Abstract
The Ccr4‐Not complex is a multisubunit complex present in all eukaryotes that contributes to regulate gene expression at all steps, from production of messenger RNAs (mRNAs) in the nucleus to their degradation in the cytoplasm. In the nucleus it influences the post‐translational modifications of the chromatin template that has to be remodeled for transcription, it is present at sites of transcription and associates with transcription factors as well as with the elongating polymerase, it interacts with the factors that prepare the new transcript for export to the cytoplasm and finally is important for nuclear quality control and influences mRNA export. In the cytoplasm it is present in polysomes where mRNAs are translated and in RNA granules where mRNAs will be redirected upon inhibition of translation. It influences mRNA translatability, and is needed during translation, on one hand for co‐translational protein interactions and on the other hand to preserve translation that stalls. It is one of the relevant players during co‐translational quality control. It also interacts with factors that will repress translation or induce mRNA decapping when recruited to the translating template. Finally, Ccr4‐Not carries deadenylating enzymes and is a key player in mRNA decay, generic mRNA decay that follows normal translation termination, co‐translational mRNA decay of transcripts on which the ribosomes stall durably or which carry a non‐sense mutation and finally mRNA decay that is induced by external signaling for a change in genetic programming. Ccr4‐Not is a master regulator of eukaryotic gene expression. WIREs RNA 2016, 7:438–454. doi: 10.1002/wrna.1332
For further resources related to this article, please visit the WIREs website.
INTRODUCTION
Cells must continuously adapt to changing environmental conditions. They face the challenge of being able to respond rapidly to dramatic alterations in the environment while buffering responses to mild environmental changes. They must achieve homeostasis in all growth conditions at a minimal energy cost. Regulation of gene expression must respond to this fundamental need of the cell. While this is evident in single cell organisms, which will strive to survive in different growth conditions, it is equally critical in multicellular organisms for appropriate development and health. For a very long time it was believed that the major control of gene expression occurred at the first step, namely transcription. However, it has becoming increasingly evident that gene expression control occurs also in a major way at all subsequent post‐transcriptional steps. Studies on messenger RNA (mRNA) export from the nucleus, localization of mRNAs, control of mRNA decay, and translational control have all risen to the forefront of the gene expression field.
The Ccr4‐Not complex is a multisubunit protein complex that is conserved in all eukaryotes and contributes to regulate RNA metabolism at all steps, from synthesis to decay. It has gradually emerged as an essential regulator of gene expression homeostasis in eukaryotes. After the three first reviews more than 10 years ago,1, 2, 3 many new reviews have been published on different aspects of this complex in the last 2 years.4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 In this review, after a brief biochemical and genetic description of the complex I will summarize the evidence for Ccr4‐Not roles at various steps of the gene expression pathway.
BIOCHEMICAL AND GENETIC DESCRIPTION OF THE Ccr4‐Not COMPLEX
The Ccr4‐Not complex consists of a scaffold protein, Not1, and a number of evolutionarily conserved core proteins that dock onto Not1 (Table 1 and Figure 1). The main core proteins are the Not2 and Not5 heterodimer, the Caf40 subunit and the Ccr4 and Caf1 catalytic subunits. The Not4 RING E3 ubiquitin ligase is a conserved protein, but it is only a stable subunit of the complex in yeast. However, its function is conserved since the human protein complements a deletion of NOT4 in yeast.19 Yeast has an additional subunit, Caf130, and it carries Not3, a protein that is homologous to Not5 and most likely originates from a gene duplication event. It is unfortunate that the ortholog in higher eukaryotes was named after Not3 rather than after Not5, since it seems that Not3 is functionally much less relevant than Not5 in yeast. In Drosophila, there is no easily recognizable ortholog of Caf130.12 Instead there are two additional subunits called Not10 and Not11. The human complex is similar in composition to the one in flies, but there are alternatively used genes encoding Caf1 and Ccr4 orthologs (Table 1). Additionally there is a mammalian‐specific protein TAB182. The subunits are known under different alternatively used names (Table 1) so I will use the core names defined in Table 1 throughout the review.
Table 1.
Name of Ccr4‐Not Genes in Yeast, Flies, and Human
| Core | Drosophila | Saccharomyces cerevisiae | Homo sapiens |
|---|---|---|---|
| Name | Gene Name | Gene Name | Gene Name |
| Not1 | Not1 | NOT1/CDC39 | CNOT1 |
| Not2 | Regena (Rga) | NOT2/CDC36 | CNOT2 |
| Not5 | Not3 | NOT5 | CNOT3 |
| Caf1 | Pop2 | CAF1/POP2 | Caf1a/CNOT7/CAF1 and Caf1b/CNOT8/POP2/CALIF |
| Ccr4 | twin | CCR4 | CCR4a/CNOT6C and CCR4b/CNOT6L |
| Caf40 | Rcd1 | CAF40 | CNOT9/Rcd1/CAF40/RQCD1 |
| Not4 | Not4 | NOT4/MOT2/SIG1 | CNOT4 |
| Not10 | Not10 | CNOT10 | |
| Not11 | Not11 | CNOT11/C2orf29 | |
| Caf130 | CAF130 | ||
| Not3 | NOT3 | ||
| TAB182 | TAB182 |
I chose Not5 as the core name for fly Not3, human CNOT3, and yeast NOT5 because yeast Not5 rather than Not3 is certainly the yeast functional ortholog of the fly and human proteins.
Figure 1.

Linear and cartoon representation of the Ccr4‐Not complex subunits. (a) The Not1 core subunit is depicted with the domains characterized as docking sites for other core subunits. The names chosen for the core subunits are those described in Table 1. DEDD and EEP refer to the signature domains of the catalytic deadenylase subunits and RING to the signature domain of the E3 ligase subunit. NOT box is a domain of homology shared by several Not proteins and RRM is a putative RNA recognition motif in Not4. Adapted from Ref 20 to include the docking of Not4 defined in Ref 22. The C‐terminal domain of Not4 binds the side surface of the first HEAT‐repeat unit of the Not1 C‐terminal domain, while Not2 together with the C‐terminal domain of Not5 bind the top and the bottom surfaces. Hence the interactions occur at largely separate surfaces of the Not1 C‐terminal domain.22 (b) Cartoon representation of the L‐shape of the Ccr4‐Not complex defined by electron microscopy23 with the expected position of the core subunits on the Not1 scaffold.20 Where Not3 and Caf130 dock onto Not1 is unknown, so we placed Not3 in contact with Not2 and Not5 because of common mutant phenotypes and the yeast specific Caf130 at the N‐terminus of Not1 where Not10 and Not11 bind in flies.
The docking of Ccr4‐Not subunits onto the Not1 scaffold has been well characterized, the most extensively in Drosophila,20 but also in yeast and human21 (Figure 1A). Several crystal structures of domains of Not1 with their interaction partners are available (Ref 22 and reviewed in Ref 8). We lack structural information about species‐specific subunits and some domains are not included in the available structures. Our current understanding of the overall architectural organization of the Ccr4‐Not complex is a low‐resolution electron microscopy reconstruction of the complex purified via Not1 from yeast after chemical cross‐linking.23 It indicates an L‐shaped structure with two arms of similar lengths of 180–190 ångström (Figure 1(b)).
The structural data available for the Ccr4‐Not complex has greatly increased very recently but we are still far from a comprehensive structural understanding of this complex. Moreover, we still do not know whether there are multiple functional forms of the complex. Core complexes of about 1 MDa, and larger complexes with all core subunits, are found in cells of different eukaryotes, but the subunits are additionally detectable in smaller complexes. It is not known whether this reflects partial dissociation of Ccr4‐Not complexes during in vitro work or instead partial complexes that truly exist and have functional relevance in vivo.
Genetic Description of the Ccr4‐Not Components
Ccr4 on one hand,24 and the five Not proteins on the other,25, 26, 27 were first identified by different genetic selections in the yeast S. cerevisiae. The Caf proteins were isolated by virtue of their interaction with Ccr4.28, 29 Caf1 has the same mutant phenotypes as Ccr4,28 but neither do Caf40 nor Caf130. Deletions of Ccr4 or Caf proteins do not pass the genetic selection that led to the identification of the Not proteins and deletions of the Not proteins do not pass the selection that led to the identification of Ccr4 (Refs 24, 26 and unpublished observations). This was early genetic evidence that the Ccr4‐Not complex is multifunctional.
