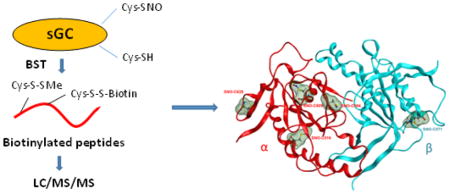
Abstract
Soluble Guanylyl Cyclase (sGC) is the main receptor for nitric oxide (NO). NO activates sGC to synthesize cGMP, triggering a plethora of signals. Recently, we discovered that NO covalently modifies select sGC cysteines via a post-translational modification termed S-nitrosation or S-nitrosylation. Earlier characterization was conducted on a purified sGC treated with S-nitrosoglutathione, and identified three S-nitrosated cysteines (SNO-Cys). Here we describe a more biologically relevant mapping of sGC SNO-Cys in cells to better understand the multi-faceted interactions between SNO and sGC. Since SNO-Cys are labile during LC/MS/MS, MS analysis of nitrosation typically occurs after a biotin switch reaction, in which a SNO-Cys is converted to a biotin-Cys. Here we report the identification of ten sGC SNO-Cys in rat neonatal cardiomyocytes using an Orbitrap MS. A majority of the SNO-Cys identified is located at the solvent-exposed surface of the sGC, and half of them in the conserved catalytic domain, suggesting biological significance. These findings provide a solid basis for future studies of the regulations and functions of diverse sGC S-nitrosation events in cells.
Keywords: Soluble guanylyl cyclase, S-nitrosation, Biotin switch, Tandem mass spectrometry
Graphical Abstract
1. Introduction
Nitric oxide (NO) signaling is a key modulator of the functions of different cells, particularly the ones involved in cardiovascular and neuronal function. NO stimulates the activity of sGC to produce cGMP, which in turn supports synaptic plasticity, relaxes smooth muscle cells (SMC) and protects from cardiac hypertrophy. Of note, sGC can produce cGMP only as an αβ heterodimer, as its catalytic domain is formed by the association of the C-terminal α and β subunits. sGC isoforms most commonly expressed are α1 and β1 subunits. NO also modulates the signaling and redox environment via S-nitrosation, a post-translational modification (PTM) of cysteines (Cys) that modifies protein localization, activity and interactions [1]. S-nitrosation was previously described as a non-enzymatic process, but there is now solid evidence that some proteins can catalyze specific transnitrosation and/or denitrosation of target proteins via protein-protein interactions [2, 3]. This emerging role of specific protein-protein interaction-driven transnitrosation is apparent in critical cellular events such as regulation of cell death (transnitrosation from thioredoxin to caspase 3, CASP3), of ubiquitination (transnitrosation between CASP3 and XIAP) and nuclear translocation (transnitrosation between GAPDH and GOSPEL) [4, 5]. Thus, dysfunction of protein-protein transnitrosation could lead to apoptosis [6] and mitochondrial dysfunction in neurodegenerative diseases.
We have previously shown that sGC, which contains more than thirty conserved Cys in its α and β subunits, is readily, yet specifically, S-nitrosated in vitro [7], and in vivo in an Angiotensin II-induced hypertensive model and during development of nitrate tolerance[8, 9]. After MALDI-TOF-TOF MS analysis of the chemical S-nitrosation of sGC purified from bovine lungs and treated with S-nitrosoglutathione (GSNO), we initially identified three sGC SNO-Cys sites, αC243 and αC516 in the α subunit and βC122 in the β subunit. Using site-directed mutagenesis, these three Cys were shown to be involved in desensitization of sGC activity to NO stimulation [7–9].
Due to SNO-Cys instability, indirect biotin switch techniques (BST) are traditionally used to identify nitrosated peptides [10]. BST indirectly detects SNO-Cys through ascorbate (Asc) reduction of SNO-Cys into free thiols for subsequent biotinylation, avidin affinity enrichment and LC/MS/MS detection. BST specificity is predicated on SNO Asc-reduction specificity and multiple controls are necessary [11–13]. Herein, we describe the more in-depth identification of nine novel SNO-Cys from rat neonatal cardiomyocytes overexpressing sGC via adenoviral infection, using a combination of the classic BST approach with the highly sensitive Orbitrap tandem mass spectrometry. We also built a model of the catalytic domain of sGC harboring SNO modified Cys. Newly discovered SNO-Cys are conserved among the mammalian species and are mostly exposed at the surface of sGC, in particular in the catalytic domain. This observation suggests that SNO-Cys in the catalytic domain could modulate sGC activity hence the NO-cGMP pathway. Since sGC SNO-Cys are solvent exposed, they could be involved in specific protein-protein interactions and possibly transnitrosation reactions to modulate other signaling pathways. Thus identifying sGC, the main NO receptor, to be itself extensively and specifically S-nitrosated in cells provides a strong rationale for further biological studies to investigate the functions of SNO-Cys in sGC.
2. Materials and methods
2.1. Materials
Phenylmethylsulfonyl fluoride (PMSF), methyl methanethiosulfonate (MMTS), protease inhibitor cocktail, copper (I) chloride, formic acid were purchased from Sigma (St. Louis, MO). N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide (Biotin-HPDP), EDTA, HEPES, C18 spin columns, trypsin and ammonium bicarbonate were purchased from Fisher Scientific (Fair Lawn, NJ). SDS was purchased from BioRad (Hercules, CA). Acetonitrile (ACN) and water were purchased from J. T. Baker Inc., (Center Valley, PA). The ICAT avidin enrichment kit was purchased from AB Sciex (Framingham, MA).
2.2. Cell culture
Neonatal cardiomyocytes (NCM) were isolated from 1–2 days old control Wistar rats (Harlan Laboratories, Somerville) by Percoll gradient centrifugation and plated overnight in cardiomyocyte culture medium with 5% horse serum and 100 μM BrdU. Plates were coated with gelatin. The medium was modified from Dulbecco’s modified Eagle’s medium (DMEM)/F-12, supplemented with sodium pyruvate, glucose, L-ascorbic acid, bovine serum albumins, sodium selenite, sodium bicarbonate, and antibiotics. Twelve hours after seeding, the cells were transferred to the medium without BrdU or horse serum. sGC was overexpressed in NCM by infecting the cells with adenovirus constructs expressing α and β subunits with a multiplicity of infection (MOI) of 5, for 48h. All animal experimentation followed the protocol approved by the Institutional Animal Care and Use Committee of Rutgers University - New Jersey Medical School.
