Abstract
Prohead RNA (pRNA) is an essential component of the powerful Φ29‐like bacteriophage DNA packaging motor. However, the specific role of this unique RNA in the Φ29 packaging motor remains unknown. This review examines pRNA as a noncoding RNA of novel structure and function. In order to highlight the reasons for exploring the structure and function of pRNA, we (1) provide an overview of Φ29‐like bacteriophage and the Φ29 DNA packaging motor, including putative motor mechanisms and structures of its component parts; (2) discuss pRNA structure and possible roles for pRNA in the Φ29 packaging motor; (3) summarize pRNA self‐assembly; and (4) describe the prospective therapeutic applications of pRNA. Many questions remain to be answered in order to connect what is currently known about pRNA structure to its novel function in the Φ29 packaging motor. The knowledge gained from studying the structure, function, and sequence variation in pRNA will help develop tools to better navigate the conformational landscapes of RNA. WIREs RNA 2016, 7:428–437. doi: 10.1002/wrna.1330
For further resources related to this article, please visit the WIREs website.
Φ29‐LIKE BACTERIOPHAGES
Bacteriophages are viruses that infect bacteria and may play an indirect role in human health and disease states.1, 2, 3 First discovered in 1915 in micrococci, phages are now known to infect a wide range of bacteria, including extreme acidophiles and thermophiles.4, 5, 6 Among the most ubiquitous phages are those that infect soil‐dwelling bacteria. These include phages hosted by members of the Bacillus genus, which are aerobic, Gram‐positive, spore‐forming eubacteria that thrive in soil and decaying plant matter. To date, all bacteriophages isolated from Bacillus species have been found to share certain features, i.e., they are all tailed phages that have double‐stranded DNA (dsDNA) genomes and prolate icosahedral heads.7 The Φ29‐like bacteriophages are phages that infect Bacillus subtilis and related species, including Bacillus pumilis, Bacillus amyloliquefaciens, and Bacillus licheniformis.7
In the order of tailed bacteriophages, Φ29‐like phages belong to the Podoviridae family, which are phages with dsDNA genomes and short, noncontractile tails.7 The Φ29‐like bacteriophage genus encompasses several species, including Φ15, B103, BS32, GA‐1, M2, Nf, and PZA, which have been split into Groups I, II, and III on the basis of serological properties, DNA physical maps, peptide maps, and partial or complete DNA sequences.7 In addition to Φ29, Group I includes species Φ15, BS32, and PZA; Group II includes B103, M2, and Nf; and Group III includes only GA‐1.7, 8 These classifications reflect not only genetic relatedness, but also geographical distribution: Members of Group I were isolated in the United States, Group II in Japan, and Group III in Europe.7 Figure 1 is a phylogenetic tree depicting the evolutionary relationships among the species in Groups I, II, and III.8
Figure 1.
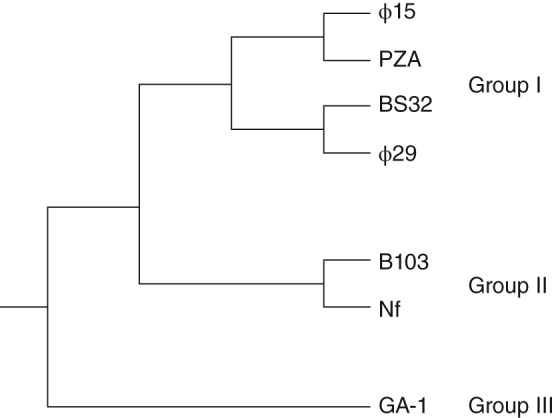
Phylogenetic tree depicting the evolutionary branching of the Φ29‐like bacteriophage genus into Groups I, II, and III. (Figure adapted from data in Ref 8.)
The Φ29‐like phages have prolate, T = 3, Q = 5 icosahedral heads with fivefold rotational symmetry. The Φ29 phage dimensions are approximately 540 Å by 450 Å, with a 16 Å‐thick protein shell.9 The Φ29 phage tail facilitates viral entry into the Bacillus host by binding and hydrolyzing the bacterial cell wall.9 It is approximately 380 Å long and exhibits pseudo‐sixfold symmetry. All Φ29‐like bacteriophages have linear, dsDNA genomes approximately 20 kb in size, with a terminal protein (TP) covalently bound to each 5′ end.8 This protein functions to prime replication via a mechanism that is also used by other phages, linear plasmids, and adenoviruses.7, 8 For this reason, Φ29 has long served as a system in which to study the mechanism of protein‐primed replication, as well as other molecular mechanisms, including transcription regulation, phage morphogenesis, and DNA packaging.7, 9, 10, 11
THE Φ29‐LIKE PHAGE PACKAGING MOTOR
Overview of the Packaging Motor
During the Φ29 phage replicative cycle, newly synthesized genomic DNA must be encapsidated to generate fully functional virus particles. This task is accomplished by the powerful Φ29 packaging motor. Single‐molecule optical tweezer experiments have shown that the Φ29 motor packages DNA against an internal force greater than 50 pN, overcoming significant enthalpic, entropic, and DNA‐bending energies to condense the 19.3 kb viral genome to near‐crystalline densities within the capsid, all within about 5.5 min.12, 13 (As a point of reference, myosin moving across actin filaments generate approximately 1–5 pN.) The high internal pressure achieved by the Φ29 motor, estimated to be approximately 60 atmospheres, propels initial DNA injection into the host cell.12, 14
The motor complex comprises three coaxial rings through which DNA is threaded during packaging.15 Proximal to the viral prohead is a ring of connector proteins, and distal to the prohead is a ring of ATPase proteins. A prohead RNA (pRNA) ring, unique to the Φ29‐like bacteriophage DNA packaging motor, forms the interface between the connector and ATPase motor proteins.16, 17 Figure 2 highlights the spatial orientation of these essential components, where the arrows indicate putative pRNA roles in the packaging motor. Motor function is driven by the ATPase (gene product, or gp, 16), which converts the chemical energy of ATP into the mechanical energy needed to power encapsidation of the viral genome. Thus, the minimal packaging motor consists of major capsid proteins (gp8), head–tail connector proteins (gp10), pRNA, ATPases (gp16), and genomic DNA linked to the TP (gp3).9 However, the Φ29 motor mechanism of DNA packaging remains to be determined.
