Abstract
Liver disease, in the form of hepatocellular carcinoma (HCC) accounts for >700,000 deaths worldwide. A major reason for this is late diagnosis of HCC. The currently used biomarker, serum alpha-fetoprotein (AFP) is elevated in 40–60% of those with HCC and other markers that can either compliment or replace AFP are desired. Our previous work has identified a number of proteins that contain altered glycans in HCC. Specifically, these altered glycans were increased levels of core and outer arm fucosylation. To determine the clinical usefulness of those identified glycoproteins, a plate based assay was developed that allowed for the detection of fucosylated glycoforms. While this method was applicable to a number of independent patient sets, it was unable to specifically detect fucosylated glycoforms in many patient samples. That is, some material was present in serum that led to non-specific signal in the lectin-fluorescence-linked immunosorbent assay (lectin-FLISA). To address this issue, a systematic process was undertaken to identify the material. This material was found to be increased levels of lectin reactive IgM. Removal of both IgG and IgM using a multi-step protein A/G incubation and filtration step removed the contaminating signal and allowed for the analysis of specific protein glycoforms. This assay was subsequently used on two sample sets, one that was shown previously to be unable to be tested via a lectin FLISA and in a larger independent sample set. The clinical usefulness of this assay in the early detection of HCC is discussed.
Keywords: biomarker, glycosylation, hepatocellular carcinoma, lectin, liver cancer
1. Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies worldwide and the incidence in the United States (USA) is increasing [1, 2]. The progression of liver disease into liver cancer has been monitored with serum levels of alpha-fetoprotein (AFP). However, AFP’s limited sensitivity and specificity has resulted in the questioning of AFP as a primary screen for HCC [3] and more sensitive biomarkers for HCC are desired.
Using fucose-specific lectins we have previously identified more than 50 glycoproteins that contained increased fucosylation with HCC [4] and have used these in plate-based assays to diagnosis HCC[5–7]. While this method was applicable to a number of independent patient sets, it was unable to specifically detect fucosylated glycoforms in certain patient samples. That is, some material was present in serum that led to non-specific signals in the lectin-FLISA[8, 9]. In the current study, we have identified the contaminating lectin reactive factors present in the serum. This lectin reactive factor was shown to be IgM and when this was removed from the serum prior to lectin-FLISA, specific glycoprotein associated lectin reactive signal could be detected. This method was used in two independent sample sets to validate the method and also to validate the performance of the fucosylated glycoforms as biomarkers of HCC. The potential use of this method as a diagnostic tool for the detection of liver cancer is discussed.
2. Material and Methods
2.1 Patient Samples
Serum samples were obtained from the University of Michigan and the University of California San Diego under a study protocol approved by the respective Institutional Review Board and written informed consent was obtained from each subject. Patients details regarding samples from the University of Michigan are found in our previous publication[10]. Detailed information regarding patients from the University of California at San Diego are found in Supplementary Table 2.
2.2 Lectin FLISA
The traditional lectin FLISA is described elsewhere[5]. The modified assay involves incubation of 1–5 μl of human serum (diluted into 45–49 μl PBS) with 20 μl of Pierce™ Protein A/G Plus (Thermo Fisher, Waltham, MA) for 1 hour prior to filtration in a 100KD Amicon Ultran 0.5 mL spin filter (EMD Millipore, Billerica, MA). Flow through was applied directly to plates and the lectin FLISA performed as before[5]. For the anti-alpha 1 anti-trypsin (A1AT) Lectin-FLISA, a polyclonal anti-A1AT (Sigma-Aldrich, St. Louis, MO) was used.
2.3. Immunoblotting
Pooled, HCC serum [11] was depleted of IgG using protein A/G coated agarose beads as described elsewhere [10] and serum incubated on 96-well plates coated with A1AT antibody for 2 hours at room temperature. Captured proteins were resolved via SDS-PAGE and either stained with colloidal Coomassie brilliant blue (Colloidal Blue Staining Kit, Thermo Fisher) or proteins transferred using western-blotting method to PVDF membranes for immunoblot analysis. Fucosylation was detected using biotin-conjugated Aleuria aurantia lectin (AAL). A1AT was detected using a polyclonal anti-A1AT (Sigma-Aldrich). IgM or IgG was detected using polyclonal antibodies (Abcam, Cambridge, MA). Bound AAL or antibody was visualized using IRDye®800-conjugated streptavidin or IRDye®700-conjugated anti-rabbit antibody.