2.3. In situ S-nitrosation and cell lysate preparations
NCM were treated with 100 μM S-nitrosocysteine (CSNO) or with control buffer (potassium phosphate buffer to evaluate basal S-nitrosation) in the culture medium and incubated at 37 °C in 5% CO2 for 30 min. The cells were then lysed in a lysis/blocking buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 2% SDS, 0.1 mM neocuproine, 0.2 mM PMSF and 20 mM MMTS) with a protease inhibitor cocktail.
2.4. Biotin switch
Biotin switch technique (BST) was performed to identify Cys-NO in sGC using a modified protocol from Jaffrey and Snyder [10]. In brief, one mg of proteins was alkylated by MMTS in the lysis/blocking buffer at 50 °C for 30 min in the dark with regular agitation. Excess MMTS was removed by cold acetone precipitation and the protein pellets were collected by centrifuging at 5,000 g for 10 min at 4 °C and washed 3X with ice cold acetone. The protein pellets were re-suspended and the SNO-Cys were biotinylated in 900 μl of the HENS2 buffer (25 mM HEPES, pH 7.7, 1 mM EDTA and 1% SDS) plus 0.7 mM biotin-HPDP, 75 mM Asc and 7 μM copper (I) chloride at room temperature for 1 hr in the dark. Excess biotin-HPDP was removed by cold acetone precipitation (Supplemental Fig. S1). Negative controls were samples without Asc treatment.
2.5. In-solution digestion
Protein pellets were dissolved in 300 μl of a buffer containing 8M urea with 100 mM Tris, at pH 7.0. The protein concentrations were measured with the BCA assay and adjusted to 2 μg/μl. Two hundred μg of proteins was diluted 10-fold with 50 mM NH4HCO3 (pH 8.3) and digested at a protein/trypsin weight ratio of 30:1, at 37 °C overnight.
2.6. Enrichment of biotinylated peptides
The resulting peptides were diluted with the AB Sciex ICAT Affinity Buffer-Load buffer. After slowly injecting the diluted sample onto the avidin cartridge, the Affinity Buffer-Load and the Affinity Buffer-Wash 1 buffers were sequentially injected onto the cartridge to remove non-biotinylated peptides, according to the manufacturer’s instructions. Subsequently, the Affinity Buffer-Wash 2 and HPLC grade water were injected onto the cartridge to further remove the nonspecifically bound peptides. The Affinity Buffer-Elute buffer was slowly injected to elute the biotinylated peptides. The resulting biotinylated peptides were desalted with Pierce C18 spin columns, completely dried in a SpeedVac and were re-suspended in 5 μl of mobile phase A (2% ACN and 0.1% formic acid in H2O) for LC/MS/MS analysis.
2.7. Identification of biotinylated-peptides by LC/MS/MS
The enriched biotinylated-peptides were analyzed by LC/MS/MS on an LTQ-Orbitrap Velos Pro mass spectrometer (Thermo Fisher Scientific) coupled with an Ultimate 3000 Chromatography System. First, the desalted peptides were trapped on a C18 pre-column (Pepmap C18, 5 mm × 300 μm, Dionex) at 2% mobile phase B (mobile phase A, 2% ACN and 0.1% formic acid; mobile phase B, 85% ACN and 0.1% formic acid) at a flow rate of 30 μl/min. Then the peptides were separated on a 15-cm C18 PepMap100 column (75-μm capillary 3 μm, 100 Å, Dionex) using an 85-min gradient (1% to 50% B) at a flow rate of 250 nL/min. A column wash program was added after each sample separation to minimize carry-overs. The eluted peptides were introduced into the mass spectrometer through a Proxeon Nanospray Flex™ Ion Source. The spray voltage was 2.15 kV and the capillary temperature was 275 °C. The spectra were acquired in a data-dependent mode. Full scans of the MS spectra (from m/z 300–2000) were acquired in the Orbitrap analyzer at a resolution of 60,000 (at m/z 400), with the lock mass option enabled. Ten most intense peptide ions with charge states of 2 to 4 were sequentially isolated and fragmented using either collision-induced dissociation (CID) with a normalized collision energy (NCE) of 30%, or higher energy collisional dissociation (HCD) with a NCE of 28%. The ion-selection threshold was set at 3000 for the MS/MS analysis.
2.8. Protein database search and bioinformatics
In order to identify biotin modified peptides and localize the modification sites, the MS/MS spectra were searched against SwissProt rat database (downloaded on January 24, 2014; ~5,379 entries) using Mascot search engine (V.2.4.1) through Proteome Discoverer (PD) platform (V. 1.4, Thermo Scientific). The search parameters were set as following: trypsin with up to two missed cleavages; precursor mass tolerance was 10 ppm and fragment mass tolerance was 0.5 Da; methionine oxidation, cysteine thiol-methylation, cysteinylation and biotin-HPDP (428.19 Da) modification were set as variable modifications. The decoy database containing all the reverse protein sequences was used to estimate the false discovery rate. The .msf files from PD were further filtered and compiled into a list of non-redundant proteins with Scaffold (v. 4.2.1, Proteome Software, Portland, OR). The protein and peptide identifications were accepted with a false discovery rate less than 1.0% based on both the Protein Prophet and the Peptide Prophet algorithms. Proteins containing the same peptides were grouped to satisfy the principles of parsimony. Biotinylated-Cys localization in MS/MS spectra was manually inspected.