Figure 2.
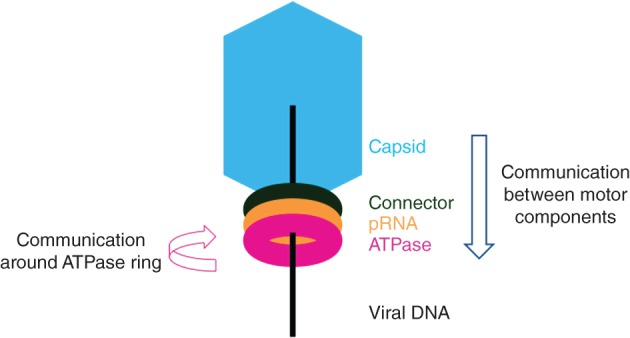
Representation of the orientation of Φ29 motor components. The viral capsid is shown in blue, the connector protein in green, the pRNA in orange, the ATPase in pink, and viral DNA as a black line. The blue vertical arrow represents the role that pRNA may play in communicating from the viral capsid and connector to the ATPase during packaging. The pink circular arrow represents the role that pRNA may play in coordinating ATPase activity around the ring.
Two very different mechanisms for Φ29 motor activity have been proposed, and each is supported by abundant experimental data. In the mechanism Liu et al. proposed,15, 18, 19, 20 each ATPase in a fivefold symmetric ring releases an ADP and binds an ATP, in turn, during a ‘dwell’ phase. In a subsequent ‘burst’ phase, the ATPs are hydrolyzed, and 2.5 bp of DNA are packaged for each of four phosphates released, totaling 10 bp of DNA translocated per burst.15, 18, 19, 20 In this model, one of the five ring ATPases plays a regulatory role, directly contacting the viral DNA, binding and hydrolyzing ATP, and releasing phosphate, without contributing to DNA translocation, per se.19, 21 The regulatory ATPase instead may play a role in initiating or concluding the ATP hydrolysis cascade,19 or it may be responsible for realigning the viral DNA, which is approximately −14° out of register following the burst phase.15, 22 It has also been suggested that this special subunit makes crucial electrostatic contacts that play a role in an underlying symmetry‐breaking mechanism that is important for DNA packaging, reconciling the fivefold rotational symmetry of the viral prohead with the 12‐fold rotational symmetry of the connector protein and sixfold symmetry of the tail.23, 24, 25, 26, 27, 28 The strengths of this model are the supporting rigorous kinetic data and single‐molecule force microscopy data. However, many questions remain to be answered about the regulatory ATPase. Most saliently, what physical or conformational features distinguish the regulatory subunit from the other subunits?
In a mechanism that Schwartz et al.29 proposed, the Φ29 viral DNA revolves, rather than rotates, during packaging.30, 31 Each ATPase in a sixfold symmetric ring binds ATP, thereby inducing a conformational change that increases ATPase affinity for dsDNA. Upon ATP hydrolysis, the ATPase undergoes another conformational change that reduces its affinity for dsDNA, thus promoting DNA handoff from one subunit to the next around the ring. Each ATP hydrolyzed moves the DNA 1.75 bp into the capsid.29 Furthermore, contrary to studies that have indicated that the ATPase binds to pRNA on the prohead prior to binding viral DNA,32 this model supports an alternative packaging motor assembly pathway in which the ATPase assembles around the viral DNA prior to docking on the prohead.12, 29 The strengths of this revolution model include the following: that it accommodates stoichiometries of either five or six ATPases; that all subunits have an equivalent, active role; and that revolution allows the viral DNA to be packaged in low‐energy, nonsupercoiled conformations.29 However, the well‐designed DNA and ATPase in vitro system used to determine stoichiometry and test the revolution mechanism may differ from the behavior of the Φ29 motor in vivo. The in vitro system will be very useful for nanotechnology applications.
Neither model yet explains the role of pRNA in the Φ29 packaging motor. More detailed structural information about the pRNA, ATPase, and the pRNA–ATPase and the ATPase–viral DNA interactions in the packaging motor may help discriminate between these two models or suggest new mechanisms.
Structures of the Packaging Motor Protein Components
In the Φ29 motor, three‐dimensional (3D) structures have been determined for the connector protein and pieces of the pRNA. The connector protein exists as a dodecameric assembly of 36 kDa gp10 protomers, forming the interface between the prohead and the rest of the packaging motor.33 The assembled, 422 kDa superstructure is cone shaped and 75 Å long, with the wide end, formed by residues 42–129 and 248–285, measuring 138 Å, and the narrow end, formed by residues 158–202, measuring just 35 Å.24, 33 A crystal structure of the connector protein assembly, refined to 2.1 Å, shows that the 35 Å‐wide central channel is largely electronegative, but it possesses two rings of lysine residues (formed by K200 and K209) spaced 20 Å apart with neighboring lysine residues in the ring spaced 10 Å apart.9, 33 This geometry would permit four points of electrostatic interactions between the phosphate backbone of the translocating DNA and the connector central channel.33 Structural comparisons between three connector protein crystal structures obtained by Simpson et al. suggest that flexibility exists in the wide end of the connector, presumably to allow for rotation within the portal during packaging.24
No structural data yet exist for the full pRNA molecule, the ATPase, or the pRNA–ATPase complex. The ATPase can be modeled from homologous bacteriophage ATPases that do not require pRNA. However, high‐resolution structural information will be essential for fully understanding the function of pRNA in the Φ29 packaging motor.