2.4 Proteomic identification of contaminating factors
For proteomic analysis well-associated proteins were digested with Trypsin Gold (Promega, Madison, WI). Samples were analysed by the Biological Mass Spectrometry Facility at Rutgers, the State University of New Jersey using a Velos LTQ Orbitrap tandem mass spectrometer coupled to a Dionex UltiMate 3000 Rapid Separation LC System (Thermo Scientific) using methods similar to previous reports[11]. The LC-MS/MS data was searched against the most up-to-date complete protein database (ensembl.org) using a local version of the Global Proteome Machine (GPM cyclone, Beavis Informatics Ltd, Winnipeg, Canada) with carbamidoethyl on cysteine as fixed modification and oxidation of methionine and tryptophan as variable modifications using a 10 ppm precursor ion tolerance and a 0.4 Da fragment ion tolerance.
Statistical analysis
Descriptive statistics for patient groups were compared by scatter plots that included the outliers. All values were reported as mean values ±SD unless otherwise stated. As data did not follow typical Gaussian distributions, a nonparametrical test (two-tailed, 95% confidence, Mann– Whitney Test) was used to determine statistical difference between groups.
3. Results
3.1 Identification of IgG and IgM as a contaminant in lectin-FLISA assays for HCC detection
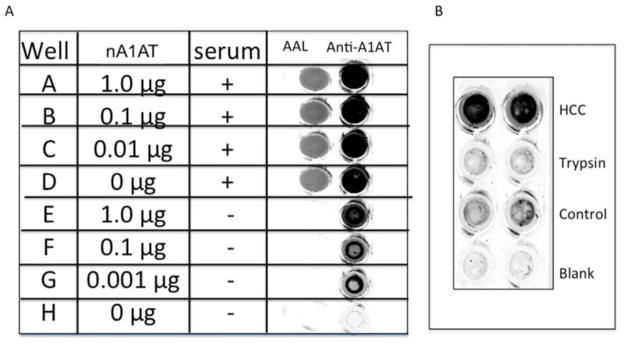
We have previously identified proteins that become hyper-fucosylated in HCC and developed a plate-based assay to assess the level of fucosylation in samples[5, 12]. This method worked well in many sample set but in others, was unable to detect a lectin reactive signal that could be attributed specifically to the protein that was captured[10]. An example of such a result is shown in Figure 1A. Here a lectin-FLISA was performed for fucosylated alpha-1-anti-trypsin (A1AT). As Figure 1A show, when such an assay is done using HCC serum, a lectin reactive signal is observed (Figure 1A; wells A–D). However, when attempts were made to compete out this signal using non fucosylated native A1AT (nA1AT), we were unable to do so, even when non fucosylated A1AT had bound to the capture antibody (Figure 1A; wells E–H). Tryptic digestion of the sample prior to analysis confirmed that the signal was in fact protein based (Figure 1B).
Figure 1. Serum contains a protein based nonspecific lectin reactive signal that confounds a plate-based lectin-FLISA.
A) Lectin-FLISA using serum from a HCC patient that was shown to be positive by Lectin-FLISA assay (well D). Attempts were made to block the signal using purchased native A1AT (nA1AT) that was not AAL reactive (wells E–H). While A1AT from both serum and exogenously sources can be detected by using an anti-A1AT antibody, an AAL signal is obtained when serum is used and cannot be competed with non-lectin reactive A1AT. B) Treatment of sample with trypsin leads to the complete loss of signal. In panel B, the HCC sample is the same as panel A, trypsin is the sample following digestion with trypsin, control is normal human sera purchased from Sigma Chemicals, blank is no sera added.
