI value for the genes was calculated using a previously published algorithm (dos Reis et al. 2004). (C) Scatterplot showing the correlation between codon adaptation index (CAI) and TE fold change upon starvation for UPP and translation genes. The Pearson correlation coefficient is shown in the plot. TE fold change for each gene was generated from Ribo-seq data. CAI values were calculated using Biopython package.
Next, we decided to question whether the differential response of UPP vs. ribosomal protein-related genes could be attributed to distinct codon adaptation in response to amino acid limitation. We found that, for both gene groups, starvation-induced changes of translation efficiency are negatively correlated with the codon adaptation index (CAI) (r = −0.6316) (Fig. 2C). The CAI value of a gene is the cumulative score of the synonymous codon usage bias of the codons that make up that gene. A higher CAI value indicates a higher frequency of common codons and a higher probability of gene expression (Sharp and Li 1987). A negative correlation between CAI values and starvation-induced TE changes suggested that in both these gene families common-codon enrichment leads to higher sensitivity to starvation (as evident in ribosomal genes). In contrast, rare codon-enriched genes become more resistant to amino acid shortage, thereby achieving selective mRNA translation (as evident in UPP genes). The correlation between starvation-induced translation efficiency and CAI does not hold for the entire genome (Supplemental Fig. S3). This is not surprising because synonymous codon usage patterns are created in the genome to reflect the action of natural selection (Plotkin and Kudla 2011). The relationship between TE and codon bias observed in UPP and ribosomal genes is perhaps an adaptive response to starvation acquired through evolution.
Rare tRNAs are enriched in elongating ribosomes during amino acid starvation
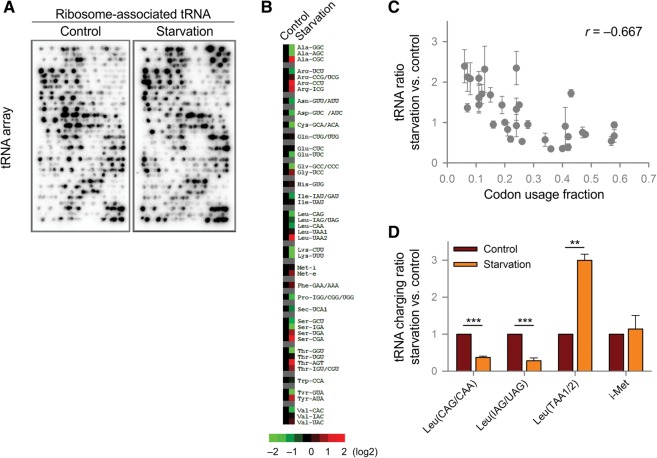
How does the presence of rare codons promote mRNA translation during amino acid starvation? We wondered if it is possible that the messages containing more non-optimal codons are less sensitive to the limited supply of charged tRNA molecules. According to the “selective tRNA charging” model, proposed and validated for E. coli (Elf et al. 2003), a general prediction is that the charged levels of tRNA isoacceptors depend on the ratios between their total concentrations and the frequencies at which their cognate codons appear in the transcriptome. When the supply of amino acids becomes rate limiting, the charged levels of certain tRNA isoacceptors decoding common codons will approach zero, but not the isoacceptors for rare codons. To provide direct evidence that this theory also holds in mammalian cells, we examined ribosome-associated tRNA levels using a microarray method. Similar to ribosome profiling, we collected RNase I-digested polyribosome samples followed by isolation of tRNA species occupying E, P, and A sites of the ribosome (Supplemental Fig. S4A). In parallel, we purified total tRNA molecules from whole cell lysates as control. Hybridization of radiolabeled tRNA samples to specific probes revealed relative ratios of charged tRNA molecules in translating ribosomes. While cell lysates contained both nuclear and mitochondrial tRNA species, ribosome samples were highly enriched with nuclear-encoded tRNAs (Supplemental Fig. S4B). Therefore, ribosome-associated tRNA species represent the pool of tRNA isoacceptors actively engaging in translation. To further validate that the tRNA microarrays are an accurate depiction of tRNAs associated with A/P/E ribosome sites, we performed the microarray experiment on RNA obtained from total cell lysate (total RNA), polysome fractions of cells treated with cycloheximide (CHX), and monosome fractions of cells treated with lactimidomycin (LTM). LTM treatment freezes ribosomes in the initiation step of translation (Lee et al. 2012). The microarray clearly shows an enrichment of initiator-tRNAMet in the LTM-treated sample (Supplemental Fig. S5).