PROHEAD RNA
Prohead RNA Primary and Secondary Structure
To date, Φ29 and related bacteriophages are the only known phages to require a nucleic acid molecule, pRNA, for packaging activity34 (Figure 2). Early experiments on the Φ29 bacteriophage packaging motor revealed sensitivity to ribonucleases (RNases) T1 and A.34 Pretreatment with RNase inhibitor restored Φ29 assembly.34, 35 Studies on the motor determined that a phage‐encoded RNA molecule, dubbed pRNA for its association with the prohead, is an essential component of the Φ29 phage packaging motor.16
Φ29 pRNA is a 174‐nucleotide (nt) transcript encoded in the viral genome from 147 to 320 bp.35, 36 It comprises two distinct domains: Domain I, spanning the first 117 bases; and Domain II, spanning bases 131–174. These domains are linked by a 13‐base stretch of unstructured, single‐stranded RNA.37 Figure 3 depicts Domains I and II in folded Φ29 pRNA.35, 37 A 120‐base construct absent Domain II packages genomic DNA with wild‐type activity in vitro and consequently is widely used for in vitro studies; however, Domain II is highly conserved among all Φ29‐like phages, suggesting that it plays an important role in vivo, potentially in phage morphogenesis.9, 35, 36 Two regions of Domain I RNA are functionally important to the packaging motor. The first is a region deemed important for prohead binding, determined by ribonuclease footprinting to be bases 22–84, and by mutagenesis to be bases 40–91.38, 39 The second region is required for DNA packaging, determined by in situ truncation and packaging assays to be bases 1–25 and furthered specified by mutagenesis studies to include bases 1–28 and 92–117.9, 38, 39, 40, 41
Figure 3.
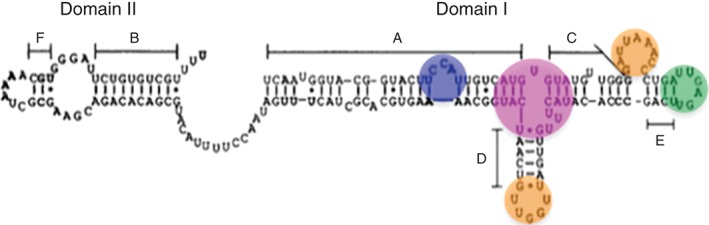
Secondary structure of Φ29 pRNA showing structured Domains I and II, an unstructured linker, and helices A through F.35, 37 The bulge loop is highlighted in blue, the 3WJ in magenta, the CE (right hand) and D (left hand) loops in orange, and the E loop in green. (Reprinted with permission from Ref 35. Copyright 1990 Elsevier)
Although the sequence similarity among all Φ29‐like bacteriophage pRNAs is only 12%, pRNA secondary structure is highly conserved.37, 42, 43 The secondary structure of pRNA comprises six helices, named A–F, all of which are found in Domain I with the exception of helices B and F37 (Figure 3). In the 120‐base Φ29 pRNA construct, there are five single‐base bulges, two stem loops, one three‐base bulge, one bifurcation bulge, and one bulge loop.44 All of the single‐base bulges appear to be nonessential; however, deletion of U5 reduces DNA packaging activity by approximately 10‐fold.41, 44, 45 The three‐base bulge (C18C19A20) in helix A is necessary for DNA packaging, and C18C19 bind the Φ29 ATPase.32 The bulge loop and two stem loops are indispensible for procapsid binding, DNA packaging, and phage assembly.44 Studies on the Φ29 pRNA three‐way junction (3WJ) have yielded conflicting results upon deletion of the polyuridine bifurcation bulge (U29, U71–73), with one study reporting loss of binding and packaging activity,45 and another reporting no loss of procapsid binding or DNA packaging activity.44 Yet another study reported that two of the four bulge uridine residues in the Φ29 3WJ (U29, U72–74) must be retained for pRNA binding to the viral prohead and for wild‐type levels of packaging activity.46 All of these studies implicate the 3WJ as an important contributor to pRNA function. It is likely that the junction acts as a hinge that imparts flexibility on the RNA, correctly placing helices in the spatial orientation necessary for packaging.44 The results of in vitro self‐assembly assays (see below) also support the hypothesis that the 3WJ is an important part of pRNA secondary structure.47
Prohead RNA Tertiary Structure and Function
The pRNA assembles on the prohead in a magnesium ion‐dependent fashion and serves as a scaffold for ring ATPase assembly.32, 39, 48, 49 Its CE and D interlocking loops mediate self‐association, while its A helices protrude from the motor like spokes, making contact with the ATPase.32, 50 In proheads isolated from infected cells, 5–6 copies of pRNA are present.36 The hairpin loop that binds the connector protein resembles ribosomal RNA protein‐binding loops.51 Crystal structures of a 71‐nt piece of Φ29 pRNA (bases 25–95) and a Φ29 3WJ construct were determined to 3.5 and 3.05 Å, respectively.49, 52 Although the 71‐nt Φ29 pRNA crystallized as a tetramer, fitting of the pRNA protomers into previous cryo‐electron microscopy (EM) reconstructions supports a pentamer model in the context of the motor.49 Electron paramagnetic resonance (EPR) spectroscopy experiments show that interhelical angles adopted by the tetramer are not representative of pRNA in a solution state and suggest the need for more complexity in modeling oligomeric pRNA.53 The Φ29 pRNA 3WJ is a multibranch loop at the intersection of helices A, C, and D (Figure 3). The 54‐nt 3WJ monomer structure revealed metal binding sites that altered the H1/H3 interhelical angle, and 3D modeling of the 3WJ structure is consistent with its existence as a hexamer in the context of the motor.52 The structure of the 5′–C18C19A20–3′ bulge loop in Φ29 pRNA has also been determined using nuclear magnetic resonance (NMR) spectroscopy. A 27‐mer RNA fragment containing the bulge showed that it introduces a 33–35° bend in the helical axis, restricting interhelical motion.54 The loops in pRNA are the flexible joints that may have multiple, dynamic conformations during the packaging process.