3.2. Identification of IgM as a potential contaminant
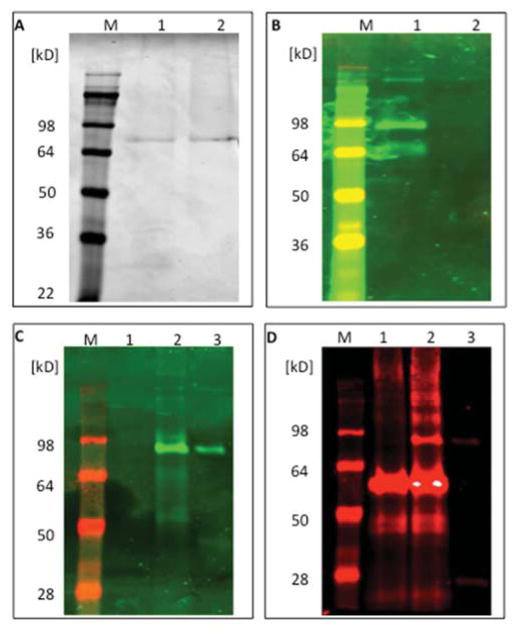
In an effort to identify the nonspecific lectin reactive material, a lectin FLISA was performed in a 96 well plate for A1AT and the captured material used for gel based lectin and proteomic analysis. As figure 2A shows, from the HCC serum incubated well (lane 1) or mock treated well (lane 2) colloidal Coomassie staining leads to the identification of only a single band, which is the capture antibody. All other bands are below the detection limit of the Coomassie stain. However, as figure 2B shows lectin blotting of this gel with a fucose-binding lectin leads to the detection of a band at ~80 kD with a weaker band observed at 65 kD (lane 1). This band was associated with the blocking buffer (data not shown). In contrasts, the ~80 kD band was not observed via staining with either Coomassie or A1AT immunoblotting. This suggested that the nonspecific lectin reactive material in these samples was a low abundant 80 kD glycoprotein that was highly fucosylated. Subsequently, proteins from wells treated identically were collected and examined via liquid chromatography tandem mass spectrometry (LC MS/MS) following digestion with trypsin. The list of glycoproteins identified in these samples is shown in Supplementary Table 1. Two proteins of ~80KD were identified, complement C1s beta chain and IgM heavy chain. Consequently, the experiment performed in Figure 2A was repeated but this time, following total protein staining and lectin blotting, immunoblotting was performed with anti-IgM antibodies and anti-complement C1s beta chain antibodies. While no evidence of complement C1s contamination was found in these samples (data not shown), IgM contamination was found. Figure 2C again shows a similar experiment to that performed in Figure 2A, where no lectin reactive material is associated with mock treated wells but is present in the serum treated well (Figure 2C, lane 2). In this gel, as a control, IgM was also added and is lectin reactive as shown in lane 3. In addition, as Figure 2C shows, in the AAL blot, the contaminating serum band runs at the same molecular weight as the IgM control (lanes 2–3) and as Figure 2D shows, overlaps with an IgM antibody following staining (lanes 2–3). As the secondary antibody in Figure 2D is anti-rabbit, the rabbit anti-A1AT IgG capture antibody that is eluted from the well also shows up in lanes 1 and 2. This result confirms that IgM from cirrhotic patients is highly lectin reactive and is a contaminating factor.
Figure 2. Identification of IgM as a potential contaminant.
A) ELISA wells coated with periodate oxidized anti-A1AT antibody were either incubated with serum from the same HCC patient as described in Figure 1 (lane 1) or mock treated (lane 2). Subsequently, captured protein was removed with SDS lysis buffer and examined by SDS-PAGE and colloidal Coomassie brilliant blue dye. B). AAL lectin blotting of samples to Panel A. Strong staining is observed from the serum incubated well with bands at 80 kD and a weak band at 50 kD. C). AAL blot from samples generated as in Panel A. Lane 1, PBS incubation, lane 2, HCC serum incubation, lane 3, IgM control. As before a lectin reactive band of 80 kD is observed from the HCC sample and this is identical to the IgM control band (lane 3). D) Same blot as in C but probed with a rabbit anti-IgM antibody. Lanes are as before.
3.3. Removal of contaminating IgM and IgG
In an effort to deplete the IgM from the serum samples, methods were developed to remove this material from serum before lectin-FLISA analysis. The first method was a simple filtration method using a centrifugal 100K spin filter. IgM is >500K and IgG, which has previously been shown to be a contaminant[5], is 150 KD and thus, this one method could remove both contaminants in one step. However, while this method could remove IgM, it was unable to remove IgG (data not shown). Figure 3 shows an experiment to test the best combination of methods and materials to remove both IgM and IgG from serum. In Figure 3, panel A, an anti-IgM immunoblot following various treatments is shown. Panel B of Figure 3 is a blot of the same samples following an immunoblot with anti-IgG antibodies and Panel C is a blot of the same samples following an immunoblot with anti-A1AT antibodies. In Lane 1, total serum was run on the gel directly, without any treatment or filtration. As this figure shows, IgM (lane 1, panel A), IgG (lane 1, panel B) and A1AT (lane 1, panel C) reactive bands are observed. While lane 2 is a spacer lane, lanes 3–6 involve incubation of serum with various forms of protein A/G or L. The logic is that IgG bound to protein A would be increased in mass enough to be retained in the 100 KD spin filter. However only treatment of serum with 20 μl of Pierce™ Protein A/G Plus agarose for one hour, prior to filter removed both IgM (lane 4 panel A) and IgG (lane 4, Panel B) from the serum with minor impacts upon the level of A1AT in the flow through (Figure 3C). Figure 3D shows that when this method was used before a lectin FLISA, in an experiment similar to that performed in Figure 1, the lectin contaminating signal observed was removed and now the signal from a positive sample could be blocked (competed) by the addition of non lectin reactive material. Thus, it appears that IgM and IgG are the contaminating lectin reactive material and their removal allows for the proper application of a plate based lectin-FLISA.
