SUMMARY
Cancers are distributed unevenly across the body, but the importance of cell intrinsic factors such as stem cell function in determining organ cancer risk is unknown. Therefore, we used Cre-recombination of conditional lineage tracing, oncogene and tumour suppressor alleles to define populations of stem and non-stem cells in mouse organs, and test their life-long susceptibility to tumourigenesis. We show that tumour incidence is determined by the life-long generative capacity of mutated cells. This relationship held true in the presence of multiple genotypes and regardless of developmental stage, strongly supporting the notion that stem cells dictate organ cancer risk. Using the liver as a model system, we further show that damage-induced activation of stem cell function markedly increases cancer risk. Therefore, we propose that a combination of stem cell mutagenesis and extrinsic factors that enhance the proliferation of these cell populations, creates a ‘perfect storm’ that ultimately determines organ cancer risk.
Graphical abstract
INTRODUCTION
Cancers are distributed unevenly across the body. Some organs are far more likely to undergo malignant change than others, and children and adults develop very different types of cancer (Howlader N et al., 2012). This temporal and topographical bias in cancer formation can be explained in part by organ-specific susceptibilities to carcinogens or inherited oncogenic mutations; but the relative contributions of these, or other factors, to organ cancer risk is unknown (Danaei et al., 2005; Futreal et al., 2004). A greater understanding of the processes that underlie tumourigenesis is crucial if we are to improve the prevention and treatment of cancer.
It was recently proposed that the number of stem cell divisions occurring in a tissue during life might dictate cancer risk (Tomasetti and Vogelstein, 2015). This so called ‘bad luck’ hypothesis states that many cancers arise following the propagation of mutations that occur by chance in highly-replicative stem cell populations, rather than following exposure to environmental carcinogens. An important implication of this hypothesis is that these cancers are unavoidable and therefore resistant to primary prevention. But the notion that ‘intrinsic’ factors such as stem cell replication are more important than ‘extrinsic’ factors in carcinogenesis has been strongly contested (Ashford et al., 2015; Gotay et al., 2015; O'Callaghan, 2015; Potter and Prentice, 2015; Song and Giovannucci, 2015; Wild et al., 2015; Wu et al., 2015). Indeed, recent mathematical modeling estimated that 70–90% of the causal factors driving the most common cancers are ‘extrinsic’ (Wu et al., 2015).
The controversy surrounding these studies of cancer risk stems largely from their use of different mathematical approaches to correlate selected human cancer incidence data with variable sources and types of stem cell proliferation metrics. While these studies are important, they do not allow direct testing of the relationship between ‘intrinsic’ factors such as stem cell proliferation and cancer risk and cannot account adequately for ‘extrinsic’ carcinogenic factors. Experiments testing these variables directly could provide crucial insights into cancer origins, but they have not been performed on an adequate scale, in appropriate experimental systems. Therefore, we performed a series of organism-wide linage tracing and tumourigenesis studies, in defined populations of cells, in neonatal and adult mice, to more directly identify cell properties that dictate organ cancer risk.
RESULTS
Prom1+ cell generative capacity varies among organs and developmental stages
As a first step to test the relationship between cell properties and cancer risk, we characterised the number, basal proliferation rate, and life-long generative capacity of defined cell populations across major organs in neonatal (postnatal day [P] 1) and adult (P60) mice. To do this we used our Prom1C-L mouse that expresses both CreER2-recombinase and LacZ from the endogenous Prom1 (Cd133) locus (Zhu et al., 2009). We focused on Prom1+ cells because Prom1 marks a variety of cell types in mouse organs (Zhu et al., 2009), as well as some normal and malignant human stem cells (Lee et al., 2005; O'Brien et al., 2007; Ricci-Vitiani et al., 2007; Singh et al., 2004; Yin et al., 1997). First, a Rosa-ZsGreen Fluorescence Protein lineage tracing allele (RosaZsG) was activated by tamoxifen-induced recombination in neonatal (n=20) and adult (n=20) Prom1C-L; RosaZsG mice. Three days later, organs were collected from 10 mice each in both age groups (‘basal’ organs; Figure 1A). The remaining mice were allowed to age for 180 days (neonatal-induced) or 600 days (adult-induced) and their organs were then collected (‘aged’ organs). Bone marrow and peripheral blood samples were also taken for GFP fluorescence activated cell sorting (FACS) of haematopoietic lineages.
