Summary
DNGR-1 is a C-type lectin receptor that binds F-actin exposed by dying cells and facilitates cross-presentation of dead cell-associated antigens by dendritic cells. Here we present the structure of DNGR-1 bound to F-actin at 7.7 Å resolution. Unusually for F-actin binding proteins, the DNGR-1 ligand binding domain contacts three actin subunits helically arranged in the actin filament, bridging over two protofilaments, as well as two neighboring actin subunits along one protofilament. Mutation of residues predicted to mediate ligand binding led to loss of DNGR-1-dependent cross-presentation of dead cell-associated antigens, formally demonstrating that the latter depends on F-actin recognition. Notably, DNGR-1 has relatively modest affinity for F-actin but multivalent interactions allow a marked increase in binding strength. Our findings shed light on modes of actin binding by cellular proteins and reveal how extracellular detection of cytoskeletal components by dedicated receptors allows immune monitoring of loss of cellular integrity.
Introduction
Damage to tissues releases damage-associated molecular patterns (DAMPs), which elicit an inflammatory response designed to maintain sterility and promote repair of the injured site. In vertebrates, DAMPs can additionally promote adaptive immune responses to foreign antigens contained within damaged cells in what may be the major pathway for initiating immunity against tumors and some viruses (Zelenay and Reis e Sousa, 2013). DNGR-1 (also known as CLEC9A) is an innate immune receptor specific for a DAMP exposed by dead cells (Sancho et al., 2009). DNGR-1 is specifically expressed by dendritic cells (DCs), a leukocyte subset responsible for initiation and regulation of immune responses (Caminschi et al., 2008; Huysamen et al., 2008; Poulin et al., 2012; Poulin et al., 2010; Sancho et al., 2008). DNGR-1 signaling in response to dead cell recognition facilitates cross-presentation of dead-cell-associated antigens by DCs and priming of cytotoxic T lymphocytes against cytopathic viruses (Iborra et al., 2012; Sancho et al., 2009; Zelenay et al., 2012). Recently, we and others reported that the DAMP recognized by DNGR-1 is the filamentous form of actin (Ahrens et al., 2012; Zhang et al., 2012), an ubiquitous and abundant intracellular component of eukaryotic cells. F-actin recognition explains how DNGR-1 can act as a universal detector of dead cells and reveals cytoskeletal exposure as a means of innate immune detection of cell damage.
DNGR-1 is a disulphide-bonded homo-dimeric type II trans-membrane protein of the C-type lectin superfamily (Huysamen et al., 2008; Sancho et al., 2008). The extracellular domain (ECD) of each DNGR-1 monomer contains a single C-type lectin-like domain (CTLD) bearing the ligand-binding site, followed by a membrane-proximal “neck” region of isoform-specific length consisting of 48 to 74 amino acids (Huysamen et al., 2008; Sancho et al., 2008). The crystal structure of the unbound CTLD of human DNGR-1 has been solved except for a missing internal segment of 5 residues (Zhang et al., 2012). The structure reveals that the CTLD of DNGR-1 is similar to that of other CTLDs in the C-type lectin superfamily. However, none of the latter have been shown to bind actin, indicating that receptor specificity can only be understood at the molecular level by solving the structure of the receptor in complex with its ligand. Here, we used electron cryomicroscopy and helical image analysis to determine the structure of DNGR-1 bound to F-actin at 7.7 Å resolution. The DNGR-1 CTLD binds to the interface between two actin protofilaments, an unusual topology among actin-binding proteins that explains the specificity of the receptor for the polymeric ligand. We have further shown that DNGR-1 affinity for F-actin is modest but is compensated by avidity to increase binding strength by at least three orders of magnitude, thus allowing efficient ligand recognition. Additionally, using mutants impaired in ligand binding, we have formally demonstrated that F-actin recognition underlies the ability of DNGR-1 to mediate cross-presentation of dead cell-associated antigens. Our data reveal how immune recognition of cell death can proceed through evolution of a CTLD optimized to detect exposed cytoskeletal components.
Results
DNGR-1 binds to the interface between actin protofilaments
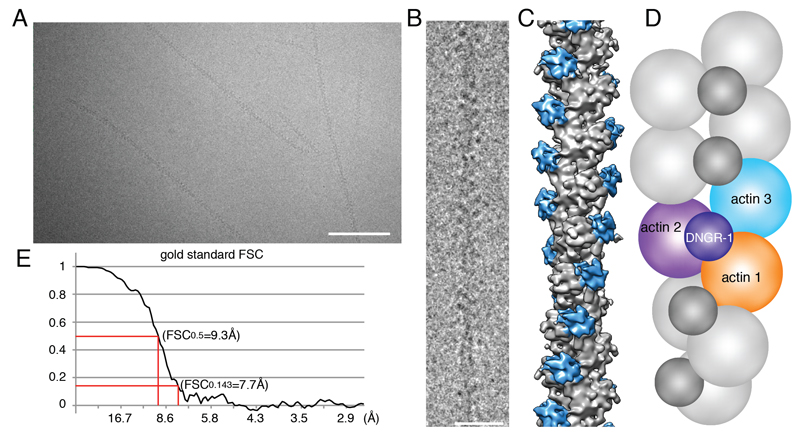
To understand the molecular basis for recognition of F-actin by DNGR-1, we set out to solve the structure of the DNGR-1 bound to actin filaments. We expressed the entire ECD of mouse DNGR-1 (long isoform; Figure S1) as a soluble disulphide-bonded dimeric protein in 293F cells (Ahrens et al., 2012). The purified ECD was then used to decorate F-actin in vitro and the complexes subjected to electron cryomicroscopy (cryoEM) and helical image analysis (Figures 1A-D). The resolution of the reconstructed three-dimensional (3D) density map was 7.7 Å as determined by the Fourier shell correlation (FSC) method at FSC = 0.143 (Figure 1E and S2). The data and analysis parameters are given in Table S1. The density map shows that the CTLD of DNGR-1 binds to actin filament subunits with 1:1 stoichiometry (Figures 1C and 2A). We could not observe densities corresponding to the neck of the bound monomer or to the other half of the dimer (Figures 1C and 2A), indicating flexibility in the neck region. Each CTLD interacts with three actin subunits that are helically arranged in F-actin, bridging over two protofilaments as well as two neighboring actin subunits along one protofilament (Figures 1C, 1D and 2A). Thus, the structure of the complex clearly explains the specificity of the receptor for polymerized actin.
Figure 1. Structure of F-actin decorated with DNGR-1.
A, B, CryoEM images of DNGR-1-decorated F-actin in a frozen-hydrated state at two different magnifications, scale bars correspond to 50nm and 20nm respectively. C, Solid surface representation of the 3D density map of DNGR-1 decorated F-actin at 7.7 Å resolution obtained by helical image reconstruction. F-actin is colored grey and DNGR-1 blue. D, Schematic representation of the data in C: DNGR-1 binds to the interface between actin protofilaments making contact with three actin filament subunits (arbitrarily numbered 1-3). E, Fourier shell correlation was performed to determine the resolution of the 3D map. See also Supplemental Figure 1 and Supplemental Table 1.
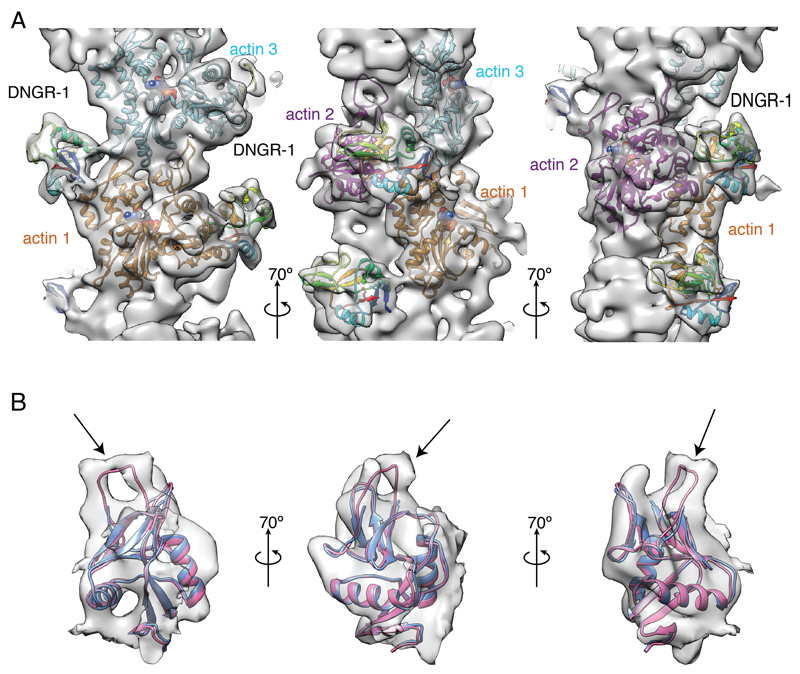
Figure 2. Helical reconstruction reveals the mode of binding of DNGR-1 to F-actin and a flexible loop missing from the crystal structure of the DNGR-1 CTLD.
