Abstract
Breast cancer is the most common cancer in women worldwide. Local invasion, metastasis, and chemotherapy resistance are the obstacles for treatment of breast cancer. In this study, we aim to investigate the role of miR-144 in breast cancer. We demonstrate that the expression of miR-144 is downregulated in breast cancer and cell lines, and lower miR-144 expression is associated with poor differentiation, higher clinical stage, and lymph node metastasis in patients with breast cancer. The rescue of miR-144 expression is able to inhibit the cell proliferation and the ability of cell migration and invasion. In addition, we show that miR-144 can directly target at 3′-untranslation region of zinc finger E-box-binding homeobox 1 and 2, that is, ZEB1 and ZEB2, and regulate their expression at transcriptional and translational levels. Moreover, we also demonstrate that ectopic expression of miR-144 can inhibit the process of epithelial mesenchymal transition in MCF-7 and MDA-MB-231 cells. Thus, we here demonstrate that miR-144 functions as a tumor suppressor in breast cancer at least partly through inhibiting ZEB1/2-mediated epithelial mesenchymal transition process. Our findings indicate that the miR-144-ZEB1/2 signaling could represent a promising therapeutic target for breast cancer treatment.
Keywords: breast cancer, miR-144, ZEB1, ZEB2, epithelial mesenchymal transition
Introduction
Breast cancer is the most common cancer in women worldwide. With the development of neoadjuvant chemotherapy, the survival of patients with breast cancer is increasing yearly.1 However, the deaths caused by breast cancer remain huge, which accounts for ~15% of all cancer deaths in women.2 Thus, it is urgently needed for understanding the involved molecular mechanisms and developing more effective treatments for breast cancer patients.
Local invasion, metastasis, and chemotherapy resistance are the obstacles for treatment of breast cancer.3 These processes are involved in many gene dysregulations, including coding RNA and noncoding RNA, such as microRNAs (miRNAs).4 miRNAs are able to silence gene expression by targeting complementary regions of messenger RNAs (mRNAs) and inhibiting protein translation, which is critical in the development and the acquisition of malignancy in breast cancer.5 For example, upregulation of miR-125b conferred a chemoresistant phenotype by targeting Bcl-2 antagonist killer 1, and maintaining cancer stem-like side population fraction.6–8 Decreased levels of miR-144 in whole blood are observed in at least 13 different cancer and noncancer diseases, representing the most nonspecific miRNAs, and these results have been validated in a cohort of 319 samples in three different centers.9 By computational analysis, Lee et al suggest that miR-144 targets a cell cycle regulator, Rb1, and also targets to PTEN, and interacts with versican 3′-untranslation region (3′-UTR).10 Various studies have showed that miR-144 was a tumor suppressive gene, which could inhibit the tumor cells proliferation and invasion.11–13 Recently, Pan et al demonstrated that overexpression of miR-144 inhibited epithelial mesenchymal transition (EMT) process evaluated by decreased vimentin and snail expression, and increased E-cadherin expression.14 However, Yu et al show that overexpression of miR-144 significantly increased radiation resistance in MDA-MB-231 and SKBR3 cells.15 Thus, the role of miR-144 in breast cancer should be further illustrated.
In this study, we investigate the expression of miR-144 and clinicopathological correlations in breast cancer tissues. We also identify two new targets of miR-144, zinc finger E-box-binding homeobox 1 and 2, that is, ZEB1 and ZEB2. We demonstrate that miR-144 acts as a tumor suppressor in breast cancer, which may be involved in the EMT process. miR-144 may be a novel target for human breast cancer treatment.
Materials and methods
Breast cancer tissue samples
Forty-four adjacent tumor tissues and 44 breast cancer tissues and another five paired cancer and adjacent tissues were collected from Xiangya Hospital of Central South University. Informed consent has been signed by all subjects. The study protocols were approved by the Medical Ethics Committee of Xiangya Hospital, Central South University. All samples were collected and identified by histopathological evaluation and stored at −80°C until used.
Cell culture
Human breast cancer cell lines Mx-1, MCF-7, MDA-MB-231, and T47D, and normal human breast epithelial cells Hs578Bst cells were obtained from ATCC (Manassas, VA, USA). All the cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 10% (v/v) fetal bovine serum and 100 units/mL penicillin/streptomycin (Thermo Fisher Scientific) at 37°C in a humidified 5% CO2 incubator.
Cell treatment
To knockdown or overexpress the expression of miR-144, the cells were transfected with miR-144 inhibitor or miR-144 mimics (RiboBio, Guangzhou, People’s Republic of China) by using Lipofectamine 3000 (Thermo Fisher Scientific) according to manufacturer’s instructions. The cells transfected with scramble sequences of inhibitor or mimics were used as negative control.
