Abstract
Background
Clinical features of epidermal growth factor receptor (EGFR) mutations: L858R, deletions in exon 19, T790M, insertions in exon 20, G719X, and L861X in non-small-cell lung cancer (NSCLC) are well-known. The clinical significance of other uncommon EGFR mutations, such as E709X, is not well understood. This study aimed to improve the understanding of E709X, and the clinical response to tyrosine kinase inhibitors (TKIs) of NSCLC patients with such an uncommon mutation.
Methods
Specimens from 3,146 patients were tested for EGFR mutations. We surveyed the clinical data and the effectiveness of TKI treatment in NSCLC patients with EGFR mutations E709X.
Results
Of the 3,146 patients, 1,689 (53.7%) had EGFR mutations. This included 726 patients with deletions in exon 19, 733 patients with L858R, and 230 (13.6%) patients with other EGFR mutations. In the 230 patients who had mutations other than single deletion in exon 19 or single L858R in exon 21, 25 (1.5%) patients had the uncommon E709X mutations. Twenty patients had complex E709X mutations and five had single E709X mutation: delE709-T710insD. Of these 25 patients, 18 received either gefitinib or erlotinib treatment. The response rate of TKIs treatment was 50.0%, and the median progression-free survival was 6.2 months. All 5 patients who had delE709-T710insD were non-responders to TKI treatments.
Conclusion
E709X EGFR mutations constituted a small part of the whole group of EGFR mutations. Most patients had complex mutations. The mutation delE709-T710insD was a single mutation and was not associated with good response to TKI treatment.
Keywords: EGFR, erlotinib, gefitinib, lung cancer, mutations
Introduction
Mutations of the epidermal growth factor receptor (EGFR) have influenced the treatment decisions in patients with non-small-cell lung cancer (NSCLC) in the recent years.1 EGFR mutations were found in 30% to 60% of Asian patients and in 10% to 20% of Caucasian patients with NSCLC.2–4 Apart from their association with ethnicity, EGFR mutations occur more frequently in NSCLC of never smokers, females, and adenocarcinoma cell type.5,6 The EGFR tyrosine kinase inhibitors (TKIs), such as erlotinib or gefitinib, are highly active against advanced NSCLC with EGFR mutations.7,8
EGFR mutations exist in exons 18 to 21, and the two major EGFR mutations are deletions in exon 19 and L858R in exon 219,10 which constitute about 80% to 90% of total EGFR mutations.2–4,11,12 In addition to deletions in exon 19 and L858R, other categories of EGFR mutations, which are also well-known, are T790M,13,14 insertions (or in-frame duplications) in exon 20,15,16 G719X and L861X.17 T790M can be a primary mutation,18,19 or a secondary mutation acquired after treatment with EGFR TKIs.13,14 Both insertions (or in-frame duplications) in exon 20 or T790M (also in exon 20) give rise to resistance to EGFR TKIs. Besides, amino acid substitutions at G719 (G719X) and L861 (L861X) are EGFR mutations that were associated with favorable effectiveness of EGFR TKIs.17
These EGFR mutations, including deletions in exon 19, L858R, G719X, L861X, T790M and insertions in exon 20, have documented clinical significance and are well clarified. In the present study, we focused on another category of EGFR mutation which is constituted of amino acid substitutions or deletions in E709 (E709X). E709X was only reported in small case numbers in the literature, and their influences on the effectiveness of EGFR TKIs have not been fully understood.17,20,21 On the contrary, the effectiveness of TKIs in NSCLC patients who harbored EGFR mutations beyond the rare mutation variant E709X has been documented. For example, in patients with deletions in exon 19 and L858R treated with TKIs, the response rate (37.5% to 82.7%),3,5,7,8 progression-free survival (PFS) (7.5 months to 12.6 months),3,4 and overall survival (16.1 months to 27.0 months)3,4 are favorable. In order to increase the understanding to the entire spectrum of EGFR mutations, we investigated the clinical features of these E709X EGFR mutations in the present study.
Materials and methods
Patient characteristics
NSCLC patients diagnosed at the National Taiwan University Hospital between January 2000 and December 2014 were included in the study. Complete cancer staging, including bronchoscopy, computed tomography (CT) of the head, chest, and abdomen, and whole-body bone scintigraphy, was performed for all patients in the hospital. The patients’ clinical data were reviewed. Never smokers were defined as those who had smoked <100 cigarettes in their lifetime. Lung cancer histology was defined on the basis of the World Health Organization pathology classification.22 Date of diagnosis, treatments received, and responsiveness to treatments were recorded. Clinical staging was decided according to the sixth edition of TNM classification of NSCLC. Tumor specimens obtained by either surgical or needle biopsy/aspiration procedures, from primary lung tumors, other distant metastases, and malignant effusion cell blocks, were sequenced for mutational analysis. This study was approved by the National Taiwan University Hospital’s Institutional Review Board. Written informed consent for use of tissue in molecular analysis was acquired from patients at the procurement of tumor specimens.