A, Three different views of main chain ribbon models of F-actin (actin subunits 1, 2, 3 from Figure 1D colored in orange, magenta, cyan, respectively) and DNGR-1 CTLD (rainbow color) fitted into the density map. B, Overlay of the crystal structure of DNGR-1 CTLD (pdb ID 3VPP; blue) and model (in magenta) of DNGR-1 CTLD including the flexible loop (R226 – A230; indicated by arrow) fitted into the observed electron density. See also Supplemental Figure 2 and Supplemental Movie 1.
A flexible loop absent in the DNGR-1 crystal structure can be visualized by cryoEM
The cryoEM density map was used to fit the crystal structure of the human DNGR-1 CTLD (PDB ID 3VPP) (Zhang et al., 2012) onto F-actin (PDB ID 3MFP) (Fujii et al., 2010) (Figure 2A). To build a reliable atomic model, we used FlexEM (Topf et al., 2008), which utilizes simulated annealing molecular dynamics with restrained stereochemical and non-bonded interaction terms. We found an unfilled density connected to the CTLD and identified it as the five residue internal loop (R226 – A230 in mouse DNGR-1) that is missing from the published structure of human DNGR-1 (Figure 2B). This loop is therefore presumably disordered in the unbound receptor but becomes ordered upon binding to F-actin. We included the loop based on the cryoEM density map to complete the model of the DNGR-1 CTLD bound to F-actin (Figure 2A and Movie S1).
DNGR-1 employs three discrete surfaces to interact with F-actin
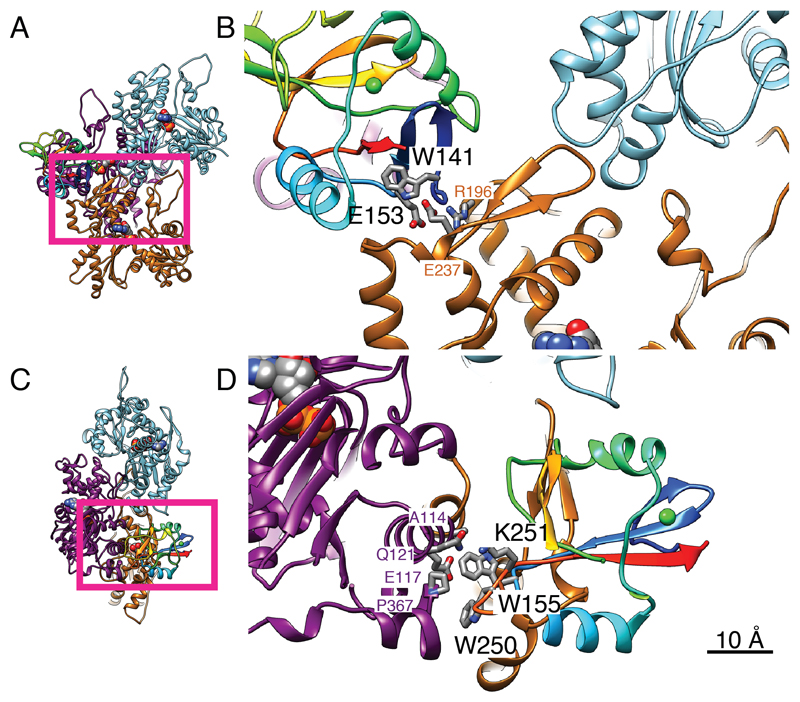
The CTLD of DNGR-1 employs three discrete surface regions to interact with three distinct actin subdomains, each belonging to a different actin filament subunit (Figures 1D, 2A, 3 and Movie S1; actin subunits in Figure 3 are numbered and colored as in Figure 2A). Although the resolution of the cryoEM density map was insufficient to discriminate individual side chains, the main chain positions were determined reliably enough to allow identification of key DNGR-1 residues putatively involved in interaction with ligand. The most extensive interactions occur with actin subunit 2 (Figure 2A right panel and Figures 3C and 3D) and, to a lesser extent, with subunit 1 (Figure 2A left panel and Figures 3A and 3B), while a weak contact is made with actin subunit 3 (Figure 2A left panel). In all, 16 putative interacting residues were identified, of which 13 are conserved between mouse and human DNGR-1 (Table S2). Notably, they include mouse DNGR-1 residues W141 and E153, which are shown in Figure 3B at the binding interface with actin subunit 1, and W155 and W250, which are shown in Figure 3D to interact with actin subunit 2. W155 and W250 of mouse DNGR-1 correspond to W131 and W227 in human DNGR-1, which were previously proposed to be important for ligand binding (Zhang et al., 2012).
Figure 3. The binding interface of DNGR-1 CTLD and F-actin.
A, B, N-terminal region of the CTLD interacting with actin 1 (orange) and actin 3 (cyan). C, D, the opposite face of the CTLD interacting with actin 2 (magenta). A and C are guides to show the portions displayed in more detail in B and D, respectively. The viewing directions of A, B and C, D correspond to the left and right panels of Figure 2A, respectively, and actin subunit numbering and coloring is as in Figure 1D. DNGR-1 CTLD is colored rainbow. Side chains of the CTLD amino acids identified by mutagenesis as important for F-actin binding are labeled in black lettering and are displayed as a stick model with oxygen in blue, nitrogen in red and carbon in grey. Actin side chains that are identified in the structural model as possible binding partners are also displayed in the same way and are labeled in the color of the corresponding actin subunit. The sequence number and identity of each amino acid are for mouse CTLD and human platelet actin. Scale bar corresponds to 10Å. See also Supplemental Table 2.
DNGR-1 residues involved in the interaction with F-actin
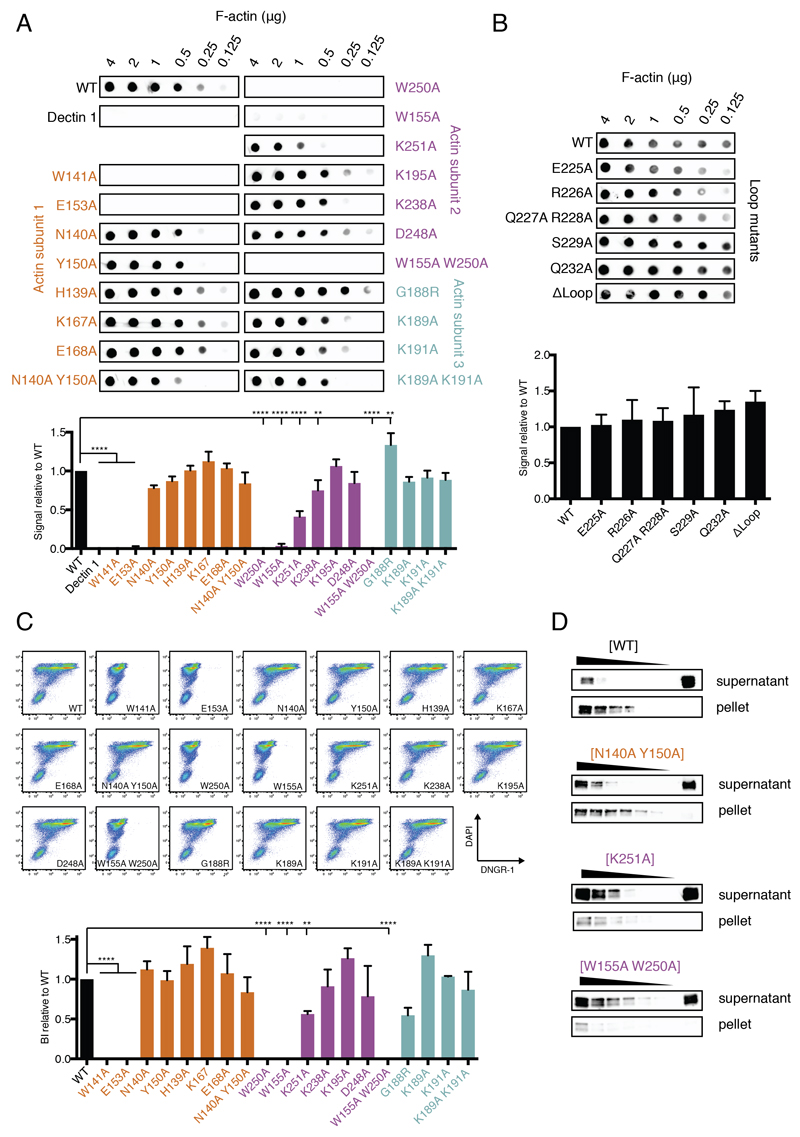
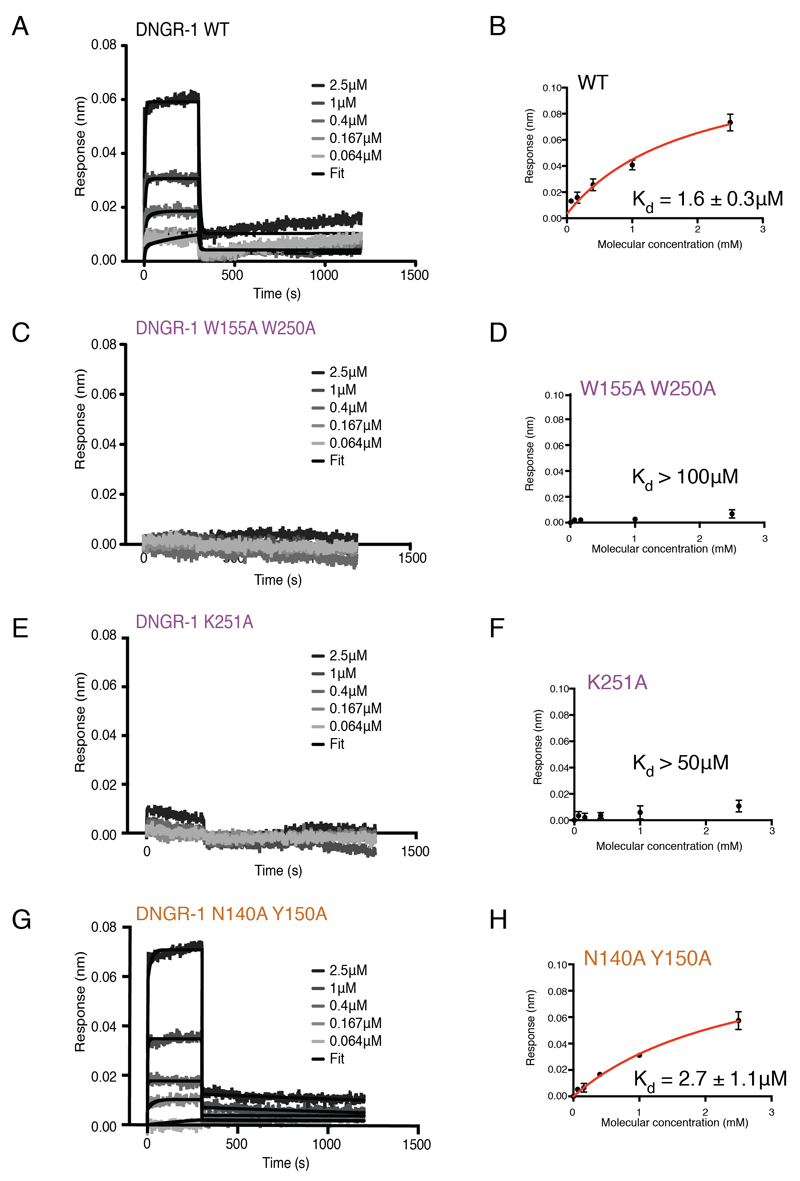
To assess the extent to which the identified residues contribute to the interaction of DNGR-1 ECD with F-actin, we mutated them either singly or in selected combinations. All mutant proteins could be expressed in 293F cells, and were secreted and accumulated in cell culture supernatants, albeit to varying amounts (Figure S3). We used Immuno blot analysis to measure ECD concentrations in supernatants and normalized them before assessing binding to in vitro polymerized actin using a dot blot assay (Ahrens et al., 2012) quantified by densitometric analysis (Figure 4A). In agreement with data for human DNGR-1 (Zhang et al., 2012), a W155A W250A double mutant mDNGR-1 ECD showed no binding to F-actin (Figure 4A). The loss in binding could largely be recapitulated by mutating W250 alone (likely interacting with A365, G366 and P367 of actin - residue numbering is for human actin; Figure 3). Single mutation of W155 (likely interacting with A114, E117 and P367 of actin; Figure 3) resulted in a pronounced decrease in binding (Figure 4A). Mutation of K251 (likely interacting with Q121 of actin; Figure 3) also had a profound effect (Figure 4A). All of these residues are predicted to be involved in the interaction between DNGR-1 and actin subunit 2 (Figure 3 and Table S2). Mutation of additional residues predicted to be involved in the same interaction surface showed only a limited effect (e.g., K238A, likely interacting with D80 of actin; Figure 3) or did not impact binding (K195, D248; Figure 4A). Point mutations (E225A, R226A, Q227A, R228A, S229A or Q232A) in the flexible loop missing from the crystal structure of human DNGR-1 CTLD (see above) did not affect DNGR-1 binding (Figure 4B). Similarly, replacing the entire loop by a string of alanine residues (ΔLoop) dramatically decreased expression efficiency of the protein (Figure S3) but not its ability to bind F-actin (Figure 4B). Taken together, these data indicate a dominant role for W155, W250 and K251 in mediating DNGR-1 contact with actin subunit 2 and suggest that the flexible loop does not impact the strength of binding.