Next we used the microarrays to compare the landscape of ribosome-associated tRNAs from cells with and without amino acid starvation (Fig. 3A). Although there was no clear tRNA pattern clustered by amino acids, we observed remarkable differences of tRNA isoacceptors decoding the same amino acid (Fig. 3B). For instance, rare tRNAs leucine isoacceptors, tRNALeu (UAA1/2), showed increased signals in starvation samples, whereas the most abundant leucine isoacceptor, tRNALeu (CAG), exhibited a concomitant decrease. The similar trend holds true for isoacceptors of tRNASer, tRNAThr, and tRNAVal. In particular, there is a strong inverse correlation between the changes of tRNA isoacceptors and their codon usage fractions (Fig. 3C). Therefore, the abundant tRNA isoacceptors are more likely depleted from translating ribosomes than the rare tRNAs under limited amino acid supply. This result is consistent with the observation in E. coli where tRNA isoacceptors that read rare codons retain high charging levels during starvation (Dittmar et al. 2005). As an additional test we measured the charging level of various tRNALeu isoacceptors in control and amino acid starved cells using a qRT-PCR-based method (Loayza-Puch et al. 2016). Our results show that the charging level of all the abundant isoacceptors decreased while the charging level of rare isoacceptors tRNALeu(UAA1/2) increased during amino acid starvation (Fig. 3D). The level of charged initiator-methionine tRNA (i-Met) remained the same upon starvation, as observed in our array data (Fig. 3B,D). The tRNA charging data further validate our hypothesis that during starvation rare tRNA isoacceptors are enriched in translating ribosomes, and this causes the translation of messages containing rare codons to be relatively resistant to amino acid deprivation.
FIGURE 3.
tRNA isoacceptors decoding rare codons are selectively enriched in translating ribosomes during amino acid starvation. (A) Representative tRNA microarray results obtained from HEK293 cells with or without amino acid starvation. (B) Heatmap showing enrichment of specific tRNAs associated with translating ribosomes during amino acid starvation. The isoacceptors for each tRNA are arranged in the decreasing order of their frequency from top to bottom. The signal obtained for each tRNA during starvation is normalized to the control. The heatmap plots the log2 of normalized tRNA enrichment values. The color change from green to red signifies an increased association with the ribosome. This heatmap is representative of two biological replicates (Supplemental Fig. S4C). (C) Scatter plot showing the correlation between tRNA isoacceptor signals after starvation and the codon usage fraction. tRNAs without isoacceptors are not included in this analysis. The Pearson correlation coefficient is shown in the plot. (D) tRNA charging levels of tRNA isoacceptors in control and starvation conditions. The assay used for this qRT-PCR-based analysis was adapted from Loayza-Puch et al. (2016). Primers used for qPCR are described in Materials and Methods. Error bar, ±SD; (*) P < 0.05, (**) P < 0.01, (***) P < 0.001.
