Abstract
Background
Activating transcription factor 5 (ATF5) is a member of the activating transcription/cAMP response element-binding protein family of basic leucine zipper proteins that plays an important role in cell survival, differentiation, proliferation, and apoptosis. The ATF5 gene generates two transcripts: ATF5 isoform 1 and ATF5 isoform 2. A number of studies indicate that ATF5 could be an attractive target for therapeutic intervention in several tumor types; however, so far, the role of ATF5 has not been investigated in papillary thyroid carcinoma (PTC).
Methods
Quantitative real-time reverse transcription polymerase chain reaction and immuno-histochemical staining were used to study ATF5 mRNA and protein expression in PTC.
Results
We report here that ATF5 is expressed more in PTC tissue than in normal thyroid tissue. Furthermore, this is the first study that describes the presence of both ATF5 isoforms in PTC.
Conclusion
These findings could provide potential applications in PTC cancer treatment.
Keywords: papillary thyroid carcinoma, ATF5, therapeutic target, qRT-PCR, IHC
Background
Thyroid cancer is the most common type of endocrine malignancy. Four thyroid tumor types have been identified based on their histopathological features: papillary (PTC), follicular (FTC), anaplastic (ATC), and medullary thyroid carcinomas (MTC).
PTC and FTC are well-differentiated cancers. Among them, PTC is the most common histotype and accounts for more than 80%–85% of all thyroid malignant neoplasms, whereas FTC accounts for 10%–15% of cases. Another 5% is accounted for by MTC, a neuroendocrine tumor, and the remaining 1% by ATC.1
Although the majority of PTC show an indolent behavior, relapses are seen in many patients. Therefore, it is important to identify new targets for PTC treatment.2,3 Several studies indicate that activating transcription factor 5 (ATF5, also known as ATFx and ATF7), a transcription factor of the ATF/CREB4 regulates differentiation,5–7 cell proliferation, and survival.8–10
Recently, we demonstrated that ATF5 plays a potential role in osteogenic differentiation.11 Previous studies showed that ATF5 supports cell survival by regulating the expression of antiapoptotic proteins MCL-112 in glioblastoma and BCL-213 in epithelial ovarian cancer.
Moreover, Liu et al14 demonstrated that Egr-1 gene was a downstream target of ATF5 that mediates ATF5-dependent cell proliferation, tumorigenic transformation, and survival.
In addition, Ishihara et al15 reported that ATF5 was one of the key molecules involved in the development of oncogenic resistance to radiotherapy, enhancing both the survival ability and malignant potential of lung cancer cells.
Therefore, ATF5 inhibition could be an attractive target for cancer therapy.15 In particular, ATF5 may be an effective method to enhance radiosensitivity in cancer cells and prevent the recurrence and progression of cancer after radiotherapy.
ATF5 is highly expressed in several cancer types, including breast, lung,16 glioma,17 B-cell chronic lymphocytic leukemia,18 rectal,19 and ovarian cancer,13 whereas it is not detectable in most normal human tissues (except the liver, where ATF5 is highly expressed).20
Recent evidence suggests that ATF5 is overexpressed in FTC.21 In particular, Barden et al21 identified the differences in expression levels of several genes, including ATF5 in follicular thyroid adenomas and FTCs.
Despite this, however, the role of ATF5 in PTC, MTC, and ATC has not yet been investigated. In addition, the ATF5 gene generates two transcripts: ATF5 isoform 1 (ATF5, transcript variant 1: NM_012068.5) and ATF5 isoform 2 (ATF5, transcript variant 2: NM_001193646.1), which encodes the same single 30 kDa protein.22 These isoforms are identical in their coding regions, but differ in the 5′ untranslated region (UTR), designated as ATF5-5′ UTRα and ATF5-5′ UTRβ. The significance of these two transcripts is presently not known and has not yet been investigated in cancer.
Recent evidence suggests that 5′ UTRα reduced the stability of ATF5 mRNA. Repression of 5′ UTRα is released by amino acid limitation or NaAsO2 exposure via eIF2α phosphorylation. On the other hand, 5′ UTRβ is insensitive to stress conditions.23–25
In the present study, ATF5 mRNA and protein expression were analyzed using quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) and immunohistochemistry (IHC) in PTC and nonneoplastic thyroid tissue.
