Abstract
Among the various subtypes of Acute Myeloid Leukemia (AML), those with chromosomal rearrangements of the MLL oncogene (AML-MLL) have a poor prognosis. AML-MLL tumor cells are resistant to current genotoxic therapies due to an attenuated response by p53, which induces cell cycle arrest and apoptosis in response to DNA damage. In addition to chemicals that damage DNA, efforts have focused on targeting DNA repair enzymes as a general chemotherapeutic approach to cancer treatment. Here, we found that inhibition of the kinase ATR, which is the primary sensor of DNA replication stress, induced chromosomal breakage and death of mouse AMLMLL cells (with an MLL-ENL fusion and a constitutively active N-RAS) independently of p53. Moreover, ATR inhibition as a single agent exhibited antitumoral activity, both reducing tumor burden after establishment and preventing tumors from growing, in an immunocompetent allograft mouse model of AMLMLL and in xenografts of a human AML-MLL cell line. We also found that inhibition of ATM, a kinase that senses DNA double-strand breaks, also promoted the survival of the AMLMLL mice. Collectively, these data indicated that ATR and ATM inhibition represent potential alternative therapeutic strategies for the treatment of AML, especially MLL-driven leukemias.
Introduction
One recurrent finding in cancer is the presence of DNA replication stress, which if persistent leads to DNA double-strand breaks (DSBs) that initiate genomic rearrangements in cancer cells (1, 2). In addition to oncogenes, many of the agents used in genotoxic chemotherapy, including antifolates, nucleotide analogs, topoisomerase inhibitors, or platinum derivatives, are potent inducers of DNA replication stress. In mammalian cells, the DNA replication stress response (RSR) is a signaling cascade that initiates with the activation of the kinase ATR and activates the kinase checkpoint kinase 1 (CHK1, encoded by CHEK1) (3, 4). The presence of DNA replication stress in cancer cells renders them particularly dependent on a proficient ATR-response for survival (5). Accordingly, low levels of ATR are particularly toxic for cells carrying oncogenic mutations (6, 7). Additionally, ATR or CHK1 inhibitors have exhibited efficacy in hematopoietic tumors, mostly in in vitro settings (6–11).
Although persistent DNA replication stress or DNA damage activate the tumor suppressor p53, encoded by TP53, which triggers cell cycle arrest and apoptosis (12), cell death caused by low ATR activity is not only p53-independent but also enhanced by p53-deficiency (13, 14). Likewise, the toxicity of chemical inhibitors of ATR is higher in cells lacking p53 (10, 15, 16). This p53-independent cell killing by ATR inhibitors is linked to their capacity to induce the accumulation of DNA replication stress and premature mitotic entry, activities that are unrelated to p53 functions (17, 18). Hence, ATR inhibitors offer an alternative for the elimination of p53-deficient tumors.
Previous studies have established that an intact p53 network is a critical determinant of to the effectiveness of chemotherapy in AML (19). MLL (mixed lineage leukemia) refers to chromosomal translocation products involving the gene KMT2A, which encodes histone-lysine N-methyltransferase 2 (20). In contrast to other oncogenic fusion proteins, acute leukemia cells with MLL fusion proteins (AMLMLL) do not mount an effective p53 response and are therefore resistant to current genotoxic treatments (19). Consistent with this, MLL rearrangements and mutations in TP53 rarely occur together in human AML (21, 22). Thus, alternative therapies are needed to overcome chemotherapy resistance associated with p53 dysfunction in AMLMLL. In addition to the need of a therapy that works on p53-deficient tumors, several lines of evidence suggested that targeting ATR could be particularly beneficial in AMLMLL. First, reduced amounts of ATR in mouse models inhibited growth of AML driven by the MLL-ENL oncogene, which encodes a fusion of KMT2A and the transcription activator ENL (7). Second, inhibitors of ATR or its target CHK1 are toxic for several lymphomas and leukemias, including p53-deficient tumors (6–9, 11, 23). Moreover, the particular efficacy of RSR inhibitors in lymphoid tumors is consistent with a preferential role for the RSR in the untransformed lymphoid compartment, exemplified by the frequent presence of anemia in mice suffering from DNA replication stress (13, 24–27). Finally, inhibition of ATR or inhibition of the related DNA damage response (DDR) kinase ATM predisposed primary stem cells infected with retroviruses expressing MLL-AF9, a fusion between the KMT2A and the transcription activator AF9, towards differentiation in vitro (28). On the basis of these data, we predicted that inhibition of ATR or ATM could have potential as a therapy for MLL-associated leukemia. Here, we report the antitumoral effects of ATR or ATM inhibitor monotherapy in an immunocompetent mouse model of AMLMLL.
Results
High levels of CHEK1 expression in lymphomas and leukemias
Since ATR inhibitors are preferentially toxic for cells experiencing DNA replication stress, tumors with high endogenous levels of this stress could be promising targets. We previously showed that increased CHK1 levels reduce DNA replication stress and improve the survival of cells expressing oncogenes or reprogramming factors (29, 30). On this basis, we predicted that high CHK1 expression could be a signature of tumors with high levels of DNA replication stress, and therefore sensitive to ATR inhibitors (29). This hypothesis recently obtained strong experimental confirmation, in work that identified an enrichment of CHEK1 gene amplifications in genomically unstable ovarian cancers, which correlated with an enhanced sensitivity to ATR inhibitors (31). Hence, CHK1 levels provide a biomarker for ATRi sensitivity.
To compare the relative levels of CHEK1 gene expression across different cancer types we analyzed data from the human Cancer Cell Line Encyclopedia (32). CHEK1 mRNA was most abundant in Burkitt lymphomas, which we previously showed are highly dependent on ATR and CHK1 for their survival (Fig. S1) (6). In addition to Burkitt, CHEK1 expression was distinctively high in various lymphomas and leukemias (Fig. S1). Moreover, recent reports have shown good efficacy of ATR or CHK1 inhibitors in hematopoietic tumors, mostly in in vitro settings (6–11). The particular efficacy of RSR inhibitors in lymphoid tumors is consistent with a preferential role for the RSR in the untransformed lymphoid compartment, exemplified by the frequent presence of anemia in mice suffering from RS (13, 24–27). Overall, these analyses identify distinctively high levels of CHEK1 expression in lymphomas and leukemias, which could be indicative of underlying DNA replication stress. Consistent with our findings, in the companion paper in this issue, David et al. (XX) report that CHEK1 expression and CHK1 abundance negatively correlate with prognosis in patients of acute myeloid leukemia.