Figure 4. Identification of DNGR-1 residues essential for binding F-actin.
A, and B, The indicated amounts of F-actin were spotted onto a nitrocellulose membrane and probed with WT or mutant DNGR-1 ECD proteins at equal concentrations. Strength of signal was quantified and plotted relative to WT (lower panel). The dot blot data depicted are from one representative experiment of 3. The quantitation is based on 3 experiments and the data represent mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; One-way ANOVA with Dunnett’s multiple comparisons test. C, HeLa cells were UV-irradiated, cultured overnight to allow secondary necrosis, and stained with WT or mutant DNGR-1 ECD proteins at equal concentrations. Binding was analyzed by flow cytometry using DAPI to identify dead cells (upper panel) and binding index was calculated (lower panel). The plots shown are from one representative experiment of 3. The binding index is from 3 pooled experiments and the data represent mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; One-way ANOVA with Dunnett’s multiple comparisons test. D, F-actin was incubated with a 2-fold dilution of WT or mutant DNGR-1 ECD proteins (decreasing protein concentration indicated by the wedge). The samples were ultracentrifuged and both pellet and supernatant fractions were tested for presence of DNGR-1 proteins by SDS-PAGE and immunoblot. Data are representative of 4 experiments for the WT and 2 experiments for each mutant. The font color used for the mutant name is meant to represent the actin subunit to which the residue normally binds, as depicted in Figure 1D. See also Supplemental Figure 3.
Among the residues potentially involved in the interaction with actin subunit 1, W141A and E153A single mutants expressed poorly (Figure S3) but, compared to other poorly expressed mutants, showed no or very weak binding, respectively (Figure 4A). In contrast, mutation of N140A and Y150A, alone or in combination, had only minimal impact on binding (possibly to the carbonyl groups of actin S233 and S234; Figure 3). There was no decrease in binding for H139A, K167A and E168A mutants (Figure 4A). Collectively, these data suggest that W141 and E153 (likely interacting with R196 or forming a carboxyl pair with E237 via Ca2+; Figure 3) provide the major contribution to the DNGR-1 interaction with actin subunit 1. Although W141 is pointing towards the inside of the CTLD in the crystal structure of human DNGR-1, it is not part of the hydrophobic core of the domain and is located instead on the hydrophilic side of the three-stranded β-sheet formed by the N- and C-terminal chains. The W141A mutant could be recognized by 2 different anti-DNGR-1 antibodies (clones 7H11 and 1F6; data not shown), suggesting that the protein is not grossly misfolded, even if it expresses poorly. We therefore hypothesize that W141 might flip out upon binding of DNGR-1 to ligand to interact with a hydrophobic patch on actin (Figure 3). This notion is supported by the observed electron density, which strongly suggests that the N-terminal β-hairpin (W141-Y150) could be unfolded to place W141 en face with actin (Figure 2A and data not shown). Finally, of the residues putatively involved in the interaction with actin subunit 3, mutation of K189 and K191, either singly or doubly, did not have any appreciable impact on binding of DNGR-1 (Figure 4A).
To confirm that the observed binding pattern of DNGR-1 ECD mutants to in vitro polymerized F-actin is representative of binding to actin in dead cells, we utilized a flow cytometry-based approach in which secondarily-necrotic HeLa cells were stained with the same proteins used in dot blot analysis (Figure 4C, upper panel). Using this assay, we saw a pattern of binding identical to the one observed using dot blot and in vitro polymerized F-actin (Figure 4C, lower panel, compare with Figure 4A, lower panel). These data indicate that DNGR-1 binding to F-actin from dead cells does not differ substantially from that to purified F-actin, suggesting that, at least under these experimental conditions, cell death does not markedly alter the structure of the polymer.
Subtle differences between mouse and human DNGR-1 CTLDs could potentially impact binding to F-actin. For example, among residues predicted to interact with actin subunit 3, glycine 188 (G188) in mouse replaces arginine (R165) in human DNGR-1. Changing G188 to arginine (G188R) appeared to increase binding of DNGR-1 to F-actin in the dot blot assay (Figure 4A) but reduce it the flow cytometry assay (Figure 4C). The differences may have been an artefact due to the fact that the mutation adversely impacted expression of the protein (Figure S3), which resulted in its concentration being limiting in the flow cytometry but not dot blot assay. We observed a similar phenomenon for the ΔLoop mutant (data not shown), which also expressed very poorly. Substituting the other non-conserved residues in mouse DNGR-1 CTLD with their human counterparts (H139N and K195D) resulted in no detectable change in binding to F-actin (data not shown).
In sum, taken together, our data suggest that W141 and E153 and, to a lesser extent N140 and Y150, play a role in the interaction of mouse DNGR-1 with actin subunit 1 while W155, W250, K251, and to a lesser extent K238 play a role in the interaction with actin subunit 2, which is dominant. The limited interaction with subunit 3, as well as the flexible loop, appear largely dispensable for binding.