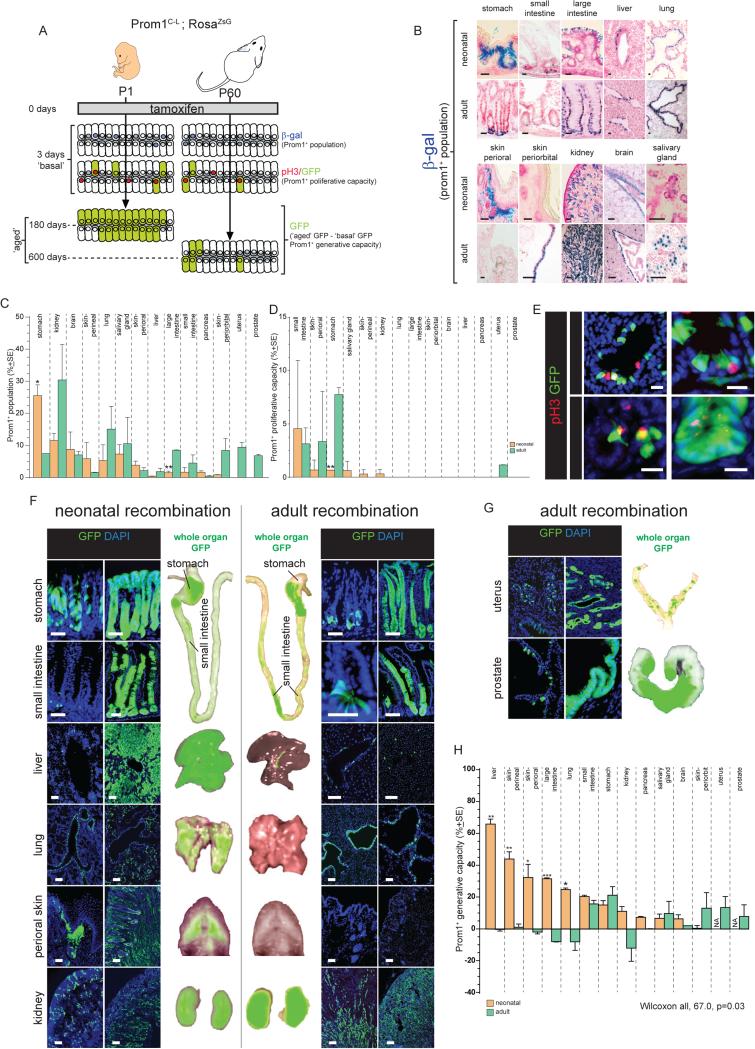
Figure 1. Prom1+ cell properties in major organs of Prom1C-L; RosaZsG mice.
A. Overall approach used the measure Prom1+ cell number, proliferative and generative capacities across organs of neonatal and adult mice. B. β-galactosidase staining of Prom1+ cells in neonatal and adult Prom1C-L mouse tissues (top panels scale bars=10μm; bottom=50μm). Percentage of Prom1+ cells (C) and percentage of proliferating Prom1+ cells (D) in indicated tissues. (E) Arrows identify proliferating Prom1+ cells in indicated tissues 1 day post tamoxifen (scale bars=10μm). (F,G) Direct GFP fluorescence microscopy of tissues at the indicated times post tamoxifen treatment. Whole organ direct GFP fluorescence images are also shown (scale bars=50μm). (E) Prom1+ cell generative capacity in indicated tissues. (*, p<0.05; **, p<0.005; ***, p<0.0005. See also Figures S1-S3 and Table S1. Data in C, D and H represent mean ±SE.