AMLMLL cells are highly sensitive to ATR inhibitors in vitro
From the different kinds of hematopoietic malignancies, we decided to focus on AML carrying MLL translocations for the following reasons. First, mouse genetic studies indicated that ATR abundance is particularly important for the viability of AML cells (7). Second, ATR-dependent phosphorylation of MLL suppresses DNA replication stress (33). Third, in vitro experiments showed that ATR inhibitors promote the differentiation MLL-AF9-infected primary stem cells towards differentiation in vitro (28). Finally, due to the p53-independent mechanism of cell killing by ATR inhibitors (18), they could potentially overcome the limitation impinged by p53 deficient responses in MLL-driven AML.
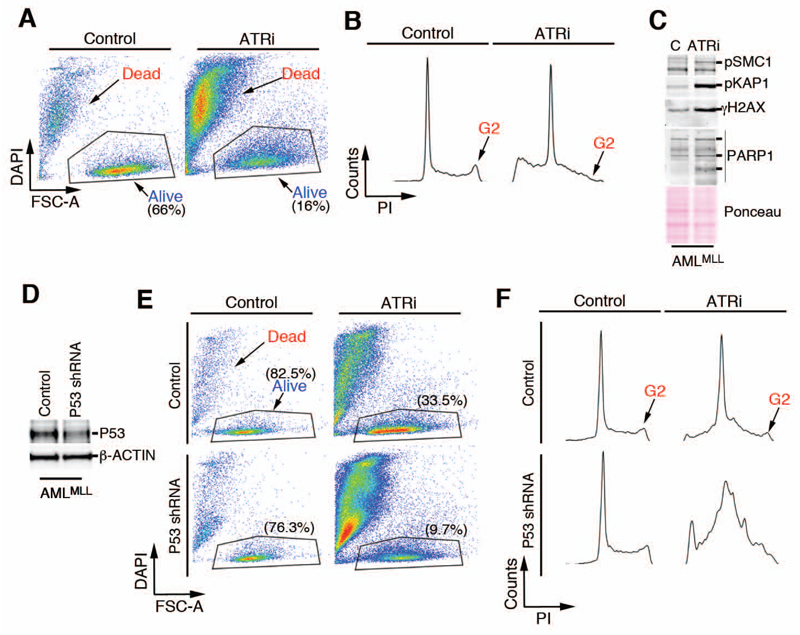
To test the efficacy of ATR inhibition in the treatment of AML in vivo, we used a previously described mouse cell line generated by transforming bone marrow cells with viruses expressing MLL-ENL and NrasG12D (AMLMLL). These cells have an activating mutation in N-Ras, which is also common in human AMLMLL patients (19), and these cells recapitulate the deficient p53 signaling and poor responses to conventional chemotherapy that are observed in the clinic with AMLMLL patients (19). A 1-day treatment of AMLMLL cells with the ATR inhibitor AZ20 (34) (hereafter referred to as ATRi) eliminated 75% of the viable cells in culture (Fig. 1A). The cytotoxicity of ATR inhibitors is mediated by forcing premature mitotic entry from G2, the phase during which DSB are generated (18). Consistently, exposure to ATRi led to the disappearance of most AMLMLL cells from the G2 phase of the cell cycle (Fig. 1B). ATRi also resulted in the accumulation of DSBs, as indicated by the phosphorylation of the DDR targets KRAB-associated protein-1 (KAP1), structural maintenance of chromosomes 1 (SMC1), and histone H2AX (Fig. 1C). Moreover, depletion of p53 with a retrovirus expressing a p53-targeting shRNA did not rescue the cells from the toxic effects of ATRi (Fig. 1D-F).
Fig. 1. p53-independent toxicity of ATRi in AMLMLL cells in culture.
(A) FACS analysis showing the percentage of viable AMLMLL cells (identified by size and DAPI exclusion) either untreated or exposed to ATRi (10μM, 24 hrs). Data are representative of 2 independent experiments. (B) FACS analysis of DNA content (PI) from the cultures used in (A) illustrating the depletion of G2 cells observed in response to ATRi. Data are representative of 2 independent experiments (C) Western blot of SMC1, KAP1, and γH2AX phosphorylation and PARP1 cleavage products in AMLMLL cells exposed to ATRi (5 μM, 6 hrs). Data are representative of 2 independent experiments. (D) WB confirming the depletion of p53 in AMLMLL cells after infection with retroviruses expressing a p53-targeting shRNA. β-ACTIN abundance is shown as a loading control. (B) FACS analysis showing the percentage of viable AMLMLL cells (identified by size and DAPI exclusion) from control or p53 shRNA-infected AMLMLL cells either untreated or exposed to ATRi (10 μM, 24 hrs). Data are representative of 2 independent experiments (C) FACS analysis of DNA content (PI) from the cultures showed in (E) illustrating the depletion of G2 cells observed in response to ATRi. Data are representative of 2 independent experiments
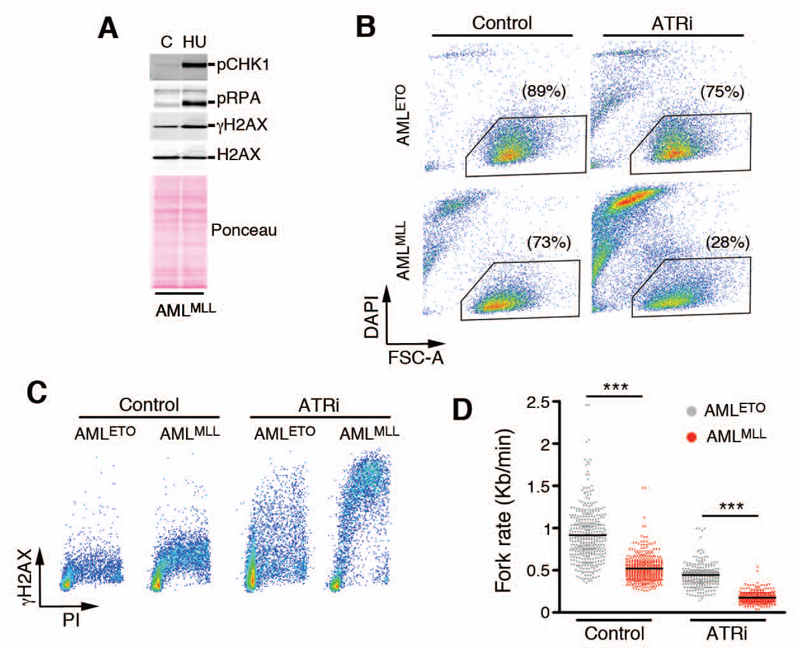
