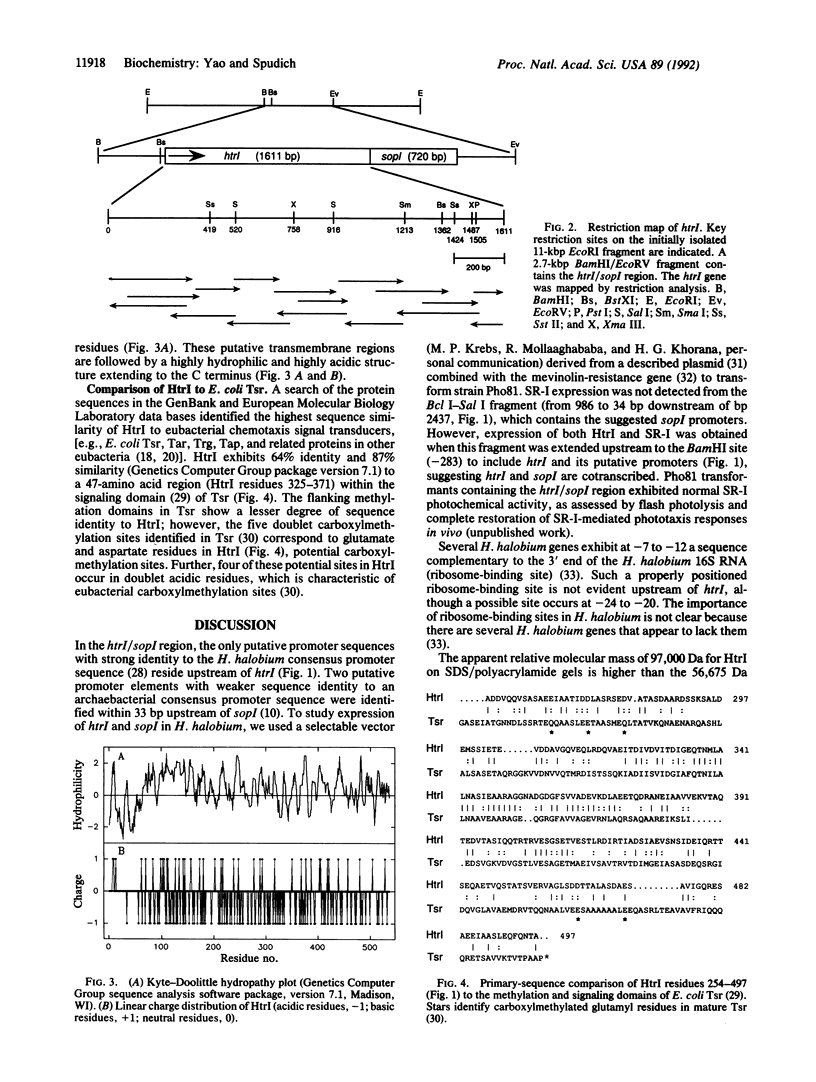
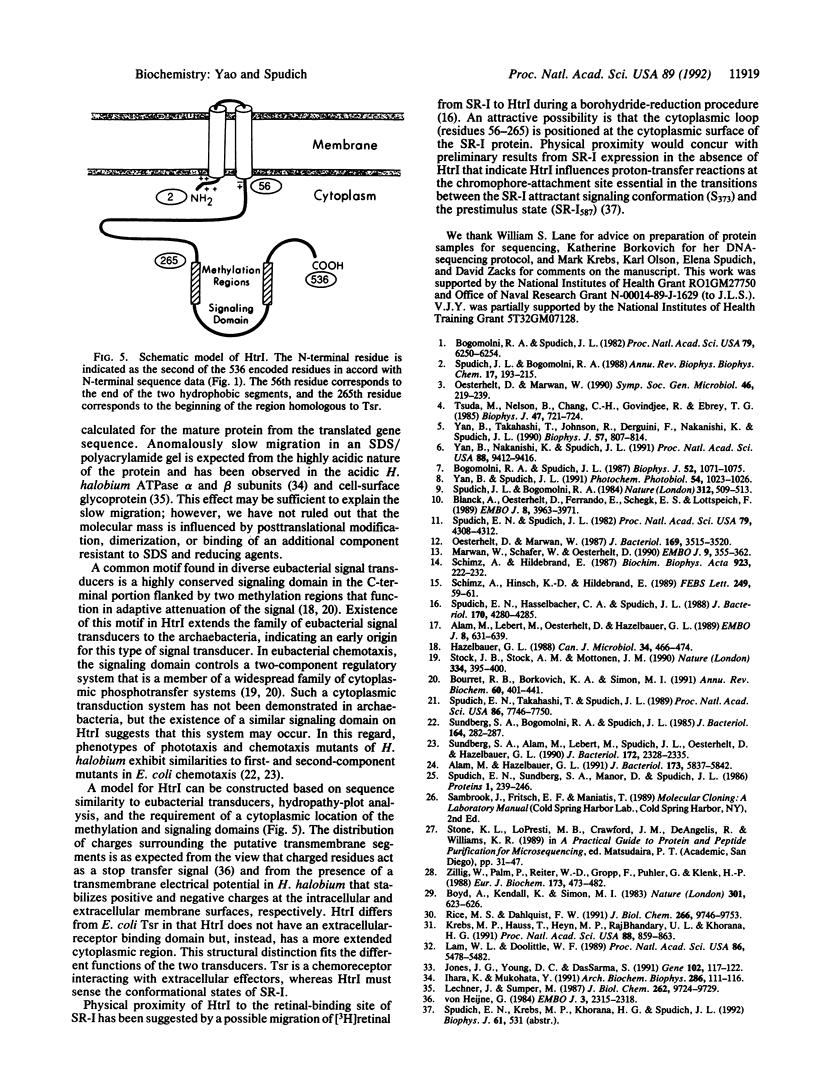
Abstract
A methylated membrane protein of 97 kDa was suggested on the basis of mutant analysis to transduce signals from the phototaxis receptor sensory rhodopsin I to the flagellar motor in Halobacterium halobium. Here we report isolation of the proposed transducer protein, cloning of its gene based on partial protein sequences, the complete gene sequence, and analysis of the encoded primary structure. The 1611-base-pair gene termination codon overlaps the initiator ATG of the sopI gene, which encodes the sensory rhodopsin I apoprotein. The predicted size of 57 kDa for the methylated protein indicates an aberrant electrophoretic migration on SDS/polyacrylamide gels, as occurs with other acidic halophilic proteins. Putative promotor elements are located in an A+T-rich region upstream of the gene. Comparison of the translated nucleotide sequence with N-terminal sequence of the purified protein shows the protein is synthesized without a processed leader peptide and the N-terminal methionine is removed in the mature protein. The deduced protein sequence predicts two transmembrane helices near the N terminal that would anchor the protein to the membrane. Beyond this hydrophobic region of 46 residues, the remainder of the protein (536-amino acid residues total) is hydrophilic. The C-terminal 270 residues contain a region homologous to the signaling domains of eubacterial transducers (e.g., Escherichia coli Tsr protein), flanked by two regions homologous to the methylation domains of the transducer family. The protein differs from E. coli Tsr in that it does not have an extramembranous-receptor binding domain but instead has a more extended cytoplasmic region. Coexpression of the methyl-accepting protein gene (designated htrI) and sopI restores sensory rhodopsin I phototaxis to a mutant (Pho81) that contains a deletion in the htrI/sopI region. These results extend the eubacterial transducer family to the archaebacteria and substantiate the proposal that the methylated membrane protein functions as a signal-transducing relay between sensory rhodopsin I and cytoplasmic sensory-pathway components.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alam M., Hazelbauer G. L. Structural features of methyl-accepting taxis proteins conserved between archaebacteria and eubacteria revealed by antigenic cross-reaction. J Bacteriol. 