β-galactosidase and GFP staining detected similar numbers and distributions of Prom1+ cells in ‘basal’ organs, indicating Prom1C-L drives efficient recombination across all major organs (Figure 1B,F and data not shown). The size of the Prom1+ cell population varied markedly among ‘basal’ organs, ranging from the greatest in adult kidney (30.4%±13.3SE) to the smallest in neonatal liver (0.4%±0.04SE; Figure 1B,C,F,G); however, within any given organ, the number and location of Prom1+ cells remained fairly constant between neonates and adults. We then quantified Prom1+ cell proliferative capacity by co-immunostaining ‘basal’ organs with GFP and the cell proliferation marker pH3. Only limited Prom1+ proliferative capacity was detected in a handful of organs at both ages (Figure 1D,E).
To determine which organs might contain Prom1+ stem cells, we calculated the generative capacity of Prom1+ cells by measuring the difference in the proportion of GFP+ cells in lineage-traced ‘aged’ organs with those in the corresponding ‘basal’ organ (Figure 1A). In select organs with high generative capacity, we also fate mapped Prom1+ cell progeny to see if these included differentiated cells. In this manner, we showed previously that adult small intestinal Prom1+ stem cells generate the entire intestinal mucosa (Zhu et al., 2009). Although the number and location of Prom1+ cells in each organ remained fairly constant between neonates and adults, those in neonates frequently displayed a greater generative capacity (Figure 1H; across all organs, p<0.05, Wilcoxon). For example, Prom1+ cells in both neonatal and adult livers were sparse and located within or adjacent to the Krt19+ bile duct epithelium; a proportion of these cells co-expressed the liver progenitor marker A6 (Figure 1B,C,F; Figure S1A,B). However, while Prom1+ cells in neonatal livers were highly generative and produced the majority of ductal cells and hepatocytes that populated the liver by P180; those in adults had no detectable generative capacity (p<0.005 Mann-Whitney; Figure 1B,F,H; Figure S1B). Other organs in which Prom1+ cell generative capacity was restricted to neonates included the perineal skin (p<0.005), perioral skin (p<0.05), large intestine (p<0.0005) and lung (p<0.05; Figure 1F,H).
In other organs, both adult and neonatal Prom1+ cells displayed similar generative capacities. Prom1+ cells were localised to the base of gastric pits in both neonates and adults – the same location as adult Lgr5+ gastric stem cells (Barker et al., 2010) – and produced mature pit (UEAI+), neck (GSII+) and parietal (DBA+) cells of the gastric epithelium (Figure 1B,F,H; Figure S2A). Prom1+ cells in the crypts of neonatal and adult small intestines also displayed similar generative capacities and produced all mature cell types of the intestinal mucosa. In contrast, Prom1+ cells in both neonatal and adult kidney, pancreas, salivary gland, and brain displayed limited or no generative capacity (Figure 1H). Similarly, no GFP+ cells were detected in the bone marrow or blood of ‘aged’ mice (data not shown). These data are in agreement with recent reports that mouse haematopoeitic stem cells cannot be isolated on the basis of Prom1 expression (Arndt et al., 2013).
Tamoxifen treatment caused involution of the neonatal prostate and uterus, precluding accurate lineage-tracing of Prom1+ cells in these organs. However, serial rounds of androgen deprivation and replacement in castrated adult male mice caused Prom1+ cells to regenerate the entire prostatic mucosa, including basal (p63+) and luminal (Androgen Receptor+) cells, while serial oestrous cycles induced Prom1+ cells to regenerate the uterine mucosa (Figure 1G,H; Figure S2 A-C; Supplementary Experimental Procedures).
