Abstract
Objective. To evaluate the impact of personal genotyping and a novel educational approach on student attitudes, knowledge, and beliefs regarding pharmacogenomics and genomic medicine.
Methods. Two online elective courses (pharmacogenomics and genomic medicine) were offered to student pharmacists at the University of Florida using a flipped-classroom, patient-centered teaching approach. In the pharmacogenomics course, students could be genotyped and apply results to patient cases.
Results. Thirty-four and 19 student pharmacists completed the pharmacogenomics and genomic medicine courses, respectively, and 100% of eligible students (n=34) underwent genotyping. Student knowledge improved after the courses. Seventy-four percent (n=25) of students reported better understanding of pharmacogenomics based on having undergone genotyping.
Conclusions. Completion of a novel pharmacogenomics elective course sequence that incorporated personal genotyping and genomic medicine was associated with increased student pharmacist knowledge and improved clinical confidence with pharmacogenomics.
Keywords: pharmacogenomics, pharmacogenetics, genomics, genetics, personalized medicine
INTRODUCTION
Tremendous scientific advances have occurred in genomic variability and its association with drug response, toxicity, disease risk, and disease prognosis. Although clinical use of pharmacogenomic and genomic data to inform patient care decisions is increasing, it is not yet routine. Coverage of these concepts in health professions education, including pharmacy education, is lacking, and most practitioners feel inadequately prepared to apply these data in clinical practice.1-3 This knowledge gap is a significant barrier to widespread implementation of genomic medicine.4 While core competencies in genomic medicine and pharmacogenomics are available for pharmacists and other health professionals, they largely emphasize scientific knowledge of genetics rather than skills needed to apply these data. Additionally, didactic lectures remain the primary teaching method for this content in health professions education, despite data showing limited benefit of this strategy in improving practitioner understanding and retention of genomic medicine concepts.5-10
Educational approaches are needed that blend knowledge-based and skills-based learning activities, incorporate real-world applications, utilize collaborative teaching methods, and are accessible by a broad audience to support use of genomic and pharmacogenomics data.11-13 Although the optimal strategy to meet this need is debated, incorporation of student personal genetic testing is proposed as one option to improve learning outcomes.11,12,14-16 Medical and graduate students enrolled in a genetics course at Stanford School of Medicine were offered personal genome-wide testing through a direct-to-consumer company.11,16 Researchers reported improved engagement, motivation, and learning outcomes in students who underwent personal genetic testing. Of the students who underwent genetic testing, 70% self-reported a better understanding of human genetics based on their participation. In addition, genotyped students demonstrated a significantly higher increase from pretest to posttest knowledge scores compared with students who did not undergo genotyping. Results of a pilot study in two interdisciplinary electives at Duke University were similar.12 Other proposed or explored approaches include development of experiential, team-based learning programs for residents;17 student performance of limited self-genotyping in laboratory exercises;18,19 and development of educational tracks or shared curricular materials for schools of pharmacy and medicine.20-22
There remains a need for development of educational models in this area. Although student genetic testing through an external laboratory provider has been employed in some models,11,12 this approach may not be scalable and has raised ethical concerns.15 Additionally, teaching strategies employed in these courses to date lack an emphasis on patient care applications of these data. They also employ traditional, residential-based teaching methods and thus do not present a solution to broad-based education in this area. Based on this need, the University of Florida (UF) College of Pharmacy, in conjunction with UF Health Personalized Medicine Program clinicians and faculty members, offered two 8-week, 1-credit hour elective courses on the clinical applications of pharmacogenomics and genomic medicine in fall 2014 for third-year doctor of pharmacy (PharmD) students. The courses were delivered in an online format. The goal of these courses was to provide students with the knowledge and skills to use pharmacogenomic and genomic medicine data in future clinical practice. The online courses incorporated a flipped-classroom model with interprofessional lectures, patient-case discussions, and role-playing exercises that required use of clinical practice guidelines, databases, and primary literature. In the pharmacogenomics course, students could have their DNA genotyped on a custom pharmacogenomics chip23 and apply their genetic information (or use de-identified genotype data) to solve patient cases.
The purpose of this study was to evaluate the impact of this educational approach and the incorporation of personal genotype evaluation on student pharmacists’ attitudes, beliefs, and knowledge of clinical applications of pharmacogenomics and genomic medicine. We hypothesized that this novel approach would engage learners in a proactive way that would stimulate their interest in the topic and cause them to consider, in more concrete ways, the decisions they would make based on their own genetic information. We hypothesized that the incorporation of personal genotype information would be feasible and positively received by students.