To determine the mechanism for the sensitivity of AMLMLL cells to ATRi, we analyzed the RSR in AMLMLL cells. Exposure to the ribonucleotide reductase inhibitor hydroxyurea (HU) stimulated the phosphorylation of the ATR targets CHK1 and replication protein A (RPA), ruling out a deficiency in activation and signaling through the DNA replication stress pathway in these cells (Fig. 2A). We also compared the response to ATRi in AMLMLL cells in an equivalent cell line that was generated by transforming bone marrow cells with viruses expressing an AML1-ETO translocation (AMLETO). This models a p53-proficient type of AML that has better prognosis than MLL-ENL AML patients (19). ATRi was significantly more toxic for AMLMLL than for AMLETO cells (Fig. 2B). The enhanced sensitivity of AMLMLL cells correlated with higher levels of DNA damage in replicating cells, as measured by monitoring γH2AX phosphorylation of cells labeled with propidium iodide by fluorescence activated cell sorting (FACS) (Fig. 2C). ATRi induced a greater γH2AX signal, indicating a greater amount of damage, in AMLMLL cells. Importantly, a structurally different ATR inhibitor, ETP-46464 (16), induced similar toxicity and accumulation of DNA damage (fig. S1). Furthermore, both ATR inhibitors induced cleavage of PARP1, an indicator of apoptosis (35), which confirms the toxicity of these inhibitors under conditions that induced DNA damage (fig. S1). Consistent with FACS data, AMLMLL and AMLETO cells exhibited differences in the rate of DNA replication fork progression when analyzed by stretched DNA fiber analyses. While replication fork progression in AMLMLL cells was slower under basal conditions than that in AMLETO cells (Fig. 2D), exposure to ATRi had a bigger effect in reducing replication fork rates in AMLMLL cells, leading to an almost complete impairment of replication fork progression in these cells (Fig. 2D). In summary, MLL-driven AML cells exhibited an intrinsically higher sensitivity to ATR inhibitors than do AMLETO cells; with ATRi inducing the accumulation of replicative DNA damage, activation of the DDR, and p53-independent death in these cells.
Fig. 2. Increased levels of ATRi-induced DNA replication stress in AMLMLL cells.
(A) WB of CHK1, RPA, and gH2AX phosphorylation in AMLMLL cells exposed to HU (2 mM, 2 hrs). Data are representative of 2 independent experiments (B) FACS analysis showing the percentage of viable AMLMLL and AMLETO cells (identified by size and DAPI exclusion) either untreated or exposed to ATRi (3 μM, 16 hrs). Data are representative of 2 independent experiments (C) FACS analysis of DNA content (PI) and H2AX phosphorylation in AMLMLL and AMLETO cells exposed to ATRi (10 μM, 5 hrs), illustrating the increased levels of ATRi-induced DNA replication stress (as measured by γH2AX in cells with an S-phase DNA content) in AMLMLL. Data are representative of 2 independent experiments. (D) Fork rates were measured in stretched DNA fibers prepared from AMLMLL and AMLETO cells exposed (or not) to ATRi (10 μM, 5 hrs). At least 200 tracks were measured per condition. ***P<0.001 by two-tailed t test.
ATR inhibitors show efficacy as single agents in a mouse model of MLL-driven AML
To examine the in vivo efficacy of ATRi, we injected AMLMLL tumor cells, which also expressed both green fluorescence protein (GFP) and luciferase for tracking, into immunocompetent mice Even in immunocompetent recipients, intravenous injection of AMLMLL cells results in a very aggressive form of AML that infiltrates multiple organs and kills mice in a few weeks (19). In contrast to previous studies (19, 36), we did not irradiate recipient animals before the transplant. Irradiation depletes the bone marrow of the recipient mice, facilitating the expansion of the transplanted tumor. Because the tumor cells proliferated even in the absence of irradiation, we preferred to avoid this treatment to further mimic the normal context of AML. Transplanted mice were treated daily through an oral gavage with a dose of 60 mg/kg of ATRi, and tumor development was followed by monitoring luciferase activity with an in vivo imaging system (IVIS). Finally, to test the efficacy of ATR inhibitors we used two protocols. In the prevention protocol, mice started receiving treatment on the day on the injection of AMLMLL cells (ATRiPr); in the therapy protocol, mice started receiving treatment after tumors were detectable by IVIS (ATRiTh).