Codon composition influences translation efficiency during amino acid starvation
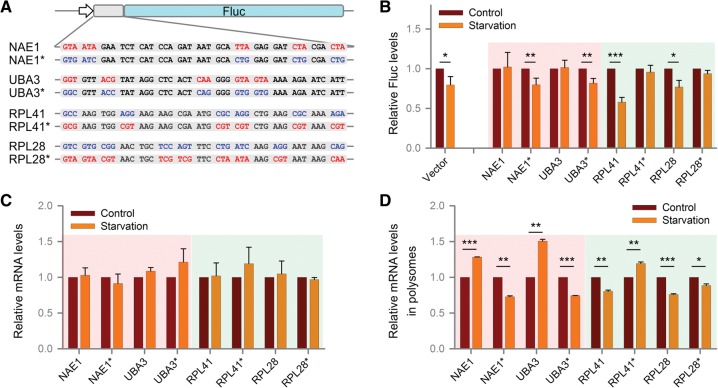
To further demonstrate the correlation between codon usage and translational potential during amino acid starvation, we constructed firefly luciferase (Fluc) reporters by fusing an extra 15-codon sequence at the NH2 terminus (Fig. 4A). Sequences containing rare codons were chosen from UPP genes (NAE1 and UBA3), whereas sequences enriched with common codons were derived from genes encoding ribosomal proteins (RPL41 and RPL28). Consistent with the behavior of endogenous genes, chimeric reporters bearing either NAE1 or UBA3 sequences maintained Fluc levels in transfected cells exposed to amino acid starvation (Fig. 4B). In contrast, reporters bearing the RPL41 or RPL28 sequences showed reduced Fluc levels during amino acid deprivation. Therefore, addition of sequences with different codon usage leads to a distinct translational response to amino acid shortage.
FIGURE 4.
Rare codons contribute to differential mRNA translation during amino acid starvation. (A) Fluc reporters were constructed by fusing a 15-codon sequence derived from UPP (UBA3 and NAE1) or ribosomal protein (RPL41, RPL28) genes in the amino termini. Certain rare codons in the UPP genes were changed to common codons while certain common codons in the ribosomal proteins were changed to rare codons without changing the encoded amino acid (mutants denoted by asterisk). The codon features are color-coded with red for rare codons and blue for common codons. (B) HEK293 cells were transfected with plasmids encoding the Fluc reporters illustrated in A followed by amino acid starvation. The relative Rluc activities under starvation are normalized by control. (C) The relative mRNA level of all the Fluc reporters was determined using qRT-PCR during control and amino acid starvation. The forward and reverse primers used for PCR covered a region of the inserted gene sequence and a region of the luciferase sequence, respectively. The primer sequences are included in Supplemental Table S1. (D) Polysome profile distribution was determined for each Fluc reporter mRNA during control and amino acid starvation. Equal volumes of fractions were analyzed for the distribution of the target mRNAs using RT-qPCR. The ratio of polyribosome-associated mRNA fraction relative to the monosome-associated fraction is presented for each gene. The primers used are the same as described in C. (B, C, D) Error bar, ±SD; (*) P < 0.05, (**) P < 0.01, (***) P < 0.001; n = 3.
We next swapped the synonymous codons in these reporters to validate the role of codon composition in starvation-induced translational regulation. Without changing the encoded amino acid, we replaced the rare codons with the common codons in reporters containing sequences derived from UPP genes (Fig. 4A). Remarkably, these mutant reporters regained the translational response after amino acid starvation by showing reduced Fluc levels in transfected cells (Fig. 4B). Furthermore, when the common codons of RPL41 or RPL28 were changed to the corresponding rare codons, these reporters maintained their translation efficiency in transfected cells upon amino acid deprivation (Fig. 4B). Additionally, we tested the total mRNA levels of all four reporters in control and starvation conditions; our data showed that the total mRNA levels for all of them remain unaltered during starvation (Fig. 4C). The polysome distribution of the reporter mRNAs during control and amino acid starvation exhibited the same pattern as observed for the native mRNAs (Fig. 4D; Supplemental Fig. S6). Together, these results firmly establish the critical role of codon optimality in translational adaptation in response to starvation.