Western blot analysis
The protein was extracted by radioimmunoprecipitation assay lysis buffer from indicated cells, and BCA Protein Assay Kit (Thermo Scientific) was used to measure the protein concentration. After being separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis gels, the protein was transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat dry milk and incubated with primary antibodies (rabbit polyclonal anti-E-cadherin [1:1,000], mouse monoclonal anti-vimentin [1:1,000], rabbit polyclonal anti-fibronectin [1:1,500], and rabbit monoclonal anti-beta Catenin [1:2,000] from Abcam, Cambridge, UK; goat polyclona anti-ZEB1 [1:500] and mouse monoclonal anti-ZEB2 [1:500] from Santa Cruz Biotechnology Inc., Dallas, TX, USA; mouse monoclonal anti-GAPDH [1:5,000] from Sigma-Aldrich Co., St Louis, MO, USA) overnight at 4°C. The membranes were washed with TBST, and then incubated with appropriate horseradish peroxidase-conjugated secondary antibody. Enhanced chemiluminescence reagent was used to detect the signal on the membrane.
Quantitative PCR analysis
Trizol (DMEM [Thermo Fisher Scientific]) was used to extract total RNA from the indicated cells according to the manufacturer’s instructions. A 2× power taq polymerase chain reaction (PCR) mastermix (Biotek, Beijing, People’s Republic of China) was used for real time PCR to detect mRNA levels. The primers are as follow: ZEB1, sense, TCCATGCTTAAGAGCGCTAGCT, antisense, ACCGTAGTTGAGTAGGTGTATGCCA; ZEB2, sense, GGCGCAAACAAGCCAATCCCA, antisense, TTCACTGGACCATCTACAGAGGCTT; β-actin, sense, AGGGGCCGGACTCGTCATACT, antisense, GGCGGCACCACCATGTACCCT. SYBR primerScript miRNA RT-PCR Kit (TaKaRa, Dalian, People’s Republic of China) was used for real time PCR to detect miR-144 levels. The primers of miR-144 (cat no CD201-0158) and U6 (cat no CD201-0145) were purchased from Tiangen (Beijing, People’s Republic of China). The expression of target genes was normalized by β-actin or U6. All the quantitative PCR data were processed using 2−ΔΔCT method.
Dual luciferase report system
Wild type and mutant 3′-UTR of ZEB1/ZEB2 were designed by GeneCopoeia (Guangzhou, People’s Republic of China), which was inserted into the downstream of dual luciferase reporter vector. For luciferase assay, 5×104 cells were plated and cultured in 24-well plates to reach ~70% confluence. MCF-7 cells were co-transfected with miR-144 mimics or miR-144 inhibitors and wild type/mutant 3′-UTR of ZEB1/ZEB2 dual luciferase reporter vector, respectively. After 48 hours transfection, the luciferase activity was detected using dual luciferase reporter gene assay kit (Beyotime, Shanghai, People’s Republic of China) on luminometer (Promega, Madison, WI, USA).
CCK-8 cell proliferation assay
Cell growth was measured by cell counting kit (CCK)-8 assay. Two thousand indicated cells were seeded in each 96-well plate for 12 hours. Following treatment, the cells were further incubated for 0, 24, 48 and 72 hours. An hour before the ending of incubation, 10 μL CCK-8 reagents (Solarbio, Beijing, People’s Republic of China) were added to each well. Optical density at 570 nm value in each well was determined by an enzyme immunoassay analyzer.
Flow cytometric analysis of apoptosis
Cells from each group were trypsinizated and washed with cold PBS and Annexin-V-FLUOS Staining Kit (Roche) was then used to stain the apoptosis cells according to the manufacturer’s instructions. The apoptosis rate was analyzed by flow cytometry (BD Biosciences, San Jose, CA, USA). The experiments were independently performed in triplicate.
Scratch assay
Cells in each group were collected and resuspended in DMEM medium. Each well of a 6-well plate was seeded with 1×105 cells and cultured for 36 hours to 90%–100% confluence. The cells were scratched with the head of a 200 μL tip, and washed with serum-free medium. These cells were further cultured for 0, 24, or 48 hours in DMEM medium containing 3% fetal bovine serum, and these cells in each group were photographed for analysis.