Efficacy evaluation of EGFR TKIs
We identified the treatments of TKIs (gefitinib or erlotinib) for stage IIIB or stage IV NSCLC from the records of the hospital’s Department of Pharmacy. The timing of different EGFR TKIs depended on the physicians’ discretion. Baseline assessments were performed before treatment. Chest radiography was routinely carried out and assessed every 2–4 weeks to evaluate the response to treatment, while a chest CT scan (including liver and adrenal glands) was performed every 2–3 months to evaluate treatment response and disease progression.
According to the criteria of the response evaluation criteria in solid tumors (RECIST) group, treatment responses were stratified as complete response, partial response, stable disease, and progressive disease.23 Responders to anti-tumor therapy were defined as patients with partial or complete response, and the rest were classed as non-responders. Disease control status comprised complete response, partial response, and stable disease.24 Disease control rate was defined as the patient number with disease control status divided by the total patient number. The cut-off date for data collection was January 10, 2016. PFS with erlotinib or gefitinib was measured from the first day of gefitinib or erlotinib treatment until the first objective or clinical sign of disease progression or death. Overall survival was measured from the first day of gefitinib or erlotinib treatment to the cut-off date or the day of death.
Mutational analysis of EGFR
Tumor specimens, including paraffin blocks or frozen tissues of surgical specimens, fine needle biopsies, and pleural effusions, were obtained for mutational analysis. Acquisition of tumor specimens for testing for EGFR mutations was performed before EGFR TKI treatments. Tissue sections were examined for adequacy by microscopy with hematoxylin and eosin staining and macrodissection was used. Some of the materials have been examined previously, and reported in studies regarding EGFR mutations.6,15,25–29
The mutational analysis of EGFR genes was performed as previously reported.6,26,30 In summary, DNA was derived from tumors embedded in paraffin blocks by using a QIAmp DNA Mini Kit (Qiagen, Valencia, CA, USA). The exons 18, 19, 20 and 21 of EGFR were amplified, while independent polymerase chain reaction (PCR) amplifications were purified and analyzed by direct dideoxynucleotide sequencing with an automatic ABI Prism 3100 or 3700 DNA Analyzer (Applied Biosystems, Foster City, CA, USA).
Frozen lung cancer tissues were procured at surgery, immediately snap frozen in liquid nitrogen, and stored until use. Malignant pleural effusion fluid was centrifuged at 250× g for 10 min, and the cell pellets were frozen in RNAlater (Qiagen).
Total mRNA was extracted from frozen cancer tissue or cell pellets from pleural effusion using TRI reagent (Molecular Research Center, Cincinnati, OH, USA) and an RNA extraction kit (RNeasy Mini Kit; Qiagen). The four exons (exons 18–21) of the EGFR gene were amplified by reverse transcription-PCR (RT-PCR) using a Qiagen One-Step RT-PCR Kit (Qiagen). The primers and RT-PCR conditions have been reported previously.26,30 The cDNA amplicons were purified and sequenced. Both the forward and reverse sequences obtained were analyzed.
Statistical analyses
All categorical variables were analyzed with chi-square tests or Fisher’s exact test. For multivariate analysis, logistic regression, using the stepwise method, was implemented. Overall survival and PFS after gefitinib or erlotinib treatment were analyzed with the Kaplan–Meier method to assess the time to death or progression. Cox-regression was used for multivariate survival analysis. A log-rank test was employed to compare cumulative survival in different groups. All P-values were two-sided and a P-value <0.05 was considered statistically significant. All analyses were performed using SPSS software (version 13.0; SPSS Inc. Chicago, IL, USA).
Results
Patient characteristics
Tumor specimens (996 surgical specimens, 1,002 fine-needle biopsies, and 1,148 pleural effusions) from a total of 3,146 NSCLC patients were examined for mutations of the EGFR tyrosine kinase domain. Of the total 3,146 patients, 696 were smokers and 2,450 were never smokers. There were 2,636 adenocarcinomas and 510 non-adenocarcinomas.
Sixteen hundred and eighty nine (53.7%) patients had EGFR mutations, and 1,457 patients had wild type EGFR (Table 1). The mutations were more frequent in never smokers than smokers (61.6% vs 26.0%, P<0.001), in adenocarcinomas than non-adenocarcinomas (61.1% vs 15.3%, P<0.001), and in females than males (63.9% vs 43.7%, P<0.001).
Table 1.