METHODS
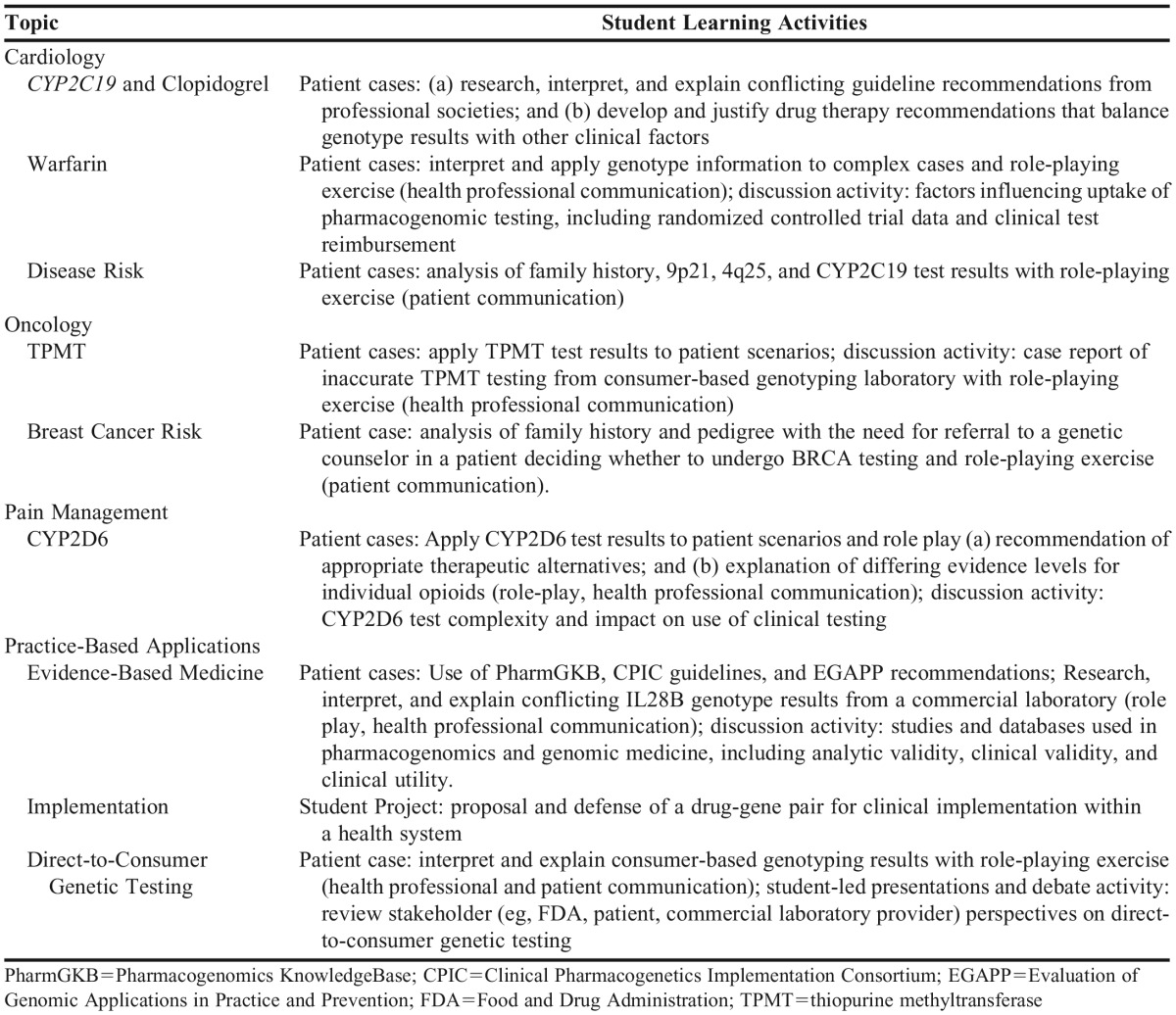
The Clinical Applications of Pharmacogenomics and Clinical Applications of Genomic Medicine courses offered by the college are represented in Figure 1. Students could enroll in either or both courses. Each course employed a combination of 1-hour weekly prerecorded lectures available on the course website and 1-hour weekly live class sessions conducted via synchronous webinar in a flipped-classroom model that used a Socratic question-and-answer teaching strategy. In addition to foundational science concepts, lecture content emphasized clinical implementation and patient care applications of pharmacogenomics and genomic medicine, such as test ordering and reimbursement, interpreting and applying test results to patient care scenarios, applying clinical guidelines, and communicating with patients and other health care professionals. Table 1 provides representative course topics and student activities.
Figure 1.
Pharmacogenomics and Genomic Medicine Elective Course Sequence in Fall 2014. Third-year pharmacy students could enroll in both courses sequentially (2 credit hours) or in one course only (1 credit hour/course) during the 16-week fall 2014 semester.
Table 1.
Representative Topics and Student Learning Activities in Pharmacogenomics and Genomic Medicine Courses
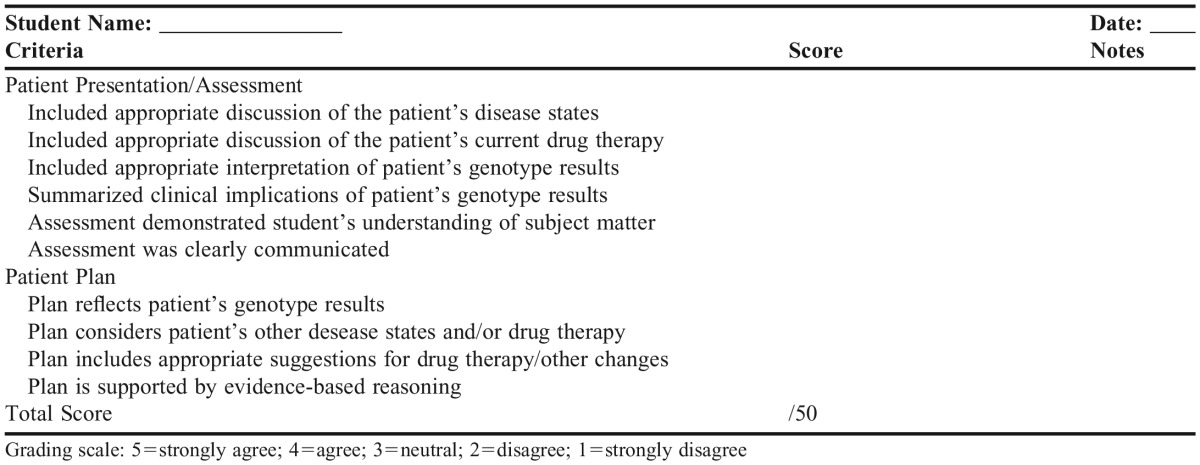
Students were assigned patient cases or learning activities after the recorded lectures were viewed. Appendix 1 shows a representative case for a clopidogrel-CYP2C19 patient. During the live class session, students were called on randomly by course faculty members to discuss their answers and rationale to individual questions (2-3 minutes/student) and graded individually on their responses using a standardized assessment rubric (Appendix 2). If the question required use of genotype information, students could use their own or de-identified data (but were not asked which they chose). Instructors used online polls and “hand-raising” webinar functionality throughout the live sessions to assess student understanding of the subject matter. An online quiz with questions based on the in-class discussion was also administered after each class to further assess students. In their final project, students reflected on drug-gene pairs discussed during the course and were required to choose two clinical implementations to defend in essay format as either being “most likely to succeed” or “least likely to succeed.” Other assessment measures included two formal examinations for each course and additional active-learning activities (eg, discussion board participation).