Transwell assay
The indicated cells were starved for 24 hours, and then resuspended in serum-free medium and added to the upper chamber of transwell (Corning Incorporated, Corning, NY, USA). The lower chamber was filled with completed medium containing 10% fetal bovine serum. Following 48 hours culture, cells attached to the bottom were fixed and stained with crystal violet for 30 minutes. The optical density at 570 nm of crystal violet dissolved by 10% acetic acid was detected by an enzyme immunoassay analyzer.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5 software (Graphpad Software, Inc., La Jolla, CA, USA) and the data are presented as the mean ± standard deviation. An unpaired two-tailed Student’s t-test or one-way analysis of variance with Bonferroni t post-test was used to analyze the data depending on conditions. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-144, ZEB1, and ZEB2 in breast cancer tissues and breast cancer cell lines
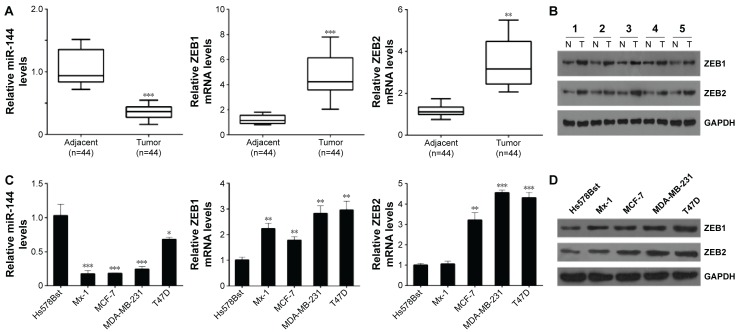
To investigate the role of miR-144 in breast cancer, we collected 44 breast cancer tissues and adjacent tissues, and detected the expression of miR-144 by using quantitative PCR. Our data showed that the average expression level of miR-144 was significantly downregulated in breast cancer tissue samples compared with adjacent controls (Figure 1A). Importantly, we analyzed the correlation of miR-144 expression and clinical features in patients with breast cancer and found that miR-144 expression was associated with differentiation status, clinical stages, and lymph node metastasis (Table 1). And we also analyzed the putative targets of miR-144, ZEB1, and ZEB2. We found that the mRNA levels of ZEB1 and ZEB2 were significantly upregulated in breast cancer tissues compared with the adjacent tissues (Figure 1A). In addition, we confirmed that the protein levels of ZEB1 and ZEB2 was also significantly upregulated in five paired breast cancer and normal adjacent tissues (Figure 1B). Moreover, the similar results were observed in the human breast cancer cell lines; the expression of miR-144 in breast cancer cell lines was significantly lower than that in Hs578Bst cells, while the mRNA and protein levels of ZEB1 and ZEB2 were higher than that in Hs578Bst cells (Figure 1C and D).
Figure 1.
The expression of miR-144, ZEB1, and ZEB2 in breast cancer tissues and cell lines.
Notes: (A) qPCR analysis for miR-144, ZEB1, and ZEB2 in breast cancer tissues and adjacent tissues. (B) Western blot analysis for ZEB1 and ZEB2 in five paired adjacent and breast cancer tissues. (C) qPCR analysis for miR-144, ZEB1, and ZEB2 in breast cancer cell lines. (D) Western blot analysis for ZEB1 and ZEB2 in breast cancer cell lines. Data are presented as mean ± SD. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: mRNA, messenger RNA; qPCR, quantitative polymerase chain reaction; SD, standard deviation; ZEB, zinc finger E-box-binding homeobox.
Table 1.