Demographics of patients screened for EGFR mutations
| All patients screened for EGFR mutations | Patients with EGFR mutations | Patients with wild type EGFR | P-valuea | Patients with E709X EGFR mutations | |
|---|---|---|---|---|---|
| Patient number | 3,146 | 1,689 | 1,457 | 25 | |
| Median age (year, range) | 66 (25–102) | 65 (27–96) | 67 (25–102) | 64 (48–85) | |
| Gender | <0.001 | ||||
| Male | 1,590 | 695 | 895 | 13 | |
| Female | 1,556 | 994 | 562 | 12 | |
| Smoking | <0.001 | ||||
| Smoker | 696 | 181 | 515 | 8 | |
| Never smoker | 2,450 | 1,508 | 942 | 17 | |
| Histology | <0.001 | ||||
| Adenocarcinoma | 2,636 | 1,611 | 1,025 | 22 | |
| Non-adenocarcinoma | 510 | 78 | 432 | 1 | |
| EGFR mutation | |||||
| Deletion in exon 19 | 726 | ||||
| L858R | 733 | ||||
| Others | 230 |
Note:
Comparison between patients with EGFR mutation and patients with wild type EGFR.
Abbreviation: EGFR, epidermal growth factor receptor.
Of the 1,689 patients with EGFR mutations, 733 (43.4%) patients had single L858R in exon 21, 726 (43.0%) patients had single deletions in exon 19, and 230 patients had other EGFR mutations (13.6%). In the 230 patients who had mutations other than single deletion in exon 19 or single L858R in exon 21, 25 (1.5%) patients had uncommon E709X mutations. The demographics of all the patients with and without EGFR mutations, and patients with E709X mutations are listed in Table 1.
In the 25 patients with EGFR E709X mutations, 5 patients had single mutation DelE709-T710insD, and the other 20 patients had complex mutations (one E709X mutation and another EGFR mutation) (Tables 2 and 3). Some of these patients had been reported in previously published studies (case 8, 10, 12, 13, 18, 19, 20, 24, 25).4,17
Table 2.
Summary of clinical information of patients treated with EGFR TKIs
| Patient no | Gender | Age (y) | Smoking | Stage | Cell | EGFR mutations | TKI | TKI response | Outcome | PFS (m) | OS (m) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 61 | No | IV | AD | DelE709-T710insD | Gefitinib | SD | Alive | 5.1 | 79.0 |
| 2 | M | 65 | Yes | IV | AD | DelE709-T710insD | Gefitinib | PD | Dead | 0.9 | 11.1 |
| 3 | F | 57 | No | IV | AD | DelE709-T710insD | Gefitinib | PD | Dead | 0.6 | 24.1 |
| 4 | M | 79 | Yes | IV | AD | DelE709-T710insD | Gefitinib | SD | Alive | 6.2 | 6.2 |
| 5 | M | 68 | Yes | IV | AD | DelE709-T710insD | Gefitinib | PD | Alive | 2.3 | 29.5 |
| 6 | F | 59 | No | IV | AD | E709A + G719C | Gefitinib | SD | Alive | 7.3 | 12.1 |
| 7 | F | 58 | No | IV | AD | E709A + G719C | Erlotinib | PR | Dead | 14.9 | 29.3 |
| 8 | M | 76 | No | IV | AD | E709A + L858R | Erlotinib | SD | Dead | 3.9 | 5.4 |
| 9 | F | 48 | No | IV | AD | E709A + L858R | Gefitinib | PR | Dead | 13.6 | 32.0 |
| 10 | M | 69 | No | IV | AD | E709G + G719C | Erlotinib | PD | Dead | 1.4 | 8.3 |
| 11 | F | 57 | No | IV | AD | E709G + Del exon19 | Gefitinib | PR | Dead | 77.4 | 104.6 |
| 12 | M | 85 | Yes | IIIB | AD | E709G + L858R | Erlotinib | PR | Dead | 8.6 | 13.2 |
| 13 | F | 48 | No | IV | AD | E709G + L858R | Gefitinib | PD | Dead | 2.4 | 6.8 |
| 14 | F | 55 | No | IV | AD | E709G + L858R | Gefitinib | PR | Dead | 18.4 | 75.3 |
| 15 | F | 64 | No | IV | AD | E709K + G719S | Gefitinib | PR | Alive | 11.1 | 11.1 |
| 16 | M | 71 | No | IV | AD | E709K + L858R | Gefitinib | PR | Alive | 6.5 | 6.5 |
| 17 | M | 69 | Yes | IV | AD | E709K + L858R | Gefitinib | PR | Alive | 8.6 | 8.6 |
| 18 | M | 66 | Yes | IV | AD | E709V + L858R | Gefitinib | PR | Dead | 9.2 | 9.5 |
Abbreviations: EGFR, epidermal growth factor receptor; AD, adenocarcinoma; TKI, tyrosine kinase inhibitor; PR, partial response; SD, stable disease; PD, progressive disease; PFS, progression-free survival; OS, overall survival; F, female; M, male.