Not1 is the only essential protein in yeast, but the deletion of Not2, Not4, or Not5 leads to a very slow growth phenotype at permissive temperature and the proteins are essential for growth at high temperature.21 The deletions of Caf1 and Ccr4 display only mild phenotypes for yeast grown on rich media in comparison to the deletions of the Not proteins (Ref 21 and unpublished observations). Deletion of Caf40 or Caf130 does not display any identified growth phenotype (unpublished observations). However mild increase or decrease of sensitivity to a variety of different conditions have been reported from global studies and are summarized on the Saccharomyces genome database (www.yeastgenome.org).
As we progress in our characterization of the Ccr4‐Not complex functionalities, the early genetic experiments in yeast should not be forgotten, in particular the phenotypes of double deletion mutants. A double mutant that grows no worse than the single mutants indicates components likely to work together. This is true for Ccr4 and Caf1, as well Not2, Not3, and Not5, but deleting either Ccr4 or Caf1 with Not2 or Not5 is lethal.21 The deletion of Not4 is lethal when combined with that of either Ccr4 or Not3, indicating a different functionality, and this is very consistent with the fact that Not4 is not a stable subunit of the complex in flies or human. This also indicates that Not3 has maintained some functional relevance in yeast.21
Cellular Localization of the Ccr4‐Not Complex
Ccr4‐Not components have been detected both in the cytoplasm and in the nucleus.26, 30, 31, 32 One study showed that the localization of a Ccr4‐Not subunit was regulated according to the stage of the cell cycle, with accumulation in the nucleus in G0 or G1, but mostly cytoplasmic distribution by the time cells are in S phase.33 In the cytoplasm, Ccr4‐Not subunits are present in polysomes where mRNAs are being translated,34, 35 and they have been detected in processing bodies (P‐bodies),36 sites to which mRNAs that undergo inhibition of translation, decapping and/or initiation of degradation, delocalize.
FUNCTIONS OF THE Ccr4‐Not COMPLEX ALONG THE GENE EXPRESSION PATHWAY
mRNA Production in the Nucleus
Chromatin Status
DNA packaging into nucleosomes and organization into chromatin fibers generates constraints for transcription. Chromatin remodelers are required for RNA polymerase II (RNAPII) to access the DNA. Post‐translational modifications of histone tails constitute a code that contributes to transcription by serving as a landing platform for protein complexes such as ATP‐dependent chromatin remodeling complexes. Two such histone marks are under the control of the Ccr4‐Not complex. Histones H3 and H4 are globally hypoacetylated in yeast cells lacking Not4 or Not5.37 Gcn5 is the histone acetyltransferase (HAT) that acetylates histones within nucleosomes when associated with other proteins in HAT complexes. Gcn5 purified from cells lacking Not5 can acetylate free histones but it fails to acetylate nucleosomes in vitro. Another chromatin mark that is affected by the Ccr4‐Not complex is histone H3 lysine four tri‐methylation. This mark is very much reduced in cells lacking Not4 or Not538, 39 probably because Not4 is responsible for the turnover of the Jhd2 demethylase that demethylates histone H3.40
Transcription Initiation
Ccr4‐Not subunits are present at sites of transcription and can be cross‐linked to promoters in yeast and in higher eukaryotes.41, 42, 43, 44, 45, 46 Tethering of Ccr4‐Not complex subunits to promoters can activate or repress transcription.47, 48 In higher eukaryotes, a couple of examples have suggested that Ccr4‐Not subunits can interact with transcription factors and repress their ability to activate transcription. For instance Not1 interacts in a ligand dependent manner with the retionoid acid X receptor49 and Caf40 binds transcriptional activators such as c‐Myb and AP‐1.50 Several transcription factors or transcriptional regulators are targets of the Not4 ubiquitin ligase in yeast or mammals. Examples are the DNA binding activator Yap1 in yeast51 or the Paf1 subunit of the PAF complex in mammalian cells.52 This suggests that transcription factors recruit Ccr4‐Not subunits to promoters but can also be regulated by Not4.
Besides its effect on specific transcription factors, the Ccr4‐Not complex is important for the relative presence of TFIID and SAGA at promoters.53, 54 TFIID and SAGA are general transcription complexes that can both recruit the TATA binding protein to promoters, but TFIID is generally present at constitutive housekeeping genes, while SAGA is mostly present at highly inducible genes. Ccr4‐Not subunit crosslinking is enriched at SAGA‐dependent genes43 and genome‐wide gene expression analyses of ccr4‐not mutants indicate that SAGA‐dependent genes are mostly affected.55 Numerous studies describe both genetic and physical interactions of Ccr4‐Not subunits with TFIID or SAGA. However, we still do not know whether presence of Ccr4‐Not at the sites of transcription is relevant for SAGA‐related phenotypes.
The connections between Ccr4‐Not and transcription initiation are summarized in Figure 2.
Figure 2.
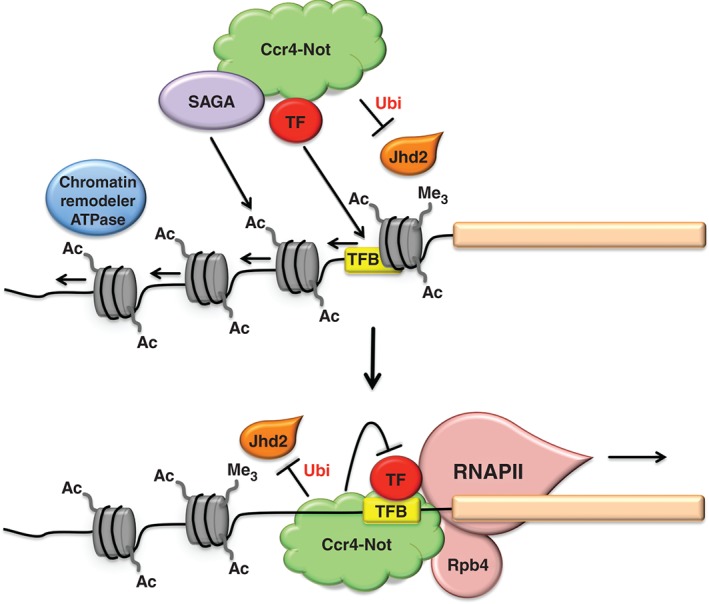
Cartoon representation of the interactions of Ccr4‐Not with components relevant for transcription initiation (a). and with components likely to recruit Ccr4‐Not to the transcription unit (b). TF, transcription factor; TFB, transcription factor binding site; Ubi, ubiquitination; Ac, acetylation; Me3, trimethylation.
Transcription Elongation
Strains harboring mutations in CCR4‐NOT genes are sensitive to inhibitors of transcription elongation, namely 6‐azauracil and mycophenolic acid.56 In addition the deletion of CCR4‐NOT genes displays a synthetically enhanced slow growth phenotype when combined mutations in transcription elongation factors. In vivo evidence for a role of the Ccr4‐Not complex in transcriptional elongation is based upon an assay, which monitors the last wave of RNAPII across a long gene after its initiation is repressed. This wave clears the gene more slowly in ccr4‐not mutants compared to the wild‐type.44 In another assay the in vivo role of Ccr4‐Not on transcription elongation was evaluated by measuring gene‐length dependent accumulation of mRNA (GLAM). Changes in GLAM were identified in ccr4‐not mutants.57 In vitro Ccr4‐Not can interact with transcription elongation complexes prepared from highly purified yeast RNAPII.44 This requires the separable Rpb4/7 module of the polymerase,58 in accord with a reported interaction between Rpb4 and both Not3 and Not5.59 Ccr4‐Not directly affects transcription elongation by rescuing backtracked RNAPII. It has no effect on the transcription rate of un‐arrested RNAPII. Interestingly, the ability of Ccr4‐Not to rescue backtracked polymerase depends upon the length of the transcript suggesting that an association of Ccr4‐Not with the new transcript contributes to promote transcription elongation. Another transcription factor that directly promotes transcription elongation and rescues backtracked RNAPII is TFIIS. It does so by stimulating intrinsic nucleolytic activity of RNAPII and transcript cleavage (reviewed in Ref 60). This mechanism is not shared by Ccr4‐Not, but Ccr4‐Not has the additional effect of improving association of TFIIS to the elongating polymerase.61 The effects of Ccr4‐Not on transcription elongation are summarized in Figure 3.