DISCUSSION
Codon usage bias is a universal feature of all genomes and has been proposed to regulate translation efficiency, accuracy, mRNA stability, and protein folding (Plotkin and Kudla 2011; Presnyak et al. 2015). Although it is widely believed that codons adapted to tRNA pools might be preferentially used in highly expressed genes, the precise nature of fitness gain associated with translational adaptation remains a topic of active debate. Despite the fact that “codon optimization” might lead to enhanced protein production, maintaining rare codons in genomes could serve beneficial purposes in gene expression. Two recent studies using Neurospora and Synechococcus elongate reported that rare codons present in circadian genes play a critical role in maintaining the circadian rhythm (Xu et al. 2013; Zhou et al. 2013). In yeast cells, genes encoding membrane and secretory proteins are enriched with rare codons at positions critical for SRP recognition (Pechmann et al. 2014). All these phenomena rely on rare codon-mediated translation slowdown that potentially influences co-translational folding (Yu et al. 2015). Our present work demonstrates a surprising role for non-optimal codons in promoting mRNA translation during amino acid starvation. Our findings not only expand the functional role of codon usage but also uncover an important cellular adaptation mechanism in response to amino acid deprivation.
Amino acids are essential cellular nutrients, hence it is imperative for cells to adapt to amino acid deprivation via multiple mechanisms. Up-regulation of the UPP pathway ensures efficient protein degradation and recycling of amino acids when the external supply is limited (He and Klionsky 2009). We have previously reported that UPP mRNAs maintained their translation potential during amino acid starvation (Gao et al. 2015), although the underlying mechanism remained elusive. It has been suggested that by sensing the presence of nutrients, activated mammalian TOR complex 1 (mTORC1) signals to various components of the translation initiation machinery to regulate cap-dependent translation (Gingras et al. 2004; Hay and Sonenberg 2004). mTOR-dependent translation reprogramming in vivo has been studied by using ribosome profiling (Hsieh et al. 2012). Treatment of pancreatic cancer cells with mTOR inhibitor PP242 and rapamycin affected the translation efficiency of several target mRNAs. UPP mRNAs were not identified in this group of mTOR responsive genes. It is possible, however, that the decreased polysome concentration and reduced TE of the ribosomal genes could be the result of diminished mTOR signaling. However, our data strongly indicate that the nature of codons present in the mRNAs can also play an important role in the translation of these mRNAs, especially during amino acid starvation.
Additionally, computational simulation studies suggest that initiation is rate-limiting under normal growth conditions but elongation becomes the limiting step under severe amino acid starvation (Firczuk et al. 2013; Racle et al. 2013; Shah et al. 2013). In E. coli, genes encoding amino acid biosynthetic enzymes preferentially use codons that are poorly adapted to the typical pool of charged tRNAs, but are well adapted to starvation-induced tRNA pools (Dittmar et al. 2005). We, for the first time, demonstrate the presence of a similar mechanism in mammalian cells, by which UPP mRNAs are selectively translated during amino acid starvation. Our results reveal a coordinated regulation between amino acid availability, tRNA charging, and selective mRNA translation. It highlights the physiological significance of codon usage bias in cellular adaptation and survival.
MATERIALS AND METHODS
Cell culture
Human embryonic kidney 293 (HEK293) cells were cultured in high glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. For amino acid starvation treatments, the cells were grown in Hank's balanced salt solution (HBSS) supplemented with 10% dialyzed fetal bovine serum. Cells were treated with 100 µg/mL cycloheximide (CHX) for 3 min prior to ribosome profiling (Ribo-seq) experiments.