Table 3.
Predictive factors associated with clinical response to TKIs
| Variables | No patients, N=296 | Responder | Response rates (%) | Univariate analysis, P-valuea | Multivariate analysis, P-valueb |
|---|---|---|---|---|---|
| Type of mutation | 0.026 | 0.035 | |||
| E709X | 18 | 9 | 50.0 | ||
| Classical | 278 | 206 | 74.1 | ||
| Gender | 0.508 | 0.611 | |||
| Female | 188 | 139 | 73.9 | ||
| Male | 108 | 76 | 70.4 | ||
| Age | 0.626 | 0.637 | |||
| ≤65 years | 153 | 113 | 73.9 | ||
| >65 years | 143 | 102 | 71.3 | ||
| Smoking status | 0.312 | 0.388 | |||
| Smoker | 65 | 44 | 67.7 | ||
| Non-smoker | 231 | 171 | 74.0 |
Notes:
Chi-square test;
logistic regression test (all 4 variables included for analysis).
Abbreviation: TKIs, tyrosine kinase inhibitors.
EGFR TKI treatment in patients with E709X mutations
Medical records from all 25 patients with these E709X EGFR mutations were reviewed in detail. Of the 25 patients, 18 had received EGFR TKI treatment (Table 2). Gefitinib was administered to 14 patients and the other 4 received erlotinib. EGFR TKIs as first-line treatment was used for 15 patients, 2 as second-line, and 1 as third-line or later lines treatment. At the start of EGFR TKI treatment, 1 was stage IIIb with malignant pleural effusion and the other 17 were stage IV.
The response rate to EGFR TKIs in patients with E709X was 50.0% (9/18), and the disease control rate was 72.2% (13/18). Median follow-up was 13 months (range, 5.4 to 104.6). The median PFS of EGFR TKIs was 6.2 months (range 0.6–77.4). The median overall survival after start of EGFR TKIs was 29.3 months (range 5.4–104.6 months).
To evaluate the effectiveness of EGFR TKI treatment in patients with uncommon EGFR mutation E709X, we compared these patients with those patients with classical EGFR mutations (L858R in exon 21 and deletions in exon 19) and patients with wild type EGFR in our previously published study with updated survival follow-up.17 These 18 patients with EGFR E709X mutation, who received TKI treatments, were compared with 278 patients with classical mutations who also received TKI treatments. Similar TKI courses were noted in these 278 patients, with most patients receiving TKI treatments as first-line therapy (158 as first-line, 52 as second-line, and 68 as third or later lines treatments). The response rate to EGFR TKIs was significantly higher in individuals with the classical EGFR mutations than in those with E709X mutations (74.1% vs 50.0%, P<0.026). Clinical factors (gender, age, smoking, and mutation type) were included for multivariate analysis by logistic regression. The mutation type was independently associated with clinical response to TKI treatment (P=0.032) (Table 3).
On the cut-off date, 16 patients (88.9%) with E709X mutations had completed follow-up, and 2 were lost to follow-up. In 278 patients with classical mutations, 243 patients (87.4%) had completed follow-up. The median PFS (9.1 months vs 6.2 months, P=0.782) and median overall survival after start of EGFR TKIs (19.1 months vs 29.3 months, P=0.370) were not statistically different between patients with classical EGFR mutations and patients with E709X mutations. We included clinical factors (gender, age, smoking, and mutation type) and performed multivariate survival analysis by Cox-regression. The mutation type was not associated with difference in survivals after TKI treatment in the multivariate analysis (Table 4).
Table 4.
Association between clinical factors and survivals after TKI treatment
| Variables | No patients, N=296 | PFS (months) | Univariate analysis, P-valuea | Multivariate analysis, P-valueb | OS (months) | Univariate analysis, P-valuea | Multivariate analysis, P-valueb |
|---|---|---|---|---|---|---|---|
| Type of mutation | 0.782 | 0.783 | 0.370 | 0.370 | |||
| E709X | 18 | 6.2 | 29.3 | ||||
| Classical | 278 | 9.1 | 19.1 | ||||
| Gender | 0.394 | 0.396 | 0.093 | 0.093 | |||
| Female | 188 | 8.4 | 21.6 | ||||
| Male | 108 | 9.8 | 17.7 | ||||
| Age | 0.675 | 0.677 | 0.934 | 0.935 | |||
| ≤65 years | 153 | 8.8 | 20.3 | ||||
| >65 years | 143 | 9.0 | 17.7 | ||||
| Smoking status | 0.099 | 0.101 | 0.877 | 0.877 | |||
| Smoker | 65 | 10.7 | 18.2 | ||||
| Non-smoker | 231 | 8.2 | 19.9 |
Notes:
Kaplan–Meier method;
Cox-regression method (all 4 variables included for analysis).