Figure 3.
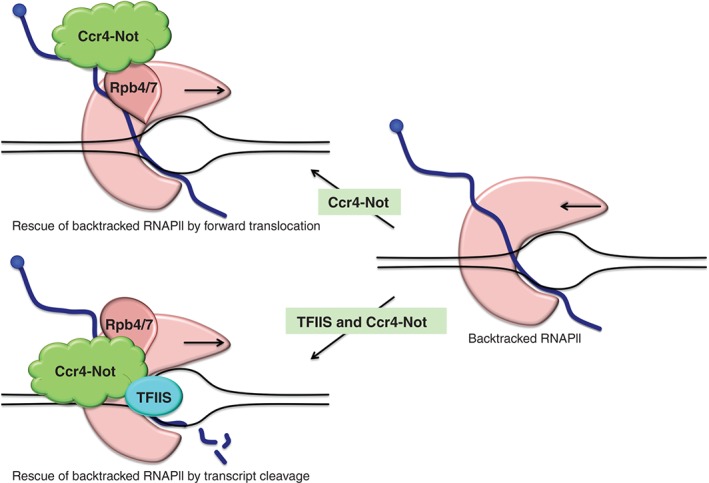
Representation of the roles of Ccr4‐Not in transcription elongation. Adapted from Refs 15, 61. On the right is depicted RNAPII that encounters a block in transcription during the productive elongation phase and that backtracks. On the upper left panel is depicted reiterative transcript binding and release by Ccr4–Not that causes realignment of the 3′ end of the RNA into the active site and promotes the resumption of elongation by RNAPII. On the lower left panel is depicted that Ccr4‐Not increases the recruitment of TFIIS to backtracked RNAPII complexes. This enhances the cleavage of the displaced transcript in backtracked RNAPII by TFIIS, allowing elongation to resume.
Quality Control and mRNA Export
Processing and maturation of newly produced transcripts depends upon many factors that assemble into heterogenous ribo‐nucleoprotein particles (hnRNPs) to prepare the mRNA for its export to the cytoplasm. The mRNPs are sensed in a quality control process to ensure that only functional mRNAs are exported. When export components are defective a number of phenotypes are observed, some of which depend upon the nuclear exosome subunit Rrp6 indicating that this exonculease is important for the mRNA quality control process.62 The Ccr4‐Not complex interacts with proteins that participate in maturation of mRNPs coupled to their export, such as the nuclear poly(A) binding protein Nab2 or Hrp1 of the THO complex, or Mlp1 of the inner basket of the nuclear pore complex (NPC).63 It is also important for appropriate interaction of Rrp6 with its co‐factor Mtr4, an RNA helicase.64 The deletion of Ccr4‐Not subunits displays synthetic growth phenotypes when combined with the deletion of export proteins such as Mft1 of the THO complex,65 or when combined with the deletion of Rrp6.64 Yet Rrp6‐dependent quality control defects observed in mft1 mutants are rescued by the co‐deletion of Ccr4‐Not subunits. This might indicate that the double mutants export defective mRNAs and hence suggests a role for Ccr4‐Not in the nuclear quality control mechanism. In line with this idea, overexpression of Not4 exacerbates the poly(A) mRNA export defect observed in an NPC mutant.63 Moreover polyadenylated snRNAs indicative of defective Rrp6‐dependent nuclear quality control accumulate in ccr4‐not mutants.64 These interactions of Ccr4‐Not related to mRNA export and nuclear quality control are depicted in Figure 4.
Figure 4.
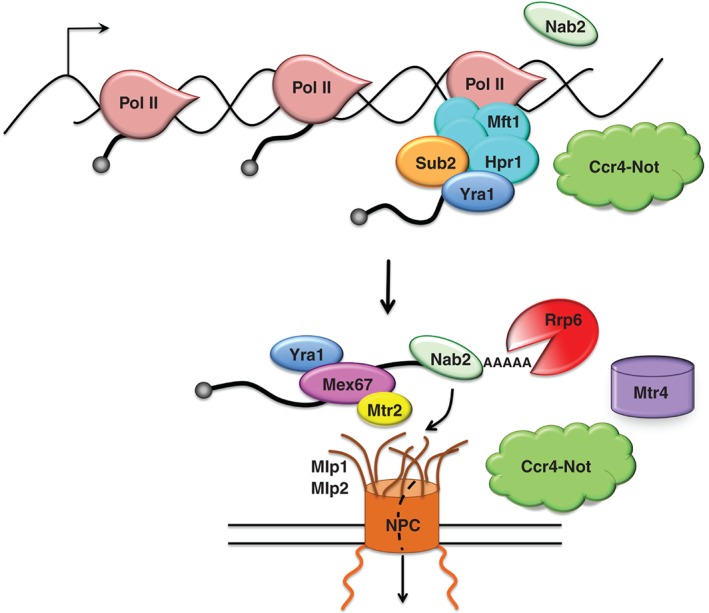
Cartoon depicting the different interactions between Ccr4‐Not and components contributing to mRNA export and nuclear quality control. Ccr4‐Not interacts with Hpr1 and Mft1 or the THO/TREX complex that includes Sub2, with the nuclear poly(A) binding protein Nab2, with Mlp1, and is necessary for the interaction of the Rrp6 exonuclease with its co‐factor the Mtr4 RNA helicase. Some other components that participate in export such as Mex67, Mtr2, Yra1, and Mlp1 are included. NPC: nuclear pore complex.
Cytoplasmic mRNA Fate
Stability and translation of mRNAs in eukaryotes depend upon the 7‐methylguanylate cap at the 5′ end of the mRNA and the poly(A) tail at the 3′ end of the mRNA, the former added co‐transcriptionally to the new message and the latter added after transcript cleavage. In mammalian cells the mRNA cap is first bound in the nucleus by the cap binding protein (CBP) that will support a pioneer round of translation in the cytoplasm. The eIF4E translation initiation factor replaces subsequently CBP and will direct steady state rounds of translation. CBP and eIF4E particles are associated with nuclear and cytoplasmic poly(A) binding proteins (PABP), respectively. CBP is not essential in yeast and newly produced mRNAs can be bound directly by eIF4E.
Cytoplasmic mRNAs cycle between polysomes and RNA granules. The mechanisms and directionality of these movements are not fully understood. Translationally repressed mRNAs will accumulate in granules and RNA granules can release transcripts to allow their translation.66 Functional mRNAs arriving in the cytoplasm are not necessarily recruited immediately into polysomes to be translated, nor are they necessarily immediately degraded after leaving polysomes.
The best‐characterized granules are the P‐bodies (PBs) and stress granules (SGs). They have features in common and specific features. PBs contain the mRNA decay machinery whereas SGs contain many translation initiation factors. SGs form in the absence of new translation. They harbor mRNAs from pre‐existing polysomes and arise in response to inhibition of translation initiation, followed by polysome elongation and a progressive loss of ribosomes. PBs are also formed from translationally repressed mRNPs onto which complexes of decapping components are recruited. mRNAs that exit translation can first go to PBs but then remodel to acquire translation initiation factors and move to SGs where translation is repaired and mRNAs can be released when they are ready to translate again. SGs may also form independently of PBs, and mRNPs may enter SGs prior to entering polysomes when produced under conditions of translation inhibition. Globally, the role of the RNA granules is still not entirely clear. They might induce a local concentration of proteins favorable for mRNA decay (PBs) or reinitiation of translation (SGs), or provide a buffering system to maintain a proper ratio of translational capacity to the pool of mRNAs that are translating (for instance to avoid titrating limiting factors), or finally serve as a reservoir of translatable mRNAs (for review see Ref 66).
The Roles of Ccr4‐Not in the Translation Process
Ccr4‐Not and mRNA Translatability
Recent data has indicated that translatability of several mRNAs, defined as the presence of polyadenylated forms of those specific mRNA in polysome fractions relative to total levels, is altered in yeast lacking Not5, some increased, others decreased.59 The mechanism is unknown. Not5 might be altering the cycling of specific mRNAs between RNA granules and polysomes, or regulating the distribution of specific newly produced mRNAs to polysomes or RNA granules, or both. Not5 has RNA binding activity67 and it is also important for integrity of the Ccr4‐Not complex that contains other proteins with RNA binding activity.