Moreover, we included some cases of ATC in our study. We clearly demonstrated that ATF5 was expressed more in PTC patients than in normal thyroid tissue, thus suggesting that ATF5 could represent a suitable tool to treat PTC.
Methods
Samples
Fourteen cases of PTC were retrieved from the archives of the Pathology Unit at the Mediterranean Institute of Oncology. Of these, frozen and nonneoplastic thyroid tissue was available for eleven patients. Histopathological data are summarized in Table 1.
Table 1.
Patient characteristics
| Age, years | Sex | Specimen type | T | N | M | Stage | Histological subtype | ATF5 IHC score in tumor | ATF5 IHC score in normal tissue |
|---|---|---|---|---|---|---|---|---|---|
| 53 | Female | Thyroid | 1 | 0 | 0 | I | PTC | 2B | 0 |
| 35 | Female | Thyroid | 1 | 0 | 0 | I | PTC | 0 | 1B |
| 48 | Female | Thyroid | 1 | 0 | 0 | I | PTC | 3B | 0 |
| 58 | Male | Thyroid | 1 (m) | 0 | 0 | I | PTC | 2B | 0 |
| 54 | Male | Thyroid | 3 | 1a | 0 | III | PTC | 2B | 0 |
| 27 | Male | Thyroid | 3 | 1 | 0 | I | PTC | 3B | 0 |
| 52 | Female | Thyroid | 1 | 0 | 0 | I | PTC | 4C | 3B |
| 80 | Female | Thyroid | 3 | 0 | 0 | III | PTC | 3B | 2A |
| 40 | Female | Thyroid | 2 | 1 | 0 | I | PTC | 3B | 2C |
| 59 | Female | Thyroid | 3 | 0 | 0 | III | PTC | 2B | 2B |
| 74 | Female | Thyroid | 3 | 0 | 0 | III | PTC | 2B | 1B |
| 31 | Male | Thyroid | 1 | 1 | 0 | I | PTC | 2A | 2A |
| 61 | Female | Thyroid | 1 | 0 | 0 | I | PTC | 4B | 4A |
| 33 | Female | Thyroid | 3 | 1 | 0 | I | PTC | 2B | 2B |
Notes: The table includes details of 14 cases of papillary thyroid carcinoma in different stages; there were 4 males and 10 females. Age range of patients was 27–80 years, with a mean age of 50 years.
Abbreviations: ATF5, activating transcription factor 5; IHC, immunohistochemistry; PTC, papillary thyroid carcinoma; T, tumor; N, lymph node; M, metastasis; m, multifocal.
In addition, two cases of ATC were evaluated using IHC, and one by qRT-PCR. This study was approved by the Institutional Review Board of Mediterranean Institute of Oncology, and written informed consent was obtained from all patients.
Total RNA extraction and reverse transcription
Total RNA was extracted from eleven normal and PTC frozen tissues and one case of ATC using the RNeasy Mini Isolation Kit (Qiagen, Valencia, CA, USA). RNA purity was assessed by measuring the ratio of the absorbance at 260 and 280 nm, considering 1.8–2 as admissible range of ratios for pure RNAs. RNA quality was analyzed using Agilent 2100 Bioanalyzer RNA assays (Agilent Technologies, Santa Clara, CA, USA) and evaluated by calculating the ratio of the 28S and 18S ribosomal RNA intensity peaks. Total RNA was stored at −80°C.
RNA samples (1 μg) were reverse transcribed using the High-Capacity cDNA Reverse Transcription (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol. Reverse transcription products were stored at −20°C.
Real-time RT-PCR
qRT-PCR was performed using EURx® SG qPCR Master Mix according to the manufacturer’s protocol on a HT7900 instrument (Applied Biosystems). Primers were designed using Primer BLAST26 and specifically recognize selected mRNAs by targeting exon–exon junctions (Table 2). ATF5 transcriptional isoforms are differentially amplified according to dissimilarities in 5′ UTRs. Specificity of the amplification products was confirmed by examination of dissociation reaction plots. A distinct single peak indicated that a single DNA sequence was amplified during PCR. Each sample was tested in triplicate, and samples were used for analysis of relative gene expression using the 2−ΔΔCT method. Reported ΔCT values are inversely proportional to target mRNA concentration.