Abbreviations: TKI, tyrosine kinase inhibitor; PFS, progression-free survival; OS, overall survival.
When compared with patients with wild type EGFR, patients with E709X mutations had a better response rate to EGFR TKIs than those with wild type EGFR (50.0% vs 16.5%, P<0.001). And the median PFS (6.2 months vs 2.0 months, P=0.003) and overall survival after start of EGFR TKIs (29.3 months vs 11.8 months, P=0.010) were also better in patients with E709X mutations than patients with wild type EGFR.
Subtypes of E709X mutations
Of the 25 patients who had uncommon E709X mutations, 20 patients had E709X complex mutations (one E709X mutation and another EGFR mutation). Thirteen out of the 20 patients received TKI treatments, and 9 of them (9/13) were responders to TKI treatment.
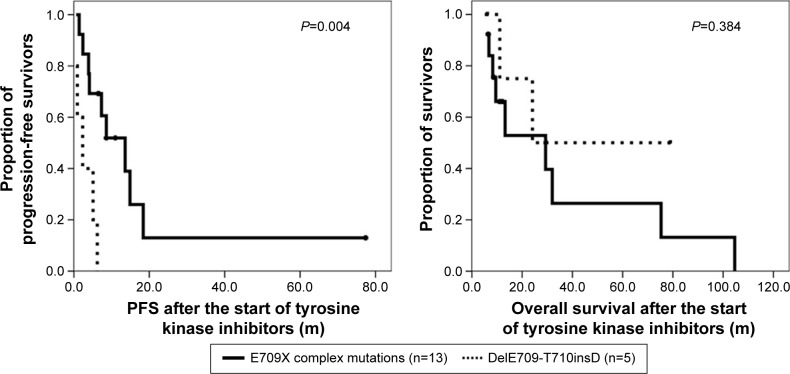
Out of the 25 patients who had uncommon E709X mutations, 5 patients had single EGFR mutation: DelE709-T710insD. All of the 5 patients, who had single DelE709-T710insD, received TKI treatments, and all were non-responders to TKI (2 had stable disease and 3 had progressive disease). Median PFS after TKI treatment of the 5 patients was only 2.3 months, which was significantly shorter than that of patients with E709 complex mutations (13.6 months, P=0.004) (Figure 1). However, the median overall survival of those patients was 24.1 months, which was not different from that of E709 complex mutation group (29.3 months, P=0.384) (Figure 1).
Figure 1.
Progression-free survival (PFS) and overall survival after the start of EGFR TKIs in patients with single DelE709-T710insD, or E709X complex mutations.
Abbreviations: EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor.
Patients who did not receive EGFR TKI treatment
Of the 25 patients who had uncommon EGFR E709X mutation, 7 did not receive TKIs in their treatment course (Table 5). One patient received treatment in another hospital after a diagnosis of cancer (case 19). Two patients (case 20 and 21) were in stage IIIA when lung cancer was diagnosed. Two patients (cases 22 and 23) had been diagnosed in early stage, and received an operation without subsequent recurrence. They underwent operation and adjuvant chemotherapy. One patient (case 24) received supportive care after her disease was diagnosed at an advanced stage. One patient received chemotherapy for his lung cancer (case 25). On the cut-off date, 4 patients were alive and the other 3 patients had expired (Table 5).
Table 5.
Summary of clinical information of patients who did not receive EGFR TKIs
| Patient no | Gender | Age (years) | Smoking | Stage | Cell | EGFR mutations | Outcome |
|---|---|---|---|---|---|---|---|
| 19 | F | 65 | No | IV | AD | E709A + G719A | Alive |
| 20 | M | 48 | No | IIIA | AD | E709K + G719A | Alive |
| 21 | M | 61 | No | IIIA | AD | E709G + L858R | Alive |
| 22 | M | 61 | Yes | IB | AD | E709G + L858R | Dead |
| 23 | F | 81 | No | IIA | AD | E709K + L858R | Alive |
| 24 | F | 72 | No | IV | SQ | E709V + L858R | Dead |
| 25 | M | 57 | Yes | IV | AD | E709V + L858R | Dead |
Abbreviations: EGFR, epidermal growth factor receptor; TKIs, tyrosine kinase inhibitors; AD, adenocarcinoma; SQ, squamous cell carcinoma; F, female; M, male.