A clinical pharmacist and a geneticist coordinated both courses and led webinar discussions. Additional instructors included a bioethics attorney to review informed consent, benefits, and risks associated with personal genotyping (pharmacogenomics course), UF Center for Pharmacogenomics faculty members, a physician with expertise in family health history, and a genetic counselor with expertise in clinical genetics. Lectures and cases were largely based on experiences with clinical implementation of pharmacogenomics and genomic medicine through the UF Health Personalized Medicine Program.24
Students participating in the Clinical Applications of Pharmacogenomics course could undergo personal genotyping of select pharmacogenomic single nucleotide polymorphisms (SNPs). Genotyping was not performed in the genomic medicine course. Collection kits for DNA, information on personal genotyping, and informed consent documents were mailed to students enrolled in the course prior to the first day of class. During the first week of class, students completed lectures on bioethics, informed consent, and benefits/risks of personal genotyping. The informed consent process for genotyping was reviewed during the first class webinar by a study coordinator not affiliated with the course. The study coordinator conducted all student communication about personal genotyping and served as a liaison between students and the lab coordinator to ensure course faculty and laboratory personnel were blinded to students’ choices regarding personal genotyping. The genotyping portion of the study was approved by the UF Institutional Review Board (IRB). Student participation was anonymous; all students gave written, informed consent.
Genotyping was conducted using a custom array on Life Technologies (Carlsbad, CA) QuantStudio 12K Flex system, with OpenArray technology.23 This custom genotyping chip included approximately 120 SNPs in genes with evidence of a pharmacogenomic effect, such as CYP2C19, CYP2C9, VKORC1, and CYP2D6. Samples of DNA were collected using ORAgene Discover OGR-500 DNA self-collection kits and isolated using the prepIT L2P kit (PT-LP2-45) (both from DNA Genotek Inc., Ontario, Canada). Genotyping costs were supported by grant funding from the National Institutes of Health. Students did not receive any compensation, benefit, or incentive to participate in personal genotyping. Students who chose not to participate in genotyping were provided with anonymous, de-identified genotyping results in an identical manner (format, method, and timeline of returning results) to students who were genotyped.
Samples of DNA were assigned an anonymous 4-digit code to ensure the laboratory was blinded to which students underwent genotyping. After genotyping was completed, the laboratory transferred the genotype data back to the study coordinator who then transferred personal genotype data (provided as untranslated SNP genotypes) back to each student via encrypted files. Students were also given access to 10 de-identified genotype data sets, which allowed students to complete patient cases using their own genotype or the de-identified data set. The laboratory destroyed any remaining DNA samples after genotyping was complete.
To ensure protection of all aspects of student confidentiality, all communication with students, faculty members, and research laboratory personnel regarding participation in personal genotyping and/or the pre/postsurvey (described below) was conducted by a nonfaculty study coordinator not associated with the course (referred to as an “honest broker”). This included all in-person or e-mail communication about the study, informed consent, mailing and receipt of DNA kits, and communication of genotyping results to students. Course faculty members were not included in these processes and study-related questions were directed to the study coordinator. In this way, faculty members were completely blinded to students’ choices to participate in any component of the study. In addition, students with questions or concerns about their genotyping results could e-mail the honest broker to obtain contact and scheduling information to meet confidentially with a health professional or genetic counselor to discuss their results at no charge to the student, with payment of any related costs or fees coordinated by the honest broker on behalf of the UF Health Personalized Medicine Program.
Students in both courses were invited to participate in pre/postcourse surveys evaluating knowledge, attitudes, and beliefs about genotyping for pharmacogenomic and genomic medicine applications. The surveys were approved by the UF IRB, and all students provided a waiver of documentation of informed consent. Surveys were developed by course faculty members, with input from researchers with expertise in survey methodology and interprofessional education, and were pilot tested prior to administration. Selected survey questions were adapted from a survey developed by researchers at Stanford School of Medicine.11,16 Surveys were administered during the first and last weeks of the courses using an online instrument (Qualtrics, LLC, Provo, UT). Students were assigned an anonymous 4-digit code to identify their surveys so that survey data remained de-identified, but pre/postsurveys could be paired for analysis. Students did not receive any compensation, benefit, or incentive to participate in the surveys and were not penalized in any way for not participating in them.
Survey results were analyzed to assess the impact of the course and personal genotyping on pharmacogenomic and genomic medicine attitudes, beliefs, and knowledge. Only students who provided a presurvey and postsurvey (paired response) were included in the analysis. Student responses to questions on attitudes were assessed on a 5-point Likert scale (strongly agree, agree, neutral, disagree, and strongly disagree). For data analysis, student responses were collapsed and reported as percentage of students who agree or strongly agree, and/or disagree or strongly disagree with the stem statement (the “neutral” response was omitted for statistical analyses). The paired precourse and postcourse responses were analyzed for change using nonparametric tests (McNemar’s test for binary responses and Wilcoxon signed-rank test for Likert scale responses). The paired precourse and postcourse knowledge questions were analyzed individually for change using McNemar’s test, and the overall knowledge score was analyzed for change by Student’s t test.