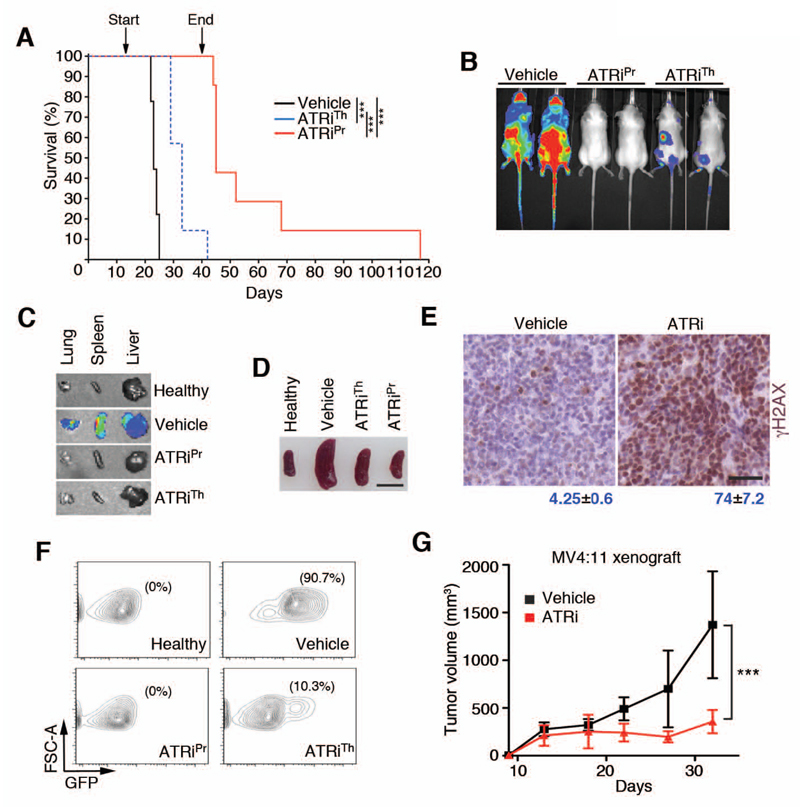
Without drug treatment, AMLMLL cells rapidly expanded leading to a lethal disease in control animals with a median survival of 23 days (Fig. 3A). The treatment of the ATRiTh group started at day 13. Both therapy and prevention groups showed a marked response as measured by IVIS at day 18 (Fig. 3B). IVIS on isolated organs confirmed that the treatment with ATRi severely limited tumor infiltration to organs including liver, spleen, and lung (Fig. 3C). The decreased tumor burden was also evident from visual analysis of spleen sizes (Fig. 3D). Moreover, and in agreement with the observations made in vitro by exposure of cells to ATR inhibitors (Fig. 2C), treatment with ATRi led to a widespread accumulation of γH2AX-positive cells in the spleens of the treated mice (Fig. 3E). On day 23, we isolated bone marrow from mice of all groups and measured the presence of tumor cells by detection of GFP (Fig. 3F). GFP-positive cells were undetectable in the prevention group, and the therapy group exhibited a 17-fold decrease in the percentage of GFP-positive AMLMLL cells in the bone marrow. Consistent with this, videos of animals recorded on day 25 showed a clear improvement of the overall health in both ATRi-treated groups (movies S1-3). Whereas all animals eventually succumbed to leukemia, both groups showed a significant increase in the median lifespan (vehicle: 23 days; ATRiTh: 33 days, ATRiPR: 45 days) (Fig. 3A). Strikingly, at 40 days, all animals from the prevention group were alive, a time at which we decided to stop the treatment to explore potential curative effects of the therapy. 40% of these mice survived for more than 50 days, and one was alive up until 117 days before succumbing to the disease.
Fig. 3. In vivo responses of AMLMLL to ATRi.
(A) Kaplan-Meyer curves of AMLMLL transplanted mice that were either treated with vehicle (n=9), ATRi from day 1 (ATRiPr; n=7) or S day 13 (ATRiTh; n=7). Treament on the prevention group stopped at day 40. The p value was calculated with the Mantel-Cox log rank test. ***; p<0.001. (B) Representative IVIS of the luciferase signals observed on mice from the groups indicated on (A) on day 18. (C) Representative examples of the luciferase signal observed by IVIS on isolated organs from the indicated groups at day 23. (D) Picture of the spleen sizes observed at day 23 of the in vivo treatment experiment. Scale bar (black) represents 1 cm. (E) Representative images of γH2AX immunohistochemistry on spleens of AMLMLL transplanted mice treated with vehicle or ATRi (60 mg/kg, 11 days). Scale bar (black) represents 50 μm. Numbers indicate the percentage of γH2AX positive cells in each case (mean ± s.d.). (F) FACS analysis from the bone marrow collected from mice at day 23 of the treatment experiment indicated in (A). GFP (x-axis) is used to monitor the presence of AMLMLL cells. y-axis indicates forward scattered light (FSC-A). The percentage of live GFP+ cells detected in each case is indicated. Data are representative of 2 independent groups. (G) Effect of ATRi as monotherapy on the growth of xenografts from the human MV4:11 cell line of MLL-driven AML. Treatment started when tumors became palpable and 8 animals were used per group. ***; p<0.001 by two-way ANOVA.
Finally, we measured the efficacy of ATRi in xenografts of the human AML cell line MV4:11, which is also driven by an MLL-AF4 translocation (37). We subcutaneously implanted MV4:11 cells into the flanks of immunodeficient SCID mice. Treatment with ATRi started when tumors became palpable and was administered daily through oral gavage at 60 mg/kg. ATRi therapy significantly limited the growth of MV4:11 xenografts (Fig. 3G). In summary, our data showed that ATR inhibition elicits antitumoral responses when used as a single agent in allografts of mouse cell AMLMLL cells and in xenografts of a human AML cell line and provide an example of antitumor activity of this class of drugs in an immunocompetent model of cancer.