As a further test of the stem cell nature of Prom1+ cells we used Gene Set Enrichment Analysis to compare the transcriptomes of Prom1+ cells (>98% purity) isolated from various adult organs as well as neonatal livers (sufficient pure Prom1+ cell isolates could not be obtained from other neonatal organs) with those of 67 published stem cell gene expression signatures. A significant and positive correlation was observed between the generative capacity of Prom1+ cells and their enrichment of stem cell signatures (R2= 0.80, p<0.005; Table S1; Figure S3). Thus, our data reveal marked variation in the generative capacity associated with Prom1+ cells among organs and provide strong evidence that Prom1+ cells in the adult small intestine, stomach, prostate and uterus and the neonatal liver are stem cells.
Prom1+ cell susceptible to transformation varies among organs and developmental stages
Having characterised the number, basal proliferation and generative capacity of Prom1+ cells in neonatal and adult organs, we next tested the susceptibility of these cells to tumourigenesis (Figure 2A). We reasoned that different tissues might be sensitive to different initiating oncogenes and tumour suppressor genes (TSGs). To characterise this variability in susceptibility we selected a broad range of conditional alleles that disrupt major signaling pathways perturbed in human cancer, including Ctnnb1flx(ex3), KrasG12D, RosaNICD1, Tp53flx/flx, Ptenflx/flx, and Cdkn2aflx/flx (Futreal et al., 2004). Mice carrying these alleles were bred with Prom1C-L; RosaZsG mice to generate a total of eight mouse lines each harboring these conditional alleles either alone or in combination (Figure 2B). To parallel our lineage tracing study, oncogene and TSG conditional alleles were recombined by tamoxifen treatment of neonatal (P1) and adult (P60) mice. Four hundred and sixty-eight mice (n=235 neonates, and 233 adults) across the eight genotypes were recombined and subjected to life-long tumour surveillance studies (Table S2). Median survival rates varied greatly among the different genotypes, ranging from 11 days post-tamoxifen survival in neonatal-induced Prom1C-L; RosaZsG; Ctnnb1flx(ex3); KrasG12D mice, to 743 days post-tamoxifen survival in neonatal-induced Prom1C-L; RosaZsG; Ptenflx/flx mice (Figure 2B). Significant differences in survival were also observed between neonatal and adult-induced mice of the same genotype (Figure 2B).
Figure 2. Prom1+ cell susceptibility to tumourigenesis in major organs of Prom1C-L; RosaZsG mice.
A. Overall approach used to measure Prom1+ cell susceptibility to tumourigenesis. B. Left, survival curves of neonatal and adult Prom1C-L ; RosaZsG mice carrying the indicated alleles. P-value=difference in adult and neonatal survival. Right, pie charts of numbers of mice autopsied containing no, single or multiple tumours. C. Incidence of tumours in the indicated tissues across all genotypes. D. Anatomical heatmaps of organ tumour incidence. Below, ratios of tumours in neonatal and adult tissues. F=Fishers test of the difference; MHC= Mantel-Haenstzel-Cochran test of the difference. See also Table S2.
To determine if the recombination of oncogene and TSG alleles in Prom1+ cells induced cancer, we performed full autopsies on 166 neonatal-induced and 114 adult-induced mice that became moribund (Figures 2B-D; Table S2). Organs from all animals were inspected both macro- and microscopically for evidence of tumour formation. A total of 316 tumours were identified across the 280 autopsies. A single tumour was identified in 46% (n=76/166) and 33% (n=38/114) of neonatal and adult-induced mice, respectively. A further 28% (n=46/166) of neonatal-induced and 30% (n=34/114) of adult-induced mice contained multiple tumours (range 2 to 5 distinct primaries; Table S2). Ninety-seven percent (n=307/316) and 3% (n=9/316) of all tumours were judged to be primary or metastatic, respectively, by the reviewing pathologists (Figure 3A; Table S2). All primary and secondary tumours were GFP+ (Figure 3A). Primary tumours included: small and large intestinal adenomas; stomach adenomas and adenocarcinomas; hepatocellular carcinomas/intrahepatic cholangiocarcinomas (HCC/ICC); hepatoblastomas; prostatic adenocarcinomas; uterine carcinomas; squamous cell skin cancers; and lung adenomas (Figure 2B-D and 3A; Table S2). Few or no tumours and no premalignant lesions were found in the salivary gland, kidney, brain, or pancreas even though some of these had large populations of Prom1+ cells and would therefore have received a significant mutational burden. Two hundred and four genotype-matched mice were also aged without tamoxifen induction to assess rates of spontaneous tumour formation. Only 2.5% (n=5/204) of these mice developed tumours that were all GFP− (Table S2). Thus, tumours in our model systems arose within recombined Prom1+ cell lineages.