Ccr4‐Not and Translation
The level of total polysomes is reduced in yeast cells lacking Not2, Not4 or Not5.35 These strains grow slowly, so it is difficult to definitively conclude that reduced polysomes are not a consequence of slow growth. However several observations in yeast are compatible with a role of the Ccr4‐Not proteins during normal translation. All Ccr4‐Not subunits are detected in polysomes35 and newly produced proteins aggregate massively in not2, not4, and not5 mutants. This suggests that folding or protein interactions of newly produced proteins are defective.
The importance of Not5 during translation is exemplified by the need for Not5 during de novo synthesis of the largest subunit of RNAPII, Rpb1.59 Rpb1 tends to aggregate and must associate with Hsp90 and a co‐chaperone called R2TP to form a soluble assembly‐competent intermediate.68 Not5 is needed for the presence of R2TP at ribosomes producing Rpb1 (Figure 5(a)), and as a consequence it is needed for efficient assembly of RNAPII. Assembly of another multisubunit complex, the proteasome, is also dependent upon the Ccr4‐Not complex, in particular upon Not4.69 Not4 is needed for assembly of the proteasome lid, and it is important for a proteasomal chaperone, Ecm29, to associate with proteasome subunits. Interestingly in fission yeast the ortholog of Ecm29 is present in polysomes producing proteasomal subunits, suggesting that Not4 may be contributing to co‐translational assembly of the proteasome with one of its chaperones (Figure 5(b)).
Figure 5.
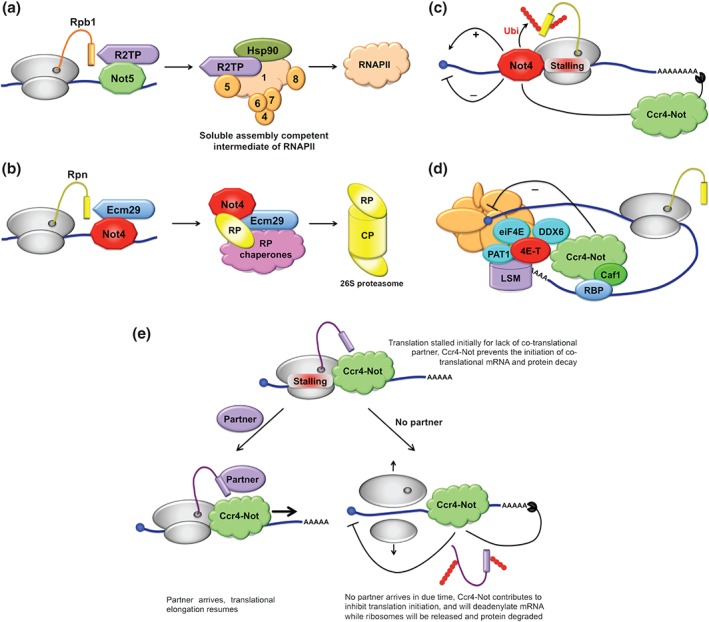
The Ccr4‐Not complex plays many roles during the translation process. (a) Not5 is needed for the presence of the R2TP co‐chaperone at polysomes producing Rpb1 to form a soluble assembly‐competent Rpb1 including Hsp90 and the Rpb4, 5, 6, 7, and 9 subunits, that will form RNAPII. (b) Not4 is needed probably co‐translationally for the interaction of the Ecm29 chaperone with regulatory particle (RP) proteasome subunits and proteasome integrity. CP: core particle. (c) Not4 is a relevant player during co‐translational quality control. It has reported positive and negative effects on translation initiation, the ability to ubiquitinate stalled peptide or finally to initiate mRNA decay by Ccr4‐Not dependent deadenylation. (d) Upon recruitment by an RNA binding protein (RBP) Ccr4‐Not mediates translational repression in a deadenylation‐independent manner via the recruitment of the 4E‐T protein. 4E‐T interacts with eIF4E and decapping components associating with the 3′ and 5′ ends of mRNAs such as the LSM, PAT1, and DDX6 proteins. DDX6 can also directly interact with Not1. (e) Cartoon with a model for both positive and negative effects of Not4 on translation.
The Not4 E3 ligase ubiquitinates a ribosomal protein Rps7A and a ribosome associated chaperone called nascent polypeptide associated complex (NAC).35, 70 This observation has fostered interest in Not4 as a possible translational regulator. Such a role for Not4 has been studied with artificial mRNA templates encoding two open reading frames (ORFs) with an intervening stretch of RNA coding for 12 subsequent lysines known to provoke ribosome stalling.34, 71, 72 Initially Not4 was reported to ubiquitinate the stalled peptides,34 but subsequently described instead to have an impact on levels of the mRNAs with stalled ribosomes.72 A third study suggested that in the absence of Not4 translation levels were reduced71 in contrast to a fourth study indicating that Not4 was needed to repress translation.73 Not4 was also described to participate in global translational repression after nutrient withdrawal in yeast, when reduced concentrations of amino acyl tRNAs are thought to promote generalized ribosome stalling.73 These different possible effects of Not4 in co‐translation quality control are summarized in Figure 5(c).
A role for the Ccr4‐Not complex in translational repression upon non‐sense mediated mRNA decay (NMD) or upon its’ tethering to mRNAs by RNA binding proteins (RBPs) or by the micro‐RNA machinery has been characterized. This translational repression ability is independent of its deadenylase function. Indeed, tethering of the Ccr4‐Not complex to mRNAs lacking a poly(A) tail in vivo 74 and in vitro 75 can induce translation arrest, and in yeast repression of translation by the Mpt5 RBP occurs via Caf1 in cells that lack the active deadenylase subunit Ccr4.76 A mechanism for translation repression has recently emerged from studies in Xenopus 77 and Drosophila.78 The eIF4E transporter protein (4E‐T), a member of the eIF4E binding proteins, can mediate translational repression of tethered Ccr4‐Not. 4E‐T interacts with eIF4E and with the mRNA decay machinery, both with components interacting with the 3′ ends of the mRNAs (such as the LSM1‐7‐PAT1 complex) and with the 5′ cap (such as the dead box protein DDX6 known as Dhh1 in yeast). 4E‐T also interacts with the Ccr4‐Not complex itself.77, 78, 79 The interaction between eIF4E and 4E‐T is necessary for deadenylation‐independent translational repression by the Ccr4‐Not complex77 and for mRNA decay.78 The proposed chain of events is that RBPs recruit Caf1 that in turn recruits Not1 to the mRNA that will then recruit 4E‐T. 4E‐T will physically link the 3′ terminal mRNA decay machinery to the 5′ cap (Figure 5(d)).
Not1 also directly interacts with DDX6 via its MIF4G domain (see Figure 1).80, 81, 82 In yeast it was shown that Dhh1 is an activator of the decapping complex, but that it also functions in translation repression.83 It accompanies slowly translocating ribosomes and when tethered to an mRNA leads to an accumulation of ribosomes on the mRNA.84 It has been suggested that translational repression after nutrient withdrawal needs Dhh1 like Not4.73 However overexpression of Dhh1 rather than deletion of Dhh1 passes the genetic selection that led to identification of the NOT genes,85 indicating that Dhh1 and Not4 might have opposite rather than similar effects on gene expression.
The described positive and negative roles of the Ccr4‐Not complex during translation seem contradictory, but they may in fact be reconcilable. The necessity for proteins to interact with partners co‐translationally could cause ribosome pausing. If partners fail to appear in a timely manner, the Ccr4‐Not complex might contribute to translational repression and mRNA decay. However, the Ccr4‐Not complex might initially preserve the ribosome‐mRNA complex to give time for partners to interact co‐translationally, and only if the delay is too long allow the co‐translational responses to proceed (Figure 5(e)). An imbalance in this mechanism would lead to the production of mis‐folded proteins.