Table 2.
List of primers used in real-time PCR analysis
| Target transcript | GenBank ID | Primer sequence | Temp Fwd/Rev (°C) | Prod size (bp) |
|---|---|---|---|---|
| ATF5 transcript variant 1 | NM_012068.5 | Fwd: CAGGAAATTCTGCAAGCAAGGAA Rev: CGGCGACACTCTTCCCTCTG |
60/62 | 220 |
| ATF5 transcript variant 2 | NM_001193646.1 | Fwd: TGTCCTCGGATCACAGTCTCT Rev: AAGTGGAAGACTCCATGGCTG |
60/60 | 201 |
| GAPDH | NM_002046.5 | Fwd: GCTCTCCAGAACATCATCCCTGCC Rev: GCGTTGTCATACCAGGAAATGAGCTT |
65/63 | 347 |
| TUBB | NM_178014.3 | Fwd: GCGCATTCCAACCTTCCAG Rev: CCCAGAACTTGGCACCGAT |
59/60 | 166 |
Abbreviations: ATF5, activating transcription factor 5; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; PCR, polymerase chain reaction; TUBB, tubulin beta chain; Prod, product; bp, base pair; Fwd, forward; Rev, reverse.
Immunohistochemistry
ATF5 expression was analyzed using IHC on 14 PTC formalin-fixed paraffin-embedded specimens and adjacent normal thyroid tissues and two ATCs. Paraffin blocks were cut as 3-μm sections. Tissue sections were dewaxed with xylene and rehydrated in descending concentrations of ethanol (100% and then 75%). Antigen retrieval was performed with a pH 8.0 EDTA solution at 95°C for 40 minutes in a Decloaking Chamber (Biocare Medical, Walnut Creek, CA, USA). After rinsing briefly in phosphate-buffered saline (PBS 1×), sections were incubated for 10 minutes with 3% H2O2 at room temperature in the dark and washed again.
Afterward, the sections were permeabilized with Triton 0.4% for 30 minutes. Subsequently, the nonspecific binding of the antibodies was blocked with 4% blocking serum (bovine serum albumin; Sigma Aldrich Co., St Louis, MO, USA) for 30 minutes. The sections were next stained with a rabbit anti-ATF5 antiserum (Novus Biologicals, Littleton, CO, USA) at a ratio of 1:500 in a humidified chamber overnight at 4°C.
The following day, sections were washed three times (5 minutes each) in PBS 1×.
Visualization was achieved by exposing the sections to Envision + Dual Link System-HRP (Dako Denmark A/S, Glostrup, Denmark) for 30 minutes at room temperature. The sections were then incubated with diaminobenzidine solution and rinsed with PBS 1×. The slides were counterstained with Harris’s hematoxylin for 1 minute at room temperature and, after dehydration, sealed with a drop of mounting medium.
Interpretation of staining was done using a semiquantitative system. The percentage of stained cells was assessed as follows: 1=1%–25%; 2=26%–50%; 3=>51%–75%; 4=76%–100%, whereas staining intensity was scored as follows: A = low; B = mid; C = strong (Table 1).
Statistical analyses
Differences in ATF5 transcripts expression between normal and tumor samples were assessed using paired Student’s t-test. ATF5 staining has been evaluated comparing both the percentage of ATF5-positive cells and the intensity of the signal in tumor tissues and adjacent area using Fisher’s exact test. Comparisons resulting in P-values <0.05 were reported as statistically significant. Statistical calculation was performed using the software R (version 3.2.1, Lucent Technologies, Murray Hill, NJ, USA).27
Results
ATF5 expression in PTC
mRNA levels of ATF5 isoforms 1 and 2 were examined by real-time PCR analysis on eleven PTC and adjacent normal samples using the primers listed in Table 2.
