Abstract
8-oxoguanine is one of the most abundant and impactful oxidative DNA lesions. However, the reasons underlying its effects, especially those not directly explained by the altered base pairing ability, are poorly understood. We report the effect of the lesion on the action of EcoRI, a widely used restriction endonuclease. Introduction of 8-oxoguanine inside, or adjacent to, the GAATTC recognition site embedded within the Drew—Dickerson dodecamer sequence notably reduced the EcoRI activity. Solution NMR revealed that 8-oxoguanine in the DNA duplex causes substantial alterations in the sugar—phosphate backbone conformation, inducing a BI→BII transition. Moreover, molecular dynamics of the complex suggested that 8-oxoguanine, although does not disrupt the sequence-specific contacts formed by the enzyme with DNA, shifts the distribution of BI/BII backbone conformers. Based on our data, we propose that the disruption of enzymatic cleavage can be linked with the altered backbone conformation and dynamics in the free oxidized DNA substrate and, possibly, at the protein—DNA interface.
Introduction
8-oxoguanine (oxoG) is a naturally abundant base lesion with a well characterized mutagenic potential [1]. It is easily generated in DNA by reactive oxygen species endogenously produced during normal aerobic metabolism as well as by environmental assaults such as ionizing radiation. Under normal circumstances, the persisting background level of oxoG in human DNA has been estimated as ~0.5–5 oxoG / 106 guanines [2]; however, oxidative stress and many diseases cause a significant increase in the amount of this lesion [3–5]. Due to its propensity to pair with adenine during replication, oxoG induces G:C→T:A transversions, which are currently regarded as the lesion’s main biological consequence [1,6].
Aside from its miscoding properties, oxoG has been shown to affect recognition of DNA by some DNA-binding proteins. The presence of oxoG in the recognition sequence has been shown to inhibit DNA cleavage by several restriction endonucleases, including HaeIII, HpaII, MspI, NheI and NgoMIV [7–10]. Oxidation of G in CpG dinucleotide substantially alters methylation of the C within the site by human DNA methyltransferases DNMT1 and DNMT3a [11,12]. The presence of oxoG in the topoisomerase I recognition site, depending on the position, may either enhance or suppress the enzyme activity [13,14]. Transcription factors AP-1 and Sp1, methyl-CpG-binding protein MeCP2, and telomere protecting proteins TERF1 and TERF2 poorly bind oxoG-containing recognition sequences [15–17], whereas binding of NF-κB to its promoter element may either decrease or increase depending on oxoG location in the sequence [18]. This interference of oxoG with normal protein—DNA interactions may potentially have biological consequences such as improper regulation of gene expression, continuing survival of the invasive foreign DNA with a damaged restriction site in a bacterial cell, etc. [19,20].
The exact reasons of the differences observed in enzymatic recognition of the DNA substrate upon introduction of the oxoG remain unclear, especially when the lesion does not form direct contacts with the protein. To explain specific changes in protein—DNA recognition, a comprehensive analysis of DNA conformation is required, including within the backbone and the base pairs [21]. Within the same helix geometry, backbone dihedral angles define different types of DNA backbone conformation (e.g. BI vs BII) [22]. In this paper, we have used restriction endonuclease EcoRI as a model to analyze possible effect of oxoG in protein—DNA interactions. EcoRI was selected for the following reasons: (i) this is a well-studied enzyme, with the activity towards many other deviations from the cognate site described experimentally [23–25]; (ii) oxoG affects the recognition of DNA by several other structurally unrelated restriction enzymes (see above); (iii) the structure of EcoRI bound to its cognate DNA is available [26], allowing the analysis of direct protein—DNA interactions; (iv) the EcoRI site -GAATTC- is a central part of extensively characterized Drew—Dickerson dodecamer [27,28], which provides an excellent reference point for comparison of the undamaged and damaged cognate site in free DNA. We report that the activity of restriction endonuclease EcoRI is inhibited by oxoGplaced within, or adjacent to, the recognition site despite the apparent conservation of the protein—DNA interface. Our solution NMR analysis of oxoG-containing DNA substrates provides a possible structural explanation (backbone alteration) of this effect.
Materials and Methods
Oligonucleotides and enzymes
Sequences of the oligonucleotides used in this study are listed in Table 1. Oligonucleotides were synthesized in-house from commercially available phosphoramidites (Glen Research, Sterling, VA). Restriction endonuclease EcoRI was from SibEnzyme (Novosibirsk, Russia). E. coli Fpg was purified as described [29].
Table 1. Sequences of oligonucleotides used in this study.
| ID | Sequence* |
|---|---|
| DDD | CGCGAATTCGCG |
| oxoG4 | CGCXAATTCGCG |
| oxoG10 | CGCGAATTCXCG |
| 8-10-G | CCTGCTTTCGCGAATTCGCGTCCACGGCAC |
| 8-10-X1 | CCTGCTTTCGCXAATTCGCGTCCACGGCAC |
| 8-10-X2 | CCTGCTTTCGCGAATTCXCGTCCACGGCAC |
| 10-8-G | GTGCCGTGGACGCGAATTCGCGAAAGCAGG |
| 10-8-X1 | GTGCCGTGGACGCXAATTCGCGAAAGCAGG |
| 10-8-X2 | GTGCCGTGGACGCGAATTCXCGAAAGCAGG |
*X = 8-oxoguanine;
DDD (Drew—Dickerson dodecamer, common for all the sequences).
Enzyme activity and kinetics
The reaction mixture included 100 mM Tris-HCl (pH 7.6), 50 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, 1.5–100 nM 32P-labeled oligonucleotide duplex, and 0.4 or 2 U/μl EcoRI (approximately 60 or 300 pM, respectively, for kinetics and single-point assay). The reaction was allowed to proceed for 20 min, terminated by adding an equal amount of formamide/EDTA and heating for 5 min at 95°C, and analyzed by electrophoresis in 20% polyacrylamide gel with 8 M urea. The gel was quantified using Molecular Imager FX with Quantity One v3.1 software (Bio-Rad, Hercules, CA).
NMR experiments
NMR data were collected utilizing 700 Bruker NMR spectrometer (1H 700 MHz) equipped with a Prodigy TCI cryoprobe and a BBO probe for the 31P-1H correlation spectra. For the NMR recordings, the duplex DNA samples were dissolved in the following buffer: 10 mM Na phosphate (pH 6.8), 50 mM NaCl, 1 mM EDTA.