Figure 3. Methods for the removal of IgM from serum.
A) Anti-IgM immunoblot following treatment of samples to remove IgM and IgG. B) Immunoblot of the same samples with anti-IgG or (C) anti-A1AT antibodies. For A–C: In Lane 1, total serum without any treatment. Lane 2, empty lane. Lane 3, serum incubated with 20 μl protein A/G agarose for one hour prior to filtration. Lane 4, treatment of serum with 20 μl of Pierce Protein A/G Plus agarose for one hour, prior to filtration. Lane 5, treatment of serum with Protein A/G dynabeads prior to filtration. Lane 6, treatment of serum with Protein L prior to filtration. D Blocking of AAL signal with non lectin reactive material. Lectin-FLISA using a mixture of serum from a patient with HCC that was shown to be positive by a Lectin-FLISA assay (well 2). Attempts were made to block the signal using nA1AT that was not AAL reactive. A1AT levels are detected in lanes 1,3,5 and 7. AAL reactivity is shown in lanes 2,4,6, and 8. The amount of nA1AT added to the serum is indicated.
3.4. Application of method in the analysis of human serum samples
We next applied this method to the analysis of fucosylated glycoforms of A1AT using a sample set where lectin-FLISA methods had failed[10]. Importantly, in this set, lectin-western data showing lectin reactive A1AT was determined and acts as the gold standard for the lectin-ELISA [10]. As figure 4A shows, without filtration, the mean value of the cirrhotic samples was 3.2 (±2.0) and the HCC samples was 2.9 (±1.8) relative units. The AUROC of this assay was 0.578 (Figure 4B). In contrast, using the filtration method, the lectin ELISA results in a mean value of 1.4 (±0.75) in the cirrhotic sample and 2.4 (±1.1) in the HCC samples (Figure 4C).
Figure 4. Impact of IgM depletion method of Lectin-FLISA.
A) Scatter plot of A1AT FLISA without depletion of IgM in 40 samples from the University of Michigan (20 cirrhotic patients and 20 cirrhotic plus HCC patients). Values are given for each sample as a relative increase over normal sera. The p value is indicated. B) AUROC for data in Panel A. C) Scatter plot of A1AT FLISA after depletion of IgM and IgG. The p value is indicated. D) AUROC for data in Panel C. E) Scatter plot of A1AT FLISA after depletion of IgM and IgG in 124 samples from the University of California at San Diego (86 cirrhotic patients and 36 cirrhotic plus HCC patients). F) AUROC for data in Panel E.
The AUROC of the comparison between the HCC and cirrhosis samples after filtration was 0.788 (Figure 4D), which was nearly identical to that observed previously by the lectin-western [10]. As a comparison, AFP had an AUROC of 0.82 in this set (data not shown[10]). Importantly, every sample that was positive for the lectin-ELISA following filtration was shown to be positive by the lectin-western and every sample shown to be negative by the lectin-ELISA following filtration was shown to be negative by the lectin-western. This assay was used to measure fucosylated A1AT in a second set of samples obtained from the University of San Diego. This set consisted of 86 patients with liver cirrhosis and 36 patients with HCC in the background of liver cirrhosis. Details on these patients are found in Supplementary Table 2. Using the filtration method, the lectin ELISA for A1AT had a mean value of 1.4 (±0.56) in the cirrhotic samples and 2.7 (±1.5) in the HCC samples (Figure 4E). The AUROC of A1AT was 0.798, which was nearly identical to that observed in the first set (Figure 4F)[10]. As a reference AFP in this set had an AUROC of 0.7490 (data not shown). At a fixed specificity of 95%, AFP had 28% sensitivity while A1AT was 58% sensitive.