2.9. Modeling and molecular dynamics simulation of the catalytic domain of SNO-sGC
Amino acid sequences for α and β subunits for rat and human sGC were obtained from a UniProt portal (access codes: human Q02108 and Q02153; rat P19686 and P20595). Human and rat sequences of sGC are highly homologues, with 89% identical residues in α subunits and 99% in β subunits. For the catalytic domain this similarity is even higher, 96% identical residues in α and 100% in β subunits. A three-dimensional model of the rat catalytic domain of sGC was made by virtual mutation of amino acid residues in a crystal structure of the catalytic domain of human sGC (PDB 4NI2) in Molecular Operating Environment (MOE v.2014.0901) [14]. A mutated protein was optimized by energy minimization with an Amber force field in MOE. S-nitrosation of selected cysteines was performed by replacement of the S-H group with S-N=O moiety. Both wild type rat sGC catalytic domain and S-nitrosated proteins were saved in pdb file format. Antechamber subroutine from AmberTools 15 package [15] was used for parameterization of non-standard SNO-Cys residues, using AM1-BCC charge method. Wild type rat catalytic domain and S-nitrosated protein structures were further refined with Molecular Dynamics (MD) in aqueous solution. After initial equilibration and minimization the production run for 20 ns with an NPT ensemble and Langevin coupling thermostat was performed in Amber 14 molecular dynamics program [15]. Changes in the position of WT Cys and Cys-NO residues were closely monitored and recorded. Average structures of proteins calculated from Amber trajectory files over all 20 ns of MD simulations were used for visual inspection and analysis.
3. Results and Discussion
3.1. Both collision-induced dissociation (CID) and high-energy collisional dissociation (HCD) approaches are effective at the identification of biotinylated-Cys from sGC expressed in cells
We previously used primary rat aortic smooth muscle cells (expressing endogenous sGC) treated with CSNO (100 μM) and showed by BST that sGC was S-nitrosated and this correlated with sGC desensitization to NO. Of note, one hour after CSNO washout, S-nitrosation of sGC was not detectable and sensitivity to NO stimulation was restored, suggesting denitrosation. However, to identify specific SNO-cysteine (s) in sGC in an earlier study, we used a purified bovine sGC (treated with S-nitrosated glutathione, GSNO) to overcome the low cellular SNO-sGC concentrations [7]. To identify sGC SNO-Cys sites in a context that is more biologically relevant than our earlier studies of S-nitrosation of purified proteins, we infected rat NCM with adenoviruses expressing both rat sGC α and β subunits. It allows us to improve signal-to-noise ratios in detection of SNO-Cys in sGC. Infected NCM were treated with either CSNO (100 μM) or with its solvent potassium phosphate buffer. The buffer treatment represents a basal endogenous level of S-nitrosation, without NCM stressed, while CSNO was used as a nitrosating agent to enhance the detection of SNO-Cys signals. After BST treatments, only the biotinylated-Cys that occurred in at least 2 independent experiments are reported here (Table 1; Supplemental Figs. S2 and S3). Overall, we identified 10 biotinylated-Cys sites in this study; 5 each in α (Figs. 1 and 2; Supplemental Fig. S2a–2e) and β sGC subunit (Supplemental Fig. S3a–3e). Of note, Cys282 in sGC-α and Cys 174, 571 in sGC-β were also potentially S-cysteinylated, albeit at low Mascot scores (data not shown).
Table 1.
Biotinylated-peptides and biotinylated-Cys sites in sGC identified by LC/MS/MS.
| Protein Description |
Gene ID | Swiss-Prot Protein Accession |
Sequence | Modification | Therotical Mass |
Actual Mass |
Observed m/z |
Charges | ΔM (ppm) |
Best Mascot Score |
|---|---|---|---|---|---|---|---|---|---|---|
| sGC alpha | Gucy1a3 | P19686 | STKPSLSPGKPQSSLVIPTSLFCK | AC282 | 2929.5479 | 2929.5427 | 977.52 | 3+ | −1.7 | 59 |
| FDQQCGELDVYK | AC516 | 1871.8256 | 1871.8293 | 936.92 | 2+ | 1.5 | 52 | |||
| YCLFGNNVTLANK | AC594 | 1883.9096 | 1883.9117 | 942.96 | 2+ | 2.4 | 68 | |||
| FESCSVPR | AC609 | 1351.6087 | 1351.608 | 676.81 | 2+ | −1.7 | 38 | |||
| DCPGFVFTPR | AC628 | 1565.7193 | 1565.7209 | 783.87 | 2+ | −0.5 | 58 | |||
| sGC beta | Gucy1b3 | P20595 | SEECDHTQFLIEEK | BC174 | 2134.9373 | 2134.94 | 1068.48 | 2+ | 1.3 | 53 |
| ISPYTFCK | BC214 | 1385.6546 | 1385.6555 | 693.84 | 2+ | 0.7 | 29 | |||
| DLVVTQCGNAIYR | BC232 | 1878.9154 | 1878.9152 | 940.46 | 2+ | −0.1 | 42 | |||
| YCLFGNTVNLTSR | BC541 | 1914.9154 | 1914.9172 | 958.47 | 2+ | 0.9 | 46 | |||
| CLMSPENSDPQFHLEHR | BC571 | 2467.0905 | 2467.092 | 823.37 | 3+ | −0.3 | 36 |
Biotinylated-Vys609 was identified in both endogenous and CSNO treated sGC
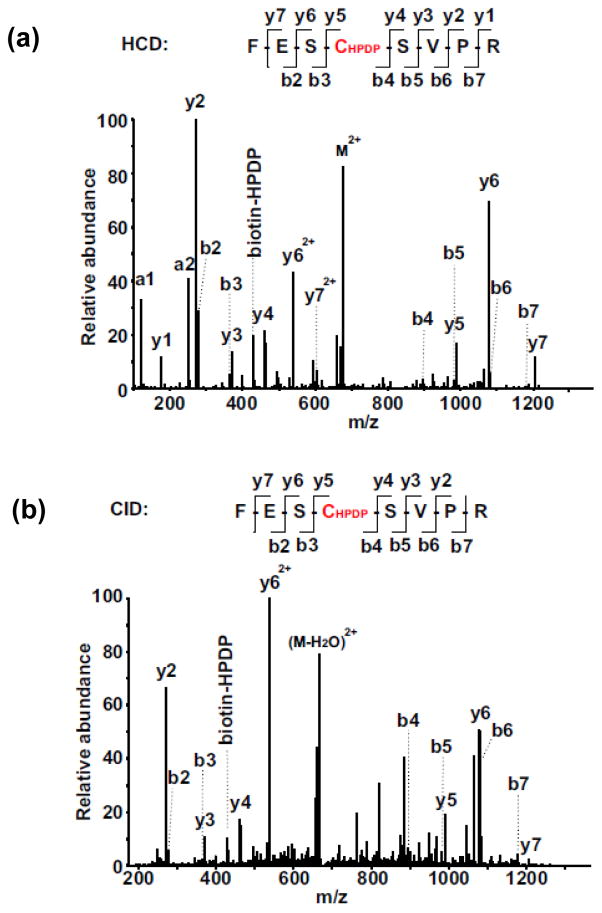
Fig. 1. Identification of biotinylated-Cys609 in FESCSVPR of sGC α subunit by LC/MS/MS.