ATM inhibitors extend the survival of leukemia-bearing mice
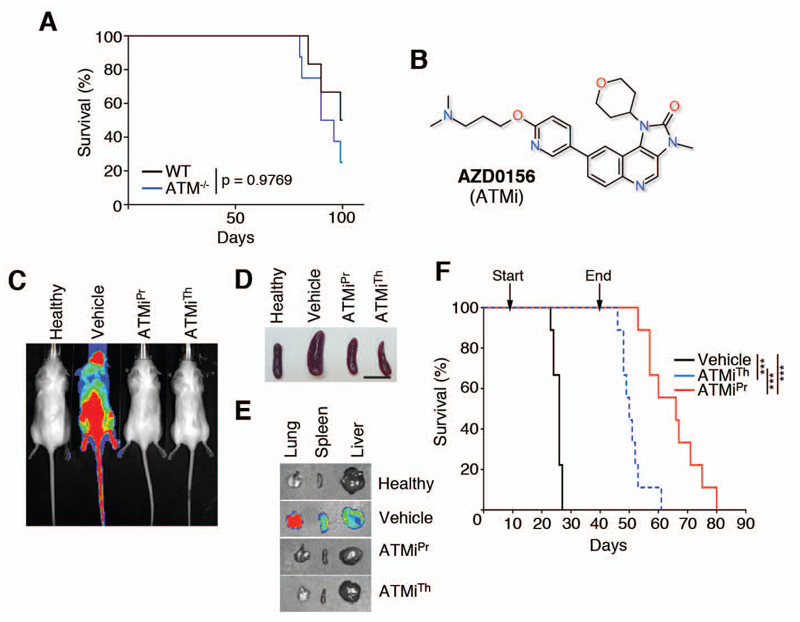
Persistent DNA replication stress leads to the breakage of replication forks and thus to DSB that trigger an ATM-dependent DDR, suggesting an active role of ATM in limiting the toxicity of replication stress. Consistently, ATM deficiency is lethal in ATR-Seckel mice that accumulate high levels of DNA replication stress (13). Moreover, previous in vitro data revealed that treatment of primary MLL-AF9 transformed cells with ATMi or transformation of ATM knockout cells with MLL-AF9 results in poor growth and increased differentiation of these leukemic cells (28). To determine whether ATM is required for MLL-leukemia in vivo, we first generated wild-type and ATM-/- AMLMLL tumors by transforming bone-marrow hematopoietic progenitor cells with retroviruses expressing MLL-AF9-IRES-neo and NrasG12D-IRES-GFP. When injected into immunodeficient NRG (Rag1null; IL2rgnull) recipient mice, both ATM wildtype and ATM-/- AMLMLL cells caused lethal leukemia with no difference in median survival (Fig. 4A). Thus, even though loss of ATM activity inhibits growth in vitro (21), we found that it did not have a detectable impact on the development of MLL-AF9 leukemia in vivo in primary transplants.
Fig. 4. In vivo responses of AMLMLL to ATMi.
(A) Kaplan-Meyer curves of mice that were transplanted with ATM wild-type (n=6) and ATM-/- (n=8) bone marrow hematopoietic progenitors that had been infected with retroviruses expressing MLL-AF9-IRES-neo and NrasG12D-IRES-GFP. The p value was calculated with the Mantel-Cox log rank test. (B) Chemical structure of the ATM inhibitor AZD0156 (ATMi). (C) Representative IVIS of the luciferase signals observed on AMLMLL transplanted mice that were either treated with vehicle, ATMi from day 1 (ATMiPr), or day 8 (ATMiTh). IVIS imaging was conducted on day 22 of the experiment. (D) Picture of the spleen sizes observed at day 22 of the in vivo treatment experiment explained in (C). Scale bar (black) represents 1 cm. (E) Representative examples of the luciferase signal observed by IVIS on isolated organs from the indicated groups at day 22. (F) Kaplan-Meyer curves of AMLMLL transplanted mice that were either treated with vehicle (n=9), ATMi from day 1 (ATMiPr; n=9), or day 8 (ATMiTh; n=9). Treament on both groups stopped at day 40. The p value was calculated with the Mantel-Cox log rank test. ***; p<0.001.
Despite the absence of an impact of ATM deletion, lack of the kinase activity of ATM is not the same as loss of ATM. For example, ATM kinase-inactivating mutations lead to embryonic lethality in mice, whereas ATM knockout mice are viable and show less genome instability than kinase-dead mutants (38, 39). To determine the effects of ATM inhibition on MLL-leukemia, we used the newly developed ATM inhibitor AZD0156 (ATMi hereafter) (40) (Fig. 4B).
To determine the efficacy of the ATMi, we repeated the same prevention and therapy protocols that we used to evaluate the ATR inhibitors. We injected AMLMLL cells through the tail vein into the mice and treated the mice daily with 20 mg/kg of the ATMi or with the vehicle. In the prevention protocol, treatment started on the day of injection ( ATMiPr); in the therapy protocol, treatment started when tumors were first detectable by IVIS (ATMiTh). The untreated cohort behaved similarly to the previously shown experiments, with a median survival of 26 days. As with ATRi, the therapy with ATMi had a notable effect with either protocol, leading to reduced overall luciferase signal measured by IVIS (Fig. 4C), smaller spleen size (Fig. 4D), reduced organ infiltration (Fig. 4E), and prolonged survival of AMLMLL-injected mice (vehicle: 23 days; ATMiTh: 50 days, ATMiPR: 66 days) (Fig. 4F). Videos of mice recorded on day 23 confirmed the improvement in overall health in ATMi-treated mice (movies S4-6).
Discussion
Current treatment of acute pediatric leukemias involves the use of broad-spectrum genotoxic approaches. However, these lines of chemotherapy are largely ineffective for treatment of leukemias that have MLL translocations. In addition, some of these therapies for AML, such as the topoisomerase II inhibitor etoposide, counterproductively promote MLL translocations and therapy-related leukemia (41, 42). Thus, treatment of this subset of leukemias would benefit from a targeted therapy that exploits a specific vulnerability in the cancer cells.
One such vulnerability is associated with the function of KMT2A, which is the protein encoded in a gene associated with MLL translocation events. KMT2A is a lysine methyltransferase that functions as an epigenetic regulator (20). Leukemias carrying MLL fusion proteins require few if any additional mutations. Rather, fusion proteins induce leukemia by deregulating transcription at MLL-fusion target genes, such as the HOXA gene cluster and MEIS1 (43, 44). Abnormal expression of these genes is associated with epigenetic changes, including alteration in DNA and histone methylation. For example, the histone 3 Lys79 (H3K79) methylase DOT1L is recruited to MLL fusion target genes, and this subtype of leukemia is dependent on DOT1L enzymatic activity (45). Similarly, MLL leukemias depend on several hematopoietic transcription factors, such as the bromodomain and extraterminal (BET) protein BRD4, to maintain their leukemic stem cell properties (31, 36, 46). Thus, one therapeutic approach for leukemogenesis resulting from MLL translocation events is to disrupt the MLL target gene expression program with drugs that target epigenetic-modifying enzymes or the products of genes that depend on such modifications (43). In this context, BET inhibition with the drug JQ1, as well as DOT1L inhibition, are currently under clinical investigation.