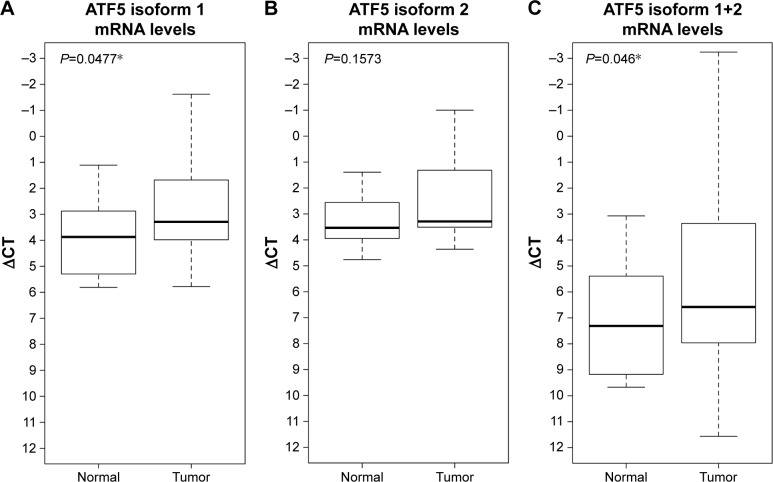
Our results indicate that ATF5 isoform 1 mRNA level was significantly higher (P=0.0477) in PTC when compared to adjacent normal tissue (ΔCT medians: 3.3 for tumor and 3.9 for normal); ATF5 isoform 2 was not statistically significant (ΔCT medians: 3.3 for tumor and 3.5 for normal). Also, there was a statistically significant difference (P=0.046) when both isoforms were considered together (ΔCT medians: 6.6 for tumor and 7.3 for normal). These data are shown in Figure 1.
Figure 1.
ATF5 isoforms mRNA levels in PTC and adjacent normal sample by qRT-PCR.
Notes: ΔCT measurements for (A) ATF5 isoform 1 (medians: 3.3 for tumor and 3.9 for normal), (B) ATF5 isoform 2 (medians: 3.3 for tumor and 3.5 for normal), and (C) both isoforms (medians: 6.6 for tumor and 7.3 for normal) in eleven PTC sample and adjacent normal sample. *indicates P<0.01.
Abbreviations: ATF5, Activating transcription factor 5; PTC, papillary thyroid carcinoma; qRT-PCR, quantitative reverse transcription polymerase chain reaction.
Such findings suggest that ATF5 is expressed more in PTC patients than in normal samples, and hence this could be an attractive therapeutic target for PTC cancer treatment.
ATF5 expression in ATC
mRNA levels of ATF5 isoforms 1 and 2 were also examined by real-time PCR analysis on one ATC and adjacent normal samples. The results showed an increased expression of both isoforms in ATC patients than in normal sample (data not shown).
IHC ATF5 expression
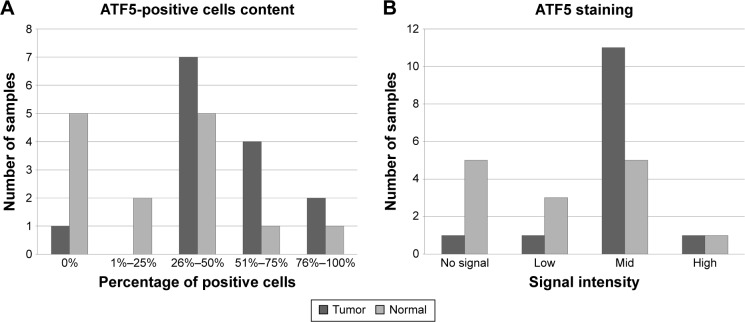
To confirm these data, we performed an anti-ATF5 immunohistochemical analysis on 14 PTC specimens. We found higher ATF5 expression in PTC when compared to adjacent normal thyroid tissue (Figure 2), confirming the data obtained from real-time PCR analysis.
Figure 2.
Increased ATF5 expression in immunohistochemical analyses performed on PTC patients.