(a) HCD spectrum of a doubly-charged ion at m/z 676.80. (b) CID spectrum of a doubly-charged ion at m/z 676.81. The biotinylated-Cys site was located on Cys609 in sGC α subunit. Both spectra contain almost complete series of the y+ and b+ ions with the biotinylated-Cys (+428.19 at Cys) found between b3 and b4, as well as between y4 and y5 ions. More y+ ions were observed from the HCD spectrum compared to the CID spectrum, due to the superior capability of HCD to detect low mass fragments.
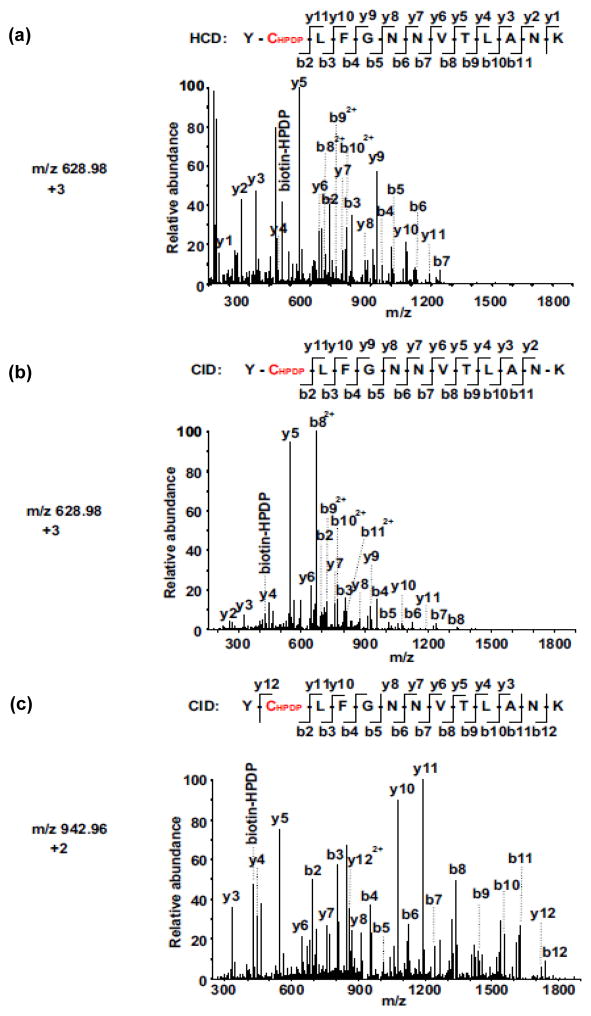
Fig. 2. Comparison of CID vs. HCD MS/MS spectra for the identification of biotinylated-Cys594 in YCLFGNNVTLANK of sGC α subunit.
(a) HCD MS/MS spectrum of the triply-charged biotinylated-peptide ion at m/z 628.97. (b) CID MS/MS spectrum of the triply-charged biotinylated-peptide ion at m/z 628.98. (c) CID MS/MS spectrum of the doubly-charged biotinylated-peptide ion at m/z 942.96. The SNO-Cys site was located on Cys594 in sGC α subunit. Both spectra (a and b) contain almost complete series of the y+ and b+ ions with the biotinylated-Cys (+428.19 at Cys) found between b1 and b2, as well as between y11 and y12 ions. The MS/MS fragmentation of the doubly-charged biotinylated-peptide ion was not observed by HCD mode. More y+ ions were observed from the HCD spectrum compared to the CID spectrum, due to the superior capability of HCD to detect low mass fragments.
We employed a highly sensitive MS approach for the detection of SNO-Cys. SNO-Cys is labile to neutral loss during both ESI and CID or HCD reactions [16, 17]. Such neutral loss gives rise to S-Cys radical-containing ions that are usually substantially more abundant than the peptide backbone ions in MS/MS spectra [17, 18], resulting in insufficient fragment ions for the localization of SNO-Cys sites. Therefore, an indirect approach that replaces SNO-Cys with a biotinylated-Cys is widely used as a surrogate method to the localization of SNO-Cys sites from biological samples. The BST was developed by Jaffrey et. al. to detect SNO-Cys (Supplemental Fig. S1). Key features include first, blockage of free thiols by MMTS (Step (a) in Supplemental Fig. S1) or N-ethylmaleimide (NEM); second, selective reduction of SNO-Cys into free cysteines by Asc (Step (b) in Supplemental Fig. S1); and third, covalent linkages of the nascent Cys thiols with biotin derivatives, including biotin-HPDP (Step (c) in Supplemental Fig. S1), for avidin-based affinity enrichment and/or downstream biotinylated-Cys (as a proxy for SNO-Cys) detection by LC/MS/MS. BST can be prone to false identifications due to either incomplete blocking of reduced thiols or non-specific thiol exchange reactions. It is essential to include negative controls. From our Asc omission step as a negative control, no biotinylation was detected. Moreover, for all 10 sGC biotinylated-Cys sites but one (αC609), we did not observe sGC nitrosation in the cells treated with only the buffer, without CSNO (Table 1), suggesting that incomplete free thiol blockage and thiol exchange were unlikely to have occurred in this experimental setup. In addition, when we mutated one of the sGC biotinylation sites (Cys 516) that we previously discovered and confirmed in this study, we observed that sGC lost the sensitivity to NO stimulation, suggesting that the S-nitrosation of Cys516 may indeed be functionally relevant [8].