1991 Sep;173(18):5837–5842. doi: 10.1128/jb.173.18.5837-5842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M., Lebert M., Oesterhelt D., Hazelbauer G. L. Methyl-accepting taxis proteins in Halobacterium halobium. EMBO J. 1989 Feb;8(2):631–639. doi: 10.1002/j.1460-2075.1989.tb03418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanck A., Oesterhelt D., Ferrando E., Schegk E. S., Lottspeich F. Primary structure of sensory rhodopsin I, a prokaryotic photoreceptor. EMBO J. 1989 Dec 20;8(13):3963–3971. doi: 10.1002/j.1460-2075.1989.tb08579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogomolni R. A., Spudich J. L. Identification of a third rhodopsin-like pigment in phototactic Halobacterium halobium. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6250–6254. doi: 10.1073/pnas.79.20.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogomolni R. A., Spudich J. L. The photochemical reactions of bacterial sensory rhodopsin-I. Flash photolysis study in the one microsecond to eight second time window. Biophys J. 1987 Dec;52(6):1071–1075. doi: 10.1016/S0006-3495(87)83301-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret R. B., Borkovich K. A., Simon M. I. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu Rev Biochem. 1991;60:401–441. doi: 10.1146/annurev.bi.60.070191.002153. [DOI] [PubMed] [Google Scholar]
- Boyd A., Kendall K., Simon M. I. Structure of the serine chemoreceptor in Escherichia coli. Nature. 1983 Feb 17;301(5901):623–626. doi: 10.1038/301623a0. [DOI] [PubMed] [Google Scholar]
- Hazelbauer G. L. The bacterial chemosensory system. Can J Microbiol. 1988 Apr;34(4):466–474. doi: 10.1139/m88-080. [DOI] [PubMed] [Google Scholar]
- Ihara K., Mukohata Y. The ATP synthase of Halobacterium salinarium (halobium) is an archaebacterial type as revealed from the amino acid sequences of its two major subunits. Arch Biochem Biophys. 1991 Apr;286(1):111–116. doi: 10.1016/0003-9861(91)90015-b. [DOI] [PubMed] [Google Scholar]
- Jones J. G., Young D. C., DasSarma S. Structure and organization of the gas vesicle gene cluster on the Halobacterium halobium plasmid pNRC100. Gene. 1991 Jun 15;102(1):117–122. doi: 10.1016/0378-1119(91)90549-q. [DOI] [PubMed] [Google Scholar]