Correlation between miR-144 expression and clinicopathological features of patients with breast cancer
| Variants | miR-144 expression
|
P-value | |
|---|---|---|---|
| High | Low | ||
| Age | 0.565 | ||
| <48 | 10 | 15 | |
| ≥48 | 6 | 13 | |
| Differentiation | 0.040 | ||
| High | 7 | 2 | |
| Moderate | 6 | 16 | |
| Poor | 3 | 10 | |
| Clinical stage | 0.035 | ||
| I–II | 11 | 10 | |
| III–IV | 5 | 18 | |
| Lymph node metastasis | 0.028 | ||
| Present | 6 | 20 | |
| Absent | 10 | 8 | |
miR-144 regulates ZEB1 and ZEB2 expression at transcriptional and translational levels by directly targeting their 3′-UTR
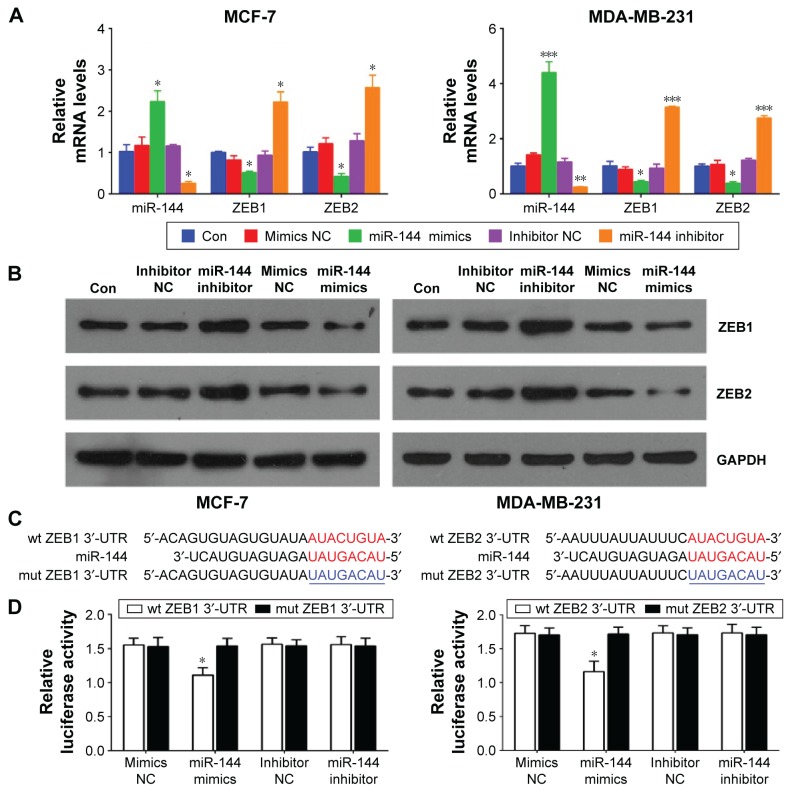
To further investigate the downstream molecules targeted by miR-144, we transfected miR-144 mimics or miR-144 inhibitor into MCF-7 and MDA-MB-231 cells to overexpress or knockdown the expression of miR-144 (Figure 2A). After miR-144 mimics or inhibitor transfection, we analyzed the expression of ZEB1 and ZEB2, two putative targets of miR-144 screened by a bioinformatic tool (Targetscan). As shown in Figure 2A and B, the mRNA and protein levels of ZEB1 and ZEB2 were markedly downregulated by miR-144 mimic transfection and upregulated by miR-144 inhibitor transfection compared with the negative control, respectively. We then wanted to know whether the 3′-UTR of ZEB1 and ZEB2 had a direct target site for miR-144. The sequences containing the wild type or mutant 3′-UTR of ZEB1 and ZEB2 (Figure 2C) were constructed into dual luciferase reporter gene. By dual luciferase reporter assay, we found that the luciferase activity was significantly repressed in the miR-144 mimics transfectant compared to the negative control transfectant. Moreover, miR-144-mediated repression of luciferase activity was abolished by the mutant type 3′-UTR of ZEB1 and ZEB2 (Figure 2D). These results determined that miR-144 directly targeted ZEB1 and ZEB2 and regulates their expression at transcriptional and translational levels.
Figure 2.
miR-144 regulates ZEB1 and ZEB2 expression at transcriptional and translational levels.
Notes: (A) qPCR analysis for miR-144, ZEB1, and ZEB2 in MCF-7 and MDA-MB-231 cells treated with miR-144 mimics or inhibitor. (B) Western blot analysis for ZEB1 and ZEB2 in MCF-7 and MDA-MB-231 cells treated with miR-144 mimics or inhibitor. (C) The target sites (red) and the mutant (mut) site (blue) at 3′-UTR of ZEB1 and ZEB2. (D) MCF-7 cells were co-transfected with miR-144 mimics/inhibitor and wt-3′-UTR of ZEB1/ZEB2 or their mut type, respectively. Compared with NC, the luciferase activity of wt-ZEB1 or ZEB2 was deceased significantly by miR-144, which was abrogated by their mut type. The luciferase activity had no significant changes in MCF-7 cells treated with miR-144 inhibitor. Data are presented as mean ± SD. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: Con, control; NC, negative control; OD, optical density; qPCR, quantitative polymerase chain reaction; SD, standard deviation; 3′-UTR, 3′-untranslation region; wt, wild type; ZEB, zinc finger E-box-binding homeobox.