RESULTS
Thirty-four and 19 pharmacy students completed the pharmacogenomics course and genomic medicine courses, respectively. Sixteen students completed both courses sequentially. All participants were PharmD students in the fall of their third professional year (P3). No students were enrolled in dual-degree or other postgraduate courses or programs.
The most common reason cited by students for enrolling in either course was the need to learn about pharmacogenomics/genomic medicine because of its importance to patient care in the future. Students who participated in both courses were asked which course they felt was more valuable in influencing their future clinical practice. Fifty-six percent of students (9 of 16) stated that the courses were equally beneficial, while 38% (6 of 16) and 6% (1 of 16) stated that the pharmacogenomics and genomic medicine course was more beneficial, respectively.
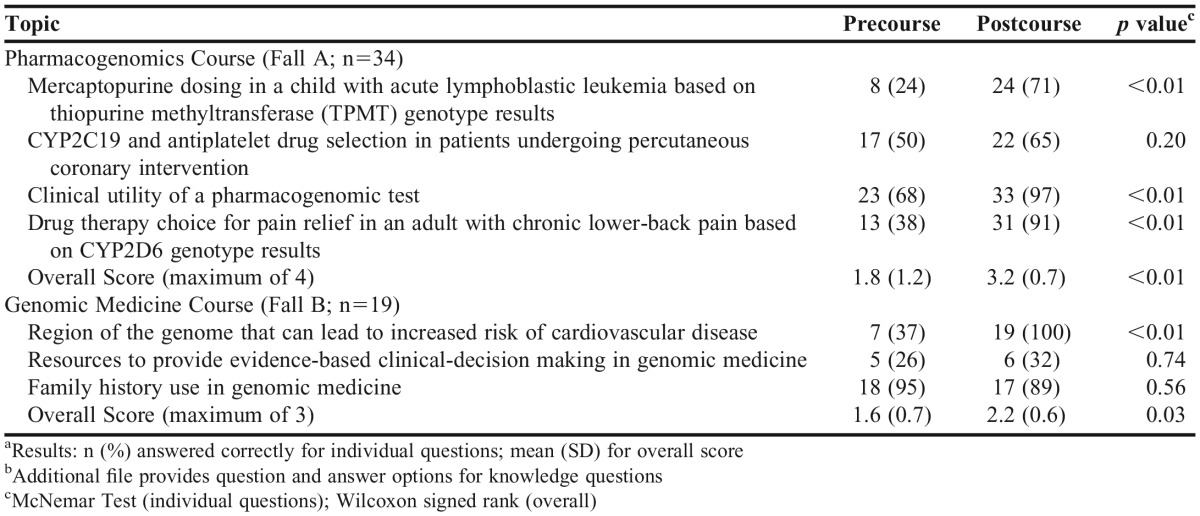
Seventy-nine percent of students (n=27) attended and participated in all live, web-based class meetings for both the pharmacogenomics and genomic medicine courses (attendance was required and incorporated into grading for both courses). Prior to the start of the pharmacogenomics course, students scored an average of 45% (1.8 of 4 questions correct) on questions related to knowledge of pharmacogenomics, which increased to 80% after completing the course (3.2 of 4 questions correct, p<0.01). Comparisons of precourse and postcourse knowledge for the pharmacogenomics and genomic medicine courses are summarized in Table 2. In the genomic medicine course, student mean scores increased from 53% (1.6 of 3 questions correct) to 74% (2.2 of 3 questions correct, p=0.03) on questions related to knowledge of genomic medicine. Notably, one of the original knowledge assessment questions for the genomic medicine course was discarded during data analysis because of a question error that invalidated student responses. An additional file provides complete question and answer options for all knowledge questions.
Table 2.
Student Knowledge Before and After Pharmacogenomics and Genomic Medicine Coursesa,b
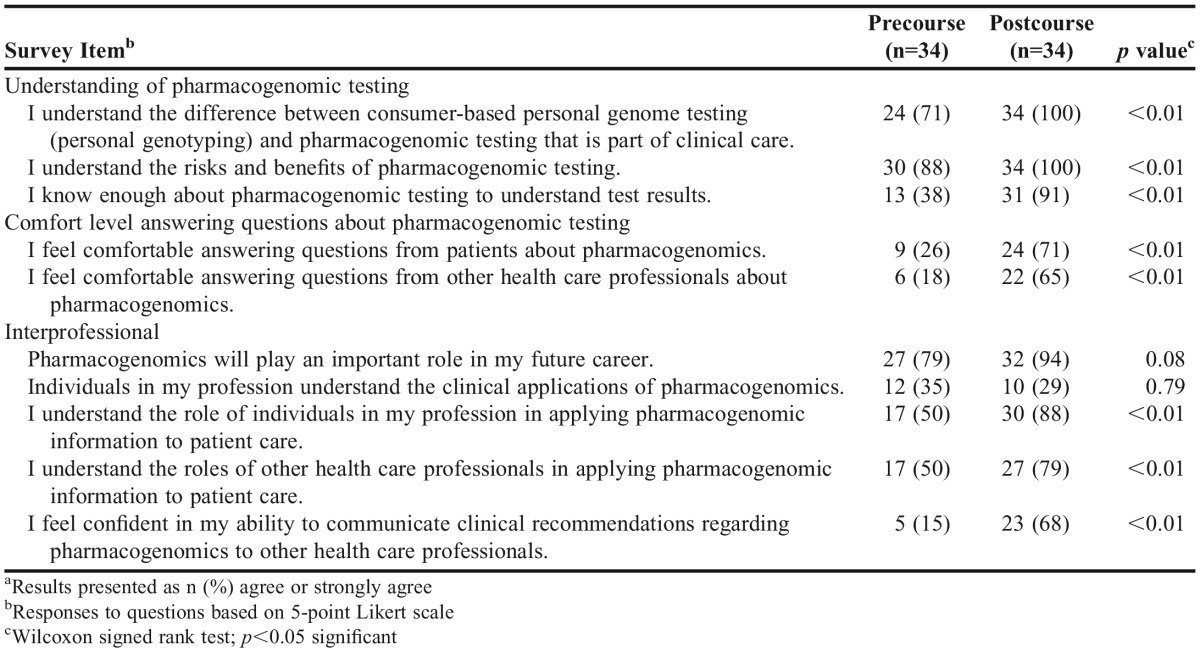
One hundred percent of students enrolled in the courses also completed the pre/postsurveys for both courses. After completing the pharmacogenomics course, students’ self-reported understanding of pharmacogenomic testing improved significantly a compared with precourse measures, with a more than 2-fold increase in percentage of students reporting they understood a patient’s pharmacogenomic test results (38% vs 91%; p<0.01). Students completing the pharmacogenomics course also reported increased comfort level and confidence in answering patient and health care professions questions about pharmacogenomics, as well as improved understanding of the roles of various health care professionals in this field after course completion. Students’ attitudes regarding pharmacogenomics before and after completing the course are summarized in Table 3.