- Krebs M. P., Hauss T., Heyn M. P., RajBhandary U. L., Khorana H. G. Expression of the bacterioopsin gene in Halobacterium halobium using a multicopy plasmid. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):859–863. doi: 10.1073/pnas.88.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam W. L., Doolittle W. F. Shuttle vectors for the archaebacterium Halobacterium volcanii. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5478–5482. doi: 10.1073/pnas.86.14.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J., Sumper M. The primary structure of a procaryotic glycoprotein. Cloning and sequencing of the cell surface glycoprotein gene of halobacteria. J Biol Chem. 1987 Jul 15;262(20):9724–9729. [PubMed] [Google Scholar]
- Marwan W., Schäfer W., Oesterhelt D. Signal transduction in Halobacterium depends on fumarate. EMBO J. 1990 Feb;9(2):355–362. doi: 10.1002/j.1460-2075.1990.tb08118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterhelt D., Marwan W. Change of membrane potential is not a component of the photophobic transduction chain in Halobacterium halobium. J Bacteriol. 1987 Aug;169(8):3515–3520. doi: 10.1128/jb.169.8.3515-3520.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice M. S., Dahlquist F. W. Sites of deamidation and methylation in Tsr, a bacterial chemotaxis sensory transducer. J Biol Chem. 1991 May 25;266(15):9746–9753. [PubMed] [Google Scholar]
- Spudich E. N., Hasselbacher C. A., Spudich J. L. Methyl-accepting protein associated with bacterial sensory rhodopsin I. J Bacteriol. 1988 Sep;170(9):4280–4285. doi: 10.1128/jb.170.9.4280-4285.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich E. N., Spudich J. L. Control of transmembrane ion fluxes to select halorhodopsin-deficient and other energy-transduction mutants of Halobacterium halobium. Proc Natl Acad Sci U S A. 1982 Jul;79(14):4308–4312. doi: 10.1073/pnas.79.14.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich E. N., Sundberg S. A., Manor D., Spudich J. L. Properties of a second sensory receptor protein in Halobacterium halobium phototaxis. Proteins. 1986 Nov;1(3):239–246. doi: 10.1002/prot.340010306. [DOI] [PubMed] [Google Scholar]
- Spudich E. N., Takahashi T., Spudich J. L. Sensory rhodopsins I and II modulate a methylation/demethylation system in Halobacterium halobium phototaxis. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7746–7750. doi: 10.1073/pnas.86.20.7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. L., Bogomolni R. A. Mechanism of colour discrimination by a bacterial sensory rhodopsin. Nature. 1984 Dec 6;312(5994):509–513. doi: 10.1038/312509a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. L., Bogomolni R. A. Sensory rhodopsins of halobacteria. Annu Rev Biophys Biophys Chem. 1988;17:193–215. doi: 10.1146/annurev.bb.17.060188.001205. [DOI] [PubMed] [Google Scholar]