The usage of the Orbitrap Velos tandem MS in place of our previously used MALDI-TOF-TOF MS is one reason why we identified additional biotinylated-Cys sites from sGC expressed in NCM compared to the three biotinylated-Cys (αC243, αC516 and βC122) identified in the purified bovine sGC treated with GSNO. Orbitrap can perform CID and HCD fragmentations and we capitalized on this ability by using both approaches to fragment biotinylated peptide ions. Both methods are able to fragment biotinylated tryptic peptides derived from sGC without fragmenting the relatively large biotinylated Cys side chains (Fig. 1 and 2). As expected, HCD MS/MS spectra are richer in the low mass regions compared with CID spectra (Fig. 1, a and b and Fig. 2, a and b). However, since we needed to identify biotinylated-Cys of sGC in the presence of biotinylated-Cys-containing peptides of all proteins isolated from NCM, we mostly employed the faster CID approach in our LC/MS/MS workflow to take advantage of its higher detection sensitivity. The resulting CID MS/MS spectra from the diverse charge states also provided complementary and unambiguous identification of the biotinylated-Cys of the sGC peptides (see an example in Fig. 2, b and c).
3.2 The SNO-Cys in sGC are found in key regulatory regions and catalytic domains of sGC
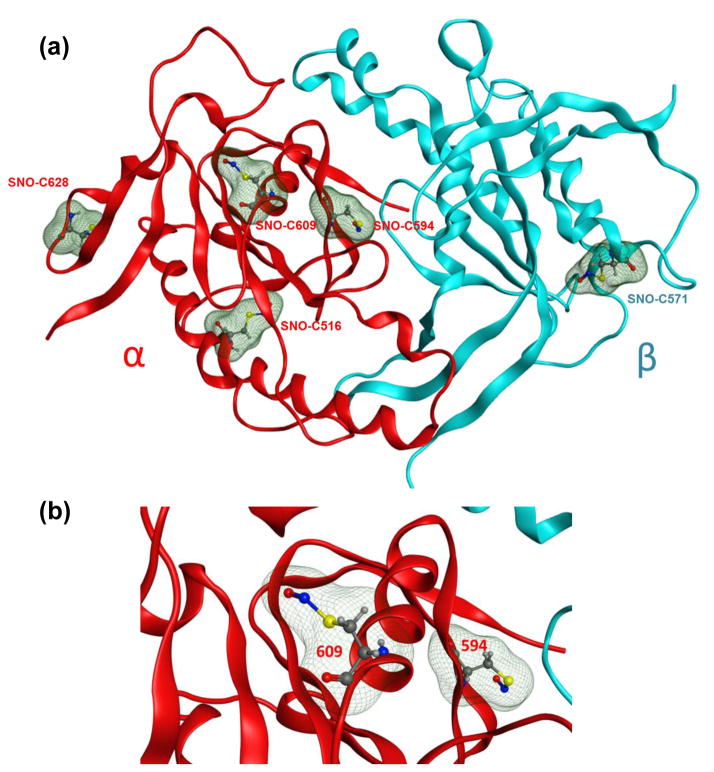
The ten identified biotinylated (SNO)-Cys are spread along the molecule with a majority in the catalytic domain formed by the association of the C-terminal parts of α and β subunits (Fig. 3a and 3b); αC516, αC594, αC609 and αC628 in the α subunit, and βC541, βC571 in the β subunit. βC541 and αC594 are key residues for the catalytic activity. βC541 is involved in the specificity of the interaction with the substrate GTP and directly located in the catalytic pocket [19], while αC594 is located in the pseudo-symmetric pocket and potentially binds inhibitory nucleotides and mediates activation of sGC by the compound YC-1 [20, 21]. Thus, S-nitrosation of these two Cys in the catalytic pocket should directly impair the catalytic activity of sGC in cells. It should be noted that βC541 is buried and thereby we could not model the SNO modification (Fig. 4). This would suggest that conformational changes are required in the catalytic site for βC541 to accommodate the PTM, yet a structural analysis of Cys S-nitrosation predicted that 35% of sulfur atoms of SNO-Cys were actually buried [22]. αC516 in the catalytic domain was one of the three original SNO-Cys sites identified in the GSNO-treated purified bovine sGC (the 2 others are αC243, and βC122). It was later shown by site-directed mutagenesis that SNO-αC516 is involved in desensitization of sGC to NO stimulation in cells treated with stress-induced angiotensin II [8]. A recent study also identified in two different families that mutation αC516 to a Tyrosine as an increased risk for the Moyamoya disease (vasculopathy), achalasia and hypertension [23]. A specific role for the three other Cys in the catalytic domain αC609, αC628 and βC571, identified as S-nitrosated in the current study, is not yet known. These three Cys are at the surface of the catalytic domain of sGC (Fig. 4). Noteworthy, αC609 and αC628 are located in the region of the catalytic domain predicted to be a regulatory surface, and a site of interaction with HNOX domain or potentially other proteins. Thus, we speculate that these two Cys could have a key regulatory role and be involved in a transnitrosation reaction of target proteins. Moreover, another group has shown that these two Cys can be modified by MMTS and could be associated with a mechanism of activation of sGC [24]. This is not surprising as a reaction with MMTS or SNO is indicative of reactive Cys. Two other reported Cys modified by MMTS, βC174 and βC214, are also found S-nitrosated in our MS analysis of sGC-infected NCM. βC174 is in the heme-binding domain, which is key for NO binding and stimulation of sGC, and βC214 together with βC232 and αC282 are located in the PAS-fold domain of sGC. This PAS-fold domain is an ancient sensory domain found also in prokaryotes that is responsible for redox sensing, and interaction with other proteins [25]. Thus, it is tempting to speculate that these Cys when S-nitrosated could regulate cellular processes via protein-protein interactions or transnitrosation reactions [26] as a function of redox environment changes. It should be noted that the surface-exposed αC609 in the catalytic domain of sGC was found S-nitrosated under basal condition and following addition of CSNO. This indicates that αC609 is endogenously S-nitrosated, which supports the idea of a physiological signaling function independent of oxidative or nitrosative stress (as one could consider that addition of CSNO to the culture medium reproduces pathophysiological conditions with excess reactive nitrogen species).