Ribosome profiling and data analysis
Ribo-seq and RNA-seq were performed based on the reported protocol (Gao et al. 2015). Briefly, cells were treated with CHX (100 µg/mL) for 3 min at 37°C and then lysed in polysome buffer (pH 7.4, 10 mM HEPES, 100 mM KCl, 5 mM MgCl2, and 100 µg/mL CHX) by vortexing for 20 sec using Lysing Matrix-D (Fisher) for five times with an interval of 40 sec on ice. The cleared lysates were separated by sedimentation through sucrose gradients (15%–45% w/v). Collected polysome fractions were digested with RNase I and the ribosome protected fragments (RPFs) were size selected and purified by gel extraction. After library construction, deep sequencing was performed using Illumina HiSEQ2000. The trimmed RPF reads were first mapped by Tophat to transcriptome (Ensembl release 70). Non-uniquely mapped reads were disregarded for further analysis due to ambiguity. The 13th position (12 nt offset from the 5′ end) of the uniquely mapped read was defined as the ribosome “P-site” position. The quantification of P-site was completed by mapping uniquely mapped reads to each individual mRNA transcript according to the NCBI Refseq gene annotation. Thirty-one and 39 million Ribo-seq reads were obtained for control and amino acid (AA) starvation, respectively, similarly 10 and 16 million reads were obtained for RNA-seq. These uniquely mapped reads were counted to calculate the mRNA and ribosome footprint RPKM values for all transcripts. For each transcript, the translation efficiency (TE) was calculated by dividing ribosome footprint RPKM by the corresponding mRNA RPKM. The fold change of translation efficiency was calculated by the ratio of TE in control vs. the TE in AA starvation. The genes with TE fold change >2 and <0.5 were used for Gene Ontology analysis (GO analysis). GO analyses were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID). GO term enrichment was summarized and visualized with REViGO.
Global codon usage calculation and other data analysis
Structures of Refseq protein-coding genes were downloaded from the UCSC Genome Browser. To create the relative synonymous codon usage (RSCU) profile for individual translation and UPP-related genes, the transcript isoform with the longest coding sequence (CDS) length was selected as the representative transcript. Each of the 61 sense codons was counted, respectively, on individual mRNA CDS, and the individual codon count was later divided by the total number of codons of CDS to obtain relative codon usage. RSCU value for every codon was calculated under the assumption of equal usage of the synonymous codons for an amino acid. tRNA adaptation index (tAI) calculations were performed according to previously published protocol (dos Reis et al. 2004). CAI values for the genes were calculated using the Biopython package. The statistical analysis throughout the manuscript was performed using Statistical Package R, and all other computation analysis was performed using in-house programming.
Isolation and labeling of tRNA from RPFs
HEK293 cells grown in control and starvation media were treated with CHX (100 µg/mL for 3 min) and ribosome-protected fragments (RPFs) were isolated as described in Ribo-seq experiments. Total RNA was obtained from RPFs using TRIzol (Invitrogen) according to the manufacturer's protocol. Total RNA (5 μg) was labeled with radioactive γ-32P-ATP and separated on 10% TBE-urea acrylamide gels. The tRNA band on the gel was visualized with a phosphor-imager and excised based on the expected size. The tRNA was then eluted in 450 µL of crush and soak buffer (50 mM KOAc, 200 mM KCl, pH 7.5) with rocking at 4°C overnight. After centrifugation, ethanol precipitation was performed to obtain the clean tRNA samples.
Hybridization and microarray analysis
Hybridization and microarray analysis was performed based on previously reported protocols (Saikia et al. 2012). Briefly, the labeled tRNA sample was first dissolved in microarray hybridization buffer (Sigma-Aldrich) containing 20 μg of salmon sperm DNA and 10 μg of poly(A). This mixture was then applied to the hybridization chamber of the GeneTAC Hyb4 station (Genomic Solutions). The following program was used for hybridization: 75°C (2 min), 60°C (probe introduction), 90°C (5 min), and 60°C hybridization for 16 h. Following hybridization, the slides were washed on the Hyb4 station twice with 2× SSC, 0.1% (w/v) SDS at 50°C, twice with 0.1× SSC, 0.1% (w/v) SDS at 42°C, and twice with 0.1× SSC at 42°C. Slides were then removed from the station, rinsed with 0.1× SSC, and dried by centrifugation. The slides were then exposed to the phosphor-imager to visualize the signals deriving from the hybridized tRNAs. Microarray slides were imaged using a FUJI BAS scanner. 32P intensities were quantified and corrected for background noise using Fuji BAS software. The median values for eight replicate spots were obtained for each tRNA and divided by the median value of the two added tRNA standards (E. coli tRNALys and yeast tRNAPhe) to obtain the normalized signal for each tRNA in a particular sample.