Notes: Immunohistochemical analysis of ATF5 expression in 14 formalin-fixed paraffin-embedded specimens from PTC patients and adjacent normal tissue. Differential immunohistochemical expression of ATF5 in normal and tumor tissue. (A) indicates the percentage of positive cells, while in (B), the staining intensity.
Abbreviations: ATF5, activating transcription factor 5; PTC, papillary thyroid carcinoma.
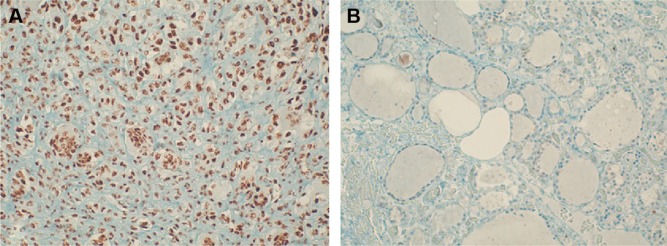
Representative staining of ATF5 protein expression in normal and PTC tissues is shown in Figure 3. Afterward, we performed an anti-ATF5 immunohistochemical analysis on two ATC specimens. Interestingly, we observed a strong expression of the protein (data not shown).
Figure 3.
Immunohistochemical expression of ATF5 in PTC.
Notes: Immunohistochemical expression of ATF5 in papillary thyroid carcinoma (A), 20×, and adjacent normal thyroid tissue (B), 20×.
Abbreviations: ATF5, activating transcription factor 5; PTC, papillary thyroid carcinoma.
Discussion
ATF5 is a stress response transcription factor whose expression is regulated by posttranscriptional, translational, and posttranslational regulation in response to cellular stresses. Although studies have reported that ATF5 is involved in the progression of several tumor types, its expression in PTC has never been studied. Our results are in line with previous data, which reports that in general, the expression level of ATF5 is significantly higher in malignant tissues than their normal counterparts. The only exception appears to be hepatocellular carcinoma cells, which express lower levels of ATF5 than normal liver cells.20
Furthermore, several studies have reported that high ATF5 expression levels may correlate with poor prognosis in cancer patients.12 In our study, however, there was no significant association between ATF5 expression and size of the tumor, local extension, lymph node involvement, or distant metastasis.
Interestingly, we observed a strong expression of the protein in some ATC included in our study. ATC is an undifferentiated carcinoma characterized by an aggressive behavior and a poor prognosis. Further analysis with real-time PCR confirmed an increased expression of both isoforms.
Moreover, more data on ATC would be necessary to confirm our data in a larger cohort of patients.
The main biological function of ATF5 is related to apoptosis, and several studies have been conducted so far to investigate the role of ATF5 as a possible therapeutic target. Interference with ATF5 function or expression in glioma cells and breast cancer cell lines caused marked apoptotic cell death.16,28 Moreover, loss of function of ATF5 in conjunction with paclitaxel treatment elicited apoptosis of pancreatic carcinoma cells.29
In addition, Karpel-Massler et al,30 demonstrated that a dominant-negative ATF5 peptide induces apoptosis in glioblastoma, triple-negative breast cancer (MDA-MB-436), hormone-refractory prostate cancer (PC3 and DU145), resistant non-small-cell lung cancer (H1975), BRAF (V600E)-mutated melanoma (A375), and pancreatic carcinoma (PANC-1). This dominant-negative ATF5 also showed in vivo efficacy in reducing the growth of a range of tumor types in xenograft models.
To our knowledge, this study provides the first evaluation of ATF5 expression and identification of its two isoforms in PTC and normal tissue. Targeting of ATF5 could potentially become a viable therapeutic approach for patients diagnosed with PTC.
Conclusion
In conclusion, our data revealed that ATF5 mRNA expression and protein was higher in PTC compared to normal thyroid tissue. Our results unambiguously demonstrate the existence of both ATF5 isoforms in PTC.
Acknowledgments
This research was partially funded by the Italian “PON Ricerca e Competitività 2007–2013, ASSE I 1078” program entitled “Identificazione di biomarcatori e sviluppo di metodi diagnostici e terapeutici nel campo dell’oncologia e della biologia vascolare”.