4. Discussions
Glycosylation changes have been observed in many cancers, including but not limited to liver[13–16], pancreas[17–20], lung [21, 22], breast [23], ovarian[24], colon[25] and prostate cancer[26, 27]. In many, proteins that contain these glycan changes have been identified. If these changes are to be used clinically, they must be examined in a rapid and in-expensive manner. To that end, several plate-based systems, have been developed. These methods all involve the capture of specific proteins followed by detection of the captured protein using a lectin[11, 28–31]. In many cases, it is assumed that the signal obtained from such an experiment is specific, without further testing.
In our previous analysis of fucosylated glycoforms, we were able to show that the lectin reactive signal was specific to the protein captured. This was done using a simple competition experiment (as in Figure 1) where non fucosylated A1AT was titrated into the lectin FLISA and could effectively block the signal from serum [5]. In those previous studies, the removal of heterophilic IgG molecules was sufficient to obtain a protein specific lectin reactive signal. However, in other sample sets, this method was unable to be used to remove non specific signals[10]. To that end, we used proteomics to identify the nonspecific lectin reactive factors present in the samples and identified IgM as the major contaminant. IgM has been shown previously to be heavily fucosylated [32, 33]. We have previously shown the anti-gal IgG from patients with liver disease have altered glycosylation and this leads to increased lectin reactivity. It is unclear if the glycosylation of IgM is altered in liver disease. However, we have shown that the level of anti-gal IgM is increased in patients with liver fibrosis[12].
In conclusion, having identified the material confounding our lectin-FLISA assays, we were able to develop a workflow that allowed for the lectin based analysis of a specific protein. Development of an assay for one protein, A1AT, allowed for near identical results from the plate based lectin-FLISA to what was observed via a lectin extraction western blot based approach[10]. Testing of this marker in a second patient set confirmed the ability of this marker and assay platform to differentiate HCC from cirrhosis. Current work is focusing on developing other markers with higher predictive ability than A1AT and incorporation of these markers into existing biomarker combinations and algorithms.
Supplementary Material
Acknowledgments
This work was supported by grants R01 CA120206 (ASM) and U01 CA168856 (ASM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Block TM, Mehta AS, Fimmel CJ, Jordan R. Molecular viral oncology of hepatocellular carcinoma. Oncogene. 2003;22:5093–5107. doi: 10.1038/sj.onc.1206557. [DOI] [PubMed] [Google Scholar]
- 2.Singal AG, El-Serag HB. Hepatocellular Carcinoma From Epidemiology to Prevention: Translating Knowledge into Practice. Clin Gastroenterol Hepatol. 2015;13:2140–2151. doi: 10.1016/j.cgh.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sherman M. Hepatocellular carcinoma: epidemiology, risk factors, and screening. Semin Liver Dis. 2005;25:143–154. doi: 10.1055/s-2005-871194. [DOI] [PubMed] [Google Scholar]
- 4.Comunale MA, Lowman M, Long RE, Krakover J, Philip R, Seeholzer S, Evans AA, Hann HWL, Block TM, Mehta AS. Proteomic analysis of serum associated fucosylated glycoproteins in the development of primary hepatocellular carcinoma. Journal of Proteome Research. 2006;6:308–315. doi: 10.1021/pr050328x. [DOI] [PubMed] [Google Scholar]