Discussion
EGFR exon 18 mutations compose a minor group of the whole EGFR mutations of lung cancer. G719X mutations, which are activating mutations, are most common exon 18 EGFR mutations. E709X mutations are second common exon 18 mutations, but the proportion of E709X was much smaller than G719X, not to mention classical mutations (deletions in exon 19 or L858R in exon 21).20 This study included a large group of patients who underwent testing for EGFR mutations, and presented patients with uncommon E709X EGFR mutations. These E709X EGFR mutations comprised 1.48% of the entire EGFR mutations, and exist in 0.79% (25/3146) of the whole NSCLC patient cohort. Generally, patients with E709X mutations who received EGFR TKI treatment had lower response rate compared to patients with deletions in exon 19 or L858R in exon 21. On the contrary, their outcomes (response rate, PFS and overall survival) were better than those with wild type EGFR. Moreover, the response of E709 mutations to EGFR TKI can be divided into two subtypes: delE709-T710insD is a unique E709X mutation, which conferred worse response and shorter PFS to TKI treatments than E709X complex mutations. To our knowledge, this study presented the largest group of E709X mutations in the literature.20
In the present study, the EGFR mutation developed at E709 is either a deletion-insertion (delE709-T710insD) or an amino acid substitution. All the amino acid substitution E709X mutations are complex mutations (combination of one E709X and another mutation). Most E709X mutations reported in the literature are complex mutations (doublets),20,31 similar to those in our study (Table 6). Even rarer exceptions are scantily reported. For example, the study of Pao et al reported a single mutation E709Q, which conferred good response to TKI treatment.13 The study of Beau-Faller et al reported a patient with delE709-T710 (without amino acid insertion), and this patient did not have a favorable response to erlotinib therapy.32
Table 6.
Response of TKI in patients with E709X EGFR mutations in the study and in literature
| Mutations | Exon | Total cases | Responder to TKI | Non-responder to TKI |
|---|---|---|---|---|
| DelE709-T710 | 18 | 1 | 1 (32) | |
| DelE709-T710insD | 18 | 6 | 1 (21) | 5 (this study) |
| E709Q | 18 | 1 | 1 (13) | |
| E709A + G719C | 18 | 5 | 1 (this study), 1 (33), 1 (34) | 1 (this study), 1 (35) |
| E709A + G719S | 18 | 1 | 1 (36) | |
| E709A + L858R | 18+21 | 2 | 1 (this study) | 1 (this study) |
| E709A + L861G | 18+21 | 1 | 1 (36) | |
| E709G + G719C | 18 | 1 | 1 (this study) | |
| E709G + Del exon19 | 18+19 | 1 | 1 (this study) | |
| E709G + L858R | 18+21 | 6 | 2 (this study), 2 (37) | 1 (this study), 1 (37) |
| E709H + G719C | 18 | 1 | 1 (5) | |
| E709K + G719A | 18 | 1 | 1 (38) | |
| E709K + G719C | 18 | 1 | 1 (20) | |
| E709K + G719S | 18 | 1 | 1 (this study) | |
| E709K + L858R | 18+21 | 4 | 2 (this study), 1 (12), 1 (20) | |
| E709V + L858R | 18+21 | 1 | 1 (this study) |
Note: Number (reference number).
Abbreviations: EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor.
Five patients had the mutation delE709-T710insD in our study. DelE709-T710insD is a rare EGFR mutation (existing in 0.16% of all 3,146 patients in our cohort), and its clinical characteristics and sensitivity to TKI treatment are not clear. All 5 of the patients in this study received gefitinib treatment for pulmonary adenocarcinoma, and all were non-responders to gefitinib. The PFS after gefitinib was 2.3 months. The study of Ackerman et al also reported an NSCLC patient who had the EGFR mutation delE709-T710insD and received gefitinib treatment. Partial response was noted in this patient.21 More clinical data and in vitro studies are needed to increase the understanding of the EGFR mutation delE709-T710insD.
In our study, we divided the E709X mutations into two groups: DelE709-T710insD and E709X complex mutations. In the E709X complex mutation group, we may postulate the E709X is simply a passenger mutation, and is possibly insignificant. The other mutation in the complex is the driver mutation. For example, response rate to TKI treatments was 77.8% (7/9) while the E709X mutation existed with a classical mutation (L858R in exon 21 or deletions in exon 19). One can simply ignore the E709X mutation in this group, and the TKI response rate for this group is totally in line with the published data on oncogenic EGFR mutations. On the contrary, DelE709-T710insD is a significant mutation, which was associated with poor response to EGFR TKI treatment.
The major limitation of this report is its retrospective nature. In addition, although the number of patients with uncommon E709X EGFR mutations in this study was relatively large compared with other studies, more clinical experience in the treatment of patients with these EGFR mutations is needed.
In conclusion, we reported on a large group of lung cancer patients with uncommon EGFR E709X mutations and their association with treatment of EGFR TKIs. These EGFR mutations comprised a small but distinct proportion of the total EGFR mutations population, and are worthy of notice. Mutations of amino acid substitutions on E709 frequently exist with another EGFR mutation, while the delE709-T710insD developed solely. Complex mutations with one E709X mutation are sensitive EGFR mutations to EGFR TKI treatment. On the contrary, delE709-T710insD conferred poor response to TKI treatment. Our report may help to guide choices for therapy in NSCLC patients.