Table 3.
Student Attitudes and Beliefs Before and After Pharmacogenomics Coursea
There was no significant difference between the percent of students who would undergo pharmacogenomic testing (100% vs 97%; p=0.32) or recommend it for patients (94% vs 97%; p=0.32) after completing the pharmacogenomics course. However, after completing the pharmacogenomics course, fewer students stated that they would recommend consumer-based personal genotyping to patients (38% vs 18%; p<0.01).
In the genomic medicine course, 85% (n=16 of 19) of students who were eligible to complete the pre/postsurveys had also taken the pharmacogenomics course (ie, completed the pharmacogenomics one week prior to starting the genomic medicine course). Because of the immediate sequential nature of the courses, presurvey data on attitudes and beliefs did not represent a true baseline measurement in the majority of students. Therefore, a valid comparison of pre/postsurvey responses was not possible for this course.
At the start of the pharmacogenomics course, 97% (n=33) of students planned to undergo personal genotype evaluation. At course completion, 100% of students reported opting in to personal genotyping. The 100% adoption rate for personal genotyping precluded us from evaluating different impact of the course based on the decision to undergo personal genotyping. When asked about their motivations for undergoing personal genotyping, student responses included: general curiosity about their genetic makeup (67%, n=22); desire to learn about genetic variations that influence drug reactions or dosing (18%, n=6); desire to understand what patients might experience (9%, n=3); and interest in providing information to family members about genetic variations that influence drug reactions or dosing (6%, n=2). Only one student reported having previously undergone consumer-based personal genotyping, genetic testing, or pharmacogenetic testing in the precourse survey.
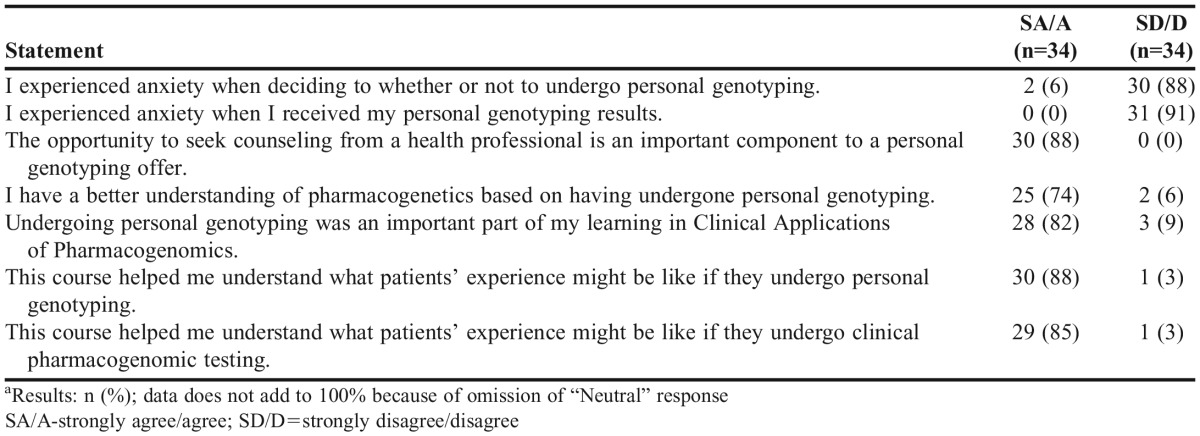
When asked to reflect on their feelings about choosing whether or not to undergo genotyping, 6% of students (n=2) agreed or strongly agreed that they felt anxiety about this decision, while no students reported feeling anxiety when they received their genotype results. Eighty-eight percent of students (n=30) strongly agreed/agreed that the opportunity to seek counseling from a health professional was an important component of a personal genotyping offer, although no students requested to meet with a health professional or genetic counselor throughout the course and genotyping processes. Table 4 summarizes students’ attitudes about personal genotyping.
Table 4.
Student Attitudes Toward Personal Genotypinga
After completing the course, 75% (n=25) of students reported being mildly or significantly pleased that they underwent genotyping, 21% (n=7) felt neutral about personal genotyping, and 3% (n=1) expressed mild regret for having undergone personal genotyping. The majority of students (74%, n=25) reported a better understanding of pharmacogenomics based on having undergone personal genotyping as part of the course. Eighty-two percent of students (n=28) stated that personal genotyping was an important part of their learning process. Eighty-eight percent (n=30) and 85% (n=29), of students, respectively, stated that the course helped them to better understand what a patient’s experience might be like if they were to undergo personal genotyping and clinical pharmacogenomic testing.