The NMR data were processed with NMRPipe software [30] and the spectra were visualized with NMRDraw [30] and NMRView [31]. The NMR resonance assignment was performed by the standard procedure [28] utilizing the sets of 2D NOESY (70, 140 and 200 ms mixing time, 100% 2H2O buffer), the 2D TOCSY (100% 2H2O buffer) as well as the 2D NOESY (90% H2O buffer) data. The assignment of 31P NMR resonances was performed utilizing the standard protocol [32].
The distance restraints were derived from the NOESY cross-peaks generated with NMRView. The volume-to-distance calibration of the NOESY peaks’ volumes was performed using the CYANA 2.1 software [33] with cytosine H5–H6 distance as the calibration standard. When reliably available from all the NOESY spectra collected, every final NOE restraint was calculated as their “averaged” distance. In all other cases, every final NOE restraint was derived from their corresponding most reliable NOESY cross-peak (“non-averaged”). All the NOE restraints were duplicated for the other (self-complimentary) strand for both samples. The obtained distance restraints were grouped according to their length: short (< 3.0 Å), medium (between 3.0 and 5.0 Å) and long (> 5.0 Å) distances as well as methyl-containing restraints. The group-specific no-penalty tolerances were employed based on the NOE distances and spectral quality (S1 Table). The standard Watson—Crick hydrogen bonding distance restraints were applied for all the base pairs based on the NOESY cross-peak patterns observed in 90% H2O. No sugar conformation restraints were used. Based on the observed 31P resonances, the backbone dihedrals were restrained to a standard BI conformation [34] for all the canonical residues, whereas the oxoG nucleotide had the backbone angle ε restrained within 215–295° and ζ within 155–205° range (BII conformation). The dihedral angles adjacent to ε and ζ (two on either side) were allowed to sample a broader range of values (by ± 20°).
Restrained molecular dynamics
Restrained molecular dynamics (rMD) simulations were conducted via a simulated annealing protocol with AMBER 12 [35] using the parm99 force field [36]. The generalized Born (GB) implicit solvation model [37] was used. The starting structures used were of A- and B- DNA type models. The oxoG-specific force field parameters, including the electric charges, were taken from [38]. The protocol for energy minimization, restraints and simulated annealing is included in S1 Document. A total of 18 structures were produced for each sample. The ensemble of structures was determined based on the lowest conformational penalty/energy values. A representative structure for each ensemble was built by averaging the lowest energy models and subjecting the obtained frames to energy minimization.
Free molecular dynamics
Free molecular dynamics (fMD) simulations were performed using PMEMD from AMBER 12. Each structure was parameterized with the AMBER ff99SB force field [39] with parmbsc0 modifications specific to the DNA backbone [40]. Additional information regarding initialization, minimization and equilibration protocols is included in S1 Document. Production molecular dynamics simulations were run for 120 ns using GPU code with the PMEMD.cuda implementation of SANDER [41] from AMBER 12. Long range interactions were calculated with PME. A 2 fs integration time step was used. Simulation coordinates were recorded every 1 ps. The first 10 ns of each simulation were removed giving 110 ns total simulation time for each sample to analyze.
Hybrid explicit/implicit solvent molecular dynamics
The starting model for the MD analysis of EcoRI bound to oxoG-containing DNA was the X-ray structure of pre-transition state EcoRI in a complex with a 13-mer duplex containing the GAATCC sequence (PDB ID 1CKQ). Both G bases in the recognition sequence were changed to oxoG. Heavy atoms missing in the structure and hydrogen atoms were restored using the Missing Atom Restoration module of BioPASED molecular modeling package [42]. Out of 302 water molecules found in the crystal unit cell, 129 tightly bound molecules were identified using the WaterShell optimization algorithm [43] and retained as explicit water, otherwise the modeling was done in EEF1 implicit solvent [44] with counter-ion correction charge scaling [45] to enhance the sampled conformational space [46]. All models were energy-minimized in 500 steps of Fletcher energy optimization followed by 100 ps of simulated annealing using the BioPASED package. Molecular dynamics simulations (12 ns) were performed using BioPASED with the AMBER ff99SB force field [39] with parmbsc0 correction [40]; the integration time step was 1 fs. The system was gradually heated from 10 K to 300 K in 50 ps and equilibrated at this temperature. Classic NVT ensemble molecular dynamics trajectories were generated with harmonic restraints of 0.25 kcal/Å2 for the terminal nucleotides and 0.001 kcal/Å2 for the rest of heavy atoms except amino acid side chains that were modeled with no restraints. Simulation coordinates were recorded every 2 ps.
Structure visualization and analysis
The structures were visualized and analyzed with VMD [47] and Chimera [48]. The molecular dynamics trajectories were analyzed using CPPTRAJ [49], Curves+ [50], 3DNA [51], MDTRA [52] and custom Python scripts.
Results
Effect of oxoG on the GAATTC sequence recognition by EcoRI
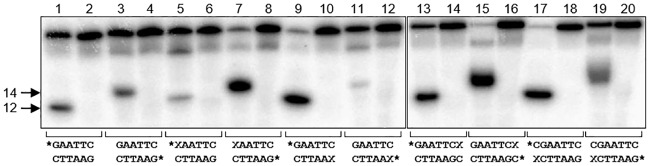
In an attempt to follow possible biological consequences of oxoG in DNA, we have compared the activity of EcoRI restriction endonuclease on its natural substrate and in several contexts containing oxoG. A 30-mer duplex contained a palindromic 12-mer CGCGAATTCGCG sequence (Drew—Dickerson dodecamer, DDD) in the middle flanked by non-palindromic shoulders. One of the guanines in the recognition site GAATTC, or a guanine immediately 3′ to this site, was replaced with oxoG (bold and underlined in the sequence above). The cleavage of either the oxidized strand or the opposite strand was followed.
In the crystal structure of EcoRI, the guanine base in the recognition sequence forms no direct hydrogen bond with the protein [26], yet its replacement with isosteric bases 7-deazaguanine or 2-aminopurine leads to ~60-fold drop in the enzyme activity [53]. In agreement with these observations, introduction of oxoG in the recognition sequence severely suppressed the cleavage of the modified strand (Fig 1, Table 2). The steady-state kinetic parameters of cleavage of the non-damaged strand were compromised, but the reaction still proceeded to completion with more enzyme (Fig 1, Table 2). In an unrelated 18-mer duplex containing the GAATTC site not embedded within the Drew—Dickerson context, the introduction of oxoG inhibited the cleavage almost completely in the modified strand and reduced it in the opposite strand (S1 Fig). When oxoG was introduced next to the GAATTC site, as in the Drew—Dickerson sequence, it increased KM and decreased Vmax of nicking of the modified strand, although the reaction was also efficient with the larger amounts of the enzyme (Fig 1, Table 2). Since EcoRI cleavage is rate-limited by the release of doubly nicked product [54], the apparent increase in the efficiency of cleavage with the concomitant decrease in the Vmax/KM ratio may be due to the change in the rate-limiting step when oxoG is present in the substrate. Regardless of the kinetic scheme, it is clear that oxoG affects the recognition or cleavage of the cognate site by EcoRI.