Acknowledgments
The authors are grateful for the support provided by the Department of Medical Research, National Taiwan University Hospital. Supported by grants MOHW103-TDU-PB-211-144002 (Department of Health, Executive Yuan, Taiwan), and 104-S2675 (National Taiwan University Hospital, Taiwan).
Abbreviations
- EGFR
epidermal growth factor receptor
- NSCLC
non-small-cell lung cancer
- PCR
polymerase chain reaction
- TKI
tyrosine kinase inhibitor
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 2.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 3.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361(10):958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 4.Wu JY, Yu CJ, Yang CH, et al. First- or second-line therapy with gefitinib produces equal survival in non-small cell lung cancer. Am J Respir Crit Care Med. 2008;178(8):847–853. doi: 10.1164/rccm.200803-389OC. [DOI] [PubMed] [Google Scholar]
- 5.Mitsudomi T, Kosaka T, Endoh H, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23(11):2513–2520. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- 6.Shih JY, Gow CH, Yu CJ, et al. Epidermal growth factor receptor mutations in needle biopsy/aspiration samples predict response to gefitinib therapy and survival of patients with advanced nonsmall cell lung cancer. Int J Cancer. 2006;118(4):963–969. doi: 10.1002/ijc.21458. [DOI] [PubMed] [Google Scholar]
- 7.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer – molecular and clinical predictors of outcome. N Engl J Med. 2005;353(2):133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 8.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non-small-cell lung cancer. J Clin Oncol. 2006;24(31):5034–5042. doi: 10.1200/JCO.2006.06.3958. [DOI] [PubMed] [Google Scholar]
- 9.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 10.Gu D, Scaringe WA, Li K, et al. Database of somatic mutations in EGFR with analyses revealing indel hotspots but no smoking-associated signature. Hum Mutat. 2007;28(8):760–770. doi: 10.1002/humu.20512. [DOI] [PubMed] [Google Scholar]
- 11.Chan SK, Gullick WJ, Hill ME. Mutations of the epidermal growth factor receptor in non-small cell lung cancer – search and destroy. Eur J Cancer. 2006;42(1):17–23. doi: 10.1016/j.ejca.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 12.Pallis AG, Voutsina A, Kalikaki AR, et al. ‘Classical’ but not ‘other’ mutations of EGFR kinase domain are associated with clinical outcome in gefitinib-treated patients with non-small cell lung cancer. Br J Cancer. 2007;97(11):1560–1566. doi: 10.1038/sj.bjc.6604068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balak MN, Gong Y, Riely GJ, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12(21):6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 15.Wu JY, Wu SG, Yang CH, et al. Lung cancer with epidermal growth factor receptor exon 20 mutations is associated with poor gefitinib treatment response. Clin Cancer Res. 2008;14(15):4877–4882. doi: 10.1158/1078-0432.CCR-07-5123. [DOI] [PubMed] [Google Scholar]
- 16.Greulich H, Chen TH, Feng W, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2(11):e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu JY, Yu CJ, Chang YC, Yang CH, Shih JY, Yang PC. Effectiveness of tyrosine kinase inhibitors on “uncommon” epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res. 2011;17(11):3812–3821. doi: 10.1158/1078-0432.CCR-10-3408. [DOI] [PubMed] [Google Scholar]
- 18.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359(4):366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosell R, Molina MA, Costa C, et al. Pre-treatment EGFR T790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clin Cancer Res. 2011;17(5):1160–1168. doi: 10.1158/1078-0432.CCR-10-2158. [DOI] [PubMed] [Google Scholar]
- 20.Cheng C, Wang R, Li Y, et al. EGFR Exon 18 mutations in east Asian patients with lung adenocarcinomas: a comprehensive investigation of prevalence, clinicopathologic characteristics and prognosis. Sci Rep. 2015;5:13959. doi: 10.1038/srep13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ackerman A, Goldstein MA, Kobayashi S, Costa DB. EGFR delE709_T710insD: a rare but potentially EGFR inhibitor responsive mutation in non-small-cell lung cancer. J Thorac Oncol. 2012;7(10):e19–e20. doi: 10.1097/JTO.0b013e3182635ab4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Travis W, Brambilla E, Müller-Hermelink HK, Harris C. Pathology and genetics of tumors of the lung, pleura, thymus and heart. Lyon: IARC Press; 2004. [Google Scholar]