The effects of miR-144 on cell proliferation, migration, and invasion in MCF-7 and MDA-MB-231 cells
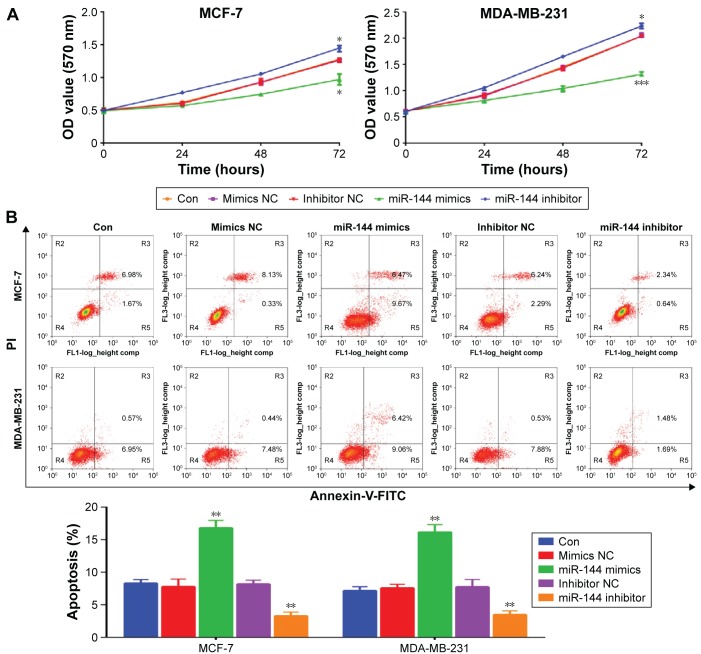
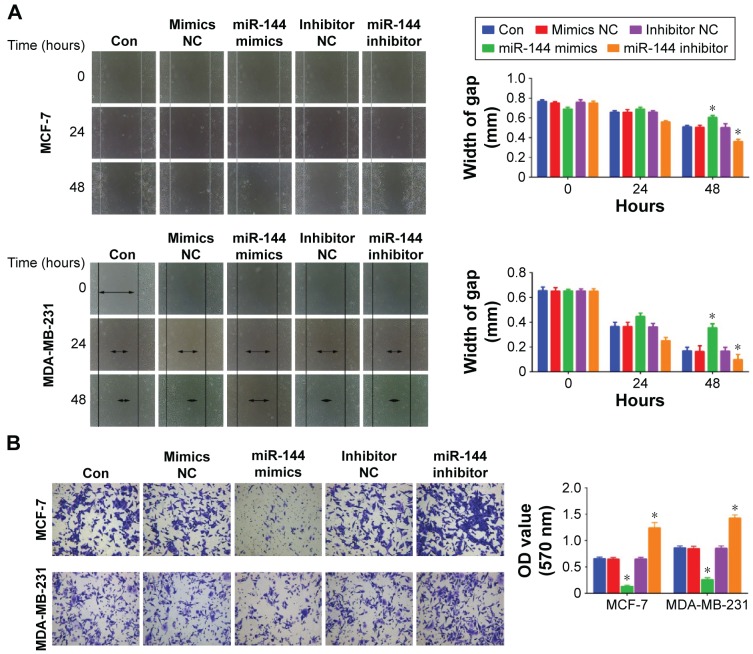
MCF-7 and MDA-MB-231 cell proliferation was measured by using CCK-8 after overexpression or knockdown of miR-144. We found that overexpression of miR-144 inhibited cell proliferation, while downregulation of miR-144 promoted cell proliferation in MCF-7 and MDA-MB-231 cell lines (Figure 3A). In addition, upregulation of miR-144 significantly increased the apoptosis rate, whereas downregulation of miR-144 exhibited an opposite role (Figure 3B). Furthermore, we also accessed the role of miR-144 in cell migration and invasion. Scratch and transwell assay were used to analyze cell migration and invasion after overexpression or knockdown of miR-144. Upregulation of miR-144 significantly reduced migration and invasion ability compared to negative control, while knockdown of miR-144 was able to enhance the ability of cell migration and invasion in MCF-7 and MDA-MB-231 cell lines (Figure 4).
Figure 3.
The effects of miR-144 on cell proliferation and apoptosis, migration, and invasion.
Notes: (A) CCK-8 assay was used to measure cell proliferation in MCF-7 and MDA-MB-231 cells treated with miR-144 mimics or inhibitor. (B) Flow cytometry analysis was used to measure cell apoptosis in MCF-7 and MDA-MB-231 cells treated with miR-144 mimics or inhibitor. Data are presented as mean ± SD. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: Con, control; NC, negative control; OD, optical density; SD, standard deviation; PI, propidium iodide; FITC, fluorescein isothiocyanate.
Figure 4.
The effects of miR-144 on cell migration and invasion.
Notes: (A) Scratch assay was used to analyze cell migration in MCF-7 and MDA-MB-231 cells treated with miR-144 mimics or inhibitor. (B) Transwell assay was used to analyze cell invasion in MCF-7 and MDA-MB-231 cells treated with miR-144 mimics or inhibitor. Magnification, ×100. Data are presented as mean ± SD. *P<0.05.
Abbreviations: Con, control; NC, negative control; OD, optical density; SD, standard deviation.