Mutation of receptor residues at the ligand interface decreases DNGR-1 affinity for F-actin
Of the ECD mutants described above, we selected for detailed analysis ones that vary in the strength of F-actin binding such as K251A, W155A W250A and N140A Y150A; we excluded W141A and E153A because of their low expression (Figure S3). First, we utilized an assay in which decreasing amounts of wild type (WT) or mutant DNGR-1 ECD proteins were incubated with a constant amount of F-actin before filaments were pelleted by ultracentrifugation (Ahrens et al., 2012). In case of the WT protein and the N140A Y150A mutant, we observed accumulation of DNGR-1 in the F-actin pellet and depletion from the supernatant. In contrast, W155A W250A and K251A mutants largely remained in the supernatant after pelleting, confirming that neither mutant binds efficiently to F-actin (Figure 4D).
To monitor binding in real-time, we developed a method based on biolayer interferometry (BLI) (Abdiche et al., 2008). We mixed biotinylated and non-biotinylated G-actin (1:4) and allowed it to polymerize directly on a streptavidin-coated biosensor. Actin polymerized in this way was stable over prolonged periods of time (Figure S4A). To confirm the applicability of the technique, we first tested binding of α-actinin, a well-characterized F-actin binding protein (Figure S4B). Real-time binding of α-actinin to the F-actin-coated sensor in a concentration-dependent fashion was observed (Figure S4B) and steady-state analysis (Figure S4C) revealed a Kd = 0.15 ± 0.04 μM, in good agreement with previously published data (Wachsstock et al., 1993). When WT mDNGR-1 ECD was tested using the same setup, binding curves displayed fast association kinetics (1.5×106 ± 4.2×105 M-1s-1), as well as a fast dissociation phase (8.48×10-1 ± 9.6×10-2 s-1) (Figure 5A), with steady state analysis revealing a Kd = 1.6 ± 0.3 μM (Figure 5B), within the range previously estimated by pelleting assay (Ahrens et al., 2012). There was no statistically significant decrease in affinity for mutants that showed little difference in the dot blot assay, such as Y150A (data not shown). In contrast, consistent with the dot blot assay, no binding of W155A W250A to F-actin was observed by BLI, indicating a Kd greater than 100 μM (Figure 5C and 5D). For the K251A mutant, only very weak binding was observed at the highest concentration used, indicating a Kd > 50 μM (Figure 5E and 5F). Finally, the N140A Y150A mutant showed only a marginal decrease in affinity when compared to the wild type protein (Kd = 2.7 ± 1.1 μM; Figure 5G and 5H). Thus, analysis by BLI, pelleting assay, flow cytometry-based assay and dot blot are all concordant and place the DNGR-1 ECD proteins in the following order of decreasing affinity for F-actin: WT = Y150A > N140A Y150A > K251A > W155A W250A.
Figure 5. Real-time measurement of DNGR-1 binding to F-actin.
F-actin was polymerized directly on a BLI sensor and binding of the indicated concentrations of A, WT, C, W155A W250A E, K251A and G, N140A Y150A DNGR-1 ECD was monitored in real time. Representative examples of binding curves are shown. Steady state analysis of binding for B, WT, D, W155A W250A F, K251A and H, N140A Y150A DNGR-1. Data points are the mean ± SD of 7 replicate experiments for WT, 3 replicate experiments for N140A Y150A mutant and 2 replicate experiments for W155A W250A and K251A mutants. Numbers represent best-fit curve values ± standard error. The font color used for the mutant name is meant to represent the actin subunit to which the residue normally binds, as depicted in Figure 1D. See also Supplemental Figure 4.
Avidity increases the strength of DNGR-1 binding to F-actin
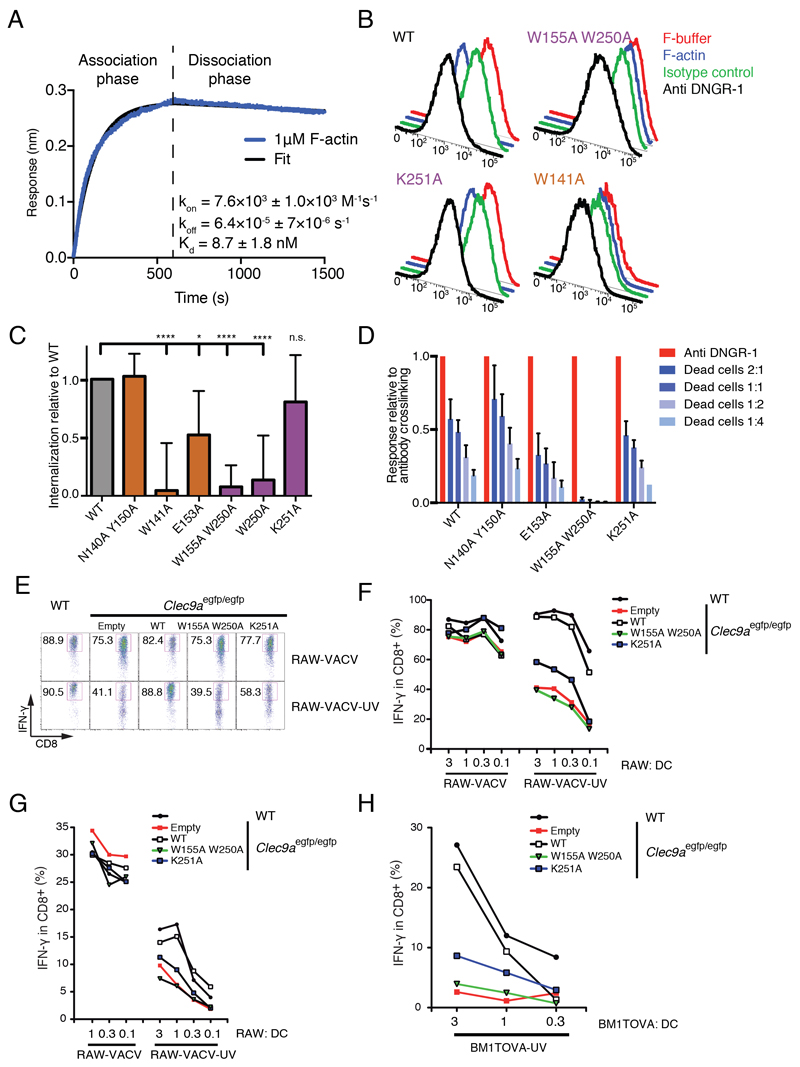
The BLI analysis of soluble DNGR-1 binding to F-actin does not take into account the avidity component of the interaction. In the case of soluble DNGR-1, the receptor is free to diffuse upon dissociation from an actin filament but, in the context of the membrane-anchored protein, the polymeric nature of the ligand means that when one molecule of DNGR-1 dissociates, the actin filament remains bound through interactions with adjacent DNGR-1 receptors. This makes the likelihood of re-association much higher, effectively decreasing the off-rate. To assess the contribution of avidity to the interaction of DNGR-1 with F-actin, we immobilized DNGR-1 ECD on the BLI biosensor and tested binding of short actin filaments capped and stabilized with gelsolin (see Experimental Procedures). Steady-state analysis was not feasible under these conditions because of the narrow range of F-actin concentrations that could be tested: lowering of concentration was limited by the fact that F-actin depolymerizes at the pointed end below the critical concentration of 0.6 μM (Pollard, 1986) while concentrations above 2 μM caused marked artifacts in the BLI signal due to large filament size (data not shown). To measure apparent affinities, we therefore used dynamic analysis of the binding curves and on- and off-rates obtained with 1 μM of actin filaments (Figure 6A). Similar to the situation where F-actin is immobilized and DNGR-1 ECD is free, we found that there is a high binding on-rate (7.6×103 ± 1.0×103 M-1s-1). However, and in contrast to the previous setup, the dissociation phase was very slow (off-rate 6.4×10-5 ± 7×10-6 s-1), which translated into a Kd of 8.7 ± 1.8 nM. Thus, the avidity component increases the strength of the DNGR-1 : F-actin interaction by as much as three orders of magnitude (Kd decrease from micromolar to nanomolar range). This is likely to be an underestimate of the true contribution of avidity as the latter is directly dependent on the size of actin filaments, which were kept deliberately short in our experimental setup.
Figure 6. Avidity increases the strength of DNGR-1 : F-actin binding.
A, WT DNGR-1 ECD was immobilized on a BLI sensor and binding of of short F-actin filaments was monitored in real time. Data shown are representative of 6 replicate experiments. Numbers are the mean value ± s.d. of all 6 experiments. B, Cells expressing full-length WT or mutant DNGR-1 trans-membrane proteins were treated with F-actin, anti-DNGR-1 antibody, F-buffer or control antibody, incubated at 37°C for 60 minutes and fixed before staining for DNGR-1 and analysis by flow cytometry. Profiles depict surface expression of DNGR-1 WT and selected mutants. The image is representative of 6 experiments. C, Data from 6 independent experiments carried out as in B, were normalized to the antibody induced internalization and expressed as mean ± s.d. internalization relative to WT. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; One-way ANOVA with Dunnett’s multiple comparisons test. In A-C, the font color used for the mutant name is meant to represent the actin subunit to which the residue normally binds, as depicted in Figure 1D. D, B3Z-Syk reporter cells expressing WT or mutant DNGR-1 receptors were incubated with decreasing numbers of UV-irradiated HeLa cells (ratio dead:reporter cells is indicated) or with plate-bound anti-DNGR-1 antibody or medium alone at 37°C overnight. Activation of the NFAT reporter was read out at the end of the incubation period. Data are normalized and expressed as mean ± s.d. of three independent experiments. E-G, RAW264.7 (RAW) cells were infected with rVACV OVA and not irradiated (RAW-VACV) or infected and UV-irradiated (RAW-VACV-UV). They were fed at the indicated ratios to DCs from WT or Clec9aegfp/egfp mice transduced or not with retroviruses encoding WT or the indicated mutant DNGR-1 constructs. OVA-specific CD8+ T cells (E, F) or VACV-specific CD8+ T cells (G) were added and IFN-γ production was measured after 6h. H, Cross-presentation of UV-treated BM1TOVA (BM1TOVA-UV) was also assessed using OVA-specific CD8+ T cells. In (E), dot plots show the production of IFN-γ by CD8+ T cells specific for OVA at the ratio 3:1 infected cells:DC. In (F-H), graphs depict the frequencies of IFN-γ+ CD8+ T cells specific for OVA (F, H) or VACV (G). E-H: one representative experiment of three performed is shown. See also Supplemental Figure 5.