Fig 1. Cleavage of native and oxoG-modified GAATTC sequence by EcoRI.
Odd lanes, enzyme added (300 pM); even lanes, no enzyme (controls). The concentration of the substrate was 100 nM. The central part of the 30-mer duplexes and the position of oxoG (X) and radioactive label (asterisk) are shown below the image. Cleavage of the top strand produces a 12-mer product; cleavage of the bottom strand produces a 14-mer product.
Table 2. Kinetics of EcoRI on oxoG-containing substrates.
| Sequence of the EcoRI site | KM, nM | Vmax, nM·min–1 | Vmax/KM, min–1 | relative to undamaged# |
|---|---|---|---|---|
|
*-GAATTCG- -CTTAAGC- |
20.0 ± 5.0 | 0.30 ± 0.03 | (15 ± 4)×10−3 | 1 |
|
*-XAATTCG- -CTTAAGC- |
n/d* | |||
|
*-GAATTCG- -CTTAAXC- |
n/d* | n/d* | (0.31 ± 0.04)×10−3 | 0.021 |
|
*-GAATTCX- -CTTAAGC- |
40.0 ± 13.0 | 0.067 ± 0.010 | (1.7 ± 0.6)×10−3 | 0.11 |
*n/d, saturation by the substrate could not be achieved; KM and Vmax were not determined individually; Vmax/KM, when possible, was determined from the linear slope of the v0 vs [S] graph.
#Based on the Vmax/KM ratio.
OxoG in the recognition sequence does not affect the structure of EcoRI—DNA complex
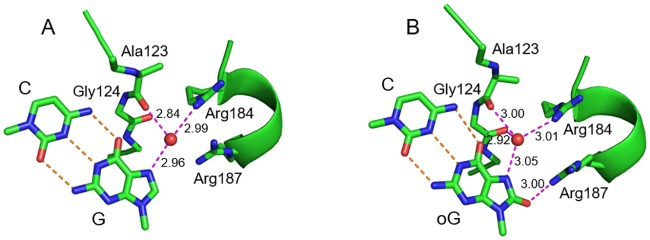
In the crystal structure of pre-transition state EcoRI—DNA complex (PDB 1CKQ), the N7 atom of the guanine base in the recognition sequence accepts an H-bonding interaction with a water molecule that, in turn, is coordinated by the carbonyl oxygen of Ala123 and Nη1 of Arg184 (Fig 2A). Since N7 in oxoG is a hydrogen bond donor rather than acceptor, we performed molecular dynamics of EcoRI bound to either native or oxoG-modified GAATTC site to assess how this change might affect the structure of the enzyme—DNA interface. No significant deviations in the occurrence or energy of bonds existing between an EcoRI dimer and the recognition site, or within base pairs in the recognition site were observed (S2 Table). The N7-bound water molecule simply re-oriented to accept a hydrogen bond from N7 of oxoG, while keeping the HOH···O[Ala123] and Nη1[Arg184]···HOH bonds (Fig 2B). Moreover, in the oxoG-modified structure, the water molecule additionally donated a hydrogen bond to the carbonyl oxygen of Gly124, and O8 of oxoG accepted a hydrogen bond from Nη1 of Arg187 (Fig 2B). We conclude that loss of activity of EcoRI on oxoG-modified recognition site is unlikely to stem from the local perturbation of the enzyme—DNA interface in the pre-incision complex by the lesion. Notably, while all non-terminal nucleotides in the G model were in BI conformation, the oxoG model spontaneously produced a significant fraction of BII in the C nucleotide opposite the lesion (S2 Fig). This backbone effect was the only significant difference observed between DNA in G and oxoG models.
Fig 2. Structures of the G:C pair in the EcoRI recognition site from molecular dynamic simulation.
The representative snapshots (10 ns) for native (A) and oxoG-modified (B) cognate site are shown. Carbon atoms and secondary structure elements are colored green, oxygens, red, nitrogens, blue. For DNA, only the G:C or oxoG:C base pair is shown. Orange dashed lines indicate Watson—Crick bonds, magenta dashed lines show hydrogen bonds with the protein formed by G or oxoG directly or through the interface water molecule (red ball). Lengths of hydrogen bonds (Å) are shown next to the lines.
NMR characterization of normal and damaged duplexes
In an attempt to uncover possible reasons for EcoRI inhibition by oxoG, we have determined solution structures of two duplex DNA samples derived from the Drew—Dickerson dodecamer sequence ([27], DDD): oxoG4 and oxoG10 (Table 1). The oxoG4 sample represents guanine oxidation within the EcoRI recognition site, whereas the oxoG10 sequence has the oxoG lesion flanking this site. In the context of our enzyme kinetics study, oxoG4 corresponds to the 8-10-X1 and 10-8-X1 oligonucleotides, whereas oxoG10 corresponds to the 8-10-X2 and 10-8-X2 counterparts (Table 1).
The composition of the peaks on all the 2D NMR spectra for oxoG4 indicates that the sample adopts a single distinct A/B-DNA conformation. The same observation was made for oxoG10. The NMR resonance assignment for oxoG4 and oxoG10 duplexes was performed by the previously reported procedure [55] utilizing the following 2D NMR data: NOESY (100% 2H2O buffer); TOCSY (100% 2H2O buffer); and NOESY (90% H2O buffer). The 2D NOESY peaks patterns for both samples are highly similar to those reported in the unmodified sequence, a de facto A/B DNA standard [28]. The main differences are localized around the oxoG sites (S3 and S4 Figs). The 2D NOESY spectra of the oxoG4 sample recorded in 100% 2H2O yielded a total of 189 unique NOEs, with 82 of them being “averaged”, 94 “non-averaged” and 13 methyl-containing restraints. The oxoG10 structure was driven by 233 unique NOEs with 120 “averaged”, 90 “non-averaged” and 23 methyl-containing restraints. The DQF-COSY H1′···H2′/H2″ cross-peak patterns, whenever unobscured, established that the sugar conformations of all the nucleotides were in the C2′-endo/C3′-exo range. The 2D NOESY recorded in 90% H2O shows the distinct set of cross-peaks linking protons from the bases on the opposite strands and confirming the Watson—Crick pattern of H-bonding within the non-terminal base pairs (S4 Fig). NMR spectral features involving oxoG protons were as expected (BMRB IDs 25369 and 25528): absence of purine H8 NOESY cross-peaks (S3 Fig), downfield shift of H2″ (> 2.33 ppm), and presence of peaks involving HN7 in spectra recorded in the 90% H2O buffer.