Although the role of pRNA in the motor remains unknown, its absence prior to prohead assembly as well as in mature Φ29 phage indicates that it performs an essential role specifically in DNA packaging.9, 17, 34, 36 Current models for genome packaging, however, often ignore pRNA or contend that it serves a purely structural function.15, 29, 55 Several roles for pRNA in the Φ29 motor are possible. One possibility is that pRNA serves as a ‘molecular washer,’ i.e., it helps absorb the stress imposed by the moving parts in the motor. Another possibility is that pRNA allows for communication between the motor components, i.e., between the viral capsid and connector and the ATPase, during packaging49 (vertical arrow in Figure 2). The Φ29 motor has been shown to change packaging rates as pressure builds within the viral head,12, 15 suggesting communication between the viral head and the ATPase. As pRNA is located at the center of the packaging motor,46 it is possible that pRNA may play a role in communicating to the ATPase when the viral head is full. Yet another possibility is that pRNA helps coordinate the activity of the ATPase subunits, resulting in their sequential firing around the ring19, 20 (circular arrow in Figure 2). It is also possible that pRNA serves both communicative functions.46
pRNA is unique to the Φ29‐like bacteriophage dsDNA packaging motor.16, 17, 37 It is speculated that in other phages, a subdomain of another motor protein has assumed the role of pRNA. The large terminase protein found in other phages, e.g., T4, is the structural equivalent of pRNA and the ATPase together.56 Furthermore, because pRNA is required for ATPase docking, it may be considered functionally analogous to a subdomain of the large terminase protein.32, 56 The idea that proteins could evolve to fulfill the role of RNA is consistent with the later stages of the RNA world hypothesis and evolution from an RNA‐only world to an RNA‐protein world.57, 58
PROHEAD RNA SELF‐ASSOCIATION
Prohead RNA Interlocking Loops
Intermolecular Watson‐Crick base pairing between nucleotides in the CE and D loops mediates self‐association of pRNA molecules in vivo on the viral prohead as well as in vitro 37, 42, 43, 50 (Figure 4). NMR studies on 19‐ and 35‐nt Φ29 pRNA heterodimers, in which each molecule contained either a CE or a D loop, showed that two intermolecular G–C base pairs form between the loops in a Mg2+‐dependent fashion.59 Disruption of the flexibility of the D loop via mutagenesis (U80C) also disrupted the G–C base pair formation and abolished heterodimer assembly, but interestingly did not affect prohead binding or DNA packaging.59
Figure 4.
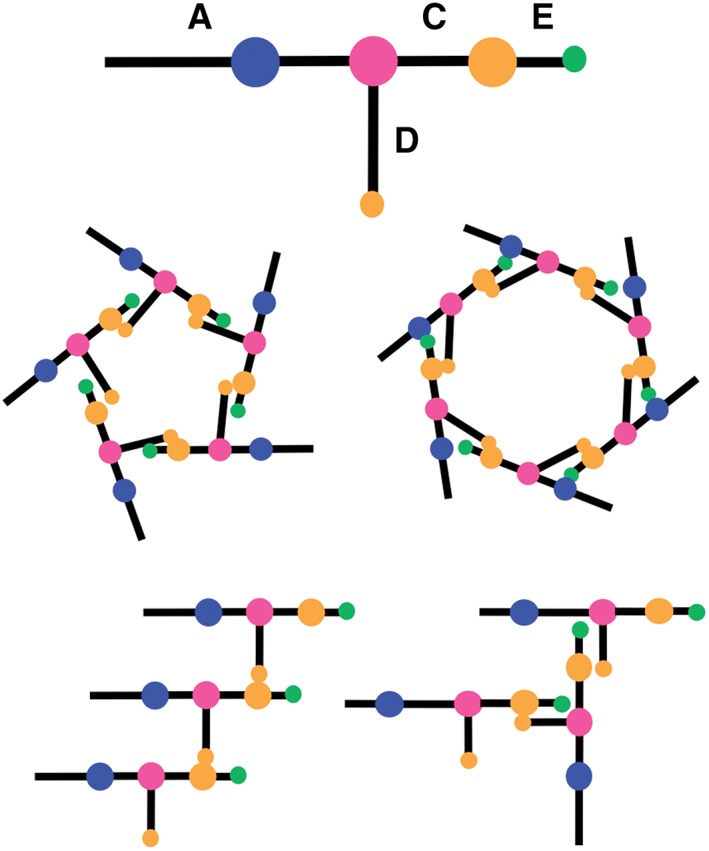
Ball‐and‐stick model of pRNA where balls are loops and sticks are helices. CE and D loops (orange) mediate self‐assembly of pRNA into nanorings (as found on the prohead) and potentially linear and/or branched structures in vitro.