Avidity can compensate for decreased in affinity in ligand-induced DNGR-1 internalization
To assess the contribution of affinity vs. avidity in a biological setting, we expressed full-length (trans-membrane) mDNGR-1 in cells as WT protein or bearing W250A, W155A W250A, K251A, W141A, E153A or N140A Y150A mutations (Figure S5A) and tested the response to F-actin. When cross-linked by either F-actin or a specific antibody, DNGR-1 underwent endocytosis, which could be read out as loss of cell surface staining (Figure 6B and S5B). Treatment of cells on ice resulted in only minimal loss of signal, excluding the possibility that the effect was due to masking of the antibody epitope by bound ligand (Figure S5C and data not shown). When treated with anti-DNGR-1 antibody, all of the mutants were able to internalize (Figure S5D), confirming that they were not misfolded or otherwise incapable of entering the endocytic pathway. In contrast, when treated with F-actin, no internalization was observed for the mutants that also showed complete loss of affinity in biochemical assays, namely W155A W250A, W250A and W141A (Figure 6B and 6C). As expected from our in vitro binding data, the N140A Y150A mutant was indistinguishable from WT receptor in the internalization assay (Figure 6C). Importantly, K251A and E153A mutants were able to internalize in response to F-actin, albeit to a lesser extent than WT (Figure 6B and 6C), lending support to the notion that the avidity component is able to compensate for a considerable decrease in receptor affinity.
Loss of DNGR-1 affinity for F-actin impairs cross-presentation of dead-cell associated antigens
To further assess the impact of affinity vs. avidity on DNGR-1 signaling through Syk, we expressed wild-type DNGR-1 or the W155A W250A, K251A, E153A or N140A Y150A mutants in B3Z-Syk reporter cells, in which Syk activation induces transcription of an NFAT-driven β-galactosidase reporter. We measured activation of the reporter in response to stimulation with dead cells and normalized the data to the response obtained by stimulation with plate-bound anti-DNGR-1 antibody in order to account for the varying surface expression of different mutants. As expected, expression of wild type DNGR-1 conferred reporter cell responsiveness to dead cell stimulation (Fig. 6D and data not shown). In contrast, expression of W155A W250A mutant led to a complete lack of response (Fig. 6D). The K251A and E153A mutants displayed a small decrease in the ability to signal via Syk in response to dead cells whereas the N140A Y150A mutant showed no impairment (Figure 6D). Thus, the ability of DNGR-1 mutants to internalize and to signal via Syk in response to dead cells largely mirror each other and indicate that a large loss of affinity such as in the case of the K251A and E153A mutants need not lead to loss of response, likely because of the avidity component of the interaction.
Finally, to assess to what extent these observations apply to DNGR-1-mediated cross-presentation of dead cell-associated antigens by DCs, we isolated bone marrow from mice in which DNGR-1 was genetically ablated (Clec9aegfp/egfp), transduced it with retroviruses encoding full-length WT, W155A W250A or K251A mutant DNGR-1 or with a control empty retrovirus, and differentiated the cells into DCs under the influence of FLT3L. Separately, we infected H-2d RAW264.7 cells with a recombinant vaccinia virus (VACV) encoding OVA, subjected them (RAW-VACV-UV) or not (RAW-VACV) to UV radiation 4 hours post-infection and left them overnight to undergo secondary necrosis. The RAW cells were then cultured with transduced Clec9aegfp/egfp or non-transduced control WT DCs at different ratios, together with OVA-specific CTL lines or vaccinia virus-specific H-2b-restricted CD8+ effector T cells purified from mice infected with VACV WR. IFN-γ production was assessed 6 hours later. In this assay, the non-irradiated RAW-VACV cells served as a source of virus that could infect DCs directly, allowing for direct antigen presentation to the CD8+ T cells (positive control). In contrast, the UV-irradiated RAW-VACV-UV cells underwent secondary necrosis and contained only inactive virus that could not infect DCs. As such, they served as a source of dead cell-associated viral antigens (including OVA) for cross-presentation.
When direct presentation was allowed (RAW-VACV), all DCs were equally competent at stimulating antigen-specific CD8+ T cells, irrespective of DNGR-1 expression (Figure 6E-G). In contrast, when restricted to cross-presentation (RAW-VACV-UV), the Clec9aegfp/egfp DCs transduced with control virus showed a pronounced defect in stimulating antigen-specific CD8+ T cells (Figure 6E-G). This defect could be rescued by reconstitution with WT DNGR-1 (Figure 6E-G), as previously reported (Sancho et al., 2009). In contrast, reconstitution with W155A W250A DNGR-1 did not rescue (Figure 6E-G), consistent with the inability of that mutant to bind F-actin. The K251A mutation showed an intermediate phenotype, restoring cross-presentation of dead cell-associated antigens to a lower extent than the WT receptor (Figure 6E-G) and lower than might have been expected from its activity in the internalization and reporter assays (see above). It is unclear whether this disparity relates to expression of the receptor or Syk in DCs vs. reporter cells, or to a greater extent of crosslinking being required for signaling for cross-presentation in DCs vs. reporter construct activation in reporter cells. Similar results were obtained using secondarily necrotic H-2bm1 fibroblasts expressing OVA (Sancho et al., 2009) as a source of dead cell-associated antigen for cross-presentation to OVA-specific CD8+ T cells (Fig. 6H). Thus, these data formally prove that the ability of DNGR-1 to bind F-actin is at the heart of its function in cross-presentation of dead cell-associated antigens.
Discussion
DNGR-1, a C-type lectin-like receptor expressed by DCs, is a sensor of cell damage and the only known transmembrane protein able to bind actin in the extracellular space. Here, we have solved the structure of DNGR-1 in complex with its ligand, F-actin, and identified the contact surfaces and residues essential for the interaction. The CTLD of DNGR-1 binds to three actin subunits that are helically arranged in F-actin, crosslinking two actin protofilaments as well as two neighboring actin subunits along the protofilament and making contacts with each actin subunit. We further defined a flexible loop, which is absent in the crystal structure and appears to be stabilized by interaction with F-actin. Notably, we observed no decrease in affinity upon removal of this loop. This is likely due to the entropy cost of folding of the loop, which cancels out the enthalpy gain of the interaction.
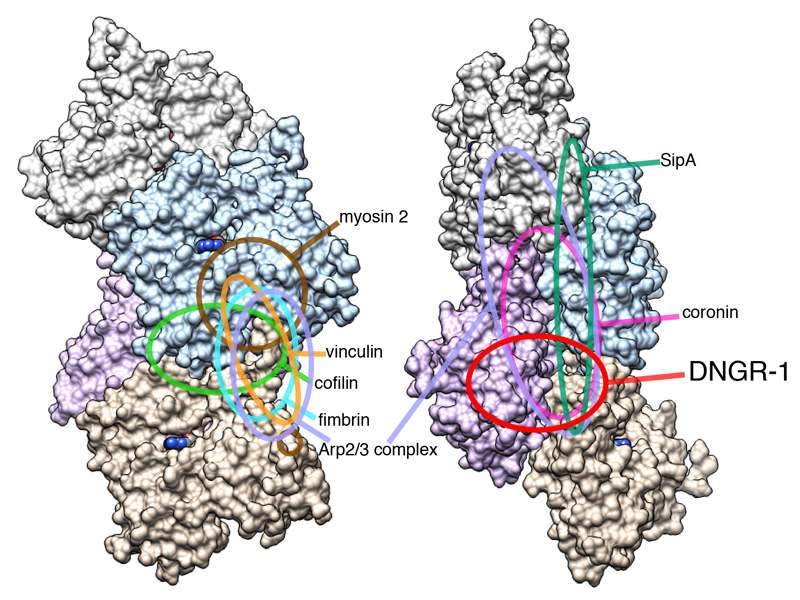
The mode of DNGR-1 binding shares some features with that of coronin (Galkin et al., 2008a; Ge et al., 2014), the Arp2/3 complex (Rouiller et al., 2008) or Salmonella SipA protein (Lilic et al., 2003) in that they all bind across the two protofilaments. However, the binding site of DNGR-1 comprises a relatively small F-actin surface and only partially overlaps with that of any of the above proteins. It is also distinct from the binding sites of other intracellular F-actin binding proteins, which generally bind to the outer surfaces of filaments (dos Remedios et al., 2003; Galkin et al., 2008b; Galkin et al., 2011; Holmes et al., 2003; Thompson et al., 2014) (Figure 7). The fact that the DNGR-1 binding site is distinctive means that it is less likely to be encumbered by other actin-binding proteins that might decorate actin filaments, which is in line with the role of DNGR-1 as a general sensor of cell damage. However, the latter function appears at odds with the rather modest affinity of the DNGR-1 CTLD for F-actin. Notably, we have uncovered a key role for avidity in the DNGR-1 : F-actin interaction that helps explain why DNGR-1 can bind tightly to its ligand under physiological conditions. The avidity component has been noted for other lectin receptors recognizing multivalent polysaccharide ligands, ranging from bacterial toxins (Kitov et al., 2000) to virus capsid glycoproteins (Connor et al., 1994) and can lead to an affinity increase of as much as six orders of magnitude (Collins and Paulson, 2004). DNGR-1 avidity is further favored by the homo-dimeric nature of the receptor, whereby binding of one CTLD facilitates binding by the second (data not shown). The importance of avidity in ligand binding has been noted for the T and B cell receptors, demonstrating a parallel between receptors of the innate and adaptive immune systems.