The authors gratefully acknowledge Dr Salvatore Scarpulla and Dr Rosario Gulino for their helpful assistance as well as Gabriele Anastasi and Giovanni Ferlito for technical assistance.
Footnotes
Author contributions
LV designed the study, analyzed the results, and wrote the manuscript. CLR performed IHC and contributed to data analysis. SF and GC performed real-time polymerase chain reaction experiments and contributed to data analysis. SS provided the surgical samples and clinical data. EA performed IHC. LM and CC analyzed IHC and critically revised the manuscript. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Cabanillas ME, McFadden DG, Durante C. Thyroid cancer. Lancet. 2016 May 27; doi: 10.1016/S0140-6736(16)30172-6. Epub. [DOI] [PubMed] [Google Scholar]
- 2.Regalbuto C, Frasca F, Pellegriti G, et al. Update on thyroid cancer treatment. Future Oncol. 2012;8(10):1331–1348. doi: 10.2217/fon.12.123. [DOI] [PubMed] [Google Scholar]
- 3.Leonardi GC, Candido S, Carbone M, et al. BRAF mutations in papillary thyroid carcinoma and emerging targeted therapies (review) Mol Med Rep. 2012;6:687–694. doi: 10.3892/mmr.2012.1016. [DOI] [PubMed] [Google Scholar]
- 4.Wei Y, Ge Y, Zhou F, et al. Identification and characterization of the promoter of human ATF5 gene. J Biochem. 2010;148(2):171–178. doi: 10.1093/jb/mvq047. [DOI] [PubMed] [Google Scholar]
- 5.Greene LA, Lee HY, Angelastro JM. The transcription factor ATF5: role in neurodevelopment and neural tumors. J Neurochem. 2009;108(1):11–22. doi: 10.1111/j.1471-4159.2008.05749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angelastro JM, Mason JL, Ignatova TN, et al. Downregulation of activating transcription factor 5 is required for differentiation of neural progenitor cells into astrocytes. J Neurosci. 2005;25(15):3889–3899. doi: 10.1523/JNEUROSCI.3447-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angelastro JM, Ignatova TN, Kukekov VG, et al. Regulated expression of ATF5 is required for the progression of neural progenitor cells to neurons. J Neurosci. 2003;23(11):4590–4600. doi: 10.1523/JNEUROSCI.23-11-04590.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dluzen D, Li G, Tacelosky D, Moreau M, Liu DX. BCL-2 is a downstream target of ATF5 that mediates the prosurvival function of ATF5 in a cell type-dependent manner. J Biol Chem. 2011;286(9):7705–7713. doi: 10.1074/jbc.M110.207639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheng Z, Ma L, Sun JE, Zhu LJ, Green MR. An activating transcription factor 5-mediated survival pathway as a target for cancer therapy. Oncotarget. 2010;1(6):457–460. doi: 10.18632/oncotarget.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Lin G, Zhang Z. ATF5 promotes cell survival through transcriptional activation of Hsp27 in H9c2 cells. Cell Biol Int. 2007;31(11):1309–1315. doi: 10.1016/j.cellbi.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Vicari L, Calabrese G, Forte S, et al. Potential role of activating transcription factor 5 during osteogenesis. Stem Cells Int. 2016:5282185. doi: 10.1155/2016/5282185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheng Z, Li L, Zhu LJ, et al. A genome-wide RNA interference screen reveals an essential CREB3L2/ATF5/MCL1 survival pathway in malignant glioma with therapeutic implications. Nat Med. 2010;16(6):671–677. doi: 10.1038/nm.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen A, Qian D, Wang B, et al. ATF5 is overexpressed in epithelial ovarian carcinomas and interference with its function increases apoptosis through the downregulation of Bcl-2 in SKOV-3 cells. Int J Gynecol Pathol. 2012;31(6):532–537. doi: 10.1097/PGP.0b013e31824df26b. [DOI] [PubMed] [Google Scholar]