The value of in vitro self‐association studies lies in providing fundamental biophysical data that inform hypotheses of in vivo mechanisms and guide rational design of RNA assemblies. In vitro self‐association studies have shown that pRNA–pRNA interaction varies by sequence and depends upon pRNA concentration as well as upon other factors, such as temperature and magnesium and sodium chloride concentration.42 Importantly, these studies indicate that different pRNAs have different self‐association properties. For example, SF5 pRNA has the highest propensity for self‐association, forming dimers, trimers, and several higher‐order multimers at room temperature.42 However, more recent studies under different conditions indicate that SF5 pRNA forms only dimers, but that M2 pRNA assembles into highly thermostable multimeric species.47 These seemingly contradictory results suggest that the self‐association behaviors of pRNA are more subtle than previously thought, and that these behaviors may be readily modulated by taking advantage of its sensitivity to salt, temperature, and other preparation conditions.47
Prohead RNA Multibranch Loop
Many studies have taken steps toward addressing why different pRNAs exhibit different self‐assembly properties. In a study by Gu et al., pRNA sequences that showed different self‐assembly properties had different base pairing stabilities in the CE and D interlocking loops but similar quaternary interaction stabilities, indicating that sequence variation outside of the interlocking loop sequences plays a role in pRNA self‐assembly energetics.42 Studies on designer pRNA sequences retaining little to no sequence similarity to wild‐type pRNA except in the paired CE and D loops and in the 3WJ indicate that helices do not contribute to pRNA assembly in a sequence‐dependent fashion.49 Investigations by Hao et al. revealed similar insights, demonstrating that mediocre CE and D loop ‘kissing’ interactions can be overcome with highly stable scaffolds.47 These results implicate the 3WJ as an important contributor to pRNA–pRNA interactions. One hypothesis is that different nucleotide sequences at the 3WJ may be responsible for the observed differences in pRNA self‐assembly properties.47 It is known that multibranch loops enable long‐range contacts by determining the angles at which helices emerge. Furthermore, multibranch loops are key determinants of structural and functional roles in other RNAs, such as hairpin ribozymes.60, 61, 62, 63 EPR spectroscopy studies on pRNA monomers and dimers reveal that the interhelical angles in pRNA vary,53, 64 and single‐molecule fluorescent resonance energy transfer (FRET) studies on the Φ29 pRNA 3WJ motif have shown that Mg2+ changes the interhelical angles.52 Thus, the sequence variation at pRNA 3WJs may confer variation in self‐assembly by way of orienting these angles or shifting the distribution of angles in a dynamic ensemble of 3WJ conformations.
APPLICATIONS AND FUTURE DIRECTIONS
Prohead RNA in Nanobiotechnology
The study of nucleic acid folding and assembly has broad implications. DNA folding and assembly properties are widely used in the de novo construction of nanostructures.65, 66, 67 Furthermore, rationally designed, self‐assembling DNA ‘nanorobots’ are capable of delivering their molecular payloads according to an AND logic gate mechanism, suggesting that nucleic acids hold promise for rational nanoparticle design of delivery vehicles.68 Relative to DNA, RNA is suspected to have more potential as a raw material due it its more diverse catalytic and recognition properties and its capacity for robust tertiary and quaternary interaction, which rival those of proteins.69, 70 Proof‐of‐concept research into RNA engineering has yielded interesting results. For example, studies exploiting RNA turn motifs demonstrated that RNA can be programmed to cotranscriptionally fold into desired nanostructures such as hexagonal lattices.71 Studies on self‐assembling RNAi microsponges showed that RNA can be programmed as a delivery vehicle comprising both carrier and cargo, simultaneously maximizing small interfering RNA transfection efficiency and minimizing cytotoxicity and degradation during delivery.72 DNA–RNA hybrid nanomotifs have also been designed to form discrete tiles, fusing the functionality of RNA with the programmability of DNA to achieve a platform with RNA modalities including siRNAs, microRNAs, and aptamers for cell targeting and gene regulation.69
The unique and variable self‐assembly properties of pRNA make it a ‘smart’ building block for rational design of 3D structures with nanotechnological and medical applications.47, 73 This has been demonstrated in several bottom‐up fabrication studies. In a proof‐of‐concept study, Φ29 pRNA dimers with a receptor‐binding moiety on one arm and a gene‐silencing molecule on another successfully targeted cancer cells.74 Additionally, thermodynamically stable X‐shaped nanostructures using the Φ29 pRNA scaffold can carry up to four siRNAs and elicit effective gene silencing compared to individual siRNA oligomers.75 Importantly, these nanostructures selectively target and remain localized in cancer cells in a mouse model.75 Corroborating these results, 14 other pRNA‐based nanoparticles have been developed with cancer‐targeting functionality.76
Unanswered Questions
Whether pRNA assembles into hexamers or pentamers on the viral prohead is still debated. Competition inhibition assays, single fluorophore imaging, atomic force microscopy, and cryo‐EM studies indicate that pRNA assembles into hexamers on the prohead.48, 50, 52, 77, 78 However, high‐resolution cryo‐EM data on complete proheads using relaxed or asymmetric reconstruction and difference maps of pRNA‐truncated proheads support a pentamer model.25, 79, 80 Crystal structures have done little to resolve this debate. Model‐building of the Φ29 pRNA 3WJ, which crystallized as a monomer, supported the hypothesis, that pRNA assembles into hexamers on the viral prohead.52 Yet, a 71‐nt pRNA construct with several deletions in the CE and D loops was shown to crystallize as a tetramer, and analysis of this structure and fitting into previously obtained cryo‐EM reconstructions supported the hypothesis that pRNA forms a pentamer on the prohead.49 EPR studies have suggested that pRNA oligomeric modeling requires more complexity.53 It is known that in vitro, pRNA is capable of forming both pentamers and hexamers.42, 50, 81 Until higher‐resolution structural data and more information about the in vivo assembly mechanism of the Φ29 motor are available, the biologically relevant stoichiometry remains to be determined.
The stoichiometry of the ATPase in the Φ29 motor is also debated. As discussed above, the numbers of subunits engaged in motor activity in the foremost models of the Φ29 motor mechanism are in conflict. Again, high‐resolution structural data of the Φ29 motor will be necessary to determine which stoichiometry is biologically relevant, as well as to distinguish between the currently proposed models.