Figure 7. Distinct DNGR-1 footprint on F-actin.
Schematic depiction of binding footprints for canonical F-actin binding proteins on F-actin compared with the footprint of the DNGR-1 CTLD.
It has been reported that DNGR-1 binds to F-actin that is complexed to the calponin homology-based actin binding domains of other cytoskeletal molecules (Zhang et al., 2012). Our data do not support the notion that the DNGR-1 binding site is specific to complexed F-actin. Nevertheless, it is conceivable that calponin homology-based actin binding proteins such as α-actinin and β-spectrin could contribute to the strength of DNGR-1 binding by crosslinking F-actin and facilitating receptor bridging of two filaments (Ahrens et al., 2012). Whether neck region flexibility is sufficient to allow the second CTLD to bind in trans to other actin filaments remains to be determined. At least under the conditions of our study, it does not appear to be the case, as we did not observe any sign of F-actin bundling upon incubation with the DNGR-1 ECD (Figure 1A). This is not to say that actin binding proteins cannot facilitate or impair DNGR-1 binding indirectly by altering the conformation of the filaments. Similarly, it is worth noting that some of the actin residues at, or near the interaction surfaces that we have mapped, are amenable to post-translational modifications including ubiquitinylation, phosphorylation and nitration (Terman and Kashina, 2013). It is therefore intriguing to speculate that decoration of F-actin by actin binding proteins or post-translational modifications of actin taking place as a result of cellular metabolism or cell death modality could contribute to obscuring or revealing the DNGR-1 binding site, thereby modulating immune responses to dead cells.
Experimental Procedures
Mice
Clec9aegfp/egfp mice (Sancho et al., 2009) were backcrossed more than 10 times to C57BL/6J-Crl and bred at CNIC in specific pathogen-free conditions. All animal experiments were performed in accordance with national and institutional guidelines for animal care and were approved by the CNIC Ethical Committee for Animal Welfare and by the Spanish Ministry for Rural and Marine Environment.
DNGR-1 ECD proteins
DNA coding for WT or mutant mouse DNGR-1 extracellular domain (ECD; K57 – I264) was cloned into p3xFLAG-CMV-9 expression vector (Sigma-Aldrich) and all the constructs were verified by sequencing. The proteins were expressed by transient transfection in 293F cells as described previously (Ahrens et al., 2012). All proteins were tagged with 3xFLAG tag at their N terminus and purified using a combination of affinity chromatography on M2 anti-FLAG gel matrix (Sigma-Aldrich) with 3xFLAG peptide elution and size-exclusion chromatography on Superdex S200 column (GE-Healthcare).
Actin and actin-binding proteins
Human platelet actin, biotinylated rabbit muscle actin, rabbit skeletal muscle α-actinin and human recombinant plasma gelsolin were purchased from Cytoskeleton. Before each experiment, lyophilized actin was reconstituted in G-buffer (5 mM Tris-HCl, pH 8.0 + 0.2 mM CaCl2). Polymerization was carried out in F-buffer (10 mM Tris-HCl, pH 7.5 + 50 mM KCl + 2 mM MgCl2 + 1 mM ATP).
Cryo-electron microscopy and analysis
Human platelet actin was polymerized in a 30 μl solution of 25 mM Hepes buffer (pH 7.5), 100 mM KCl, 1 mM MgCl2 and 1 mM ATP. F-actin and DNGR-1 were mixed in final concentrations of 1.8 μM and 7.6 μM, respectively. A 2.4 μl aliquot was applied onto a holey carbon molybdenum EM grid (R0.6/1.0, Quantifoil), blotted and plunge-frozen into liquid ethane by Vitrobot (FEI). The grid was observed at temperatures of ~80 K using a JEOL JEM3200FSC electron cryomicroscope equipped with an Ω-type energy filter and a field-emission electron gun operated at 200 kV. Zero energy-loss images, with a slit setting to remove electrons of an energy-loss larger than 10 eV, were recorded on a 4k × 4k 15 μm/pixel slow-scan CCD camera (TemCam-F415MP, TVIPS) at a magnification of 109,489× (1.37 Å/pixel), a defocus range of 1.0 – 2.0 μm and an electron dose of ~20 electrons/Å2. Helical image analysis and 3D image reconstruction was carried out as previously described (Fujii et al., 2010) by using the iterative helical real-space refine method (Egelman, 2000). The resolution of the reconstructed 3D image was determined by the Fourier shell correlation (FSC) method, in which the filament images were randomly divided into two sets, image analysis was carried out independently for the two sets of data to produce two 3D images, and the FSC was calculated from these two 3D images.
DNGR-1 mutagenesis
Mutants were prepared using QuikChange II XL kit (Agilent Technologies) or QuikChange Lightning kit (Agilent Technologies) with the following primers:
5’-GACTGCAGCCCTTGTCCAGCCAACTGGATTCAGAATGG-3’ for H139A,
5’-GCAGCCCTTGTCCACACGCCTGGATTCAGAATGGAAAAAG-3’ for N140A,
5’-GCAGCCCTTGTCCACACAACGCCATTCAGAATGGAAAAAGTTG-3’ for W141A,
5’-GATTCAGAATGGAAAAAGTTGTTACGCCGTCTTTGAACGCTGGGAAATGTGG-3’ for Y150A,
5’-GGAAAAAGTTGTTACTATGTCTTTGCCCGCTGGGAAATGTGGAACATCAG-3’ for E153A,
5’-GTTGTTACTATGTCTTTGAACGCGCCGAAATGTGGAACATCAGTAAGAAG-3’ for W155A,
5’-CATCAGTAAGAAGAGCTGTTTAGCCGAGGGCGCTAGTCTCTTTC-3’ for K167A,
5’-CATCAGTAAGAAGAGCTGTTTAAAAGCCGGCGCTAGTCTCTTTCAAATAGAC-3’ for E168A,
5’-GAAGAAATGGAGTTCATCAGCAGTATAAGGAAACTCAAAGGAGGAAATAAATATTGGG-3’ for G188R,
5’-GGAGTTCATCAGCAGTATAGGGGCCCTCAAAGGAGGAAATAAATATTGGG-3’ for K189A,
5’-CATCAGCAGTATAGGGAAACTCGCCGGAGGAAATAAATATTGGGTG-3’ for K191A,
5’-GGAGTTCATCAGCAGTATAGGGGCCCTCGCCGGAGGAAATAAATATTGGG-3’ for K189A K191A,
5’-GGGAAACTCAAAGGAGGAAATGCCTATTGGGTGGGAGTGTTTC-3’ for K195A,
5’-GACTTGTTGCCAGCAGCGAGACAGCGATCAGCCGGCC-3’ for E225A,
5’-GACTTGTTGCCAGCAGAAGCGCAGCGATCAGCCGGCC-3’ for R226A,
5’-GACTTGTTGCCAGCAGAAAGAGCCGCCTCAGCCGGCCAGATCTGTGG-3’ for Q227A R228A
5’-GTTGCCAGCAGAAAGACAGCGAGCCGCCGGCCAGATCTGTGGATACC-3’ for S229A
5’-GCAGAAAGACAGCGATCAGCCGGCGCCATCTGTGGATACCTCAAAGATTC-3’ for Q232A
5’-CTGAGATGAGAGTAGAATCGGCGAGGTATCCACAGATCTGGCC-3’ for K238A,
5’-CTCACAGATAAAATATTTCCAGCTGGCGCACTTATCTGAGATGAGAGTAGAATC-3’ for D248A,
5’-CATCTCAGATAAGTGCGATAGCGCCAAATATTTTATCTGTGAGAAGAAGGC-3’ for W250A,
5’-CTCAGATAAGTGCGATAGCTGGGCCTATTTTATCTGTGAGAAGAAGG-3’ for K251.
5’-GAATCTTTGAGGTATCCACAGATGGCGGCGGCGGCGGCGGCGGCTGCTGGCA
ACAAGTCAGAGAG-3’ for ΔLoop
All constructs were verified by sequencing.