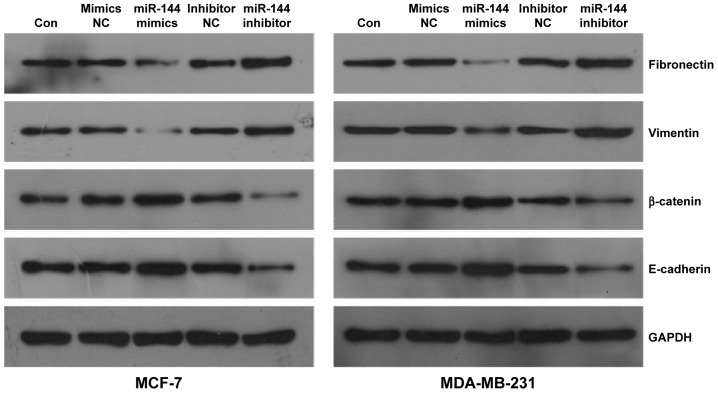
miR-144 regulates EMT-related gene expression
To explore potential downstream molecular signaling underlying miR-144 targeting to ZEB1 and ZEB2, we tested the expression of EMT markers, including fibronectin, vimentin, β-catenin, and E-cadherin, by Western blot in MCF-7 and MDA-MB-231 cells after miR-144 knockdown or overexpression. We found that a significant increase of β-catenin and E-cadherin and a significant decrease of fibronectin and vimentin were observed in the cells with upregulated miR-144. Inversely, knockdown of miR-144 significantly reduced the expression of β-catenin and E-cadherin, but increased the expression of fibronectin and vimentin in MCF-7 and MDA-MB-231 cells compared with that in control (Figure 5). These results suggest that upregulation of miR-144 is able to repress the process of EMT.
Figure 5.
The effect of miR-144 on the expression of epithelial mesenchymal transition-related genes.
Note: Western blot analysis for fibronectin, vimentin, β-catenin, and E-cadherin in MCF-7 and MDA-MB-231 cells treated with miR-144 mimics or inhibitor.
Abbreviations: Con, control; NC, negative control.
Discussion
In this study, we demonstrate that the expression of miR-144 is downregulated in breast cancer and cell lines, and the rescue of its expression is able to inhibit the cell proliferation and the ability of cell migration and invasion. In addition, lower miR-144 expression is associated with poor differentiation, higher clinical stage, and lymph node metastasis in patients with breast cancer. Downregulation of miR-144 has been observed in many malignancies, such as lung cancer, glioblastoma, and gastric cancer. miR-144 is able to inhibit proliferation, enhance apoptosis, and increase autophagy by targeting the p53-induced glycolysis and apoptosis regulator in lung cancer cell lines.16 Lan et al demonstrate that the loss of miR-144 effectively predicts the decreased overall survival in glioma patients, and overexpression of miR-144 potently represses glioma cell proliferation and invasion via suppressing mesenchyal epithelial transition in vitro and in vivo.17 And miR-144 is also significantly downregulated in both highly metastatic gastric cancer cell lines and tissues and acts as a tumor suppressor in gastric cancer by targeting mesenchyal epithelial transition.11 In addition, miR-144 functions as a tumor suppressor in bladder cancer cells by regulating cell cycle-related genes.18 Thus, miR-144 is able to target several genes that are involved in cell proliferation, apoptosis, and cell cycle, which subsequently results in inhibition of tumor growth. Therefore, miR-144 can control many targets and signaling pathways to promote epithelial gene expression and suppress cell invasion and metastasis by regulating a network of genes.
ZEB1 and ZEB2 are two members of the ZEB family of transcription factors characterized by the presence of two zinc finger clusters and a centrally located homeodomain.19 The expression of ZEB1/2 is regulated by multiple signaling pathways, including WNT, transforming growth factor beta, and miRNAs. In human breast cancer cells, knockdown of ZEB1/2 results in aberrant expression of many genes, most of which are determinants of epithelial differentiation and cell–cell adhesion.20,21 Previous studies showed a critical role of ZEB in developmental EMT. And recently, accumulating evidence suggests that ZEB can promote migration and invasion by inducing EMT in cancer cells.22 Aberrant expression of ZEB1 has been observed in many human cancers, such as pancreatic cancer, liver cancer, colon cancer, and breast cancer.23–25 Overexpression of ZEB1 can induce EMT and promote metastasis in breast cancer cell lines, and there is the relevance of ZEB1 induction of EMT and tumor progression in clinical cancers.26,27 Recently, Guan et al demonstrated that miR-144 as a tumor suppressor suppressed the expression of ZEB1 and ZEB2 by directly binding to their 3′-UTRs in thyroid cancer.28 We now also show that miR-144 can directly target to 3′-UTR of ZEB1 and ZEB2 and regulate their expression at transcriptional and translational levels. In non-small-cell lung cancers, miR-144 interacts with the oncogene ZEB1 at its 3′-UTR, and a decrease in miR-144 results in increased ZEB1 expression and an EMT phenotype;14 reduced expression of miR-144 may have diagnostic value for malignant solitary pulmonary nodule.12 Moreover, we also demonstrated that ectopic expression of miR-144 can inhibit the process of EMT in MCF-7 and MDA-MB-231 cells. ZEB1 and ZEB2 are critically involved in EMT and cell invasion in human breast cancer.29 And ZEB1/2 are also regulated by other factors to promote EMT, such as the forkhead box A2 and miR-200 family.21,30 Thus, we here demonstrated that miR-144 functions as a tumor suppressor in breast cancer at least partly through inhibiting ZEB1/2-mediated EMT process.