Fig. 3. Sequence of rat sGC α subunit (a) and sGC β subunit (b).
Biotinylated-Cys residues identified are in bold and colored in red. The sequences of the tryptic peptides where S-nitrosation was detected in the sGC protein by MS/MS are highlighted in yellow. α subunit: Swiss-Prot Accession No.: P19686; β subunit Swiss-Prot Accession No.: P20595.
Fig. 4. Model of the S-nitrosated catalytic domain of rat sGC.
(a) α subunit of rat sGC is shown as red ribbons and β subunit is as light blue ribbons. S-nitrosated Cys residues are depicted as balls-and-sticks with their molecular volumes added as green mesh surfaces for visual purpose. The structure presented here is an averaged structure obtained after 20 ns of MD simulation. (b) A close-up of S-nitrosated cysteine 609 and 594. The color scheme is the same as in (a).
3.3 Implications of multiple SNO-Cys in sGC
Different Cys were identified in our previous study of the purified sGC treated with GSNO [7] compared to cellular overexpressed sGC treated with CSNO. This difference could reflect different mechanisms and S-nitrosation specificities for GSNO and CSNO. In a purified protein system with addition of GSNO, the Cys site that is targeted is one that can accommodate specific docking of GSNO to go through the transnitrosation process [27]. Indeed, the specificity of S-nitrosation is apparently dependent upon the NO donor used and its concentration [22]. In addition, S-nitrosation specificity is expected to be different in a cellular context where Cys could be S-nitrosated by additional available mechanisms, in particular protein-protein driven transnitrosation or NO oxidation, which will be discussed in depth below [22, 28]. However, it is important to mention that other groups have suggested that treatment with CSNO, which could induce nitrosative stress, can induce thiol oxidation other than S-nitrosation [29–31].
The model system used in this study is biologically relevant, based on our previous studies. We previously used CSNO at 100 μM to determine the effect of S-nitrosation on sGC activity, in particular the SNO-dependent desensitization to NO stimulation [32]. In these previous studies, we also showed that S-nitrosation and sGC desensitization to NO stimulation were functions of both CSNO concentrations and time of exposure. The 10 putative SNO-Cys sites reported in this study may be on different sGC molecules with different functions. Multi-site S-nitrosation may impact sGC enzymatic activity and other biological functions by propagating different downstream signaling events with diverse interacting proteins, which may bind to distinct SNO-Cys-containing domains in the sGC molecules. The identification of 10 distinct SNO-Cys sites in this study can provide a basis for future studies to clarify whether the increased SNO signal in sGC is due to an increased number of S-nitrosated sGC molecules or an increased number of SNO sites per molecule of sGC. For such quantitative proteomics studies, one can use either ICAT [33] or iodoTMT-based BST in the place of biotin-HPDP [34], in order to obtain accurate quantitative information to distinguish Cys more sensitive to S-nitrosation than others.
In future MS experiments based on the current system (e.g. adenoviruses overexpressing sGC), we will vary both time and concentrations of CSNO treatments and other cellular stimuli to assay which of the 10 Cys identified in the current study are more prone to S-nitrosation under more biologically relevant setups. These 10 sites may be modified in cells by SNOs derived from different NOS pathways. Different concentrations of endogenous NO donors may be produced upon the stimulation of cellular NO synthases via biologically relevant cellular stressors. For example, we previously studied a few physiological and pathophysyiological inducers of S-nitrosation in the vascular system, including acetylcholine, vascular endothelial growth factor (VEGF) [32], nitroglycerin[9] and angiotensin II [8]. Angiotensin II and nitroglycerin probably increased S-nitrosation through oxidative stress [35, 36], and acetylcholine and VEGF via the stimulation of endothelial NOS (eNOS). Colocalization of the NO source and its target is believed to contribute to the specificity of S-nitrosation [37]; interestingly sGC is associated with both eNOS [38] and neuronal NOS (nNOS) [39]. NCM used in the present study express constitutively all 3 isoforms of NOS (eNOS, nNOS and iNOS) with eNOS and nNOS highly compartmentalized in NCM [40]. However, because we used CSNO as the NO donor, we can only speculate that sGC could be differentially S-nitrosated with derivatives of NO produced by different NOS, as a function of its localizations in different cellular compartments in the cardiomyocytes.
4. Conclusion
In the current work, we used 100 μM CSNO to treat NCM with overexpressed sGC (via adenoviral infection) to amplify the signal and ensure detection of 10 SNO-Cys sites in sGC expressed in heart cells by a highly sensitive Orbitrap LC/MS/MS approach. Our study shows that both CID and HCD approaches are effective for the localization of biotin-HPDP-modified cysteines in tryptic peptides, as a surrogate marker for SNO-Cys. The reason we identified a higher number of SNO-Cys sites and mostly different from the previous study is probably due to a combination of the higher sensitivity of the Orbitrap MS and the different mechanisms of nitrosation between GSNO-treated purified sGC and CSNO-treated cardiomyocytes. It could be argued that by using CSNO as an NO donor, what we potentially lost in sGC S-nitrosation specificity is compensated with what we gained in signal-to-noise ratios. However, one of the Cys identified in this setup (C516) was shown in a previous study [8] by mutational analysis to be key in the mechanism of sGC desensitization by S-nitrosation, suggesting that this approach can identify biologically relevant SNO-Cys in sGC. Mutational analysis of the other cysteines found in this study is one of our future goals. Overall, the identification of 10 SNO-Cys here provides a sound foundation for future studies of the function of sGC nitrosation.
Supplementary Material
Significance.