Dot Blot
F-actin stabilized with 5 μM phalloidin (Life Technologies) was spotted onto a nitrocellulose membrane (Whatman) pre-soaked in PBS in twofold dilution series starting at 10 μg/ml. The membrane was blocked in 5% milk in PBS + 0.05% Tween-20 overnight and incubated with supernatants containing relevant proteins. Binding of DNGR-1 WT or mutants was tested using HRP conjugated M2 anti-FLAG antibody (Sigma-Aldrich). Signal was revealed by SuperSignal West Pico Chemiluminescent substrate (Thermo Fisher). Signal in blots was quantified using ImageJ software and compared to WT. Each experiment was repeated 3 times and statistical analysis (one-way ANOVA with Dunnett’s multiple comparisons test) was performed in GraphPad Prism version 6.0 for Mac OS X (GraphPad Software).
Dead cell staining
HeLa cells were UV-irradiated (UVC 240 mJ/cm2) and left overnight to undergo secondary necrosis. Following incubation, the cells were recovered, washed in FACS buffer and stained with supernatants containing a fixed concentration of WT or mutant DNGR-1 proteins. Binding of DNGR-1 was revealed by Cy3 anti-FLAG antibody (Sigma-Aldrich) and data were acquired on LSR Fortessa flow cytometer (BD Biosciences). DAPI was used to discriminate dead cells, and strength of DNGR-1 staining was compared to cells stained with secondary antibody only. Data were analyzed using FlowJo version 9.6.2 software (Treestar). To compare between mutant and WT proteins, binding index was calculated as the mean fluorescence intensity of DNGR-1+ cells multiplied by their frequency in the DAPI+ population and normalized to WT. Each experiment was repeated 3 times and statistical analysis (one-way ANOVA with Dunnett’s multiple comparisons test) was performed in GraphPad Prism version 6.0 for Mac OS X (GraphPad Software).
Pelleting assay
2 μg of F-actin in 10 μl of F-buffer was mixed with different amounts of purified WT or mutant DNGR-1 ECD proteins (2-fold dilution series starting at 5 μg) in 30 μl of PBS. As a control, 5 μg of DNGR-1 ECD was mixed with F-buffer only. Mixtures were incubated at room temperature for 1 hour and pelleted at 120 000 x g for 90 minutes. Supernatants and pellets were separated by SDS-PAGE and immunoblotted with HRP-conjugated M2 anti-FLAG antibody (Sigma-Aldrich). Signal was revealed using the SuperSignal West Pico Chemiluminescent substrate (Thermo Fisher).
Biolayer Interferometry
BLI experiments were carried out using Octet RED96 System (ForteBio). For experiments with DNGR-1 ECD in solution, streptavidin (SA) biosensors (ForteBio) were pre-wetted in water for 20 minutes and equilibrated in F-buffer for 30 seconds. Biotinylated and non-biotinylated G-actin were mixed in 1:4 ratio, diluted to final concentration of 1μM in 200μl of F-buffer and allowed to polymerize directly on the SA biosensors. Free binding sites on SA were blocked using 10μg/ml biocytin (Sigma-Aldrich) in F-buffer + 0.1% (w/v) BSA + 0.02% (v/v) Tween-20. Baseline was determined in the interaction buffer (F-buffer + 0.1% (w/v) BSA + 0.02% (v/v) Tween-20) and association with DNGR-1 WT or mutants at various concentrations (2.5 μM, 1 μM, 0.4 μM, 0.167 μM and 0.064 μM) was monitored for 300s. Dissociation was monitored for 900s. Data analysis was performed in Data Analysis 4.1 software (ForteBio). Non-linear regression (curve fit, one site – specific binding) for the steady state analysis was performed using GraphPad Prism version 6.0 for Mac OS X (GraphPad Software). The experiment was repeated 7 times for WT and at least 2 times for each of the tested mutant proteins. For experiments with F-actin in solution, 23 μM actin was polymerized in the presence of 0.23 μM gelsolin (ratio 100:1) to obtain short F-actin filaments because large filaments caused artifacts in the BLI signal. Anti-FLAG (FLG) biosensors (ForteBio) were pre-wetted in water for 20 minutes and equilibrated in PBS for 60 seconds. 3xFLAG-tagged DNGR-1 ECD was diluted to 0.15 μM in PBS and immobilized on the sensor for 180 seconds. Baseline was determined in interaction buffer (F-buffer + 0.1% (w/v) BSA + 0.02% (v/v) Tween-20) containing 5 μM phalloidin. Association of 1µM short F-actin filaments was monitored for 600 seconds in phalloidin-free interaction buffer. Dissociation phase was carried out for 900 seconds in interaction buffer with 5 μM phalloidin to prevent depolymerization of F-actin bound to DNGR-1. The experiment was repeated 6 times.
Cells
All cells were cultured in RPMI 1640 medium supplemented with glutamine, penicillin, streptomycin, 2-mercaptoethanol (all from Invitrogen), and 10% heat-inactivated fetal calf serum (Source BioScience). For Flt3L DC cultures, medium was additionally supplemented with 150 ng/ml Flt3L (R&D Systems). For the B3Z-Syk reporter experiments, the cells were switched to AIM-V medium (Invitrogen).
Internalization assay
Full-length DNGR-1 WT and mutant constructs were cloned into pMSCV retroviral vector, verified by sequencing and used to produce retroviral vectors. Phoenix cells retrovirally transduced with WT or mutant DNGR-1 constructs were treated with 1 μM F-actin, F-buffer, 5 μg/ml anti-DNGR-1 (clone 7H11) or 5 μg/ml isotype control antibody, and incubated at 37°C for the indicated times. The cells were washed in ice-cold 5mM EDTA, harvested on ice and fixed in 2% formaldehyde. Fixed cells were washed twice in FACS buffer (5mM EDTA, 1% FCS, 0.0125% NaN3) and surface-stained with PE-conjugated anti-DNGR-1 antibody (clone 1F6). Staining of membrane DNGR-1 was analyzed using LSR Fortessa flow cytometer (BD Biosciences). Data analysis was carried out in FlowJo version 9.6.2 software (Treestar). The extent of internalization was determined by comparing the Median Fluorescence Intensity (MFI) of F-actin treated samples with F-buffer treated samples, and anti-DNGR-1 treated samples with isotype control treated samples. The data were normalized to the maximal internalization induced by antibody treatment by using formula (MFI (F-buffer) – MFI (F-actin)) / (MFI (isotype control) – MFI (anti-DNGR-1)) and plotted relative to the internalization of WT. The experiment was repeated 6 times and statistical analysis (one-way ANOVA with Dunnett’s multiple comparisons test) was performed in GraphPad Prism version 6.0 for Mac OS X (GraphPad Software). To check if 7H11 binding interfered with 1F6 binding, the cells were treated with 7H11 and incubated on ice to prevent internalization before being fixed and analyzed as described above.
B3Z-Syk reporter assay
The DNGR-1 reporter assay was described previously (Sancho et al., 2009). Briefly, 1x105 B3Z-Syk reporter cells transduced with WT or indicated mutant DNGR-1 proteins were incubated with previously UV-irradiated (UVC, 480 mJ/cm2) HeLa cells at the indicated ratios, or with plate-bound anti-DNGR-1 antibody (clone 7H11), or in medium alone for 16 – 18 hours in a 96-well plate at 37°C and 5% CO2. Following incubation, the cells were washed in PBS and LacZ activity was determined by lysing the cells in CPRG (Roche)-containing buffer and measuring O.D. 595 (using O.D. 655 as a reference) at multiple timepoints. Each experiment was repeated 3 times with all treatments in duplicate. The mean response to plate-bound antibody in each experiment was taken as a reference to which responses to other treatments were normalized. Normalized data were plotted as mean ± s.d. using GraphPad Prism version 6.0 for Mac OS X (GraphPad Software).
Antigen presentation assays
Mouse Flt3L DCs were generated by culturing BM cells in complete RPMI 1640 medium (with 5mM glutamine, penicillin, streptomycin, 2-mercaptoethanol (all from Invitrogen) and 10% heat-inactivated fetal bovine serum (Sigma)) in the presence of 150 ng/ml Flt3L (R&D) as previously described (Iborra et al., 2012). For generation of retrovirally transduced Flt3L DCs, bone marrow cells from DNGR-1-deficient mice were seeded in 6-well plates (2x106 cells per well) and spin-infected with supernatants containing pMSCV-based retroviral vectors encoding GFP and WT or mutant DNGR-1 on days 0, 1 and 2. Flt3L 150 ng/ml (R&D) was added to cultures on day 0 and 3. GFP and DNGR-1 expression was assessed at day 8 and routinely indicated a 50% transduction efficiency. On day 9, most cells had a typical DC morphology and a CD8α-like phenotype (MHC class II+, CD11c+, CD24hi, CD11blo, B220-) and were harvested and used for antigen presentation studies. Sources of antigen were RAW264.7 cells (cultured in DMEM-based complete medium) infected with rVACV OVA (kind gift from Jonathan W. Yewdell and Jack R. Bennink, National Institutes of Health, Bethesda, MD) for 4h and then treated with UVC (250 mJ/cm2) to inactivate the virus (RAW-VACV-UV) or left non-irradiated (RAW-VACV) and further cultured for 16h before use, as described (Iborra et al 2012). Alternatively, H-2bm1 fibroblasts expressing OVA (Sancho et al., 2009) irradiated and cultured as above were used. Responder T cells were either CD8+ T cells purified from spleens of mice infected intradermally 7d before with rVACV OVA or CD8+ T cell lines specific for OVA. DCs, antigen sources and responder T cells were co-cultured for 6h in complete medium additionally supplemented with non-essential amino acids, hepes and sodium-pyruvate (Invitrogen). Brefeldin A (Sigma, 5 μg/ml) added for the last 4h of culture and cells were then stained with PE-anti-CD8α, fixed with 4% PFA and incubated with APC-anti-IFN-γ during permeabilization with 0.1% saponin. An average of 5,000 T cells was analyzed in each sample to assess % positive for IFN-γ.