- 5.Wang M, Long RE, Comunale MA, Junaidi O, Marrero J, Di Bisceglie AM, Block TM, Mehta AS. Novel fucosylated biomarkers for the early detection of hepatocellular carcinoma. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2009;18:1914–1921. doi: 10.1158/1055-9965.EPI-08-0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comunale MA, Wang M, Hafner J, Krakover J, Rodemich L, Kopenhaver B, Long RE, Junaidi O, Bisceglie AM, Block TM, Mehta AS. Identification and development of fucosylated glycoproteins as biomarkers of primary hepatocellular carcinoma. J Proteome Res. 2009;8:595–602. doi: 10.1021/pr800752c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comunale MA, Rodemich-Betesh L, Hafner J, Wang M, Norton P, Di Bisceglie AM, Block T, Mehta A. Linkage Specific Fucosylation of Alpha-1-Antitrypsin in Liver Cirrhosis and Cancer Patients: Implications for a Biomarker of Hepatocellular Carcinoma. PLoS ONE. 2010;5:e12419. doi: 10.1371/journal.pone.0012419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehta AS, Long RE, Comunale MA, Wang M, Rodemich L, Krakover J, Philip R, Marrero JA, Dwek RA, Block TM. Increased levels of galactose-deficient anti-Gal immunoglobulin G in the sera of hepatitis C virus-infected individuals with fibrosis and cirrhosis. Journal of virology. 2008;82:1259–1270. doi: 10.1128/JVI.01600-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balagopal A, Philp FH, Astemborski J, Block TM, Mehta A, Long R, Kirk GD, Mehta SH, Cox AL, Thomas DL, Ray SC. Human immunodeficiency virus-related microbial translocation and progression of hepatitis C. Gastroenterology. 2008;135:226–233. doi: 10.1053/j.gastro.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comunale MA, Wang M, Anbarasan N, Betesh L, Karabudak A, Moritz E, Devarajan K, Marrero J, Block TM, Mehta A. Total serum glycan analysis is superior to lectin-FLISA for the early detection of hepatocellular carcinoma. Proteomics Clinical applications. 2013;7:690–700. doi: 10.1002/prca.201200125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comunale MA, Wang M, Hafner J, Krakover J, Rodemich L, Kopenhaver B, Long RE, Junaidi O, Bisceglie AM, Block TM, Mehta AS. Identification and development of fucosylated glycoproteins as biomarkers of primary hepatocellular carcinoma. Journal of Proteome Research. 2009;8:595–602. doi: 10.1021/pr800752c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamontagne A, Long RE, Comunale MA, Hafner J, Rodemich-Betesh L, Wang M, Marrero J, Di Bisceglie AM, Block T, Mehta A. Altered functionality of antibacterial antibodies in patients with chronic hepatitis C virus infection. PLoS ONE. 2013;8:e64992. doi: 10.1371/journal.pone.0064992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aoyagi Y, Isokawa O, Suda T, Watanabe M, Suzuki Y, Asakura H. The fucosylation index of alpha-fetoprotein as a possible prognostic indicator for patients with hepatocellular carcinoma. Cancer. 1998;83:2076–2082. [PubMed] [Google Scholar]
- 14.Naitoh A, Aoyagi Y, Asakura H. Highly enhanced fucosylation of serum glycoproteins in patients with hepatocellular carcinoma. J Gastroenterol Hepatol. 1999;14:436–445. doi: 10.1046/j.1440-1746.1999.01882.x. [DOI] [PubMed] [Google Scholar]
- 15.Goldman R, Ressom HW, Varghese RS, Goldman L, Bascug G, Loffredo CA, Abdel-Hamid M, Gouda I, Ezzat S, Kyselova Z, Mechref Y, Novotny MV. Detection of hepatocellular carcinoma using glycomic analysis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:1808–1813. doi: 10.1158/1078-0432.CCR-07-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanda M, Pompach P, Brnakova Z, Wu J, Makambi K, Goldman R. Quantitative liquid chromatography-mass spectrometry-multiple reaction monitoring (LC-MS-MRM) analysis of site-specific glycoforms of haptoglobin in liver disease. Molecular & cellular proteomics : MCP. 2013;12:1294–1305. doi: 10.1074/mcp.M112.023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terao N, Takamatsu S, Minehira T, Sobajima T, Nakayama K, Kamada Y, Miyoshi E. Fucosylation is a common glycosylation type in pancreatic cancer stem cell-like phenotypes. World J Gastroenterol. 2015;21:3876–3887. doi: 10.3748/wjg.v21.i13.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarrats A, Saldova R, Pla E, Fort E, Harvey DJ, Struwe WB, de Llorens R, Rudd PM, Peracaula R. Glycosylation of liver acute-phase proteins in pancreatic cancer and chronic pancreatitis. Proteomics Clin Appl. 2010;4:432–448. doi: 10.1002/prca.200900150. [DOI] [PubMed] [Google Scholar]
- 19.Zhao J, Qiu W, Simeone DM, Lubman DM. N-linked glycosylation profiling of pancreatic cancer serum using capillary liquid phase separation coupled with mass spectrometric analysis. J Proteome Res. 2007;6:1126–1138. doi: 10.1021/pr0604458. [DOI] [PubMed] [Google Scholar]