Students were also asked whether they perceived that faculty members had any knowledge of whether individual students underwent genotyping and/or the effects that this knowledge may have on grading. Ninety-one percent of students (n=30) did not feel that course faculty members knew whether they had undergone personal genotype evaluation in the course, and 94% of students (n=31) stated that they did not feel required to divulge their personal genotype information to ask questions of faculty members in the course. Accordingly, the majority of students (94%, 31) reported that they did not feel they were at a disadvantage in the course because of any perceived faculty knowledge of student participation in personal genotyping.
DISCUSSION
This is the first study of educational outcomes among pharmacy students given the opportunity to undergo array-based personal genotyping of pharmacogenomic SNPs as a component of an academic course in pharmacogenomics. All eligible students opted to undergo personal genotyping, and the majority of these students agreed that genotyping was an important part of learning in the course and that it helped them understand a patient’s experience with genotyping (>80% of students for all measures). Students’ knowledge of pharmacogenomics and genomic medicine increased significantly after completing the respective courses, with a nearly 2-fold increase in knowledge assessment. These results are consistent with previous studies that incorporated personal genome evaluation into medical and/or graduate education,11,12,14,16 although our model differs from these in its focus on clinically actionable pharmacogenomic SNPs.
One unexpected finding of this study was the decreased percentage of students stating that they would recommend consumer-based personal genotyping to patients after completing the pharmacogenomics course. Although we were not able to explore this finding further, based on our interactions with students, we anticipate that this finding reflects increased understanding and discernment among students regarding the complexity of pharmacogenetic and genomic testing overall, and the need for a health professional’s guidance in interpreting test results. During the course, the importance of interpreting test results correctly for patient care applications, in light of emerging evidence and advancing genotyping procedures, was discussed extensively. For example, one TPMT (thiopurine methyltransferase) discussion activity required students to role play explaining an erroneous laboratory report delivered by a consumer-based genotyping company for TPMT assessment to a patient and prescriber.25
The use of a clinician-led, flipped-classroom, online teaching approach focusing on clinical implementation and communication with patients and other health care professionals is unique to this study. This emphasis on applying knowledge to clinical practice was reinforced throughout all course activities (eg, patient cases, role-playing exercises) and incorporated into student assessment rubrics. We believe that patient-centered teaching approaches such as these will be essential to prepare student pharmacists for future practice activities that include the routine use pharmacogenomic data. These teaching and learning strategies are consistent with other applications-based, pharmacotherapy content students are learning during their third year of pharmacy school and support student achievement of baseline clinical knowledge and skills in pharmacogenomics as defined by professional competency statements.22 This approach was associated with increased student confidence in answering questions about pharmacogenomics from patients and health care professionals and in communicating clinical recommendations to other health care professionals. These findings support the need to further develop and disseminate educational strategies for pharmacogenomics and genomic medicine that equip students with necessary clinical skills, such as the ability to incorporate data into existing patient information, analyze and apply emerging evidence, use publicly available databases, evaluate clinical validity and utility of genomic tests, counsel patients about test results, and navigate clinical reimbursement issues.7,10,12,14
The distinction between scientific knowledge and clinical skills and an emphasis on active-learning, patient-centered teaching approaches are important considerations when charting the future course of education in pharmacogenomics and genomic medicine. Coverage of pharmacogenomics and genomic medicine is increasing in health professions education, but teaching strategies are dominated by traditional didactic lectures that emphasize basic science concepts,6,8,20,21 an approach that is insufficient for clinical skill development.7,26 Significant barriers exist to widespread adoption of an active-learning approach that emphasizes clinical skills in pharmacogenomics and genomic medicine, including a shortage of faculty members with knowledge and clinical experience in this area and the need to stay current with a developing evidence base for clinical validity, utility, and reimbursement for tests across broad therapeutic areas.7,8,10,21 Strategies proposed to overcome these challenges include curricular integration,8 creation of shared curricula across institutions and/or professional societies,14,21,27 implementation of educational tracks within professional curricula to enhance exposure to genetics and genomics,20 development of “train-the-trainer” programs for teaching faculty members,17,21 and use of novel online educational approaches such as courses that use an open, wiki platform and crowd-sourcing from the medical community.10 Future exploration of these and other novel strategies is needed to advance health professional education in this area on a large scale.
Study findings also contribute to the growing body of evidence that thoughtful genotyping processes can help overcome potential ethical questions with student personal genotype evaluation, including concerns about student vulnerability, exploitation, or coercion, the possibility of returning test results with unclear or shifting clinical implications for disease risk, and a blurred line between educational and human subject research.14,15,28,29 We employed measures to protect students that included the use of an honest broker intermediary for all study-related communication, faculty blinding to student genotyping choice, coverage of bioethics and informed consent for genetic testing prior to genotyping offer, offer of genetic counseling at no charge, and use of specific verbiage during question-and-answer sessions to avoid revealing students’ genotyping choice. Although there is minimal risk of incidental findings or disease associations with the pharmacogenetic SNP data, we also conducted searches of the database of Genotypes and Phenotypes (dbGaP), and the National Human Genome Research Institute Genome-Wide Association Study (NHGRI GWAS) Catalog on all SNPs prior to the start of the course and again before returning any student genotyping data to identify and remove any SNPs that could be associated with disease risk. To ensure a clear delineation between educational and human subject research, we did not retain, report, or use student genotype data in any form, although students could opt to share their data anonymously into dbGaP before it was destroyed by the laboratory. These measures are consistent with those previously used or recommended by ethicists11,12,16,28,29 and were associated with no or minimal student anxiety in the decision to undergo genotyping and student confidence that course instructors were unaware of their choice. However, scalability of these protective measures will be an important consideration for future educational efforts in this area. Additional research is needed to determine the appropriate level of student protection needed and strategies to support educators incorporating student genotyping into teaching and learning models on a larger scale.