Supplementary Material
Acknowledgements
We thank all members of the Immunobiology Laboratory for helpful discussions and suggestions. We thank the Flow Cytometry, Equipment Park and Structural Biology Science Technology Platforms for assistance. This work was funded by Cancer Research UK, a prize from Fondation Bettencourt-Schueller and a grant from the European Research Council (ERC Advanced Researcher Grant AdG- 2010-268670) to CRS, and by JSPS KAKENHI Grant number 25000013 to KN and 25711010 to TF. Work in the DS laboratory is funded by the CNIC and grants from the Spanish Ministry of Economy and Competitiveness (SAF-2013-42920R) and the European Research Council (ERC Starting Independent Researcher Grant StG-2010-260414).
Footnotes
Accession numbers
The reconstructed density was deposited to Electron Microscopy Data Bank with accession code EMD-6102 and the atomic coordinate to Protein Data Bank with RCSB ID code rcsb160368 and PDB ID code 3J82.
Author Contributions
P.H. produced all DNGR-1 proteins, analyzed their binding to F-actin and carried out the internalization and reporter assays. T.F. and K.N. generated and analyzed electron cryomicroscopy data with assistance from Y.Y. and performed protein docking. S.I. and D.S. carried out all antigen presentation experiments. J.H., O.S., S.A., S.K., M.W. and D.S. provided advice and contributed to design of experiments. P.H. and C.R.S. designed the study, analyzed data and wrote the manuscript with assistance from K.N. and D.S.
References
- Abdiche Y, Malashock D, Pinkerton A, Pons J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Analytical biochemistry. 2008;377:209–217. doi: 10.1016/j.ab.2008.03.035. [DOI] [PubMed] [Google Scholar]
- Ahrens S, Zelenay S, Sancho D, Hanc P, Kjaer S, Feest C, Fletcher G, Durkin C, Postigo A, Skehel M, et al. F-actin is an evolutionarily conserved damage-associated molecular pattern recognized by DNGR-1, a receptor for dead cells. Immunity. 2012;36:635–645. doi: 10.1016/j.immuni.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Caminschi I, Proietto AI, Ahmet F, Kitsoulis S, Shin Teh J, Lo JC, Rizzitelli A, Wu L, Vremec D, van Dommelen SL, et al. The dendritic cell subtype-restricted C-type lectin Clec9A is a target for vaccine enhancement. Blood. 2008;112:3264–3273. doi: 10.1182/blood-2008-05-155176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins BE, Paulson JC. Cell surface biology mediated by low affinity multivalent protein-glycan interactions. Current opinion in chemical biology. 2004;8:617–625. doi: 10.1016/j.cbpa.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiological reviews. 2003;83:433–473. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Brieher W, Kueh HY, Mitchison TJ, Egelman EH. Coronin-1A stabilizes F-actin by bridging adjacent actin protomers and stapling opposite strands of the actin filament. Journal of molecular biology. 2008a;376:607–613. doi: 10.1016/j.jmb.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Cherepanova O, Lebart MC, Egelman EH. High-resolution cryo-EM structure of the F-actin-fimbrin/plastin ABD2 complex. Proceedings of the National Academy of Sciences of the United States of America. 2008b;105:1494–1498. doi: 10.1073/pnas.0708667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Kudryashov DS, Solodukhin A, Reisler E, Schroder GF, Egelman EH. Remodeling of actin filaments by ADF/cofilin proteins. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20568–20572. doi: 10.1073/pnas.1110109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge P, Durer ZA, Kudryashov D, Zhou ZH, Reisler E. Cryo-EM reveals different coronin binding modes for ADP- and ADP-BeFx actin filaments. Nature structural & molecular biology. 2014;21:1075–1081. doi: 10.1038/nsmb.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KC, Angert I, Kull FJ, Jahn W, Schroder RR. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature. 2003;425:423–427. doi: 10.1038/nature02005. [DOI] [PubMed] [Google Scholar]
- Huysamen C, Willment JA, Dennehy KM, Brown GD. CLEC9A is a novel activation C-type lectin-like receptor expressed on BDCA3+ dendritic cells and a subset of monocytes. The Journal of biological chemistry. 2008;283:16693–16701. doi: 10.1074/jbc.M709923200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra S, Izquierdo HM, Martinez-Lopez M, Blanco-Menendez N, Reis e Sousa C, Sancho D. The DC receptor DNGR-1 mediates cross-priming of CTLs during vaccinia virus infection in mice. The Journal of clinical investigation. 2012;122:1628–1643. doi: 10.1172/JCI60660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitov PI, Sadowska JM, Mulvey G, Armstrong GD, Ling H, Pannu NS, Read RJ, Bundle DR. Shiga-like toxins are neutralized by tailored multivalent carbohydrate ligands. Nature. 2000;403:669–672. doi: 10.1038/35001095. [DOI] [PubMed] [Google Scholar]
- Lilic M, Galkin VE, Orlova A, VanLoock MS, Egelman EH, Stebbins CE. Salmonella SipA polymerizes actin by stapling filaments with nonglobular protein arms. Science. 2003;301:1918–1921. doi: 10.1126/science.1088433. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. The Journal of cell biology. 1986;103:2747–2754. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin LF, Reyal Y, Uronen-Hansson H, Schraml BU, Sancho D, Murphy KM, Hakansson UK, Moita LF, Agace WW, Bonnet D, Reis e Sousa C. DNGR-1 is a specific and universal marker of mouse and human Batf3-dependent dendritic cells in lymphoid and nonlymphoid tissues. Blood. 2012;119:6052–6062. doi: 10.1182/blood-2012-01-406967. [DOI] [PubMed] [Google Scholar]
- Poulin LF, Salio M, Griessinger E, Anjos-Afonso F, Craciun L, Chen JL, Keller AM, Joffre O, Zelenay S, Nye E, et al. Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8alpha+ dendritic cells. The Journal of experimental medicine. 2010;207:1261–1271. doi: 10.1084/jem.20092618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouiller I, Xu XP, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, Hanein D. The structural basis of actin filament branching by the Arp2/3 complex. The Journal of cell biology. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho D, Joffre OP, Keller AM, Rogers NC, Martinez D, Hernanz-Falcon P, Rosewell I, Reis e Sousa C. Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature. 2009;458:899–903. doi: 10.1038/nature07750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho D, Mourao-Sa D, Joffre OP, Schulz O, Rogers NC, Pennington DJ, Carlyle JR, Reis e Sousa C. Tumor therapy in mice via antigen targeting to a novel, DC-restricted C-type lectin. The Journal of clinical investigation. 2008;118:2098–2110. doi: 10.1172/JCI34584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terman JR, Kashina A. Post-translational modification and regulation of actin. Current opinion in cell biology. 2013;25:30–38. doi: 10.1016/j.ceb.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Tolbert CE, Shen K, Kota P, Palmer SM, Plevock KM, Orlova A, Galkin VE, Burridge K, Egelman EH, et al. Identification of an actin binding surface on vinculin that mediates mechanical cell and focal adhesion properties. Structure. 2014;22:697–706. doi: 10.1016/j.str.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topf M, Lasker K, Webb B, Wolfson H, Chiu W, Sali A. Protein structure fitting and refinement guided by cryo-EM density. Structure. 2008;16:295–307. doi: 10.1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachsstock DH, Schwartz WH, Pollard TD. Affinity of alpha-actinin for actin determines the structure and mechanical properties of actin filament gels. Biophysical journa. 1993;l65:205–214. doi: 10.1016/S0006-3495(93)81059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenay S, Keller AM, Whitney PG, Schraml BU, Deddouche S, Rogers NC, Schulz O, Sancho D, Reis e Sousa C. The dendritic cell receptor DNGR-1 controls endocytic handling of necrotic cell antigens to favor cross-priming of CTLs in virus-infected mice. The Journal of clinical investigation. 2012;122:1615–1627. doi: 10.1172/JCI60644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenay S, Reis e Sousa C. Adaptive immunity after cell death. Trends in immunology. 2013;34:329–335. doi: 10.1016/j.it.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Zhang JG, Czabotar PE, Policheni AN, Caminschi I, Wan SS, Kitsoulis S, Tullett KM, Robin AY, Brammananth R, van Delft MF, et al. The dendritic cell receptor Clec9A binds damaged cells via exposed actin filaments. Immunity. 2012;36:646–657. doi: 10.1016/j.immuni.2012.03.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.