- 23.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organisation for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92(3):205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 24.Lara PN, Jr, Redman MW, Kelly K, et al. Disease control rate at 8 weeks predicts clinical benefit in advanced non-small-cell lung cancer: results from Southwest Oncology Group randomized trials. J Clin Oncol. 2008;26(3):463–467. doi: 10.1200/JCO.2007.13.0344. [DOI] [PubMed] [Google Scholar]
- 25.Yang CH, Yu CJ, Shih JY, et al. Specific EGFR mutations predict treatment outcome of stage IIIB/IV patients with chemotherapy-naive non-small-cell lung cancer receiving first-line gefitinib monotherapy. J Clin Oncol. 2008;26(16):2745–2753. doi: 10.1200/JCO.2007.15.6695. [DOI] [PubMed] [Google Scholar]
- 26.Wu SG, Gow CH, Yu CJ, et al. Frequent epidermal growth factor receptor gene mutations in malignant pleural effusion of lung adenocarcinoma. Eur Respir J. 2008;32(4):924–930. doi: 10.1183/09031936.00167407. [DOI] [PubMed] [Google Scholar]
- 27.Wu SG, Chang YL, Hsu YC, et al. Good response to gefitinib in lung adenocarcinoma of complex epidermal growth factor receptor (EGFR) mutations with the classical mutation pattern. Oncologist. 2008;13(12):1276–1284. doi: 10.1634/theoncologist.2008-0093. [DOI] [PubMed] [Google Scholar]
- 28.Wu JY, Yu CJ, Shih JY, Yang CH, Yang PC. Influence of first-line chemotherapy and EGFR mutations on second-line gefitinib in advanced non-small cell lung cancer. Lung Cancer. 2010;67(3):348–354. doi: 10.1016/j.lungcan.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 29.Wu JY, Wu SG, Yang CH, et al. Comparison of gefitinib and erlotinib in advanced NSCLC and the effect of EGFR mutations. Lung Cancer. 2011;72(2):205–212. doi: 10.1016/j.lungcan.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 30.Shiao TH, Chang YL, Yu CJ, et al. Epidermal growth factor receptor mutations in small cell lung cancer. J Thorac Oncol. 2011;6(1):195–198. doi: 10.1097/JTO.0b013e3181f94abb. [DOI] [PubMed] [Google Scholar]
- 31.Chen Z, Feng J, Saldivar JS, Gu D, Bockholt A, Sommer SS. EGFR somatic doublets in lung cancer are frequent and generally arise from a pair of driver mutations uncommonly seen as singlet mutations: one-third of doublets occur at five pairs of amino acids. Oncogene. 2008;27(31):4336–4343. doi: 10.1038/onc.2008.71. [DOI] [PubMed] [Google Scholar]
- 32.Beau-Faller M, Prim N, Ruppert AM, et al. Rare EGFR exon 18 and exon 20 mutations in non-small-cell lung cancer on 10,117 patients: a multicentre observational study by the French ERMETIC-IFCT network. Ann Oncol. 2014;25(1):126–131. doi: 10.1093/annonc/mdt418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hijiya N, Miyawaki M, Kawahara K, et al. Phosphorylation status of epidermal growth factor receptor is closely associated with responsiveness to gefitinib in pulmonary adenocarcinoma. Hum Pathol. 2008;39(3):316–323. doi: 10.1016/j.humpath.2007.10.027. [DOI] [PubMed] [Google Scholar]
- 34.De Pas T, Toffalorio F, Manzotti M, et al. Activity of epidermal growth factor receptor-tyrosine kinase inhibitors in patients with non-small cell lung cancer harboring rare epidermal growth factor receptor mutations. J Thorac Oncol. 2011;6(11):1895–1901. doi: 10.1097/JTO.0b013e318227e8c6. [DOI] [PubMed] [Google Scholar]
- 35.Yip PY, Yu B, Cooper WA, et al. Patterns of DNA mutations and ALK rearrangement in resected node negative lung adenocarcinoma. J Thorac Oncol. 2013;8(4):408–414. doi: 10.1097/JTO.0b013e318283558e. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi S, Canepa HM, Bailey AS, et al. Compound EGFR mutations and response to EGFR tyrosine kinase inhibitors. J Thorac Oncol. 2013;8(1):45–51. doi: 10.1097/JTO.0b013e3182781e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keam B, Kim DW, Park JH, et al. Rare and complex mutations of epidermal growth factor receptor, and efficacy of tyrosine kinase inhibitor in patients with non-small cell lung cancer. Int J Clin Oncol. 2014;19(4):594–600. doi: 10.1007/s10147-013-0602-1. [DOI] [PubMed] [Google Scholar]
- 38.Han SW, Kim TY, Hwang PG, et al. Predictive and prognostic impact of epidermal growth factor receptor mutation in non-small-cell lung cancer patients treated with gefitinib. J Clin Oncol. 2005;23(11):2493–2501. doi: 10.1200/JCO.2005.01.388. [DOI] [PubMed] [Google Scholar]