Luciferase assay
Cells were transfected with the luciferase reporter for 24 h followed by splitting equally in two wells of a six-well plate for another 24 h incubation. The cells were then subjected to either complete (control) or amino acid depleted media (starvation) for 1 h. Cell pellets were lysed in reporter lysis buffer (Promega) followed by centrifugation to clear the lysates. Luminescence reactions were initiated with Promega DLR (100 μL; Promega) added to the lysates (30 μL). Luciferase activities were measured using a Synergy 2 Luminescence Plate Reader (Biotek).
Proteasome activity assay
HEK293 cells were plated equally (10,000 cells per well) in 96-well white-walled plates. The wells were subjected to amino acid starvation conditions for various time points (0, 0.5, 1, 2, and 3 h, respectively). Proteasome-Glo Cell-Based Reagent (Promega, catalog # G8660) was prepared as per manufacturer's protocol and an equal volume was added to each well. The content of the plate was mixed at 700 RPM for 2 min and then incubated at room temperature for 10 min. Luminescence was read using a Synergy 2 Luminescence Plate Reader (Biotek).
tRNA-charging assay
The tRNA-charging assay protocol was adapted from a recent publication (Loayza-Puch et al. 2016). Briefly, RNA was isolated using acetate-saturated phenol/CHCl3 (pH 4.8). Precipitated RNA was resuspended in 10 mM NaOAc/HOAc (pH 4.8). Samples were split in two, one half (5 μg) was oxidized with 50 mM NaIO4 for 30 min at room temperature and the other half (5 μg) was incubated in 50 mM NaCl. Samples were quenched with 100 mM glucose for 5 min at room temperature, purified in G25 columns (GE Healthcare), and then ethanol precipitated. tRNAs were deacylated in 50 mM Tris–HCl (pH 9) for 30 min at 37°C. RNA was precipitated and then ligated to the 3′adaptor (5′-/5rApp/TGGAATTCTCGGGTGCCAAGG/3ddC/-3′) using T4 RNA ligase 2 (NEB) for 4 h at 37°C. Relative aminoacylation levels were calculated by qRT-PCR using tRNA-specific primers. Due to similar sequences in some cases, multiple tRNA isoacceptors were detected using the same primer. Primer sequences are as follows: reverse primer, GCCTTGGCACCCGAGAATTCCA; tRNA Leu(CAG/CAA) primer, GTCAGGATGGCCGAGCGGTC; tRNA Leu(IAG/UAG), GGTAGCGTGGCCGAGCGGTC; tRNA Leu(TAA1/2), ACCAGGATGGCCGAGTGGT; i-Met, AGCAGAGTGGCGCAGCG.
Additional methods
Total RNAs from fractions of polyribosome analysis were prepared using TRIzol (Invitrogen). mRNA levels were monitored by RT-qPCR using Power SYBR Green PCR Master Mix (Applied Biosystems). The primer sets used for qPCR are included in Supplemental Table S1.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank the Qian laboratory members for insightful discussions. We thank Cornell University Life Sciences Core Laboratory Center for performing the deep sequencing. This work was supported by a postdoctoral fellowship from the American Heart Association (14POST20100022) to M.S., and grants from the National Institutes of Health to T.P. (DP1GM105386) and S.-B.Q. (R01AG042400).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.058180.116.