CONCLUSION
The Φ29 bacteriophage DNA packaging motor is a unique and powerful biomolecular motor that uses pRNA, a viral RNA of novel structure and function, to package its genome during replication. However, the specific indispensible function of pRNA is yet unknown. Outside of the context of the motor, pRNA displays robust self‐assembly behavior that is highly dependent on sequence and assembly conditions, with some species of pRNA capable of forming thermodynamically stable multimers. It has been shown that RNA, like DNA, can serve as a raw material in the rational design of functional 3D structures. The pRNA is uniquely positioned to serve as a platform for developing supramolecular structures with targeting and delivery functions due to its stability and self‐association properties. However, much remains to be learned about pRNA, and elucidating its role in the Φ29 viral molecular machine will hinge on high‐resolution structural data to help inform new structure–function hypotheses.
Conflict of interest: The authors have declared no conflicts of interest for this article.
REFERENCES
- 1. Norman Jason M, Handley Scott A, Baldridge Megan T, Droit L, Liu Catherine Y, Keller Brian C, Kambal A, Monaco Cynthia L, Zhao G, Fleshner P, et al. Disease‐specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015, 160:447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Denou E, Bruttin A, Barretto C, Ngom‐Bru C, Brussow H, Zuber S. T4 phages against Escherichia coli diarrhea: potential and problems. Virology 2009, 388:21–30. [DOI] [PubMed] [Google Scholar]
- 3. Summers WC. Bacteriophage therapy. Annu Rev Microbiol 2001, 55:437–451. [DOI] [PubMed] [Google Scholar]
- 4. Tapia P, Flores F, Covarrubias P, Acuna L, Holmes D, Quatrini R. Complete genome sequence of temperate bacteriophage AcaML1 from the extreme acidophile Acidithiobacillus caldus ATCC 51756. J Virol 2012, 86:12452–12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sakaki Y, Oshima T. Isolation and characterization of a bacteriophage infectious to an extreme thermophile, Thermus thermophilus HB8. J Virol 1975, 15:1449–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Twort F. An investigation on the nature of ultra‐microscopic viruses. Bacteriophage 2011, 1:127–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meijer W, Horcajadas J, Salas M. Phi29 family of phages. Microbiol Mol Biol Rev 2001, 65:261–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pecenkova T, Paces V. Molecular phylogeny of phi29‐like phages and their evolutionary relatedness to other protein‐primed replicating phages and other phages hosted by gram‐positive bacteria. J Mol Evol 1999, 48:197–208. [DOI] [PubMed] [Google Scholar]
- 9. Morais M. Viral molecular machines chapter 23: the dsDNA packaging motor in bacteriophage phi29. Adv Exp Med Biol 2012, 726:511–547. [DOI] [PubMed] [Google Scholar]
- 10. Grimes S, Jardine PJ, Anderson D. Bacteriophage Φ29 DNA packaging In: Advances in Virus Research, vol. 58 New York, NY: Academic Press; 2002, 255–294. [DOI] [PubMed] [Google Scholar]
- 11. Guo P, Grimes S, Anderson D. A defined system for in vitro packaging of DNA‐gp3 of the Bacillus subtilis bacteriophage phi 29. Proc Natl Acad Sci 1986, 83:3505–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smith D, Tans S, Smith S, Grimes S, Anderson D, Bustamante C. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature 2001, 413:748–752. [DOI] [PubMed] [Google Scholar]
- 13. Cao S, Saha M, Zhao W, Jardine P, Zhang W, Grimes S, Morais M. Insights into the structure and assembly of the bacteriophage phi29 double‐stranded DNA packaging motor. J Virol 2014, 88:3986–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. González‐Huici V, Salas M, Hermoso JM. The push–pull mechanism of bacteriophage Ø29 DNA injection. Mol Microbiol 2004, 52:529–540. [DOI] [PubMed] [Google Scholar]
- 15. Liu S, Chistol G, Hetherington C, Tafoya S, Aathavan K, Schnitzbauer J, Grimes S, Jardine P, Bustamante C. A viral packaging motor varies its DNA rotation and step size to preserve subunit coordination as the capsid fills. Cell 2014, 157:702–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guo P, Erickson S, Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage phi 29 DNA. Science 1987, 236:690–694. [DOI] [PubMed] [Google Scholar]
- 17. Hendrix RW, Bacteriophage DNA. Packaging: RNA gears in a DNA transport machine. Cell 1998, 94:147–150. [DOI] [PubMed] [Google Scholar]
- 18. Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Mechanism of force generation of a viral DNA packaging motor. Cell 2005, 122:683–692. [DOI] [PubMed] [Google Scholar]
- 19. Chistol G, Liu S, Hetherington Craig L, Moffitt Jeffrey R, Grimes S, Jardine Paul J, Bustamante C. High degree of coordination and division of labor among subunits in a homomeric ring ATPase. Cell 2012, 151:1017–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moffitt J, Chemla Y, Aathavan K, Grimes S, Jardine P, Anderson D, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature 2009, 457:446–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aathavan K, Politzer A, Kaplan A, Moffitt J, Chemla Y, Grimes S, Jardine P, Anderson D, Bustamante C. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature 2009, 461:669–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang J. Helical repeat of DNA in solution. Proc Natl Acad Sci 1979, 76:200–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chistol G. Dissecting the operating mechanism of a biological motor one molecule at a time. 2013. Available at: http://www.escholarship.org/uc/item/8x6526d1.