In addition to lineage-specific transcriptional circuits, our experiments suggested that a second point of vulnerability in MLL-driven AML is the DNA replication stress response. Whereas we found that the amount of DNA replication stress markers (γH2AX) is not distinctively high in these tumors, we propose that this is because these cells have increased levels of RSR factors, such as CHK1, which help to buffer the levels of replication stress. Accordingly, the accompanying manuscript from David et al (XX) reveals increased CHK1 levels in cells from human AMLMLL patients. We propose that while this increase in RSR factors limits the basal toxicity of DNA replication stress and thus facilitates tumor growth, it also becomes a vulnerability since tumor cells become particularly dependent on a proficient RSR. To which extent MLL translocations are responsible for the DNA replication stress in AMLMLL cells, which have other defects such as N-RAS hyperactivity, remains to be established. Despite the hope for new targeted therapies, mechanisms for resistance to inhibitors of BET (31, 47, 48) or ATR (18) have been recently uncovered. Interestingly, perturbing the chromatin-related functions of MLL proteins has been shown to lead to DNA replication stress (33, 49). In this context, a combination of RSR inhibitors, leading to p53-independent toxicity, and epigenetic inhibitors that may both interfere with the transcriptional properties of the MLL fusion protein and further increase the levels of replication stress, could help overcome the resistance of MLL-driven AML to chemotherapy.
Materials and Methods
Cells and reagents
AML cells carrying the MLL-ENL translocation (plus IRES-GFP) and oncogenic N-RAS (Luciferase-IRES-N-RASG12D), referred in the text as AMLMLL, were developed as previously described (19). The retroviral plasmid with a p53-targeting shRNA was provided by Mariano Barbacid and transduced using standard protocols. MV4:11 cells were obtained from ATCC. Cells were cultured in standard conditions (5% CO2 and 20% O2) in RPMI-1640 (Euro Clone) medium supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich) and 10 μg/mL peniciline/streptomycin (Pen/Strep; Life Technologies). ATRi (AZ20, synthesized by GVK-BIO), ATMi (AZD0156, Astra Zeneca) and HU (Sigma-Aldrich) were used as indicated.
Flow cytometry
To measure viability, cells were collected, washed once with PBS (pH 7.4), stained in a DAPI solution (0.2 μg/mL DAPI in PBS) and analysed by flow cytometry in a FACS Canto II (Becton-Dickinson) machine. For cell cycle profiles, cells were collected, washed with PBS, and fixed in suspension in ice-cold 70% (v/v) ethanol in PBS. After washing in PBS, cells were stained in PBS containing propidium iodide (10 μg/ml) and RNase A (0.5 mg/ml) and collected in a Becton-Dickinson FACS Calibur machine. For DNA content and γH2AX analysis, p-Ser139 H2AX (Millipore) antibodies were used as previously described (18). Data was analyzed by using FACS Diva (BD Biosciences) and FlowJo (Treestar) softwares.
Protein extracts and Western blotting
For total protein extract preparation, AMLMLL cells were collected upon treatment with ATRi (5 μM for 6 h) or HU (2 mM for 2 h), washed once with PBS and lysed in UREA buffer (8 M urea, 1 % Chaps, Tris-HCl 50 mM, pH 8.0) for 30 min at 4°C with agitation. Samples were resolved by SDS-PAGE and analysed by standard western blotting techniques. The following primary antibodies were used: p-Ser345 Chk1 (Cell Signaling), p-Ser4/Ser8 RPA32 (Bethyl), p-Ser139 H2AX (Millipore), H2AX (Abcam), p-SMC1 (Monoclonal Antibody Unit, CNIO), p-Ser824 KAP-1 (Bethyl) and PARP1 (Cell Signaling). Alexa Fluor 680- or 800- conjugated secondary antibodies (Life Technologies) were used for detection with a LI-COR Odyssey infrared imaging system (LI-COR Biosciences).
Transplantation and in vivo treatment studies
For tumour induction and treatment studies, 105 AMLMLL cells were transplanted by tail vein injection into 8-12 weeks old immunocompetent albino recipient mice (C57BL/6/BrdCrHsd-Tyrc). In the case of xenografts, 1.4x105 MV4:11 cells were injected subcutaneously into the flanks of SCID mice and tumor growth was measured with a caliper. Mice were treated daily with ATRi (50 mg/kg; dissolved in 10% N-Methyl-2-Pyrrolidone/90% Polyethylen Glycol 300) or ATMi (20 mg/kg; dissolved in 10% DMSO/90% Captisol (30%)) and the corresponding vehicles by oral gavage. Health status of treated mice was monitored daily. Mice were maintained at the Spanish National Cancer Research Centre (CNIO) under standard housing conditions with free access to chow diet and water, in recommendation of the Federation of European Laboratory Animal Science Association (FELASA). All mouse work was performed in accordance with the Guidelines for Humane Endpoints for Animals Used in Biomedical Research, and under the supervision of the Ethics Committee for Animal Research of the Instituto de Salud Carlos III.
Monitoring of leukemias
To monitor tumour formation, AMLMLL-transplanted mice were monitored every 3-4 days, starting 5 days after the injection of tumor cells, by bioluminescent imaging using an IVIS spectrum imaging system. Mce were intraperitoneally injected with 150 mg/kg of D-Luciferin (Perkin-Elmer), anesthetized with isofluorane, and imaged for 30 sec after 5 min following luciferin injection. For bioluminescent analysis of organs, mice were injected with D-Luciferin, euthanized by CO2, and organs were collected and imaged for 5 seconds. To measure the persistence of AMLMLL cells, bone marrow from vehicle- or ATRi-treated animals was isolated by flushing femurs and tibias (RPMI-1640 medium supplemented with 10% FBS and 10 μg/mL Pen/Strep). Erythrocyte lysis was performed by treating bone marrow cells with a commercial ACK red lysis buffer (Lonza) for 5 min at room temperature.Cells were stained with c-KIT APC-H7 antibody (BD Biosciences) and analyzed in a FACS Canto II (BD Biosciences; FACS Diva software). Data were analyzed with FlowJo (Treestar) software.