Both 1D and 2D 31P NMR spectra for the two samples show the majority of 31P resonances clustered within 0.5 ppm as previously reported for the unmodified sequence [56] and a single peak from oxoG phosphate group, shifted downfield by ~0.3 ppm (S5 Fig). The chemical shift value of the oxoG 31P resonance indicates a more rare BII backbone conformation [22] unlike the common BI conformation of all other nucleotides.
Solution NMR structure of oxoG-containing duplexes
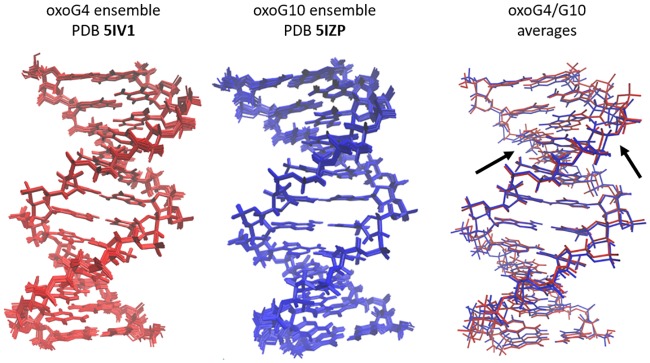
An ensemble of lowest-energy structures was derived from rMD simulations: 13 models for oxoG4 and 14 for oxoG10 (Fig 3). The greatest NOE restraint violation for any model was 0.10 Å and dihedral violations did not exceed 11°. Within each ensemble, the largest pairwise all-atom RMSD value was less than 0.92 Å. The main structural differences between samples oxoG4 and oxoG10 were localized at their respective modification sites (Fig 3).
Fig 3. Ensembles and representative structures for oxoG4 and oxoG10.
The entirely red ensemble (panel on the left) shows the 13 lowest energy structures and the averaged minimized structure for oxoG4, PDB ID 5IV1. The entirely blue ensemble (middle panel) shows the 14 lowest energy structures and the averaged minimized structure for oxoG10, PDB ID 5IZP. The panel on the right shows an overlay of the averaged minimized structures for both oxoG4 and oxoG10, in red and blue, respectively. In this last panel, the arrows show the general location of the two modified base pairs, C3:oxoG4 and C9:oxoG10.
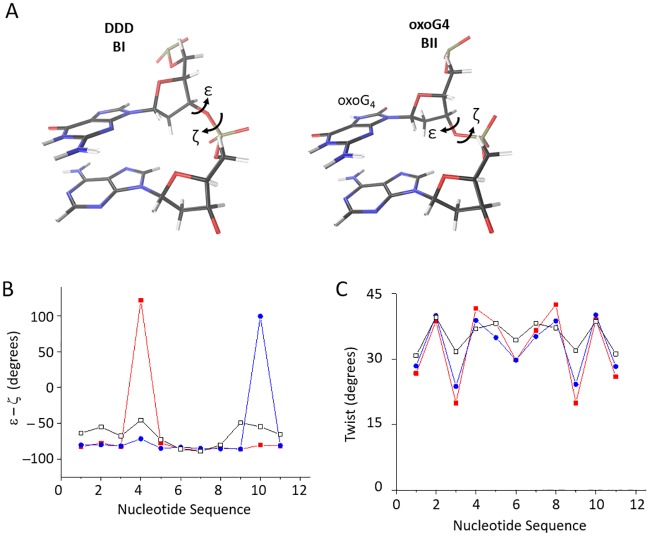
Overall, the structures of oxoG4 and oxoG10 are very similar to each other (backbone RMSD of 1.2 Å between the representative oxoG4 and oxoG10 structures, Fig 3) as well as to known DDD structures (PDB 1BNA, 355D and 1NAJ; backbone RMSD of 0.9–1.4 Å). The major structural perturbation of our oxidized samples with respect to B-DNA is the switch of the backbone to BII conformation 3' to oxoG (Fig 4A and 4B). In addition, the twist helical parameter adopts unusually low values for the base-pairs adjacent to oxoG nucleotides from the 5′ side (Fig 4C). This unwinding effect was previously predicted based on the free molecular dynamics modeling [57] and is also demonstrated in our free MD analysis below.
Fig 4. Results from the NMR-driven restrained molecular dynamics simulation of oxoG4 and oxoG10.
(A) The difference in backbone conformation between BI and BII conformations. ε and ζ are highlighted in accordance to their torsion angle definition, as BI and BII are defined by the ε–ζ. For both DDD BI and oxoG4 BII structures, the G4 and A5 are shown and rotated to highlight the backbone differences. The DDD BI conformation (left panel) is from the PDB ID INAJ structure. The BII conformation induced by oxoG4 (right panel) is from our averaged minimized structure, PDB ID 5IV1. Carbons are shown in black, oxygen is shown in red, nitrogen in blue and hydrogens in white. The additional oxygen at C8 and hydrogen at N7 in the oxoG4 base are color-coded in the same way. (B) The ε–ζ shows BII directly 3' of the modification site for oxoG4 and oxoG10. BI is defined as ε–ζ less than 20°, with over 20° defined as BII. The nucleotide sequence numbers correspond to the steps in the sequence, with ε–ζ for nucleotide step 1 corresponding to the torsion angles between C1 and G2 in C1G2C3G4A5A6T7T8C9G10C11G12. In black with open square markers, are the ε–ζ for 1NAJ (DDD). In red with closed square markers are ε–ζ for oxoG4 and blue with closed circle markers are oxoG10. (C) oxoG4 and oxoG10 cause significant unwinding near the modification site. Color schemes and data origins are the same as in panel B.
Free molecular dynamics of oxoG4 and oxoG10
The coordinates of the last 110 ns of the free dynamics were used to build a representative, averaged structure for each sample. The structures of oxoG4 and oxoG10 are very similar to each other (backbone RMSD of 0.89 Å) as well as to the unmodified DDD (backbone RMSD of 1.4 Å with respect to 1NAJ). The base pair parameters and backbone torsion angles of these average structures were similar to those of our solution NMR structures. Any significant conformational differences were observed only at or around the oxoG sites (S6 and S7 Figs). Introduction of oxoG causes a decrease in twist value for the oxoG-containing CpG base pairs (S6 Fig). The simulations also yielded a decrease in the ζ backbone torsion angle and an increase in the ε angle values for the oxoGpA and oxoGpC steps (in oxoG4 and oxoG10 respectively). As a result, the backbone adopts the BII conformation 3′ to oxoG as opposed to the BI state seen elsewhere in the duplex (S7 Fig).