- 24. Simpson AA, Leiman PG, Tao Y, He Y, Badasso MO, Jardine PJ, Anderson DL, Rossmann MG. Structure determination of the head–tail connector of bacteriophage ϕ29. Acta Crystallogr D Biol Crystallogr 2001, 57:1260–1269. [DOI] [PubMed] [Google Scholar]
- 25. Morais M, Tao Y, Olson N, Grimes S, Jardine P, Anderson D, Baker T, Rossmann M. Cryoelectron‐microscopy image reconstruction of symmetry mismatches in bacteriophage phi29. J Struct Biol 2001, 135:38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tang J, Olson N, Jardine PJ, Grimes S, Anderson DL, Baker TS. DNA poised for release in bacteriophage ø29. Structure 2008, 16:935–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xiang Y, Morais MC, Battisti AJ, Grimes S, Jardine PJ, Anderson DL, Rossmann MG. Structural changes of bacteriophage Φ29 upon DNA packaging and release. EMBO J 2006, 25:5229–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tao Y, Olson NH, Xu W, Anderson DL, Rossmann MG, Baker TS. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell 1998, 95:431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schwartz C, De Donatis GM, Zhang H, Fang H, Guo P. Revolution rather than rotation of AAA+ hexameric phi29 nanomotor for viral dsDNA packaging without coiling. Virology 2013, 443:28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao Z, Khisamutdinov E, Schwartz C, Guo P. Mechanism of one‐way traffic of hexameric phi29 DNA packaging motor with four electropositive relaying layers facilitating antiparallel revolution. ACS Nano 2013, 7:4082–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guo P. Biophysical studies reveal new evidence for one‐way revolution mechanism of bacteriophage Φ29 DNA packaging motor. Biophys J 2014, 106:1837–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koti J, Morais M, Rajagopal R, Owen B, McMurray C, Anderson D. DNA packaging motor assembly intermediate of bacteriophage phi29. J Mol Biol 2008, 381:1114–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guasch A, Pous J, Ibarra B, Gomis‐Rüth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. Detailed architecture of a DNA translocating machine: the high‐resolution structure of the bacteriophage Φ29 connector particle. J Mol Biol 2002, 315:663–676. [DOI] [PubMed] [Google Scholar]
- 34. Guo P, Peterson C, Anderson D. Initiation events in in vitro packaging of bacteriophage phi 29 DNA‐gp3. J Mol Biol 1987, 197:219–228. [DOI] [PubMed] [Google Scholar]
- 35. Anderson D, Bodley J. Role of RNA in bacteriophage phi29 DNA packaging. J Struct Biol 1990, 104:70–74. [DOI] [PubMed] [Google Scholar]
- 36. Wichitwechkarn J, Bailey S, Bodley J, Anderson D. Prohead RNA of bacteriophage phi29: size, stoichiometry and biological activity. Nucleic Acids Res 1989, 17:3459–3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bailey S, Wichitwechkarn J, Johnson D, Reilly B, Anderson D, Bodley J. Phylogenetic analysis and secondary structure of the Bacillus subtilis bacteriophage RNA required for DNA packaging. J Biol Chem 1990, 265:22365–22370. [PubMed] [Google Scholar]
- 38. Reid R, Bodley J, Anderson D. Identification of bacteriophage phi 29 prohead RNA domains necessary for in vitro DNA‐gp3 packaging. J Biol Chem 1994, 269:9084–9089. [PubMed] [Google Scholar]
- 39. Reid R, Bodley J, Anderson D. Characterization of the prohead‐pRNA interaction of bacteriophage phi 29. J Biol Chem 1994, 269:5157–5162. [PubMed] [Google Scholar]
- 40. Zhang C, Lee C, Guo P. The proximate 5′ and 3′ ends of the 120‐base viral RNA (pRNA) are crucial for the packaging of bacteriophage phi 29 DNA. Virology 1994, 201:77–85. [DOI] [PubMed] [Google Scholar]
- 41. Zhang C, Tellinghuisen T, Guo P. Confirmation of the helical structure of the 5′/3′ termini of the essential DNA packaging pRNA of phage phi 29. RNA 1995, 1:1041–1050. [PMC free article] [PubMed] [Google Scholar]
- 42. Gu X, Schroeder S. Different sequences show similar quaternary interaction stabilities in prohead viral RNA self‐assembly. J Biol Chem 2011, 286:14419–14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen C, Zhang C, Guo P. Sequence requirement for hand‐in‐hand interaction in formation of RNA dimers and hexamers to gear phi29 DNA translocation motor. RNA 1999, 5:805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang C, Tellinghuisen T, Guo P. Use of circular permutation to assess six bulges and four loops of DNA‐packaging pRNA of bacteriophage phi29. RNA 1997, 3:315–323. [PMC free article] [PubMed] [Google Scholar]
- 45. Reid R, Zhang F, Benson S, Anderson D. Probing the structure of bacteriophage phi29 prohead RNA with specific mutations. J Biol Chem 1994, 269:18656–18661. [PubMed] [Google Scholar]
- 46. Zhao W, Saha M, Ke A, Morais M, Jardine P, Grimes S. A three‐helix junction is the interface between two functional domains of prohead RNA in phi29 DNA packaging. J Virol 2012, 86:11625–11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hao Y, Kieft J. Diverse self‐association properties within a family of phage packaging RNAs. RNA 2014, 20:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Trottier M, Guo P. Approaches to determine stoichiometry of viral assembly components. J Virol 1997, 71:487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ding F, Lu C, Zhao W, Rajashankar K, Anderson D, Jardine P, Grimes S, Ke A. Structure and assembly of the essential RNA ring component of a viral DNA packaging motor. Proc Natl Acad Sci 2011, 108:7357–7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Guo P, Zhang C, Chen C, Garver K, Trottier M. Inter‐RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell 1998, 2:149–155. [DOI] [PubMed] [Google Scholar]
- 51. Harris S, Schroeder S. Nuclear magnetic resonance structure of the prohead RNA E‐loop hairpin. Biochemistry 2010, 49:5989–5997. [DOI] [PubMed] [Google Scholar]