- 14.Liu DX, Qian D, Wang B, Yang JM, Lu Z. p300-dependent ATF5 acetylation is essential for Egr-1 gene activation and cell proliferation and survival. Mol Cell Biol. 2011;31(18):3906–3916. doi: 10.1128/MCB.05887-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishihara S, Yasuda M, Ishizu A, Ishikawa M, Shirato H, Haga H. Activating transcription factor 5 enhances radioresistance and malignancy in cancer cells. Oncotarget. 2015;6(7):4602–4614. doi: 10.18632/oncotarget.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monaco SE, Angelastro JM, Szabolcs M, Greene LA. The transcription factor ATF5 is widely expressed in carcinomas, and interference with its function selectively kills neoplastic, but not nontransformed, breast cell lines. Int J Cancer. 2007;120(9):1883–1890. doi: 10.1002/ijc.22469. [DOI] [PubMed] [Google Scholar]
- 17.Arias A, Lamé MW, Santarelli L, Hen R, Greene LA, Angelastro JM. Regulated ATF5 loss-of-function in adult mice blocks formation and causes regression/eradication of gliomas. Oncogene. 2012;31(6):739–751. doi: 10.1038/onc.2011.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mittal AK, Hegde GV, Aoun P, et al. Molecular basis of aggressive disease in chronic lymphocytic leukemia patients with 11q deletion and trisomy 12 chromosomal abnormalities. Int J Mol Med. 2007;20(4):461–469. [PubMed] [Google Scholar]
- 19.Kong X, Meng W, Zhou Z, et al. Overexpression of activating transcription factor 5 in human rectal cancer. Exp Ther Med. 2011;2(5):827–831. doi: 10.3892/etm.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y, Wu B, Chen R, Zheng Y, Huang Z. High ATF5 expression is a favorable prognostic indicator in patients with hepatocellular carcinoma after hepatectomy. Med Oncol. 2014;31:269. doi: 10.1007/s12032-014-0269-0. [DOI] [PubMed] [Google Scholar]
- 21.Barden CB, Shister KW, Zhu B, et al. Classification of follicular thyroid tumors by molecular signature: results of gene profiling. Clin Cancer Res. 2003;9(5):1792–1800. [PubMed] [Google Scholar]
- 22.Hatano M, Umemura M, Kimura N, et al. The 5′-untranslated region regulates ATF5 mRNA stability via nonsense-mediated mRNA decay in response to environmental stress. FEBS J. 2013;280(18):4693–4707. doi: 10.1111/febs.12440. [DOI] [PubMed] [Google Scholar]
- 23.Watatani Y, Ichikawa K, Nakanishi N, et al. Stress-induced translation of ATF5 mRNA is regulated by the 5-untranslated region. J Biol Chem. 2008;283(5):2543–2553. doi: 10.1074/jbc.M707781200. [DOI] [PubMed] [Google Scholar]
- 24.Zhou D, Palam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC. Phosphorylation of eIF2 directs ATF5 translational control in response to Diverse stress conditions. J Biol Chem. 2008;283(11):7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]
- 25.Watatani Y, Kimura N, Shimizu YI, et al. Amino acid limitation induces expression of ATF5 mRNA at the post-transcriptional level. Life Sci. 2007;80(9):879–885. doi: 10.1016/j.lfs.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 26.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13(134):1–11. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Accessed August 6, 2016]. Available at https://www.R-project.org/ [Google Scholar]
- 28.Angelastro JM, Canoll PD, Kuo J, et al. Selective destruction of glioblastoma cells by interference with the activity or expression of ATF5. Oncogene. 2006;25(6):907–916. doi: 10.1038/sj.onc.1209116. [DOI] [PubMed] [Google Scholar]
- 29.Hu M, Wang B, Qian D, et al. Interference with ATF5 function enhances the sensitivity of human pancreatic cancer cells to paclitaxel-induced apoptosis. Anticancer Res. 2012;32(10):4385–4394. [PubMed] [Google Scholar]
- 30.Karpel-Massler G, Horst BA, Shu C, et al. A synthetic cell-penetrating dominant-negative ATF5 peptide exerts anti-cancer activity against a broad spectrum of treatment resistant cancers. Clin Cancer Res. 2016 Apr 28; doi: 10.1158/1078-0432.CCR-15-2827. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]