Since the Ccr4‐Not complex is important for control of poly(A) tail length, it is to be expected that it will play an important role in the movement of mRNAs from the translating pool to the pool directed to RNA granules. In HeLa cells the depletion of Not1 suppresses largely the formation of PBs.86 Ccr4‐Not subunits localize to PBs in stressed yeast36 but they do not seem to be a major component of PBs in flies.12
The Roles of Ccr4‐Not in mRNA Degradation
Since stability and translation of mRNAs depend upon the cap and the poly(A) tail of the mRNA, translation and decay are intimately connected and generally in competition with each other. The major mRNA decay pathway in eukaryotes involves removal of the cap followed by 5′→3′ exonucleolytic degradation. The initial and rate‐limiting step in this pathway is deadenylation performed mainly by Caf1 and Ccr4, the major eukaryotic deadenylases.
Initiation of mRNA degradation is intimately connected to the process of translation.87 Different types of mRNA decay can be distinguished: generic decay of mRNAs that have undergone normal cycles of translation, quality control induced mRNA decay, which occurs upon ribosomal stalling without regular translational termination, and finally induced mRNA decay brought about by tethering of the Ccr4‐Not complex to specific mRNAs in response to specific signaling.
Generic mRNA Deadenylation
Not much is known about how the Ccr4‐Not complex participates in generic decay of all mRNAs upon translation termination. Either Ccr4‐Not is present during translation of all mRNAs in an inactive form, and then is modified upon translation termination to induce deadenylation, or it is recruited upon translation termination to initiate shortening of the poly(A) tail. One study has described a series of coupled events starting with the recruitment of the translation termination factors eRF1 and eRF3, recruitment of the minor deadenylation complex Pan2‐Pan3, and finally recruitment of Ccr4‐Not by the BTG/TOB family of proteins via their interaction with PABP.88 BTG/TOB proteins are absent in budding and fission yeast and they are not constitutively expressed in mammalian cells (for reviews see Ref 89). Thus, while this mechanism (Figure 6(a)) might be a response that leads to a general mRNA deadenylation response in metazoans, it cannot be the mechanism for generic mRNA decay.
Figure 6.
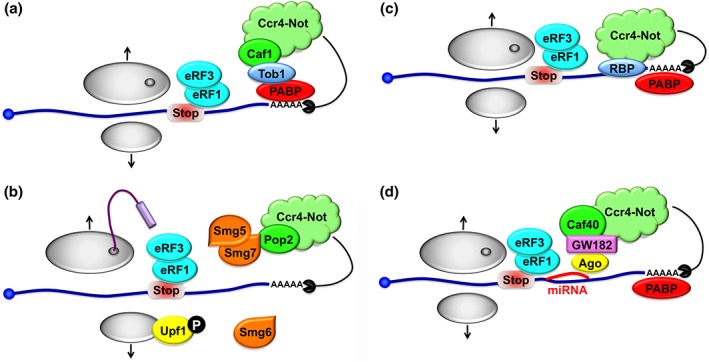
The Ccr4‐Not is the major eukaryotic deadenylase. (a) Ccr4‐Not can be recruited to mRNAs via an interaction between Caf1 and Tob1 itself binding to PABP. This can cause a generalized deadenylation of mRNAs upon Tob1 expression. (b) During NMD in verterbrates, phosphorylated Upf1 can recruit the Smg5, 6, and 7 proteins. Smg6 is an endonuclease that acts redundantly to the Ccr4‐Not complex recruited by the Smg5/7 heterodimer, via the interaction of Smg7 and Pop2, to bring about mRNA decay. (c) RBPs can tether the Ccr4‐Not complex to target mRNAs and lead to deadenylation probably after normal translation termination. (d) The micro‐RNA machinery can tether Ccr4‐Not and bring about mRNA deadenylation after normal translation termination.
Non‐sense Mediated mRNA Decay (NMD)
NMD results from the presence of an aberrant stop codon within the open reading frame. The recruitment of the Ccr4‐Not complex to initiate deadenylation of NMD targets has been characterized in vertebrates (Figure 6(b)). During the pioneer round of translation, CBP interacts with the conserved Upf1 protein that will become phosphorylated. This promotes recruitment of the Smg 5, 6, and 7 proteins.90 Smg6 is an endonuclease and the Smg5‐Smg7 heterodimer recruits the Ccr4‐Not complex via interactions between Smg7 and Pop2/CNOT8 (but not its homolog Caf1/CNOT7). The recruitment of Ccr4‐Not by Smg7 is critical for NMD in cells lacking Smg6, indicating that Smg6 and the Ccr4‐Not complex act redundantly to promote mRNA decay of NMD substrates. In yeast, Upf1 is not phosphorylated and there are no orthologs of Smg5, 6, and 7. NMD targets are rapidly decapped without prior need for deadenylation. A specific role for Ccr4‐Not in NMD in yeast is uncertain.
Targeted mRNA Decay
Certainly the most well established function of the Ccr4‐Not complex is its ability to repress gene expression upon its targeted recruitment to the 3′ untranslated regions (UTRs) of mRNAs. The complex is recruited by RBPs or the micro‐RNA machinery that associate with specific sequences at the 3′ end of the mRNAs. Experiments in yeast have demonstrated that the tethering of the Ccr4‐Not complex is sufficient to destabilize an mRNA in vivo. This has established that Ccr4‐Not is constitutively active to deadenylate an mRNA provided that the deadenylase subunits are recruited to the mRNA (Figure 6(c)).
Many sequence specific RBPs that recruit Ccr4‐Not have been described. They either bind Not1 directly, or they recruit Not1 by binding to other subunits of the Ccr4‐Not complex. They can also interact with proteins such as the BTG/TOB proteins that in turn associate with a Ccr4‐Not subunit. Three binding partners have been characterized structurally. Tristetraprolin (TTP) interacts with Not191 and associates with ARE elements in the 3′ UTRs of genes such as TNFα, c‐Fos, and c‐Myc. Nanos1 is important for germ cell line maintenance and survival and interacts also with Not1.92 Finally, BTG/TOB proteins interact with Caf1 homologs to mediate their antiproliferative activity.93
The interaction of the miRNA machinery with the Ccr4‐Not complex and mRNA targets has been well studied. It is a multilayered process starting with base pairing of the Argonaute (Ago1) miRNPs to the target mRNA. This leads to recruitment of the GW182 protein that in turn will recruit deadenylases including Ccr4‐Not. This recruitment involves multiple regions in GW182, a 400 amino acid C‐terminal region called CED being the most important one. It can recruit Not1 by interaction with a central domain of Not1 in three different ways, one being mediated by Caf4080 (Figure 6(d)). These interactions involve tryptophan residues in GW182 and Trp‐binding pockets in Not1 and Caf40. In turn Not1 is associated with Caf1 and Ccr4 that will deadenylate the mRNA, and it can bind DDX6 that has the effects discussed above (reviewed in Ref 94).
Deadenalyation Independent Decapping
Deadenylation‐independent decapping of certain mRNAs has been reported for Not2, Not4, and Not5 in yeast.95 As mentioned above, Dhh1 associates with the Not1 MIF4G domain and can function as a decapping activator. It could be that specific mRNAs need Ccr4‐Not to be efficiently decapped because Ccr4‐Not interacts with Dhh1.
CROSS‐TALK BETWEEN DIFFERENT LEVELS OF GENE EXPRESSION VIA Ccr4‐Not
The different functions of the Ccr4‐Not complex at different stages of gene expression described above are compatible with the presence of the subunits both in the nucleus and in the cytoplasm. Moreover the reported change in localization of a Ccr4‐Not subunit in response to the stage of the cell cycle is compatible with the ability of the Ccr4‐Not complex to differentially modify its role along the gene expression pathway in response to the need of the cell. In addition the Ccr4‐Not subunits harbor a multitude of post‐transcriptional modifications that have not yet been functionally characterized.
One can imagine that the Ccr4‐Not complex acts independently at different steps along the gene expression pathway, or instead that its functions at the different steps are physically connected (Figure 7). It has been proposed that gene‐specific regulators could recruit specific Ccr4‐Not subunits to promoters for subsequent post‐transcriptional regulation.45 Moreover the Ccr4‐Not complex can interact with the transcription elongation complex and with the nascent transcripts. Hence the Ccr4‐Not complex may associate with some mRNAs as they are synthesized and then remain bound within the mRNPs exported to the cytoplasm. This may not concern all mRNAs, but only those whose synthesis depends upon TFs that can recruit Ccr4‐Not or those synthesized from genes whose transcription elongation is particularly prone to mobilize Ccr4‐Not. If mRNAs are bound in the nucleus by Ccr4‐Not, probably this is a form of the complex that is not immediately active for induction of deadenylation, translational repression, and decapping.