Soluble Guanylyl Cyclase (sGC) is the main receptor for nitric oxide (NO), an important signaling molecule in mammalian cardiovascular system. NO activates sGC via its heme to synthesize cGMP and stimulates the downstream cGMP-dependent kinase, triggering a plethora of signals. Recently, we discovered that NO can be covalently added onto select sGC cysteines via a post-translational modification (PTM) termed S-nitrosation or S-nitrosylation. A previous study was conducted on purified sGC treated with S-nitrosoglutathione (GSNO); has identified three S-nitrosated cysteines (SNO-Cys). However, identification of sGC S-nitrosation sites in cells has never been done and is likely more biologically relevant. We report in this study the use of a highly sensitive Orbitrap LC/MS/MS approach to identify ten SNO-Cys sites in sGC expressed in heart cells, including nine novel sGC SNO-Cys sites. Our study shows that both CID and HCD approaches are effective for the localization of biotin-HPDP-modified cysteines in tryptic peptides, as a surrogate marker for SNO-Cys. Our findings provide the basis for future studies of the diverse sGC S-nitrosation events, and provide a more thorough understanding of the multi-faceted interactions between NO and sGC.
Highlights.
Ten SNO-Cys PTM sites, including nine novel sites, were identified in sGC expressed in cardiac myocytes using nano- LC/MS/MS approach following a biotin switch reaction.
The newly discovered SNO-Cys are conserved in mammalian sGC, and most of them are solvent-exposed, e.g. on the surface of the sGC molecule, suggesting novel functions in specific protein-protein interactions.
Acknowledgments
The project described was supported by a grant from the National Institute of General Medical Sciences (R01GM112415 to HL and AB), RO1 GM067640 to AB, and the instrument used is supported by a grant (P30NS046593) from the National Institute of Neurological Disorders and Stroke. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors report no conflict of interest. We appreciate the help from Dr. Junichi Sadoshima’ group for the preparation of the NCM.
Abbreviations
- Asc
ascorbate
- BCA
the bicinchoninic acid assay
- BST
biotin switch technique
- CID
collision-induced dissociation
- CSNO
S-nitrosocysteine
- GSNO
S-nitrosoglutathione
- HCD
higher-energy collisional dissociation
- MMTS
methyl methanethiosulfonate
- NCM
neonatal cardiomyocytes
- PTM
post-translational modification
- sGC
soluble guanylyl cyclase
- SMC
smooth muscle cells
Footnotes
Conflict of interest
The authors declare that there is no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benhar M, Forrester MT, Stamler JS. Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat Rev Mol Cell Biol. 2009;10:721–32. doi: 10.1038/nrm2764. [DOI] [PubMed] [Google Scholar]
- 3.Wu C, Parrott AM, Fu C, Liu T, Marino SM, Gladyshev VN, et al. Thioredoxin 1-mediated post-translational modifications: reduction, transnitrosylation, denitrosylation, and related proteomics methodologies. Antioxid Redox Signal. 2011;15:2565–604. doi: 10.1089/ars.2010.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kornberg MD, Sen N, Hara MR, Juluri KR, Nguyen JV, Snowman AM, et al. GAPDH mediates nitrosylation of nuclear proteins. Nat Cell Biol. 2010;12:1094–100. doi: 10.1038/ncb2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamura T, Lipton SA. Emerging role of protein-protein transnitrosylation in cell signaling pathways. Antioxid Redox Signal. 2013;18:239–49. doi: 10.1089/ars.2012.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–74. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 7.Sayed N, Baskaran P, Ma X, van den Akker F, Beuve A. Desensitization of soluble guanylyl cyclase, the NO receptor, by S-nitrosylation. Proceedings of the National Academy of Sciences. 2007;104:12312–7. doi: 10.1073/pnas.0703944104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crassous PA, Couloubaly S, Huang C, Zhou Z, Baskaran P, Kim DD, et al. Soluble guanylyl cyclase is a target of angiotensin II-induced nitrosative stress in a hypertensive rat model. American journal of physiology Heart and circulatory physiology. 2012;303:H597–604. doi: 10.1152/ajpheart.00138.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sayed N, Kim DD, Fioramonti X, Iwahashi T, Duran WN, Beuve A. Nitroglycerin-induced S-nitrosylation and desensitization of soluble guanylyl cyclase contribute to nitrate tolerance. Circ Res. 2008;103:606–14. doi: 10.1161/CIRCRESAHA.108.175133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;2001:PL1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 11.Bechtold E, King SB. Chemical methods for the direct detection and labeling of S-nitrosothiols. Antioxid Redox Signal. 2012;17:981–91. doi: 10.1089/ars.2012.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forrester MT, Foster MW, Stamler JS. Assessment and Application of the Biotin Switch Technique for Examining Protein S-Nitrosylation under Conditions of Pharmacologically Induced Oxidative Stress. J Biol Chem. 2007;282:13977–83. doi: 10.1074/jbc.M609684200. [DOI] [PubMed] [Google Scholar]
- 13.Landino LM, Koumas MT, Mason CE, Alston JA. Ascorbic acid reduction of microtubule protein disulfides and its relevance to protein S-nitrosylation assays. Biochem Biophys Res Commun. 2006;340:347–52. doi: 10.1016/j.bbrc.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 14.Molecular Operating Environment (MOE) Chemical Computing Group, Inc; 2015. 2013.08 ed. [Google Scholar]
- 15.Case DAJTB, Betz RM, Cerutti DS, Cheatham TE, III, Darden TA, Duke RE, Giese TJ, Gohlke H, Goetz AW, Homeyer N, Izadi S, Janowski P, Kaus J, Kovalenko A, Lee TS, LeGrand S, Li P, Luchko T, Luo R, Madej B, Merz KM, Monard G, Needham P, Nguyen H, Nguyen HT, Omelyan I, Onufriev A, Roe DR, Roitberg A, Salomon-Ferrer R, Simmerling CL, Smith W, Swails J, Walker RC, Wang J, Wolf RM, Wu X, York DM, Kollman PA. AMBER 2015. University of California; San Francisco: 2015. [Google Scholar]