- 20.Singh S, Pal K, Yadav J, Tang H, Partyka K, Kletter D, Hsueh P, Ensink E, Kc B, Hostetter G, Xu HE, Bern M, Smith DF, Mehta AS, Brand R, Melcher K, Haab BB. Upregulation of glycans containing 3′ fucose in a subset of pancreatic cancers uncovered using fusion-tagged lectins. J Proteome Res. 2015;14:2594–2605. doi: 10.1021/acs.jproteome.5b00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruhaak LR, Taylor SL, Stroble C, Nguyen UT, Parker EA, Song T, Lebrilla CB, Rom WN, Pass H, Kim K, Kelly K, Miyamoto S. Differential N-Glycosylation Patterns in Lung Adenocarcinoma Tissue. J Proteome Res. 2015;14:4538–4549. doi: 10.1021/acs.jproteome.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wikoff WR, Grapov D, Fahrmann JF, DeFelice B, Rom WN, Pass HI, Kim K, Nguyen U, Taylor SL, Gandara DR, Kelly K, Fiehn O, Miyamoto S. Metabolomic markers of altered nucleotide metabolism in early stage adenocarcinoma. Cancer Prev Res (Phila) 2015;8:410–418. doi: 10.1158/1940-6207.CAPR-14-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abbott KL, Aoki K, Lim JM, Porterfield M, Johnson R, O’Regan RM, Wells L, Tiemeyer M, Pierce M. Targeted glycoproteomic identification of biomarkers for human breast carcinoma. J Proteome Res. 2008;7:1470–1480. doi: 10.1021/pr700792g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abbott KL, Lim JM, Wells L, Benigno BB, McDonald JF, Pierce M. Identification of candidate biomarkers with cancer-specific glycosylation in the tissue and serum of endometrioid ovarian cancer patients by glycoproteomic analysis. Proteomics. 2010;10:470–481. doi: 10.1002/pmic.200900537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sethi MK, Kim H, Park CK, Baker MS, Paik YK, Packer NH, Hancock WS, Fanayan S, Thaysen-Andersen M. In-depth N-glycome profiling of paired colorectal cancer and non-tumorigenic tissues reveals cancer-, stage- and EGFR-specific protein N-glycosylation. Glycobiology. 2015;25:1064–1078. doi: 10.1093/glycob/cwv042. [DOI] [PubMed] [Google Scholar]
- 26.Drake RR, Jones EE, Powers TW, Nyalwidhe JO. Altered glycosylation in prostate cancer. Adv Cancer Res. 2015;126:345–382. doi: 10.1016/bs.acr.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Nyalwidhe JO, Betesh LR, Powers TW, Jones EE, White KY, Burch TC, Brooks J, Watson MT, Lance RS, Troyer DA, Semmes OJ, Mehta A, Drake RR. Increased bisecting N-acetylglucosamine and decreased branched chain glycans of N-linked glycoproteins in expressed prostatic secretions associated with prostate cancer progression. Proteomics Clin Appl. 2013;7:677–689. doi: 10.1002/prca.201200134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haab BB. Antibody-lectin sandwich arrays for biomarker and glycobiology studies. Expert Rev Proteomics. 2010;7:9–11. doi: 10.1586/epr.09.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu J, Lin Z, Wu J, Yin H, Dai J, Feng Z, Marrero J, Lubman DM. Analysis of serum haptoglobin fucosylation in hepatocellular carcinoma and liver cirrhosis of different etiologies. J Proteome Res. 2014;13:2986–2997. doi: 10.1021/pr500128t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Zolotarevsky E, Thompson I, Anderson MA, Simeone DM, Casper JM, Mullenix MC, Lubman DM. A multiplexed bead assay for profiling glycosylation patterns on serum protein biomarkers of pancreatic cancer. Electrophoresis. 2011;32:2028–2035. doi: 10.1002/elps.201000693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, He J, Li C, Benitez R, Fu S, Marrero J, Lubman DM. Identification and confirmation of biomarkers using an integrated platform for quantitative analysis of glycoproteins and their glycosylations. J Proteome Res. 2010;9:798–805. doi: 10.1021/pr900715p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu Rev Immunol. 2007;25:21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- 33.Arnold JN, Wormald MR, Suter DM, Radcliffe CM, Harvey DJ, Dwek RA, Rudd PM, Sim RB. Human serum IgM glycosylation: identification of glycoforms that can bind to mannan-binding lectin. J Biol Chem. 2005;280:29080–29087. doi: 10.1074/jbc.M504528200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.