- 52. Zhang H, Endrizzi J, Shu Y, Haque F, Sauter C, Shlyakhtenko L, Lyubchenko Y, Guo P, Chi Y. Crystal structure of 3WJ core revealing divalent ion‐promoted thermostability and assembly of the Phi29 hexameric motor pRNA. RNA 2013, 19:1226–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang X, Tung C‐S, Sowa GZ, Hatmal MM, Haworth IS, Qin PZ. Global structure of a three‐way junction in a Phi29 packaging RNA dimer determined using site‐directed spin labeling. J Am Chem Soc 2012, 134:2644–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Harjes E, Kitamura A, Zhao W, Morais M, Jardine P, Grimes S, Matsuo H. Structure of the RNA claw of the DNA packaging motor of bacteriophage Phi29. Nucleic Acids Res 2012, 40:9953–9963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guo P, Grainge I, Zhao Z, Vieweger M. Two classes of nucleic acid translocation motors: rotation and revolution without rotation. Cell Biosci 2014, 4:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rao V, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet 2008, 42:647–681. [DOI] [PubMed] [Google Scholar]
- 57. Atkins J, Gesteland R, Cech T. RNA Worlds: From Life's Origins to Diversity in Gene Regulation. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2010. [Google Scholar]
- 58. Strulson CA, Molden RC, Keating CD, Bevilacqua PC. RNA catalysis through compartmentalization. Nat Chem 2012, 4:941–946. [DOI] [PubMed] [Google Scholar]
- 59. Kitamura A, Jardine PJ, Anderson DL, Grimes S, Matsuo H. Analysis of intermolecular base pair formation of prohead RNA of the phage ø29 DNA packaging motor using NMR spectroscopy. Nucleic Acids Res 2008, 36:839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wilson TJ, Lilley DM. Do the hairpin and VS ribozymes share a common catalytic mechanism based on general acid–base catalysis? A critical assessment of available experimental data. RNA 2011, 17:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Walter N, Burke J, Millar D. Stability of hairpin ribozyme tertiary structure is governed by the interdomain junction. Nat Struct Biol 1999, 6:544–549. [DOI] [PubMed] [Google Scholar]
- 62. Tan E, Wilson T, Nahas M, Clegg R, Lilley D, Ha T. A four‐way junction accelerates hairpin ribozyme folding via a discrete intermediate. Proc Natl Acad Sci USA 2003, 100:9308–9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Müller S, Appel B, Krellenberg T, Petkovic S. The many faces of the hairpin ribozyme: structural and functional variants of a small catalytic RNA. IUBMB Life 2012, 64:36–47. [DOI] [PubMed] [Google Scholar]
- 64. Lescoute A, Westhof E. Topology of three‐way junctions in folded RNAs. RNA 2006, 12:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Seeman N. DNA in a material world. Nature 2003, 421:427–431. [DOI] [PubMed] [Google Scholar]
- 66. Lin C, Liu Y, Rinker S, Yan H. DNA tile based self‐assembly: building complex nano‐architectures. Chemphyschem 2006, 7:1641–1647. [DOI] [PubMed] [Google Scholar]
- 67. Feldkamp U, Niemeyer C. Rational design of DNA nanoarchitectures Angewandte Chemie International Edition 2006, 45:1856–1876. [DOI] [PubMed] [Google Scholar]
- 68. Douglas S, Bachelet I, Church G. A logic‐gated nanorobot for targeted transport of molecular payloads. Science 2012, 335:831–834. [DOI] [PubMed] [Google Scholar]
- 69. Ko S, Su M, Zhang C, Rbbe A, Jiang W, Mao C. Synergystic self‐assembly of RNA and DNA molecules. Nat Chem 2010, 2:1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ishikawa J, Furuta H, Ikawa Y. RNA tectonics (tectoRNA) for RNA nanostructure design and its application in synthetic biology. Wiley Interdiscip Rev RNA 2013, 4:651–664. [DOI] [PubMed] [Google Scholar]
- 71. Geary C, Rothemund P, Andersen E. A single‐stranded architecture for cotranscriptional folding of RNA nanostructures. Science 2014, 345:799–804. [DOI] [PubMed] [Google Scholar]
- 72. Lee JB, Hong J, Bonner DK, Poon Z, Hammond PT. Self‐assembled RNA interference microsponges for efficient siRNA delivery. Nat Mater 2012, 11:316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shu D, Moll W, Deng Z, Mao C, Guo P. Bottom‐up assembly of RNA arrays and superstructures as potential parts in nanotechnology. Nano Lett 2004, 4:1717–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Guo S, Tschammer N, Mohammed S, Guo P. Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Hum Gene Ther 2005, 16:1097–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Haque F, Shu D, Shu Y, Shlyakhtenko L, Rychahou P, Evers B, Guo P. Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nanotoday 2012, 7:245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Shu Y, Haque F, Shu D, Li W, Zhu Z, Kotb M, Lyubchenko Y, Guo P. Fabrication of 14 different RNA nanoparticles for specific tumor targeting without accumulation in normal organs. RNA 2013, 19:767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shu D, Zhang H, Jin J, Guo P. Counting of six pRNAs of phi29 DNA‐packaging motor with customized single‐molecule dual‐view system. EMBO J 2007, 26:527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ibarra B, Caston J, Llorca O, Valle M, Valpuesta J, Carrascosa J. Topology of the components of the DNA packaging machinery in the phage phi29 prohead. J Mol Biol 2000, 298:807–815. [DOI] [PubMed] [Google Scholar]
- 79. Morais M, Koti J, Bowman V, Reyes‐Aldrete E, Anderson D, Rossman M. Defining molecular and domain boundaries in the bacteriophage phi29 DNA packaging motor. Structure 2008, 16:1267–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, et al. Structure of the bacteriophage ϕ29 DNA packaging motor. Nature 2000, 408:745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang F, Lemieux S, Wu X, St.‐Arnaud D, McMurray CT, Major F, Anderson D. Function of hexameric RNA in packaging of bacteriophage Φ29 DNA in vitro . Mol Cell 1998, 2:141–147. [DOI] [PubMed] [Google Scholar]