REFERENCES
- Adams J. 2003. The proteasome: structure, function, and role in the cell. Cancer Treat Rev 29Suppl 1: 3–9. [DOI] [PubMed] [Google Scholar]
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D. 2003. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci 100: 3889–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlanga JJ, Santoyo J, De Haro C. 1999. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2α kinase. Eur J Biochem 265: 754–762. [DOI] [PubMed] [Google Scholar]
- Bulmer M. 1987. Coevolution of codon usage and transfer RNA abundance. Nature 325: 728–730. [DOI] [PubMed] [Google Scholar]
- Dever TE, Feng L, Wek RC, Cigan AM, Donahue TF, Hinnebusch AG. 1992. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68: 585–596. [DOI] [PubMed] [Google Scholar]
- Dittmar KA, Sorensen MA, Elf J, Ehrenberg M, Pan T. 2005. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep 6: 151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Savva R, Wernisch L. 2004. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res 32: 5036–5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. 2009. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet 10: 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf J, Nilsson D, Tenson T, Ehrenberg M. 2003. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science 300: 1718–1722. [DOI] [PubMed] [Google Scholar]
- Firczuk H, Kannambath S, Pahle J, Claydon A, Beynon R, Duncan J, Westerhoff H, Mendes P, McCarthy JE. 2013. An in vivo control map for the eukaryotic mRNA translation machinery. Mol Syst Biol 9: 635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wan J, Liu B, Ma M, Shen B, Qian SB. 2015. Quantitative profiling of initiating ribosomes in vivo. Nat Methods 12: 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, Christophersen NS, Christensen LL, Borre M, Sorensen KD, et al. 2014. A dual program for translation regulation in cellular proliferation and differentiation. Cell 158: 1281–1292. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. 2004. mTOR signaling to translation. Curr Top Microbiol Immunol 279: 169–197. [DOI] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. 2004. Upstream and downstream of mTOR. Genes Dev 18: 1926–1945. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. 2009. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43: 67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA. 2008. Selection on codon bias. Annu Rev Genet 42: 287–299. [DOI] [PubMed] [Google Scholar]
- Heyer EE, Moore MJ. 2016. Redefining the translational status of 80S monosomes. Cell 164: 757–769. [DOI] [PubMed] [Google Scholar]
- Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. 2012. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485: 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. 2012. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci 109: E2424–E2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Qian SB. 2014. Translational reprogramming in cellular stress response. Wiley Interdiscip Rev RNA 5: 301–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza-Puch F, Rooijers K, Buil LC, Zijlstra J, Oude Vrielink JF, Lopes R, Ugalde AP, van Breugel P, Hofland I, Wesseling J, et al. 2016. Tumour-specific proline vulnerability uncovered by differential ribosome codon reading. Nature 530: 490–494. [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J. 2009. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10: 307–318. [DOI] [PubMed] [Google Scholar]
- Mizushima N. 2010. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol 22: 132–139. [DOI] [PubMed] [Google Scholar]
- Murata S, Yashiroda H, Tanaka K. 2009. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol 10: 104–115. [DOI] [PubMed] [Google Scholar]
- Pechmann S, Chartron JW, Frydman J. 2014. Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat Struct Mol Biol 21: 1100–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin JB, Kudla G. 2011. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet 12: 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR, et al. 2015. Codon optimality is a major determinant of mRNA stability. Cell 160: 1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racle J, Picard F, Girbal L, Cocaign-Bousquet M, Hatzimanikatis V. 2013. A genome-scale integration and analysis of Lactococcus lactis translation data. PLoS Comput Biol 9: e1003240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia M, Krokowski D, Guan BJ, Ivanov P, Parisien M, Hu GF, Anderson P, Pan T, Hatzoglou M. 2012. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem 287: 42708–42725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342. [DOI] [PubMed] [Google Scholar]
- Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB. 2013. Rate-limiting steps in yeast protein translation. Cell 153: 1589–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. 1986. An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol 24: 28–38. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li WH. 1987. The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 15: 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood R, Porter AC, Olsen DA, Cavener DR, Wek RC. 2000. A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2α. Genetics 154: 787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE. 2010. Translational regulation of gene expression during conditions of cell stress. Mol Cell 40: 228–237. [DOI] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. 2010. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141: 344–354. [DOI] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG. 2006. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans 34: 7–11. [DOI] [PubMed] [Google Scholar]
- Xu Y, Ma P, Shah P, Rokas A, Liu Y, Johnson CH. 2013. Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature 495: 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CH, Dang Y, Zhou Z, Wu C, Zhao F, Sachs MS, Liu Y. 2015. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol Cell 59: 744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Guo J, Cha J, Chae M, Chen S, Barral JM, Sachs MS, Liu Y. 2013. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature 495: 111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. 2011. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12: 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.