Discussion
8-Oxoguanine is a pro-mutagenic base constantly generated in DNA and removed through base excision repair. Chemically, it deviates from guanine only by two positions, which do not affect the Watson—Crick edge: oxygen replaces hydrogen at C8, and N7 is protonated. However, the effect of these changes is propagated to other physicochemical properties, some of which are used by DNA repair enzymes to facilitate oxoG recognition. Compared with G, oxoG thermodynamically destabilizes DNA [58], carries a differently oriented dipole [59], has pKa of N1 lower by ~1 pH unit [60], and is more easily oxidized [61]. Moreover, oxoG can perturb the DNA structure. One example is oxoG:A mispair where oxoG adopts a syn conformation possibly due to steric repulsion involving O8 [55]. The BII conformation of the backbone 3' of oxoG in Watson—Crick base pairs is another major oxoG-related structural alteration, which was predicted computationally [57], present (but not described) in a crystal DNA structure [62] and is shown for the first time in solution NMR structures in this work.
The presence of oxoG in DNA interferes with its cleavage by several restriction endonucleases [7–10] and binding by several regulatory proteins [15–18]. Our work shows that oxidation of guanine within the EcoRI recognition site suppresses the endonuclease activity on the modified strand (Fig 1, S1 Fig). Although in some cases, such as MspI and NgoMIV endonucleases [63,64] or AP-1 Jun—Fos heterodimer [65], this effect can be directly attributed to the loss of interactions at the protein—DNA interface, EcoRI seems to conserve the full set of interactions with oxoG-containing recognition sequence as indicated by our MD simulations (Fig 2). Based on our structure and function data, we suggest that conformational changes effected by oxoG on DNA, particularly on the sugar—phosphate backbone, may be responsible for this reduction in EcoRI activity. Our solution NMR spectra and structures as well as free Molecular Dynamics data show that oxoG causes dramatic unwinding of the DNA helix 5′ to oxoG and pushes the backbone into the BII conformation 3′ to oxoG (Fig 4A and 4B and S5 and S7 Figs). The local unwinding (lower twist, Fig 4C, S6 Fig) could hinder either the recognition step or the following DNA kinking necessary for EcoRI activity. Both phosphates flanking G1 (G4 in our DDD sequence) in the EcoRI recognition sequence (G1A2A3T4T5C6) are part of the “primary clamp”, the set of phosphates identified by interference experiments that are required for proper formation of the catalytically competent complex [53] [66]. A similar observation of backbone conformation affecting EcoRI activity was made for a series of 2′-substituted G1 nucleotides, where the gauche-gauche to gauche-trans shift in the GpA phosphodiester bond rendered it non-scissile [67]. Therefore, we suggest that the oxoG lesion could affect backbone-mediated and indirect DNA recognition by other sequence-dependent proteins.
Supporting Information
(PDF)
Odd lanes, no enzyme; even lanes, EcoRI. The asterisk marks the 32P-labeled chain. X = oxoG. The sequence of the EcoRI site-bearing duplex is shown.
(TIF)
(TIF)
The dodecamer sequence was confirmed to be symmetric, right handed helix within this region. The cross-peaks between the base proton H6/H8 and the H1’ of current or next bases (the “walk”) lead to the NMR resonance assignment (base protons labeled). The number of peaks shown confirms the loss of the guanine H8 at the appropriate modification site for both oxoG4 and oxoG10.
(TIF)
(A) 2D NOESY spectra highlighting the Watson-Crick base pairing interactions in oxoG4 and oxoG10. Three separate CG base pairs are observed for both samples, with the resonances from terminal base pairs not showing due to their decreased thermal stability. Two distinct AT base pairs are also observed for both samples. (B) The imino proton regions of the 1D spectra. The modified residues contain an additional hydrogen at N7 (HN7) that is not seen in canonical guanines. The peaks marked with arrows correspond to the HN7 in both samples.
(TIF)
Labels are provided to highlight the residue whose H3' corresponds to that specific peak, all phosphates belong to the residue on the 3' end of the labelled residue. In both spectra, a significant shift is seen in the 31P dimension of the modified residue, demonstrating evidence for that the backbone on the 3' end of the oxidized guanine residue is in BII, and all others are in the canonical BI form.
(TIF)
The nucleotide sequence numbers correspond to the steps in the sequence, with the twist for nucleotide step 1 corresponding to the twist of the first CG base pair with respect to the next one. In black with open square markers, are the DDD twist for the unmodified DDD. In red with closed square markers are the twist for oxoG4 and blue with closed circle markers are oxoG10.
(TIF)
The nucleotide sequence numbers correspond to the steps in the sequence, with the twist for nucleotide step 1 corresponding to the backbone step between the C1 and G2 in DDD sequence. In black with open square markers, are the DDD twist for the unmodified DDD. In red with closed square markers are the twist for oxoG4 and blue with closed circle markers are oxoG10.
(TIF)
(PDF)
(PDF)
Acknowledgments
This research activity of SLS, DRG, JJH, HLM was supported by the United States National Science Foundation (NSF Award 1244641 to SLS). Enzyme kinetic studies and protein molecular dynamics were supported by Russian Science Foundation (14-24-00093 to DOZ). ASK and AVY acknowledge the Russian Science Foundation [grant No. 15-13-20035] for the NMR data collection. AVE is a recipient of the Russian Presidential Fellowship (SP- 673.2015.4). D.O.Z. acknowledges the State Program of Research Support by the Russian Ministry of Education and Science. Molecular graphics and analyses were performed with the UCSF Chimera developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311). Computations were performed in part on the NKS-30T cluster at the SB RAS Supercomputing Center. Special thanks to Brennan Vanden Bos for help with the restrained molecular dynamics and Russell McFarland for assistance with proofreading the manuscript and figures.
Abbreviations
- oxoG
8-oxoguanine
Data Availability
Most of the relevant data are within the paper. The solution NMR and duplex DNA structure are available via RCSB repository, PDB accession numbers 5IV1 and 5IZP as described in the publication. The respective solution NMR data for the duplex DNA samples are available via BMRB repository, accession numbers 25369 and 25528. The PDB/BMRB data is publicly available as of August 5, 2016.