In summary, our findings indicate that miR-144 is an important tumor suppressor that is inactivated during malignant transformation in breast cancer, and the miR-144-ZEB1/2 signaling could represent a promising therapeutic target for breast cancer treatment.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Zhang HF, Xu LY, Li EM. A family of pleiotropically acting microR-NAs in cancer progression, miR-200: potential cancer therapeutic targets. Curr Pharm Des. 2014;20(11):1896–1903. doi: 10.2174/13816128113199990519. [DOI] [PubMed] [Google Scholar]
- 2.Chapman CH, Jagsi R. Postmastectomy radiotherapy after neoadjuvant chemotherapy: A review of the evidence. Oncology (Williston Park) 2015;29(9):657–666. [PubMed] [Google Scholar]
- 3.Fedele P, Orlando L, Schiavone P, et al. Recent advances in the treatment of hormone receptor positive HER2 negative metastatic breast cancer. Crit Rev Oncol Hematol. 2015;94(3):291–301. doi: 10.1016/j.critrevonc.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Kaboli PJ, Rahmat A, Ismail P, Ling KH. MicroRNA-based therapy and breast cancer: A comprehensive review of novel therapeutic strategies from diagnosis to treatment. Pharmacol Res. 2015;97:104–121. doi: 10.1016/j.phrs.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 5.Kotiyal S, Bhattacharya S. Breast cancer stem cells, EMT and therapeutic targets. Biochem Biophys Res Commun. 2014;453(1):112–116. doi: 10.1016/j.bbrc.2014.09.069. [DOI] [PubMed] [Google Scholar]
- 6.Luo Y, Wang X, Wang H, et al. High Bak expression is associated with a favorable prognosis in breast cancer and sensitizes breast cancer cells to paclitaxel. PLoS One. 2015;10(9):e138955. doi: 10.1371/journal.pone.0138955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang HJ, Guo YQ, Tan G, et al. miR-125b regulates side population in breast cancer and confers a chemoresistant phenotype. J Cell Biochem. 2013;114(10):2248–2257. doi: 10.1002/jcb.24574. [DOI] [PubMed] [Google Scholar]
- 8.Zhou M, Liu Z, Zhao Y, et al. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J Biol Chem. 2010;285(28):21496–21507. doi: 10.1074/jbc.M109.083337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen LL, Mitscher LA, Sharma PN, et al. Mechanism of inhibition of DNA gyrase by quinolone antibacterials: a cooperative drug–DNA binding model. Biochemistry. 1989;28(9):3886–3894. doi: 10.1021/bi00435a039. [DOI] [PubMed] [Google Scholar]
- 10.Lee DY, Jeyapalan Z, Fang L, et al. Expression of versican 3′-untranslated region modulates endogenous microRNA functions. PLoS One. 2010;5(10):e13599. doi: 10.1371/journal.pone.0013599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Xue H, Zhang J, et al. MicroRNA-144 inhibits the metastasis of gastric cancer by targeting MET expression. J Exp Clin Cancer Res. 2015;34:35. doi: 10.1186/s13046-015-0154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang G, An H, Fang X. MicroRNA-144 regulates proliferation, invasion, and apoptosis of cells in malignant solitary pulmonary nodule via zinc finger E-box-binding homeobox 1. Int J Clin Exp Pathol. 2015;8(5):5960–5967. [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y, Ying L, Tian Y, et al. miR-144 downregulation increases bladder cancer cell proliferation by targeting EZH2 and regulating Wnt signaling. FEBS J. 2013;280(18):4531–4538. doi: 10.1111/febs.12417. [DOI] [PubMed] [Google Scholar]
- 14.Pan HL, Wen ZS, Huang YC, et al. Down-regulation of microRNA-144 in air pollution-related lung cancer. Sci Rep. 2015;5:14331. doi: 10.1038/srep14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu L, Yang Y, Hou J, et al. MicroRNA-144 affects radiotherapy sensitivity by promoting proliferation, migration and invasion of breast cancer cells. Oncol Rep. 2015;34(4):1845–1852. doi: 10.3892/or.2015.4173. [DOI] [PubMed] [Google Scholar]