Figure 7.

Ccr4‐Not connects the nuclear and cytoplasmic phases of gene expression. Adapted from Ref 96. Ccr4‐Not is present at sites of transcription by interaction with transcription factors (TFs) and with the elongating RNAPII via Rpb4. It might associate with newly produced mRNAs and be exported to the nucleus with the mRNPs. In the cytoplasm it will contribute to regulate translation and bring about mRNA degradation. But it will also promote assembly of new RNAPII that can re‐enter the nucleus to start new rounds of transcription.
The Rpb4 subunit of RNAPII was reported to associate with newly produced mRNAs under stress and to be exported with them out to the cytoplasm, where it will contribute to their translation, targeting to PBs and/or decay.97 Interestingly, Rpb4 is essential for association of Ccr4‐Not with elongating RNAPII and Rpb4 export to the cytoplasm depends upon Not5.59 Hence an intriguing possibility is that Rpb4 could be necessary in the nucleus for the Ccr4‐Not complex to associate with new transcripts, allowing Ccr4‐Not to module their function post‐transcriptionally (Figure 7).
What might be the advantage of imprinting mRNAs in the nucleus with Ccr4‐Not? The post‐transcriptional fate of such mRNAs may be more immediately determined. Moreover, it provides a mean for the eukaryotic cell to connect physically the nuclear and cytoplasmic stages of gene expression. Changes in transcription levels may be compensated for by opposite changes in mRNA degradation via the Ccr4‐Not complex to maintain similar steady state levels of mRNAs, but also by changes in translation to produce similar levels of protein. Such gene expression buffering is known to exist. Rpb4 and the Ccr4‐Not complex have been connected to this forward buffering (reviewed in Ref 98). Reciprocally, Ccr4‐Not is important during translation for production of the transcription machine (RNAPII), and this provides Ccr4‐Not with the ability to communicate the translation status of the cell back to the nucleus.
CONCLUSION
The Ccr4‐Not complex intervenes at all stages of the gene expression pathway, helping to produce mRNAs and contributing positively to translation, but also initiating translation repression and mRNA degradation. A major challenge facing our understanding of this master regulator is to determine in which forms the Ccr4‐Not complex acts for each of these functions and how it is modified to change roles. Characterizing the flow of Ccr4‐Not between the nucleus and the cytoplasm is an essential part of these questions.
Finally, one has to wonder why such a complex regulator as Ccr4‐Not was conserved across the eukaryotic kingdom. The most obvious explanation is that a regulator such as this one is the most efficient to coordinate the different steps of gene expression. It is ideally suited to mediate changes in programs of gene expression, but also ideally designed to buffer undesirable changes in a given step along the gene expression pathway. Not surprisingly, evidence for essential roles of Ccr4‐Not in cellular programing and gene expression buffering is increasing.
FURTHER READING
Many reviews have been written on the Ccr4‐Not complex, the first ones about 10 years ago,1, 2, 3 and recently many more, some of which cover aspects not discussed in this review, such as for instance more detailed structural information on the complex or the physiological roles of Ccr4‐Not in different species or genetic programming, that are exciting additional reading.4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18
ACKNOWLEDGMENTS
I would like to thank Olesya Panasenko for help with the illustrations and Virginie Ribaud, Zoltan Villanyi, Sari Kassem, and Ravish Rashpa for a critical reading of the manuscript. This work was supported by grant 31003a_135794 from the Swiss National Science Foundation.
Conflict of interest: The author has declared no conflicts of interest for this article.
The copyright line in this article was changed on 25 February 2016 after online publication.
REFERENCES
- 1. Collart MA, Timmers HT. The eukaryotic Ccr4‐not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog Nucleic Acid Res Mol Biol 2004, 77:289–322. [DOI] [PubMed] [Google Scholar]
- 2. Denis CL, Chen J. The CCR4‐NOT complex plays diverse roles in mRNA metabolism. Prog Nucleic Acid Res Mol Biol 2003, 73:221–250. [DOI] [PubMed] [Google Scholar]
- 3. Collart MA. Global control of gene expression in yeast by the Ccr4‐Not complex. Gene 2003, 313:1–16. [DOI] [PubMed] [Google Scholar]
- 4. Collart MA, Panasenko OO. The Ccr4‐‐not complex. Gene 2012, 492:42–53. [DOI] [PubMed] [Google Scholar]
- 5. Collart MA, Panasenko OO, Nikolaev SI. The Not3/5 subunit of the Ccr4‐Not complex: a central regulator of gene expression that integrates signals between the cytoplasm and the nucleus in eukaryotic cells. Cell Signal 2013, 25:743–751. [DOI] [PubMed] [Google Scholar]
- 6. Miller JE, Reese JC. Ccr4‐Not complex: the control freak of eukaryotic cells. Crit Rev Biochem Mol Biol 2012, 47:315–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Panepinto JC, Heinz E, Traven A. The cellular roles of Ccr4‐NOT in model and pathogenic fungi‐implications for fungal virulence. Front Genet 2013, 4:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu K, Bai Y, Zhang A, Zhang Q, Bartlam MG. Insights into the structure and architecture of the CCR4‐NOT complex. Front Genet 2014, 5:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Inada T, Makino S. Novel roles of the multi‐functional CCR4‐NOT complex in post‐transcriptional regulation. Front Genet 2014, 5:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Panasenko OO. The role of the E3 ligase Not4 in cotranslational quality control. Front Genet 2014, 5:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wahle E, Winkler GS. RNA decay machines: deadenylation by the Ccr4‐not and Pan2‐Pan3 complexes. Biochim Biophys Acta 1829, 2013:561–570. [DOI] [PubMed] [Google Scholar]
- 12. Temme C, Simonelig M, Wahle E. Deadenylation of mRNA by the CCR4‐NOT complex in Drosophila: molecular and developmental aspects. Front Genet 2014, 5:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shirai YT, Suzuki T, Morita M, Takahashi A, Yamamoto T. Multifunctional roles of the mammalian CCR4‐NOT complex in physiological phenomena. Front Genet 2014, 5:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chapat C, Corbo L. Novel roles of the CCR4‐NOT complex. WIREs RNA 2014, 5:883–901. [DOI] [PubMed] [Google Scholar]
- 15. Reese JC. The control of elongation by the yeast Ccr4‐not complex. Biochim Biophys Acta 2013, 1829:127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doidge R, Mittal S, Aslam A, Winkler GS. Deadenylation of cytoplasmic mRNA by the mammalian Ccr4‐Not complex. Biochem Soc Trans 2012, 40:896–901. [DOI] [PubMed] [Google Scholar]
- 17. Winkler GS, Balacco DL. Heterogeneity and complexity within the nuclease module of the Ccr4‐Not complex. Front Genet 2013, 4:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Collart MA. The NOT4 RING E3 ligase: a relevant player in co‐translational quality control. ISRN Mol Biol 2013, doi: 10.1155/2013/548359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Albert TK, Lemaire M, van Berkum NL, Gentz R, Collart MA, Timmers HT. Isolation and characterization of human orthologs of yeast CCR4‐NOT complex subunits. Nucleic Acids Res 2000, 28:809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bawankar P, Loh B, Wohlbold L, Schmidt S, Izaurralde E. NOT10 and C2orf29/NOT11 form a conserved module of the CCR4‐NOT complex that docks onto the NOT1 N‐terminal domain. RNA Biol 2013, 10:228–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maillet L, Tu C, Hong YK, Shuster EO, Collart MA. The essential function of Not1 lies within the Ccr4‐Not complex. J Mol Biol 2000, 303:131–143. [DOI] [PubMed] [Google Scholar]
- 22. Bhaskar V, Basquin J, Conti E. Architecture of the ubiquitylation module of the yeast Ccr4‐Not complex. Structure 2015, 23:921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nasertorabi F, Batisse C, Diepholz M, Suck D, Bottcher B. Insights into the structure of the CCR4‐NOT complex by electron microscopy. FEBS Lett 2011, 585:2182–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Denis CL. Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics 1984, 108:833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Collart MA, Struhl K. NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global‐negative regulator of transcription that differentially affects TATA‐element utilization. Genes Dev 1994, 8:525–537. [DOI] [PubMed] [Google Scholar]