Funding Statement
This research activity of SLS, DRG, JJH, HLM was supported by the United States National Science Foundation (NSF Award 1244641 to SLS). Enzyme kinetic studies and protein molecular dynamics were supported by Russian Science Foundation (14-24-00093 to DOZ). ASK and AVY acknowledge the Russian Science Foundation [grant No. 15-13-20035] for the NMR data collection. AVE is a recipient of the Russian Presidential Fellowship (SP- 673.2015.4). DOZ acknowledges the State Program of Research Support by the Russian Ministry of Education and Science. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kamiya H (2003) Mutagenic potentials of damaged nucleic acids produced by reactive oxygen/nitrogen species: Approaches using synthetic oligonucleotides and nucleotides. Nucleic Acids Research 31: 517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ESCODD (European Standards Committee on Oxidative DNA Damage) (2002) Comparative analysis of baseline 8-oxo-7,8-dihydroguanine in mammalian cell DNA, by different methods in different laboratories: An approach to consensus. Carcinogenesis 23: 2129–2133. [DOI] [PubMed] [Google Scholar]
- 3.Olinski R, Gackowski D, Rozalski R, Foksinski M, Bialkowski K (2003) Oxidative DNA damage in cancer patients: A cause or a consequence of the disease development? Mutation Research—Fundamental and Molecular Mechanisms of Mutagenesis 531: 177–190. [DOI] [PubMed] [Google Scholar]
- 4.Loft S, Danielsen PH, Mikkelsen L, Risom L, Forchhammer L, Møller P (2008) Biomarkers of oxidative damage to DNA and repair. Biochemical Society Transactions 36: 1071–1076. 10.1042/BST0361071 [DOI] [PubMed] [Google Scholar]
- 5.Jacob KD, Hooten NN, Trzeciak AR, Evans MK (2013) Markers of oxidant stress that are clinically relevant in aging and age-related disease. Mechanisms of Ageing and Development 134: 139–157. 10.1016/j.mad.2013.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grollman AP, Moriya M (1993) Mutagenesis by 8-oxoguanine: An enemy within. Trends in Genetics 9: 246–249. [DOI] [PubMed] [Google Scholar]
- 7.Allinson SL, Dianova II, Dianov GL (2001) DNA polymerase β is the major dRP lyase involved in repair of oxidative base lesions in DNA by mammalian cell extracts. EMBO Journal 20: 6919–6926. 10.1093/emboj/20.23.6919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turk PW, Weitzman SA (1995) Free radical DNA adduct 8-OH-deoxyguanosine affects activity of HpaII and MspI restriction endonucleases. Free Radical Research 23: 255–258. [DOI] [PubMed] [Google Scholar]
- 9.Wood ML, Dizdaroglu M, Gajewski E, Essigmann JM (1990) Mechanistic studies of ionizing radiation and oxidative mutagenesis: Genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry 29: 7024–7032. [DOI] [PubMed] [Google Scholar]
- 10.Le Page F, Randrianarison V, Marot D, Cabannes J, Perricaudet M, Feunteun J, et al. (2000) BRCA1 and BRCA2 are necessary for the transcription-coupled repair of the oxidative 8-oxoguanine lesion in human cells. Cancer Research 60: 5548–5552. [PubMed] [Google Scholar]
- 11.Turk PW, Laayoun A, Smith SS, Weitzman SA (1995) DNA adduct 8-hydroxyl-2'-deoxyguanosine (8-hydroxyguanine) affects function of human DNA methyltransferase. Carcinogenesis 16: 1253–1255. [DOI] [PubMed] [Google Scholar]
- 12.Maltseva DV, Baykov AA, Jeltsch A, Gromova ES (2009) Impact of 7,8-dihydro-8-oxoguanine on methylation of the CpG site by Dnmt3a. Biochemistry 48: 1361–1368. 10.1021/bi801947f [DOI] [PubMed] [Google Scholar]
- 13.Pourquier P, Ueng L-M, Fertala J, Wang D, Park H-J, Essigmann JM, et al. (1999) Induction of reversible complexes between eukaryotic DNA topoisomerase I and DNA-containing oxidative base damages: 7,8-dihydro-8-oxoguanine and 5-hydroxycytosine. Journal of Biological Chemistry 274: 8516–8523. [DOI] [PubMed] [Google Scholar]
- 14.Lesher D-TT, Pommier Y, Stewart L, Redinbo MR (2002) 8-Oxoguanine rearranges the active site of human topoisomerase I. Proceedings of the National Academy of Sciences USA 99: 12102–12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh R, Mitchell DL (1999) Effect of oxidative DNA damage in promoter elements on transcription factor binding. Nucleic Acids Research 27: 3213–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valinluck V, Tsai H-H, Rogstad DK, Burdzy A, Bird A, Sowers LC (2004) Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Research 32: 4100–4108. 10.1093/nar/gkh739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Opresko PL, Fan J, Danzy S, Wilson DM III, Bohr VA (2005) Oxidative damage in telomeric DNA disrupts recognition by TRF1 and TRF2. Nucleic Acids Research 33: 1230–1239. 10.1093/nar/gki273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hailer-Morrison MK, Kotler JM, Martin BD, Sugden KD (2003) Oxidized guanine lesions as modulators of gene transcription. Altered p50 binding affinity and repair shielding by 7,8-dihydro-8-oxo-2'-deoxyguanosine lesions in the NF-κB promoter element. Biochemistry 42: 9761–9770. 10.1021/bi034546k [DOI] [PubMed] [Google Scholar]
- 19.Lu T, Pan Y, Kao S-Y, Li C, Kohane I, Chan J, et al. (2004) Gene regulation and DNA damage in the ageing human brain. Nature 429: 883–891. 10.1038/nature02661 [DOI] [PubMed] [Google Scholar]
- 20.Du Y, Wooten MC, Gearing M, Wooten MW (2009) Age-associated oxidative damage to the p62 promoter: Implications for Alzheimer disease. Free Radical Biology and Medicine 46: 492–501. 10.1016/j.freeradbiomed.2008.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wecker K, Bonnet MC, Meurs EF, Delepierre M (2002) The role of the phosphorus BI—BII transition in protein—DNA recognition: the NF-κB complex. Nucleic Acids Research 30: 4452–4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heddi B, Foloppe N, Bouchemal N, Hantz E, Hartmann B (2006) Quantification of DNA BI/BII backbone states in solution. Implications for DNA overall structure and recognition. Journal of the American Chemical Society 128: 9170–9177. 10.1021/ja061686j [DOI] [PubMed] [Google Scholar]
- 23.Brennan CA, Van Cleve MD, Gumport RI (1986) The effects of base analogue substitutions on the cleavage by the EcoRI restriction endonuclease of octadeoxyribonucleotides containing modified EcoRI recognition sequences. J Biol Chem 261: 7270–7278. [PubMed] [Google Scholar]