- 16.Chen YJ, Ku WC, Lin PY, Chou HC, Khoo KH, Chen YJ. S-alkylating labeling strategy for site-specific identification of the s-nitrosoproteome. J Proteome Res. 2010;9:6417–39. doi: 10.1021/pr100680a. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Liu T, Wu C, Li H. A strategy for direct identification of protein S-nitrosylation sites by quadrupole time-of-flight mass spectrometry. J Am Soc Mass Spectrom. 2008;19:1353–60. doi: 10.1016/j.jasms.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao G, Gross SS. Electrospray tandem mass spectrometry analysis of S- and N-nitrosopeptides: facile loss of NO and radical-induced fragmentation. J Am Soc Mass Spectrom. 2006;17:1725–30. doi: 10.1016/j.jasms.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 19.Sunahara RK, Beuve A, Tesmer JJ, Sprang SR, Garbers DL, Gilman AG. Exchange of substrate and inhibitor specificities between adenylyl and guanylyl cyclases. J Biol Chem. 1998;273:16332–8. doi: 10.1074/jbc.273.26.16332. [DOI] [PubMed] [Google Scholar]
- 20.Chang FJ, Lemme S, Sun Q, Sunahara RK, Beuve A. Nitric oxide-dependent allosteric inhibitory role of a second nucleotide binding site in soluble guanylyl cyclase. J Biol Chem. 2005;280:11513–9. doi: 10.1074/jbc.M412203200. [DOI] [PubMed] [Google Scholar]
- 21.Friebe A, Koesling D. Mechanism of YC-1-induced activation of soluble guanylyl cyclase. Mol Pharmacol. 1998;53:123–7. doi: 10.1124/mol.53.1.123. [DOI] [PubMed] [Google Scholar]
- 22.Marino SM, Gladyshev VN. Structural analysis of cysteine S-nitrosylation: a modified acid-based motif and the emerging role of trans-nitrosylation. J Mol Biol. 2010;395:844–59. doi: 10.1016/j.jmb.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallace S, Guo DC, Regalado E, Mellor-Crummey L, Banshad M, Nickerson DA, et al. Disrupted Nitric Oxide Signaling due to GUCY1A3 Mutations Increases Risk for Moyamoya Disease, Achalasia and Hypertension. Clinical genetics. 2016 doi: 10.1111/cge.12739. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernhoff NB, Derbyshire ER, Marletta MA. A nitric oxide/cysteine interaction mediates the activation of soluble guanylate cyclase. Proc Natl Acad Sci U S A. 2009;106:21602–7. doi: 10.1073/pnas.0911083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiology and molecular biology reviews: MMBR. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marozkina NV, Gaston B. S-Nitrosylation signaling regulates cellular protein interactions. Biochim Biophys Acta. 2012;1820:722–9. doi: 10.1016/j.bbagen.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Craven PA, DeRubertis FR. Effects of thiol inhibitors on hepatic guanylate cylase activity. Biochim Biophys Acta. 1978;524:231–44. doi: 10.1016/0005-2744(78)90121-3. [DOI] [PubMed] [Google Scholar]
- 28.Smith BC, Marletta MA. Mechanisms of S-nitrosothiol formation and selectivity in nitric oxide signaling. Current opinion in chemical biology. 2012;16:498–506. doi: 10.1016/j.cbpa.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lancaster JR., Jr Nitroxidative, nitrosative, and nitrative stress: kinetic predictions of reactive nitrogen species chemistry under biological conditions. Chem Res Toxicol. 2006;19:1160–74. doi: 10.1021/tx060061w. [DOI] [PubMed] [Google Scholar]
- 30.Riego JA, Broniowska KA, Kettenhofen NJ, Hogg N. Activation and inhibition of soluble guanylyl cyclase by S-nitrosocysteine: Involvement of amino acid transport system L. Free Radic Biol Med. 2009 doi: 10.1016/j.freeradbiomed.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang YT, Piyankarage SC, Williams DL, Thatcher GR. Proteomic profiling of nitrosative stress: protein S-oxidation accompanies S-nitrosylation. ACS Chem Biol. 2014;9:821–30. doi: 10.1021/cb400547u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sayed N, Baskaran P, Ma X, van den Akker F, Beuve A. Desensitization of soluble guanylyl cyclase, the NO receptor, by S-nitrosylation. Proc Natl Acad Sci U S A. 2007;104:12312–7. doi: 10.1073/pnas.0703944104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu C, Parrott AM, Liu T, Beuve A, Li H. Functional proteomics approaches for the identification of transnitrosylase and denitrosylase targets. Methods. 2013;62:151–60. doi: 10.1016/j.ymeth.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray CI, Uhrigshardt H, O’Meally RN, Cole RN, Van Eyk JE. Identification and quantification of S-nitrosylation by cysteine reactive tandem mass tag switch assay. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.013441. M111 013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi H, Tostes RC, Webb RC. Thioredoxin reductase inhibition reduces relaxation by increasing oxidative stress and s-nitrosylation in mouse aorta. J Cardiovasc Pharmacol. 2011;58:522–7. doi: 10.1097/FJC.0b013e31822d80a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munzel T, Daiber A, Mulsch A. Explaining the phenomenon of nitrate tolerance. Circ Res. 2005;97:618–28. doi: 10.1161/01.RES.0000184694.03262.6d. [DOI] [PubMed] [Google Scholar]
- 37.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–66. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 38.Venema RC, Venema VJ, Ju H, Harris MB, Snead C, Jilling T, et al. Novel complexes of guanylate cyclase with heat shock protein 90 and nitric oxide synthase. Am J Physiol Heart Circ Physiol. 2003;285:H669–78. doi: 10.1152/ajpheart.01025.2002. [DOI] [PubMed] [Google Scholar]
- 39.Russwurm M, Wittau N, Koesling D. Guanylyl cyclase/PSD-95 interaction: targeting of the nitric oxide- sensitive alpha2beta1 guanylyl cyclase to synaptic membranes. J Biol Chem. 2001;276:44647–52. doi: 10.1074/jbc.M105587200. [DOI] [PubMed] [Google Scholar]
- 40.Waldman SaMF. Cyclic GMP synthesis and function. Pharmacological Reviews. 1987;39:163–96. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.