Histopathology
Spleens were collected from vehicle- and ATRi-treated mice, fixed in formalin and embedded in paraffin/formalin blocks. Sections and immunohistochemistry staining against γH2AX were performed following standard procedures. Slides where digitalized with a Mirax Scan (Zeiss) and γH2AX positive cells were automatically quantified from digitalized slides using AxioVision 4.6.3 software (Zeiss).
DNA fiber analyses
AML and AML_ETO cells were pulse-labeled with 50 μM CldU (20 min) followed by 250 μM IdU (20 min). Labeled cells were collected and DNA fibers were spread in buffer containing 0.5% SDS, 200 mM Tris pH 7.4 and 50 mM EDTA. For immunodetection of labeled tracks, fibers were incubated with primary antibodies (for CldU, rat anti-BrdU; for IdU, mouse anti-BrdU) for 1 hour at room temperature and developed with the corresponding secondary antibodies for 30 minutes at room temperature. Mouse anti-ssDNA antibody was used to assess fiber integrity. Slides were examined with a Leica DM6000 B microscope, as described previously (51). The conversion factor used was 1 μm=2.59 kb (52).
Supplementary Material
Acknowledgments
We thank Joannes Zuber for reagents and discussions, Andrew L. Kung for help with human AML cells and Mariano Barbacid for providing the plasmid containing the p53-targeting shRNA.
Funding: Work in O.F. laboratory was supported by Fundación Botín, by Banco Santander through its Santander Universities Global Division and by grants from MINECO (SAF2011-23753, SAF2014-57791-REDC and SVP-2013-068072), Howard Hughes Medical Institute and the European Research Council (ERC-617840). The A.N. laboratory was supported by the Intramural Research Program of the NIH, the National Cancer Institute, the Center for Cancer Research, an Ellison Medical Foundation Senior Scholar in Aging, and the Alex Lemonade Stand Foundation Award.
Footnotes
Author Contributions: I.M., A.D., M.M., V.L., M.E.A., A.T., H.C., A.E., R.A., A.B., X.W. contributed to the experiments; K.G.P., B.B., E.E. and A.J.P. worked in defining the experimental conditions for ATM and ATR inhibitors; S.A. helped in the analysis of the in vivo data; O.F. and A.N. designed the project and wrote the paper.
Competing interests: AZD0156 was provided by AstraZeneca to the investigators under terms of an MCRADA agreement with the NCI. This compound has been patented by AstraZeneca.
Data and materials availability: AZD0156 is available to third party investigators by AstraZeneca through an Open Innovation Portal request (http://openinnovation.astrazeneca.com/).
References and Notes
- 1.Bartkova J, et al. ATM activation in normal human tissues and testicular cancer. Cell Cycle. 2005;4:838–845. doi: 10.4161/cc.4.6.1742. [DOI] [PubMed] [Google Scholar]
- 2.Gorgoulis VG, et al. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 3.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez-Contreras AJ, Fernandez-Capetillo O. The ATR barrier to replication-born DNA damage. DNA Repair (Amst) 2010;9:1249–1255. doi: 10.1016/j.dnarep.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lecona E, Fernandez-Capetillo O. Replication stress and cancer: It takes two to tango. Exp Cell Res. 2014 doi: 10.1016/j.yexcr.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murga M, et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol. 2011;18:1331–1335. doi: 10.1038/nsmb.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schoppy DW, et al. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. J Clin Invest. 2012;122:241–252. doi: 10.1172/JCI58928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cottini F, et al. Synthetic lethal approaches exploiting DNA damage in aggressive myeloma. Cancer Discov. 2015 doi: 10.1158/2159-8290.CD-14-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derenzini E, et al. Constitutive activation of the DNA damage response pathway as a novel therapeutic target in diffuse large B-cell lymphoma. Oncotarget. 2015;6:6553–6569. doi: 10.18632/oncotarget.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwok M, et al. ATR inhibition induces synthetic lethality and overcomes chemoresistance in TP53- or ATM-defective chronic lymphocytic leukemia cells. Blood. 2016;127:582–595. doi: 10.1182/blood-2015-05-644872. [DOI] [PubMed] [Google Scholar]
- 11.Sarmento LM, et al. CHK1 overexpression in T-cell acute lymphoblastic leukemia is essential for proliferation and survival by preventing excessive replication stress. Oncogene. 2015;34:2978–2990. doi: 10.1038/onc.2014.248. [DOI] [PubMed] [Google Scholar]
- 12.Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25:304–317. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murga M, et al. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat Genet. 2009;41:891–898. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruzankina Y, et al. Tissue regenerative delays and synthetic lethality in adult mice after combined deletion of Atr and Trp53. Nat Genet. 2009;41:1144–1149. doi: 10.1038/ng.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reaper PM, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol. 2011;7:428–430. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 16.Toledo LI, et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol. 2011;18:721–727. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buisson R, Boisvert JL, Benes CH, Zou L. Distinct but Concerted Roles of ATR, DNA-PK, and Chk1 in Countering Replication Stress during S Phase. Mol Cell. 2015;59:1011–1024. doi: 10.1016/j.molcel.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz S, et al. A Genome-wide CRISPR Screen Identifies CDC25A as a Determinant of Sensitivity to ATR Inhibitors. Mol Cell. 2016;62:307–313. doi: 10.1016/j.molcel.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuber J, et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 2009;23:877–889. doi: 10.1101/gad.1771409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao RC, Dou Y. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat Rev Cancer. 2015;15:334–346. doi: 10.1038/nrc3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haferlach C, et al. Mutations of the TP53 gene in acute myeloid leukemia are strongly associated with a complex aberrant karyotype. Leukemia. 2008;22:1539–1541. doi: 10.1038/leu.2008.143. [DOI] [PubMed] [Google Scholar]