Study limitations include single-site design, small sample size with no control group, and the potential for social desirability bias in survey responses. Although we anticipated that some students would select to receive de-identified genotype data and thereby serve as internal controls, this did not occur. Because of the elective nature of the course, students who enrolled may have entered the course with a higher interest in or enthusiasm for pharmacogenomics and/or genomic medicine. It is possible that observed changes may have been caused by factors outside the course, and no additional follow up was conducted to measure the persistence of observed changes in students’ knowledge, attitudes, or beliefs. Additionally, the large percentage (85%) of students who enrolled in both courses sequentially, which invalidated baseline measures of student attitudes and beliefs in the genomic medicine course presurvey, was unanticipated. We incorporated changes into the design of the fall 2015 course and research protocol to address this issue.
The course was offered for the second time in fall 2015, with a number of changes incorporated to both the educational and research content and design. The course structure was revised to combine the individual 8-week courses to form a 16-week course titled Clinical Applications of Personalized Medicine. The 16-week format allowed faculty members to combine coverage of pharmacogenomics and genomic medicine content by disease state or organ system (eg, cardiology, hematology/oncology) to streamline teaching and avoid potential content duplication between the courses. In the second offering, personal genotyping results were returned to students in a “clinical lab report” format with star-allele and phenotype designations rather than as raw SNP data to better mimic clinical practice. In addition, the online teaching model was expanded to enable increased availability of this course to students in other health professions and at other universities, irrespective of geographic location, and additional active-learning activities (eg, journal club and discussion board) were incorporated into the course. Revisions were also made to the research plan, including designation of a control group, development of a more efficient online research enrollment and consenting process for students, use of a larger number of pre/postcourse knowledge assessment questions mapped to individual course objectives, and revision of the pre/postsurveys to allow valuation of specific teaching strategies and assessment of student readiness to implement clinical pharmacogenomics in practice.
CONCLUSION
This novel and interactive approach to pharmacogenomics (including personal genotype evaluation) and genomic medicine education in a subset of students was associated with increased student learning and improved student confidence in communicating clinical recommendations to other health professionals and patients about pharmacogenomics. The opportunity to use personal genotype information was an important part of the learning process and helped students better understand a patient’s experience with genetic testing.
ACKNOWLEDGMENTS
The authors thank Dr. Taimour Langaee for laboratory assistance with genotyping, Drs. Allyson G. Hall and Amy Blue for assistance with survey development, Dr. Reggie Frye for input on course content, and Dr. Rhonda Cooper-Dehoff for assistance with study design. The UF Health Personalized Medicine Program has been funded to date by NIH grants U01 GM074492 and U01 HL105198 (as part of TPP project in NIH PGRN); NIH/NCATS UF CTSA UL1 TR000064, IGNITE Network grant U01 HG007269 and substantial institutional support from the University of Florida. The authors have no conflicts of interest.
Appendix 1. Sample CYP2C19-Clopidogrel Patient Case
HPI: RF is a 67-year-old male (wt. 110 kg) with a history of obesity, diabetes mellitus, gout, and prostate carcinoma s/p XRT, who has had substernal chest pain for 6 months. The pain is exacerbated by exertion and relieved by rest. He underwent a treadmill stress test approximately 1 month ago and exercised 4 minutes. No ischemic appearing EKG changes were observed but he did have significant chest pain. He denies having any bleeding problems or upcoming surgeries. Patient presented to the catheterization lab for unstable angina today and underwent PCI. Blood was sent to lab for CYP2C19 genotype today.
Procedure Summary: LHC revealed high grade obstructive LAD lesion successfully treated with one DES in the mid-LAD
Insurance: Information pending
Medications:
Aspirin 81mg daily; Clopidogrel 75mg daily; Atorvastatin 80mg daily; Allopurinol 300mg daily; Glyburide-metformin 5-500mg daily; Metformin 500mg daily; Metoprolol XL 50mg daily; Sitagliptin 50mg daily
Using the genotype assigned to you (or your own CYP2C19 genotype if desired), answer the following questions:
1. What are the treatment options for RF’s dual antiplatelet therapy? (See antiplatelet recommendations in the 2012 ACCF/AHA Focused Update of the Guideline for the Management of Patients With Unstable Angina/Non–ST-Elevation Myocardial Infarction).
2. What additional information do you need for this patient?
3. What patient-specific (nongenotype) factors will influence the choice of antiplatelet therapy for RF?
4. Based on this patient’s presentation, genotype, and other patient-specific factors, provide a drug therapy recommendation for this patient’s antiplatelet therapy below, including drug, dose, and duration of therapy.