- 26. Collart MA, Struhl K. CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters. EMBO J 1993, 12:177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oberholzer U, Collart MA. Characterization of NOT5 that encodes a new component of the Not protein complex. Gene 1998, 207:61–69. [DOI] [PubMed] [Google Scholar]
- 28. Draper MP, Salvadore C, Denis CL. Identification of a mouse protein whose homolog in Saccharomyces cerevisiae is a component of the CCR4 transcriptional regulatory complex. Mol Cell Biol 1995, 15:3487–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen J, Rappsilber J, Chiang YC, Russell P, Mann M, Denis CL. Purification and characterization of the 1.0 MDa CCR4‐NOT complex identifies two novel components of the complex. J Mol Biol 2001, 314:683–694. [DOI] [PubMed] [Google Scholar]
- 30. Tucker M, Valencia‐Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae . Cell 2001, 104:377–386. [DOI] [PubMed] [Google Scholar]
- 31. Bogdan JA, Adams‐Burton C, Pedicord DL, Sukovich DA, Benfield PA, Corjay MH, Stoltenborg JK, Dicker IB. Human carbon catabolite repressor protein (CCR4)‐associative factor 1: cloning, expression and characterization of its interaction with the B‐cell translocation protein BTG1. Biochem J 1998, 336(Pt 2):471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, Simonelig M, Wahle E. Subunits of the Drosophila CCR4‐NOT complex and their roles in mRNA deadenylation. RNA 2010, 16:1356–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Morel AP, Sentis S, Bianchin C, Le Romancer M, Jonard L, Rostan MC, Rimokh R, Corbo L. BTG2 antiproliferative protein interacts with the human CCR4 complex existing in vivo in three cell‐cycle‐regulated forms. J Cell Sci 2003, 116:2929–2936. [DOI] [PubMed] [Google Scholar]
- 34. Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent peptide‐dependent translation arrest leads to Not4p‐mediated protein degradation by the proteasome. J Biol Chem 2009, 284:10343–10352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Panasenko OO, Collart MA. Presence of Not5 and ubiquitinated Rps7A in polysome fractions depends upon the Not4 E3 ligase. Mol Microbiol 2012, 83:640–653. [DOI] [PubMed] [Google Scholar]
- 36. Teixeira D, Parker R. Analysis of P‐body assembly in Saccharomyces cerevisiae . Mol Biol Cell 2007, 18:2274–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peng W, Togawa C, Zhang K, Kurdistani SK. Regulators of cellular levels of histone acetylation in Saccharomyces cerevisiae . Genetics 2008, 179:277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mulder KW, Brenkman AB, Inagaki A, van den Broek NJ, Timmers HT. Regulation of histone H3K4 tri‐methylation and PAF complex recruitment by the Ccr4‐Not complex. Nucleic Acids Res 2007, 35:2428–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laribee RN, Shibata Y, Mersman DP, Collins SR, Kemmeren P, Roguev A, Weissman JS, Briggs SD, Krogan NJ, Strahl BD. CCR4/NOT complex associates with the proteasome and regulates histone methylation. Proc Natl Acad Sci USA 2007, 104:5836–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mersman DP, Du HN, Fingerman IM, South PF, Briggs SD. Polyubiquitination of the demethylase Jhd2 controls histone methylation and gene expression. Genes Dev 2009, 23:951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Deluen C, James N, Maillet L, Molinete M, Theiler G, Lemaire M, Paquet N, Collart MA. The Ccr4‐not complex and yTAF1 (yTaf(II)130p/yTaf(II)145p) show physical and functional interactions. Mol Cell Biol 2002, 22:6735–6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Swanson MJ, Qiu H, Sumibcay L, Krueger A, Kim SJ, Natarajan K, Yoon S, Hinnebusch AG. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol Cell Biol 2003, 23:2800–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck‐Walsh C, et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces . Mol Cell 2011, 41:480–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kruk JA, Dutta A, Fu J, Gilmour DS, Reese JC. The multifunctional Ccr4‐Not complex directly promotes transcription elongation. Genes Dev 2011, 25:581–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome‐wide RNAi screen identifies a new transcriptional module required for self‐renewal. Genes Dev 2009, 23:837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Venturini G, Rose AM, Shah AZ, Bhattacharya SS, Rivolta C. CNOT3 is a modifier of PRPF31 mutations in retinitis pigmentosa with incomplete penetrance. PLoS Genet 2012, 8:e1003040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zwartjes CG, Jayne S, van den Berg DL, Timmers HT. Repression of promoter activity by CNOT2, a subunit of the transcription regulatory Ccr4‐not complex. J Biol Chem 2004, 279:10848–10854. [DOI] [PubMed] [Google Scholar]
- 48. Benson JD, Benson M, Howley PM, Struhl K. Association of distinct yeast Not2 functional domains with components of Gcn5 histone acetylase and Ccr4 transcriptional regulatory complexes. EMBO J 1998, 17:6714–6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Winkler GS, Mulder KW, Bardwell VJ, Kalkhoven E, Timmers HT. Human Ccr4‐Not complex is a ligand‐dependent repressor of nuclear receptor‐mediated transcription. EMBO J 2006, 25:3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Haas M, Siegert M, Schurmann A, Sodeik B, Wolfes H. c‐Myb protein interacts with Rcd‐1, a component of the CCR4 transcription mediator complex. Biochemistry 2004, 43:8152–8159. [DOI] [PubMed] [Google Scholar]
- 51. Gulshan K, Thommandru B, Moye‐Rowley WS. Proteolytic degradation of the Yap1 transcription factor is regulated by subcellular localization and the E3 ubiquitin ligase Not4. J Biol Chem 2012, 287:26796–26805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sun HY, Kim N, Hwang CS, Yoo JY. Protein degradation of RNA polymerase II‐association factor 1(PAF1) is controlled by CNOT4 and 26S proteasome. PLoS One 2015, 10:e0125599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lenssen E, James N, Pedruzzi I, Dubouloz F, Cameroni E, Bisig R, Maillet L, Werner M, Roosen J, Petrovic K, et al. The Ccr4‐Not complex independently controls both Msn2‐dependent transcriptional activation‐‐via a newly identified Glc7/Bud14 type I protein phosphatase module‐‐and TFIID promoter distribution. Mol Cell Biol 2005, 25:488–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. James N, Landrieux E, Collart MA. A SAGA‐independent function of SPT3 mediates transcriptional deregulation in a mutant of the Ccr4‐not complex in Saccharomyces cerevisiae . Genetics 2007, 177:123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cui Y, Ramnarain DB, Chiang YC, Ding LH, McMahon JS, Denis CL. Genome wide expression analysis of the CCR4‐NOT complex indicates that it consists of three modules with the NOT module controlling SAGA‐responsive genes. Mol Genet Genomics 2008, 279:323–337. [DOI] [PubMed] [Google Scholar]