- 24.Lesser DR, Kurpiewski MR, Jen-Jacobson L (1990) The energetic basis of specificity in the Eco RI endonuclease—DNA interaction. Science 250: 776–786. [DOI] [PubMed] [Google Scholar]
- 25.Jen-Jacobson L (1995) Structural-perturbation approaches to thermodynamics of site-specific protein-DNA interactions. Methods Enzymol 259: 305–344. [DOI] [PubMed] [Google Scholar]
- 26.Kim Y, Grable JC, Love R, Greene PJ, Rosenberg JM (1990) Refinement of Eco RI endonuclease crystal structure: A revised protein chain tracing. Science 249: 1307–1309. [DOI] [PubMed] [Google Scholar]
- 27.Drew HR, Wing RM, Takano T, Broka C, Tanaka S, Itakura K, et al. (1981) Structure of a B-DNA dodecamer: Conformation and dynamics. Proceedings of the National Academy of Sciences USA 78: 2179–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hare DR, Wemmer DE, Chou S-H, Drobny G, Reid BR (1983) Assignment of the non-exchangeable proton resonances of d(C-G-C-G-A-A-T-T-C-G-C-G) using two-dimensional nuclear magnetic resonance methods. Journal of Molecular Biology 171: 319–336. [DOI] [PubMed] [Google Scholar]
- 29.Gilboa R, Zharkov DO, Golan G, Fernandes AS, Gerchman SE, Matz E, et al. (2002) Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. Journal of Biological Chemistry 277: 19811–19816. 10.1074/jbc.M202058200 [DOI] [PubMed] [Google Scholar]
- 30.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: A multidimensional spectral processing system based on UNIX pipes. Journal of Biomolecular NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- 31.Johnson BA (2004) Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods in Molecular Biology 278: 313–352. 10.1385/1-59259-809-9:313 [DOI] [PubMed] [Google Scholar]
- 32.Sklenar V, Miyashiro H, Zon G, Miles HT, Bax A (1986) Assignment of the 31P and 1H resonances in oligonucleotides by two-dimensional NMR spectroscopy. FEBS Lett 208: 94–98. [DOI] [PubMed] [Google Scholar]
- 33.Güntert P (2004) Automated NMR structure calculation with CYANA. Methods in Molecular Biology 278: 353–378. 10.1385/1-59259-809-9:353 [DOI] [PubMed] [Google Scholar]
- 34.Trieb M, Rauch C, Wellenzohn B, Wibowo F, Loerting T, Liedl KR (2004) Dynamics of DNA: BI and BII phosphate backbone transitions. Journal of Physical Chemistry B Condensed Matter, Materials, Surfaces, Interfaces and Biophysical 108: 2470–2746. [Google Scholar]
- 35.Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, et al. (2012) AMBER 12. University of California, San Francisco. [Google Scholar]
- 36.Cheatham TE III, Cieplak P, Kollman PA (1999) A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. Journal of Biomolecular Structure and Dynamics 16: 845–862. 10.1080/07391102.1999.10508297 [DOI] [PubMed] [Google Scholar]
- 37.Tsui V, Case DA (2000) Theory and applications of the generalized Born solvation model in macromolecular simulations. Biopolymers 56: 275–291. [DOI] [PubMed] [Google Scholar]
- 38.Miller JH, Fan-Chiang C-CP, Straatsma TP, Kennedy MA (2003) 8-Oxoguanine enhances bending of DNA that favors binding to glycosylases. Journal of the American Chemical Society 125: 6331–6336. 10.1021/ja029312n [DOI] [PubMed] [Google Scholar]
- 39.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins: Structure, Function, and Bioinformatics 65: 712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pérez A, Marchán I, Svozil D, Sponer J, Cheatham TE III, Laughton CA, et al. (2007) Refinement of the AMBER force field for nucleic acids: Improving the description of α/γ conformers. Biophysical Journal 92: 3817–3829. 10.1529/biophysj.106.097782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pierce LCT, Salomon-Ferrer R, de Oliveira CAF, McCammon JA, Walker RC (2012) Routine access to millisecond time scale events with accelerated molecular dynamics. Journal of Chemical Theory and Computation 8: 2997–3002. 10.1021/ct300284c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Popov AV, Vorob'ev YN (2010) GUI-BioPASED: A program for molecular dynamics simulations of biopolymers with a graphical user interface. Molecular Biology (Moscow) 44: 648–654. [PubMed] [Google Scholar]
- 43.Popov AV, Vorobjev YN, Zharkov DO (2015) MDTRA: Unified approach to molecular dynamics data preparation and analysis. Journal of Biomolecular Structure and Dynamics 33: 37–38. 10.1080/07391102.2015.1032671 [DOI] [Google Scholar]
- 44.Lazaridis T, Karplus M (1999) Effective energy function for proteins in solution. Proteins: Structure, Function, and Bioinformatics 35: 133–152. [DOI] [PubMed] [Google Scholar]
- 45.Ravishanker G, Auffinger P, Langley DR, Jayaram B, Young MA, Beveridge DL (1997) Treatment of counterions in computer simulations of DNA. Reviews in Computational Chemistry 11: 317–372. [Google Scholar]
- 46.Vorobjev YN (2011) Advances in implicit models of water solvent to compute conformational free energy and molecular dynamics of proteins at constant pH. Advances in Protein Chemistry and Structural Biology 85: 281–322. 10.1016/B978-0-12-386485-7.00008-9 [DOI] [PubMed] [Google Scholar]
- 47.Humphrey W, Dalke A, Schulten K (1996) VMD: Visual molecular dynamics. Journal of Molecular Graphics 14: 33–38. [DOI] [PubMed] [Google Scholar]
- 48.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. (2004) UCSF Chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry 25: 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 49.Roe DR, Cheatham TE III (2013) PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. Journal of Chemical Theory and Computation 9: 3084–3095. 10.1021/ct400341p [DOI] [PubMed] [Google Scholar]
- 50.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K (2009) Conformational analysis of nucleic acids revisited: Curves+. Nucleic Acids Research 37: 5917–5929. 10.1093/nar/gkp608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu X-J, Olson WK (2003) 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Research 31: 5108–5121. 10.1093/nar/gkg680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Popov AV, Vorobjev YN, Zharkov DO (2013) MDTRA: A molecular dynamics trajectory analyzer with a graphical user interface. Journal of Computational Chemistry 34: 319–325. 10.1002/jcc.23135 [DOI] [PubMed] [Google Scholar]