- 16.Chen S, Li P, Li J, et al. MiR-144 inhibits proliferation and induces apoptosis and autophagy in lung cancer cells by targeting TIGAR. Cell Physiol Biochem. 2015;35(3):997–1007. doi: 10.1159/000369755. [DOI] [PubMed] [Google Scholar]
- 17.Lan F, Yu H, Hu M, Xia T, Yue X. miR-144-3p exerts anti-tumor effects in glioblastoma by targeting c-Met. J Neurochem. 2015;135(2):274–286. doi: 10.1111/jnc.13272. [DOI] [PubMed] [Google Scholar]
- 18.Matsushita R, Seki N, Chiyomaru T, et al. Tumour-suppressive microRNA-144-5p directly targets CCNE1/2 as potential prognostic markers in bladder cancer. Br J Cancer. 2015;113(2):282–289. doi: 10.1038/bjc.2015.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang P, Sun Y, Ma L. ZEB1: at the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle. 2015;14(4):481–487. doi: 10.1080/15384101.2015.1006048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Si W, Huang W, Zheng Y, et al. Dysfunction of the reciprocal feedback loop between GATA3- and ZEB2-nucleated repression programs contributes to breast cancer metastasis. Cancer Cell. 2015;27(6):822–836. doi: 10.1016/j.ccell.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z, Yang C, Gao W, et al. FOXA2 attenuates the epithelial to mesenchymal transition by regulating the transcription of E-cadherin and ZEB2 in human breast cancer. Cancer Lett. 2015;361(2):240–250. doi: 10.1016/j.canlet.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 22.Agajanian M, Runa F, Kelber JA. Identification of a PEAK1/ZEB1 signaling axis during TGFbeta/fibronectin-induced EMT in breast cancer. Biochem Biophys Res Commun. 2015;465(3):606–612. doi: 10.1016/j.bbrc.2015.08.071. [DOI] [PubMed] [Google Scholar]
- 23.Sureban SM, May R, Lightfoot SA, et al. DCAMKL-1 regulates epithelial-mesenchymal transition in human pancreatic cells through a miR-200a-dependent mechanism. Cancer Res. 2011;71(6):2328–2338. doi: 10.1158/0008-5472.CAN-10-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sureban SM, May R, Mondalek FG, et al. Nanoparticle-based delivery of siDCAMKL-1 increases microRNA-144 and inhibits colorectal cancer tumor growth via a Notch-1 dependent mechanism. J Nanobiotechnology. 2011;9:40. doi: 10.1186/1477-3155-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perdigao-Henriques R, Petrocca F, Altschuler G, et al. miR-200 promotes the mesenchymal to epithelial transition by suppressing multiple members of the Zeb2 and Snail1 transcriptional repressor complexes. Oncogene. 2016;35(2):158–172. doi: 10.1038/onc.2015.69. [DOI] [PubMed] [Google Scholar]
- 26.Yu JM, Sun W, Hua F, et al. BCL6 induces EMT by promoting the ZEB1-mediated transcription repression of E-cadherin in breast cancer cells. Cancer Lett. 2015;365(2):190–200. doi: 10.1016/j.canlet.2015.05.029. [DOI] [PubMed] [Google Scholar]
- 27.Jang MH, Kim HJ, Kim EJ, Chung YR, Park SY. Expression of epithelial-mesenchymal transition-related markers in triple-negative breast cancer: ZEB1 as a potential biomarker for poor clinical outcome. Hum Pathol. 2015;46(9):1267–1274. doi: 10.1016/j.humpath.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 28.Guan H, Liang W, Xie Z, et al. Down-regulation of miR-144 promotes thyroid cancer cell invasion by targeting ZEB1 and ZEB2. Endocrine. 2015;48(2):566–574. doi: 10.1007/s12020-014-0326-7. [DOI] [PubMed] [Google Scholar]
- 29.Qiao Y, Shiue CN, Zhu J, et al. AP-1-mediated chromatin looping regulates ZEB2 transcription: new insights into TNFalpha-induced epithelial-mesenchymal transition in triple-negative breast cancer. Oncotarget. 2015;6(10):7804–7814. doi: 10.18632/oncotarget.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhodes LV, Martin EC, Segar HC, et al. Dual regulation by microRNA-200b-3p and microRNA-200b-5p in the inhibition of epithelial-to-mesenchymal transition in triple-negative breast cancer. Oncotarget. 2015;6(18):16638–16652. doi: 10.18632/oncotarget.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]