- Stock J. B., Stock A. M., Mottonen J. M. Signal transduction in bacteria. Nature. 1990 Mar 29;344(6265):395–400. doi: 10.1038/344395a0. [DOI] [PubMed] [Google Scholar]
- Sundberg S. A., Alam M., Lebert M., Spudich J. L., Oesterhelt D., Hazelbauer G. L. Characterization of Halobacterium halobium mutants defective in taxis. J Bacteriol. 1990 May;172(5):2328–2335. doi: 10.1128/jb.172.5.2328-2335.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg S. A., Bogomolni R. A., Spudich J. L. Selection and properties of phototaxis-deficient mutants of Halobacterium halobium. J Bacteriol. 1985 Oct;164(1):282–287. doi: 10.1128/jb.164.1.282-287.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M., Nelson B., Chang C. H., Govindjee R., Ebrey T. G. Characterization of the chromophore of the third rhodopsin-like pigment of Halobacterium halobium and its photoproduct. Biophys J. 1985 May;47(5):721–724. doi: 10.1016/S0006-3495(85)83969-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B., Nakanishi K., Spudich J. L. Mechanism of activation of sensory rhodopsin I: evidence for a steric trigger. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9412–9416. doi: 10.1073/pnas.88.21.9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B., Spudich J. L. Evidence that the repellent receptor form of sensory rhodopsin I is an attractant signaling state. Photochem Photobiol. 1991 Dec;54(6):1023–1026. doi: 10.1111/j.1751-1097.1991.tb02125.x. [DOI] [PubMed] [Google Scholar]
- Yan B., Takahashi T., Johnson R., Derguini F., Nakanishi K., Spudich J. L. All-trans/13-cis isomerization of retinal is required for phototaxis signaling by sensory rhodopsins in Halobacterium halobium. Biophys J. 1990 Apr;57(4):807–814. doi: 10.1016/S0006-3495(90)82600-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillig W., Palm P., Reiter W. D., Gropp F., Pühler G., Klenk H. P. Comparative evaluation of gene expression in archaebacteria. Eur J Biochem. 1988 May 2;173(3):473–482. doi: 10.1111/j.1432-1033.1988.tb14023.x. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Analysis of the distribution of charged residues in the N-terminal region of signal sequences: implications for protein export in prokaryotic and eukaryotic cells. EMBO J. 1984 Oct;3(10):2315–2318. doi: 10.1002/j.1460-2075.1984.tb02132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]