- 53.Lesser DR, Kurpiewski MR, Jen-Jacobson L (1990) The energetic basis of specificity in the EcoRI endonuclease-DNA interaction. Science 250: 776–786. [DOI] [PubMed] [Google Scholar]
- 54.Wright DJ, Jack WE, Modrich P (1999) The kinetic mechanism of EcoRI endonuclease. Journal of Biological Chemistry 274: 31896–31902. [DOI] [PubMed] [Google Scholar]
- 55.Kouchakdjian M, Bodepudi V, Shibutani S, Eisenberg M, Johnson F, Grollman AP, et al. (1991) NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H-dG) opposite deoxyadenosine in a DNA duplex. 8-Oxo-7H-dG(syn)•dA(anti) alignment at lesion site. Biochemistry 30: 1403–1412. [DOI] [PubMed] [Google Scholar]
- 56.Gorenstein DG, Schroeder SA, Fu JM, Metz JT, Roongta V, Jones CR (1988) Assignments of 31P NMR resonances in oligodeoxyribonucleotides: Origin of sequence-specific variations in the deoxyribose phosphate backbone conformation and the 31P chemical shifts of double-helical nucleic acids. Biochemistry 27: 7223–7237. [DOI] [PubMed] [Google Scholar]
- 57.Dršata T, Kara M, Zacharias M, Lankaš F (2013) Effect of 8-oxoguanine on DNA structure and deformability. Journal of Physical Chemistry B Condensed Matter, Materials, Surfaces, Interfaces and Biophysical 117: 11617–11622. 10.1021/jp407562t [DOI] [PubMed] [Google Scholar]
- 58.Plum GE, Grollman AP, Johnson F, Breslauer KJ (1995) Influence of the oxidatively damaged adduct 8-oxodeoxyguanosine on the conformation, energetics, and thermodynamic stability of a DNA duplex. Biochemistry 34: 16148–16160. [DOI] [PubMed] [Google Scholar]
- 59.Banerjee A, Yang W, Karplus M, Verdine GL (2005) Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature 434: 612–618. 10.1038/nature03458 [DOI] [PubMed] [Google Scholar]
- 60.Cho BP (1993) Structure of oxidatively damaged nucleic acid adducts: pH dependence of the 13C NMR spectra of 8-oxoguanosine and 8-oxoadenosine. Magnetic Resonance in Chemistry 31: 1048–1053. [Google Scholar]
- 61.Goyal RN, Dryhurst G (1982) Redox chemistry of guanine and 8-oxyguanine and a comparison of the peroxidase-catalyzed and electrochemical oxidation of 8-oxyguanine. Journal of Electroanalytical Chemistry and Interfacial Electrochemistry 135: 75–91. [Google Scholar]
- 62.Lipscomb LA, Peek ME, Morningstar ML, Verghis SM, Miller EM, Rich A, et al. (1995) X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine. Proceedings of the National Academy of Sciences USA 92: 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu QS, Roberts RJ, Guo H-C (2005) Two crystal forms of the restriction enzyme MspI—DNA complex show the same novel structure. Protein Science 14: 2590–2600. 10.1110/ps.051565105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deibert M, Grazulis S, Sasnauskas G, Siksnys V, Huber R (2000) Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA. Nature Structural and Molecular Biology 7: 792–799. [DOI] [PubMed] [Google Scholar]
- 65.Glover JNM, Harrison SC (1995) Crystal structure of the heterodimeric bZIP transcription factor c-Fos—c-Jun bound to DNA. Nature 373: 257–261. 10.1038/373257a0 [DOI] [PubMed] [Google Scholar]
- 66.Rosati O, Srivastava TK, Katti SB, Alves J (2002) Importance of phosphate contacts for sequence recognition by EcoRI restriction enzyme. Biochemical and Biophysical Research Communications 295: 198–205. [DOI] [PubMed] [Google Scholar]
- 67.Gohda K, Matsuo N, Oda Y, Ikehara M, Uesugi S (1997) Effects of 2'-substituents of the first deoxyguanosine residue in the recognition sequence of EcoRI restriction endonuclease activity. Journal of Biochemistry 121: 219–224. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Odd lanes, no enzyme; even lanes, EcoRI. The asterisk marks the 32P-labeled chain. X = oxoG. The sequence of the EcoRI site-bearing duplex is shown.
(TIF)
(TIF)
The dodecamer sequence was confirmed to be symmetric, right handed helix within this region. The cross-peaks between the base proton H6/H8 and the H1’ of current or next bases (the “walk”) lead to the NMR resonance assignment (base protons labeled). The number of peaks shown confirms the loss of the guanine H8 at the appropriate modification site for both oxoG4 and oxoG10.
(TIF)
(A) 2D NOESY spectra highlighting the Watson-Crick base pairing interactions in oxoG4 and oxoG10. Three separate CG base pairs are observed for both samples, with the resonances from terminal base pairs not showing due to their decreased thermal stability. Two distinct AT base pairs are also observed for both samples. (B) The imino proton regions of the 1D spectra. The modified residues contain an additional hydrogen at N7 (HN7) that is not seen in canonical guanines. The peaks marked with arrows correspond to the HN7 in both samples.
(TIF)
Labels are provided to highlight the residue whose H3' corresponds to that specific peak, all phosphates belong to the residue on the 3' end of the labelled residue. In both spectra, a significant shift is seen in the 31P dimension of the modified residue, demonstrating evidence for that the backbone on the 3' end of the oxidized guanine residue is in BII, and all others are in the canonical BI form.
(TIF)
The nucleotide sequence numbers correspond to the steps in the sequence, with the twist for nucleotide step 1 corresponding to the twist of the first CG base pair with respect to the next one. In black with open square markers, are the DDD twist for the unmodified DDD. In red with closed square markers are the twist for oxoG4 and blue with closed circle markers are oxoG10.
(TIF)
The nucleotide sequence numbers correspond to the steps in the sequence, with the twist for nucleotide step 1 corresponding to the backbone step between the C1 and G2 in DDD sequence. In black with open square markers, are the DDD twist for the unmodified DDD. In red with closed square markers are the twist for oxoG4 and blue with closed circle markers are oxoG10.
(TIF)
(PDF)
(PDF)
Data Availability Statement
Most of the relevant data are within the paper. The solution NMR and duplex DNA structure are available via RCSB repository, PDB accession numbers 5IV1 and 5IZP as described in the publication. The respective solution NMR data for the duplex DNA samples are available via BMRB repository, accession numbers 25369 and 25528. The PDB/BMRB data is publicly available as of August 5, 2016.