5. How does this patient’s genotype affect your drug therapy recommendation?
Appendix 2. Content of Patient Case Student Assessment Rubric
Clinical Applications of Pharmacogenomics Grading Rubric
REFERENCES
- 1.Haga SB, Burke W, Ginsburg GS, Mills R, Agans R. Primary care physicians’ knowledge of and experience with pharmacogenetic testing. Clin Genet. 2012;82(4):388–394. doi: 10.1111/j.1399-0004.2012.01908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stanek EJ, Sanders CL, Taber KA, et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clin Pharmacol Ther. 2012;91(3):450–458. doi: 10.1038/clpt.2011.306. [DOI] [PubMed] [Google Scholar]
- 3.McCullough KB, Formea CM, Berg KD, et al. Assessment of the pharmacogenomics educational needs of pharmacists. Am J Pharm Educ. 2011;75(3):Article 51. doi: 10.5688/ajpe75351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manolio TA, Chisholm RL, Ozenberger B, et al. Implementing genomic medicine in the clinic: the future is here. Genet Med. 2013;15(4):258–267. doi: 10.1038/gim.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy JE, Green JS, Adams LA, Squire RB, Kuo GM, McKay A. Pharmacogenomics in the curricula of colleges and schools of pharmacy in the United States. Am J Pharm Educ. 2010;74(1):Article 7. doi: 10.5688/aj740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green JS, O’Brien TJ, Chiappinelli VA, Harralson AF. Pharmacogenomics instruction in US and Canadian medical schools: implications for personalized medicine. Pharmacogenomics. 2010;11(9):1331–1340. doi: 10.2217/pgs.10.122. [DOI] [PubMed] [Google Scholar]
- 7.Salari K. The dawning era of personalized medicine exposes a gap in medical education. PLoS Med. 2009;6(8):e1000138. doi: 10.1371/journal.pmed.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plunkett-Rondeau J, Hyland K, Dasgupta S. Training future physicians in the era of genomic medicine: trends in undergraduate medical genetics education. Genet Med. 2015;17(11):927–934. doi: 10.1038/gim.2014.208. [DOI] [PubMed] [Google Scholar]
- 9.Formea CM, Nicholson WT, McCullough KB, et al. Development and evaluation of a pharmacogenomics educational program for pharmacists. Am J Pharm Educ. 2013;77(1):Article 10. doi: 10.5688/ajpe77110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patay BA, Topol EJ. The unmet need of education in genomic medicine. Am J Med. 2012;125(1):5–6. doi: 10.1016/j.amjmed.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Salari K, Karczewski KJ, Hudgins L, Ormond KE. Evidence that personal genome testing enhances student learning in a course on genomics and personalized medicine. PLoS One. 2013;8(7):e68853. doi: 10.1371/journal.pone.0068853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katsanis SH, Dungan JR, Gilliss CL, Ginsburg GA. Educating future providers of personalized medicine. N C Med J. 2013;74(6):491–492. [PubMed] [Google Scholar]
- 13.Boguski MS, Boguski RM, Berman MR. Personal genotypes are teachable moments. Genome Med. 2013;5(3):22. doi: 10.1186/gm426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanderson SC, Linderman MD, Kasarskis A, et al. Informed decision-making among students analyzing their personal genomes on a whole genome sequencing course: a longitudinal cohort study. Genome Med. 2013;5(12):113. doi: 10.1186/gm518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salari K, Pizzo PA, Prober CG. Commentary: to genotype or not to genotype? Addressing the debate through the development of a genomics and personalized medicine curriculum. Acad Med. 2011;86(8):925–927. doi: 10.1097/ACM.0b013e3182223acf. [DOI] [PubMed] [Google Scholar]
- 16.Vernez SL, Salari K, Ormond KE, Lee SS. Personal genome testing in medical education: student experiences with genotyping in the classroom. Genome Med. 2013;5(3):24. doi: 10.1186/gm428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haspel RL. Teaching residents genomic pathology: a novel approach for new technology. Adv Anat Pathol. 2013;20(2):125–129. doi: 10.1097/PAP.0b013e31828629b2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krynetskiy E, Calligaro I. Introducing pharmacy students to pharmacogenomic analysis. Am J Pharm Educ. 2009;73(4):Article 71. doi: 10.5688/aj730471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knoell DL, Johnston JS, Bao S, Kelley KA. A genotyping exercise for pharmacogenetics in pharmacy practice. Am J Pharm Educ. 2009;73(3):Article 43. doi: 10.5688/aj730343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhar SU, Alford RL, Nelson EA, Potocki L. Enhancing exposure to genetics and genomics through an innovative medical school curriculum. Genet Med. 2012;14(1):163–167. doi: 10.1038/gim.0b013e31822dd7d4. [DOI] [PubMed] [Google Scholar]
- 21.Lee KC, Hudmon KS, Ma JD, Kuo GM. Evaluation of a shared pharmacogenomics curriculum for pharmacy students. Pharmacogenomics. 2015;16(4):315–322. doi: 10.2217/pgs.14.181. [DOI] [PubMed] [Google Scholar]
- 22.ASHP statement on the pharmacist’s role in clinical pharmacogenomics. Am J Health Syst Pharm. 2015;72(7):579–581. doi: 10.2146/sp150003. [DOI] [PubMed] [Google Scholar]
- 23.Johnson JA, Burkley BM, Langaee TY, Clare-Salzler MJ, Klein TE, Altman RB. Implementing personalized medicine: development of a cost-effective customized pharmacogenetics genotyping array. Clin Pharmacol Ther. 2012;92(4):437–439. doi: 10.1038/clpt.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weitzel KW, Elsey AR, Langaee TY, et al. Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am J Med Genet. 2014;166C(1):56–67. doi: 10.1002/ajmg.c.31390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brownstein CA, Margulies DM, Manzi SF. Misinterpretation of TPMT by a DTC genetic testing company. Clin Pharmacol Ther. 2014;95(6):598–600. doi: 10.1038/clpt.2014.60. [DOI] [PubMed] [Google Scholar]
- 26.Kassirer JP. Teaching clinical reasoning: case-based and coached. Acad Med. 2010;85(7):1118–1124. doi: 10.1097/acm.0b013e3181d5dd0d. [DOI] [PubMed] [Google Scholar]
- 27.Haspel RL, Arnaout R, Briere L, et al. A call to action: training pathology residents in genomics and personalized medicine. Am J Clin Pathol. 2010;133(6):832–834. doi: 10.1309/AJCPN6Q1QKCLYKXM. [DOI] [PubMed] [Google Scholar]
- 28.Callier SL. Swabbing students: should universities be allowed to facilitate educational DNA testing? Am J Bioeth. 2012;12(4):32–40. doi: 10.1080/15265161.2012.656803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parker LS, Grubs R. Ethical considerations regarding classroom use of personal genomic information. J Microbiol Biol Ed. 2014;15(2):191–196. doi: 10.1128/jmbe.v15i2.856. [DOI] [PMC free article] [PubMed] [Google Scholar]