- 22.Megonigal MD, Rappaport EF, Nowell PC, Lange BJ, Felix CA. Potential role for wild-type p53 in leukemias with MLL gene translocations. Oncogene. 1998;16:1351–1356. doi: 10.1038/sj.onc.1201637. [DOI] [PubMed] [Google Scholar]
- 23.Kwok M, et al. Synthetic lethality in chronic lymphocytic leukaemia with DNA damage response defects by targeting the ATR pathway. Lancet. 2015;385(Suppl 1):S58. doi: 10.1016/S0140-6736(15)60373-7. [DOI] [PubMed] [Google Scholar]
- 24.Austin WR, et al. Nucleoside salvage pathway kinases regulate hematopoiesis by linking nucleotide metabolism with replication stress. J Exp Med. 2012;209:2215–2228. doi: 10.1084/jem.20121061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flach J, et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014;512:198–202. doi: 10.1038/nature13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alvarez S, et al. Replication stress caused by low MCM expression limits fetal erythropoiesis and hematopoietic stem cell functionality. Nat Commun. 2015;6:8548. doi: 10.1038/ncomms9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farres J, et al. PARP-2 sustains erythropoiesis in mice by limiting replicative stress in erythroid progenitors. Cell Death Differ. 2015;22:1144–1157. doi: 10.1038/cdd.2014.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santos MA, et al. DNA-damage-induced differentiation of leukaemic cells as an anti-cancer barrier. Nature. 2014;514:107–111. doi: 10.1038/nature13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez-Contreras AJ, Gutierrez-Martinez P, Specks J, Rodrigo-Perez S, Fernandez-Capetillo O. An extra allele of Chk1 limits oncogene-induced replicative stress and promotes transformation. J Exp Med. 2012;209:455–461. doi: 10.1084/jem.20112147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruiz S, et al. Limiting replication stress during somatic cell reprogramming reduces genomic instability in induced pluripotent stem cells. Nat Commun. 2015;6:8036. doi: 10.1038/ncomms9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krajewska M, et al. ATR inhibition preferentially targets homologous recombination-deficient tumor cells. Oncogene. 2015;34:3474–3481. doi: 10.1038/onc.2014.276. [DOI] [PubMed] [Google Scholar]
- 32.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H, et al. Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint. Nature. 2010;467:343–346. doi: 10.1038/nature09350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foote KM, et al. Discovery of 4-{4-[(3R)-3-Methylmorpholin-4-yl]-6-[1-(methylsulfonyl)cyclopropyl]pyrimidin-2-y l}-1H-indole (AZ20): a potent and selective inhibitor of ATR protein kinase with monotherapy in vivo antitumor activity. J Med Chem. 2013;56:2125–2138. doi: 10.1021/jm301859s. [DOI] [PubMed] [Google Scholar]
- 35.Tallis M, Morra R, Barkauskaite E, Ahel I. Poly(ADP-ribosyl)ation in regulation of chromatin structure and the DNA damage response. Chromosoma. 2014;123:79–90. doi: 10.1007/s00412-013-0442-9. [DOI] [PubMed] [Google Scholar]
- 36.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersson A, et al. Gene expression profiling of leukemic cell lines reveals conserved molecular signatures among subtypes with specific genetic aberrations. Leukemia. 2005;19:1042–1050. doi: 10.1038/sj.leu.2403749. [DOI] [PubMed] [Google Scholar]
- 38.Daniel JA, et al. Loss of ATM kinase activity leads to embryonic lethality in mice. J Cell Biol. 2012;198:295–304. doi: 10.1083/jcb.201204035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto K, et al. Kinase-dead ATM protein causes genomic instability and early embryonic lethality in mice. J Cell Biol. 2012;198:305–313. doi: 10.1083/jcb.201204098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Degorce SL, et al. Discovery of Novel 3-Quinoline Carboxamides as Potent, Selective, and Orally Bioavailable Inhibitors of Ataxia Telangiectasia Mutated (ATM) Kinase. J Med Chem. 2016;59:6281–6292. doi: 10.1021/acs.jmedchem.6b00519. [DOI] [PubMed] [Google Scholar]
- 41.Blanco JG, Edick MJ, Relling MV. Etoposide induces chimeric Mll gene fusions. FASEB J. 2004;18:173–175. doi: 10.1096/fj.03-0638fje. [DOI] [PubMed] [Google Scholar]
- 42.Libura J, Slater DJ, Felix CA, Richardson C. Therapy-related acute myeloid leukemia-like MLL rearrangements are induced by etoposide in primary human CD34+ cells and remain stable after clonal expansion. Blood. 2005;105:2124–2131. doi: 10.1182/blood-2004-07-2683. [DOI] [PubMed] [Google Scholar]
- 43.Bernt KM, Armstrong SA. Targeting epigenetic programs in MLL-rearranged leukemias. Hematology Am Soc Hematol Educ Program. 2011;2011:354–360. doi: 10.1182/asheducation-2011.1.354. [DOI] [PubMed] [Google Scholar]
- 44.Somervaille TC, Cleary ML. Grist for the MLL: how do MLL oncogenic fusion proteins generate leukemia stem cells? Int J Hematol. 2010;91:735–741. doi: 10.1007/s12185-010-0579-8. [DOI] [PubMed] [Google Scholar]
- 45.Chen CW, Armstrong SA. Targeting DOT1L and HOX gene expression in MLL-rearranged leukemia and beyond. Exp Hematol. 2015;43:673–684. doi: 10.1016/j.exphem.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR. BET Bromodomain Inhibition Suppresses the Function of Hematopoietic Transcription Factors in Acute Myeloid Leukemia. Mol Cell. 2015;58:1028–1039. doi: 10.1016/j.molcel.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rathert P, et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015;525:543–547. doi: 10.1038/nature14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fong CY, et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015;525:538–542. doi: 10.1038/nature14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kantidakis T, et al. Mutation of cancer driver MLL2 results in transcription stress and genome instability. Genes Dev. 2016;30:408–420. doi: 10.1101/gad.275453.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lecona E, Fernandez-Capetillo O. Replication stress and cancer: it takes two to tango. Exp Cell Res. 2014;329:26–34. doi: 10.1016/j.yexcr.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacome A, et al. NSMCE2 suppresses cancer and aging in mice independently of its SUMO ligase activity. EMBO J. 2015;34:2604–2619. doi: 10.15252/embj.201591829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.