- 56. Denis CL, Chiang YC, Cui Y, Chen J. Genetic evidence supports a role for the yeast CCR4‐NOT complex in transcriptional elongation. Genetics 2001, 158:627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gaillard H, Tous C, Botet J, Gonzalez‐Aguilera C, Quintero MJ, Viladevall L, Garcia‐Rubio ML, Rodriguez‐Gil A, Marin A, Arino J, et al. Genome‐wide analysis of factors affecting transcription elongation and DNA repair: a new role for PAF and Ccr4‐not in transcription‐coupled repair. PLoS Genet 2009, 5:e1000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Babbarwal V, Fu J, Reese JC. The Rpb4/7 module of RNA polymerase II is required for carbon catabolite repressor protein 4‐negative on TATA (Ccr4‐Not) complex to promote elongation. J Biol Chem 2014, 289:33125–33130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Villanyi Z, Ribaud V, Kassem S, Panasenko OO, Pahi Z, Gupta I, Steinmetz L, Boros I, Collart MA. The not5 subunit of the ccr4‐not complex connects transcription and translation. PLoS Genet 2014, 10:e1004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wind M, Reines D. Transcription elongation factor SII. Bioessays 2000, 22:327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dutta A, Babbarwal V, Fu J, Brunke‐Reese D, Libert DM, Willis J, Reese JC. Ccr4‐Not and TFIIS function cooperatively to rescue arrested RNA polymerase II. Mol Cell Biol 2015, 35:1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Libri D, Dower K, Boulay J, Thomsen R, Rosbash M, Jensen TH. Interactions between mRNA export commitment, 3′‐end quality control, and nuclear degradation. Mol Cell Biol 2002, 22:8254–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kerr SC, Azzouz N, Fuchs SM, Collart MA, Strahl BD, Corbett AH, Laribee RN. The Ccr4‐Not complex interacts with the mRNA export machinery. PLoS One 2011, 6:e18302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Azzouz N, Panasenko OO, Colau G, Collart MA. The CCR4‐NOT complex physically and functionally interacts with TRAMP and the nuclear exosome. PLoS One 2009, 4:e6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Assenholt J, Mouaikel J, Saguez C, Rougemaille M, Libri D, Jensen TH. Implication of Ccr4‐Not complex function in mRNA quality control in Saccharomyces cerevisiae . RNA 2011, 17:1788–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Decker CJ, Parker R. P‐bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 2012, 4:a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bhaskar V, Roudko V, Basquin J, Sharma K, Urlaub H, Seraphin B, Conti E. Structure and RNA‐binding properties of the Not1‐Not2‐Not5 module of the yeast Ccr4‐Not complex. Nat Struct Mol Biol 2013, 20:1281–1288. [DOI] [PubMed] [Google Scholar]
- 68. Boulon S, Bertrand E, Pradet‐Balade B. HSP90 and the R2TP co‐chaperone complex: building multi‐protein machineries essential for cell growth and gene expression. RNA Biol 2012, 9:148–154. [DOI] [PubMed] [Google Scholar]
- 69. Panasenko OO, Collart MA. Not4 E3 ligase contributes to proteasome assembly and functional integrity in part through Ecm29. Mol Cell Biol 2011, 31:1610–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Panasenko O, Landrieux E, Feuermann M, Finka A, Paquet N, Collart MA. The yeast Ccr4‐Not complex controls ubiquitination of the nascent‐associated polypeptide (NAC‐EGD) complex. J Biol Chem 2006, 281:31389–31398. [DOI] [PubMed] [Google Scholar]
- 71. Halter D, Collart MA, Panasenko OO. The Not4 E3 ligase and CCR4 deadenylase play distinct roles in protein quality control. PLoS One 2014, 9:e86218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bengtson MH, Joazeiro CA. Role of a ribosome‐associated E3 ubiquitin ligase in protein quality control. Nature 2010, 467:470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Preissler S, Reuther J, Koch M, Scior A, Bruderek M, Frickey T, Deuerling E. Not4‐dependent translational repression is important for cellular protein homeostasis in yeast. EMBO J 2015, 34:1905–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cooke A, Prigge A, Wickens M. Translational repression by deadenylases. J Biol Chem 2010, 285:28506–28513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jeske M, Meyer S, Temme C, Freudenreich D, Wahle E. Rapid ATP‐dependent deadenylation of nanos mRNA in a cell‐free system from Drosophila embryos. J Biol Chem 2006, 281:25124–25133. [DOI] [PubMed] [Google Scholar]
- 76. Hook BA, Goldstrohm AC, Seay DJ, Wickens M. Two yeast PUF proteins negatively regulate a single mRNA. J Biol Chem 2007, 282:15430–15438. [DOI] [PubMed] [Google Scholar]
- 77. Waghray S, Williams C, Coon JJ, Wickens M. Xenopus CAF1 requires NOT1‐mediated interaction with 4E‐T to repress translation in vivo. RNA 2015, 21:1335–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Nishimura T, Padamsi Z, Fakim H, Milette S, Dunham WH, Gingras AC, Fabian MR. The eIF4E‐binding protein 4E‐T is a component of the mRNA decay machinery that bridges the 5′ and 3′ termini of target mRNAs. Cell Rep 2015, 11:1425–1436. [DOI] [PubMed] [Google Scholar]
- 79. Ozgur S, Basquin J, Kamenska A, Filipowicz W, Standart N, Conti E. Structure of a human 4E‐T/DDX6/CNOT1 complex reveals the different interplay of DDX6‐binding proteins with the CCR4‐NOT complex. Cell Rep 2015, 13:703–711. [DOI] [PubMed] [Google Scholar]
- 80. Mathys H, Basquin J, Ozgur S, Czarnocki‐Cieciura M, Bonneau F, Aartse A, Dziembowski A, Nowotny M, Conti E, Filipowicz W. Structural and biochemical insights to the role of the CCR4‐NOT complex and DDX6 ATPase in microRNA repression. Mol Cell 2014, 54:751–765. [DOI] [PubMed] [Google Scholar]
- 81. Chen Y, Boland A, Kuzuoglu‐Ozturk D, Bawankar P, Loh B, Chang CT, Weichenrieder O, Izaurralde E. A DDX6‐CNOT1 complex and W‐binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell 2014, 54:737–750. [DOI] [PubMed] [Google Scholar]
- 82. Rouya C, Siddiqui N, Morita M, Duchaine TF, Fabian MR, Sonenberg N. Human DDX6 effects miRNA‐mediated gene silencing via direct binding to CNOT1. RNA 2014, 20:1398–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell 2005, 122:875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sweet T, Kovalak C, Coller J. The DEAD‐box protein Dhh1 promotes decapping by slowing ribosome movement. PLoS Biol 2012, 10:e1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Maillet L, Collart MA. Interaction between Not1p, a component of the Ccr4‐not complex, a global regulator of transcription, and Dhh1p, a putative RNA helicase. J Biol Chem 2002, 277:2835–2842. [DOI] [PubMed] [Google Scholar]
- 86. Ito K, Takahashi A, Morita M, Suzuki T, Yamamoto T. The role of the CNOT1 subunit of the CCR4‐NOT complex in mRNA deadenylation and cell viability. Protein Cell 2011, 2:755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co‐translational mRNA decay in Saccharomyces cerevisiae . Nature 2009, 461:225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Funakoshi Y, Doi Y, Hosoda N, Uchida N, Osawa M, Shimada I, Tsujimoto M, Suzuki T, Katada T, Hoshino S. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev 2007, 21:3135–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Winkler GS. The mammalian anti‐proliferative BTG/Tob protein family. J Cell Physiol 2010, 222:66–72. [DOI] [PubMed] [Google Scholar]
- 90. Loh B, Jonas S, Izaurralde E. The SMG5‐SMG7 heterodimer directly recruits the CCR4‐NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev 2013, 27:2125–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Fabian MR, Frank F, Rouya C, Siddiqui N, Lai WS, Karetnikov A, Blackshear PJ, Nagar B, Sonenberg N. Structural basis for the recruitment of the human CCR4‐NOT deadenylase complex by tristetraprolin. Nat Struct Mol Biol 2013, 20:735–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bhandari D, Raisch T, Weichenrieder O, Jonas S, Izaurralde E. Structural basis for the Nanos‐mediated recruitment of the CCR4‐NOT complex and translational repression. Genes Dev 2014, 28:888–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Doidge R, Mittal S, Aslam A, Winkler GS. The anti‐proliferative activity of BTG/TOB proteins is mediated via the Caf1a (CNOT7) and Caf1b (CNOT8) deadenylase subunits of the Ccr4‐not complex. PLoS One 2012, 7:e51331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Filipowicz W, Sonenberg N. The long unfinished march towards understanding microRNA‐mediated repression. RNA 2015, 21:519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Muhlrad D, Parker R. The yeast EDC1 mRNA undergoes deadenylation‐independent decapping stimulated by Not2p, Not4p, and Not5p. EMBO J 2005, 24:1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Villanyi Z, Collart MA. Ccr4‐Not is at the core of the eukaryotic gene expression circuitry. Biochem Soc Trans 2015, 43:1253–1258. [DOI] [PubMed] [Google Scholar]
- 97. Lotan R, Bar‐On VG, Harel‐Sharvit L, Duek L, Melamed D, Choder M. The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes Dev 2005, 19:3004–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Collart MA, Reese JC. Gene expression as a circular process: cross‐talk between transcription and mRNA degradation in eukaryotes; International University of Andalusia (UNIA) Baeza, Spain. RNA Biol 2014, 11:320–323. [DOI] [PMC free article] [PubMed] [Google Scholar]