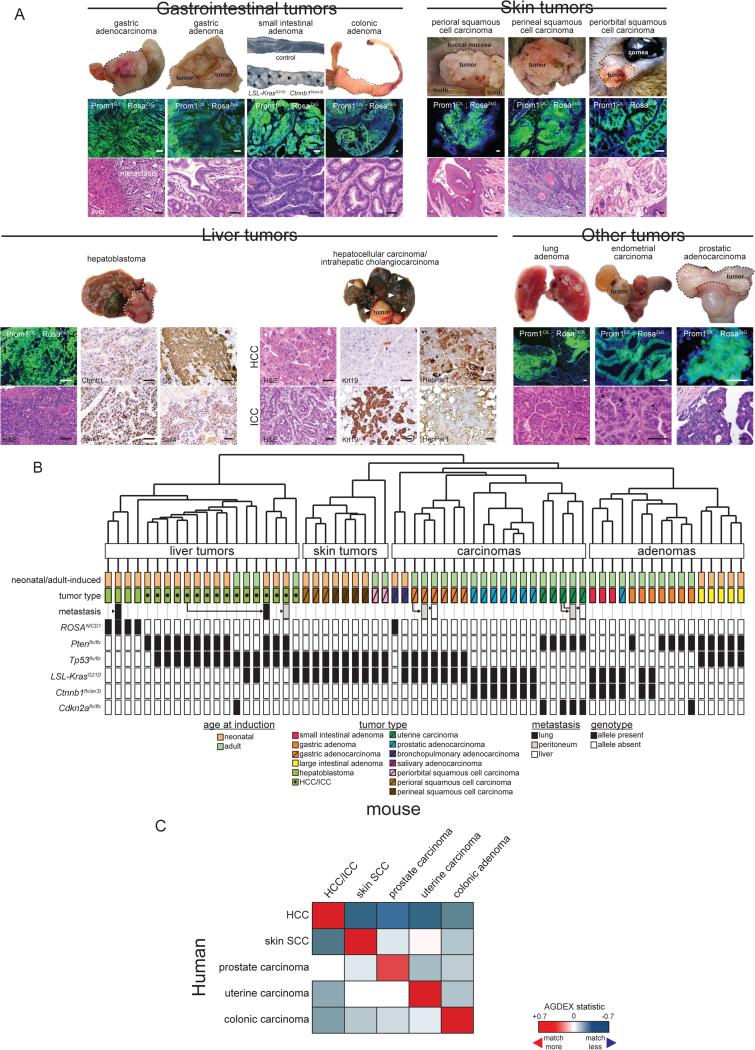
Figure 3. Histology and transcriptomic analysis of Prom1+ cell derived tumours in mice.
A. Top: gross specimens (arrows and dotted lines denote tumours), and photomicrographs of direct GFP fluorescence, liver immunohistochemistry (HCC and hepatoblastoma marker expression) and H&E stains (arrows denote mitoses) of exemplary tumours (scale bars=50μm). B. Unsupervised hierarchical clustering of mouse tumours. Details of age at recombination, tumour histology and genotype are shown below. C. AGDEX comparison of histologically matched human and mouse tumours. See also Tables S3 and S4.
Since almost one third of mice developed multiple tumours, of which some appeared to be metastatic, we sought to more rigorously assess the accuracy of our histological diagnoses. To do this we subjected 58 primary and seven metastatic tumours to gene expression profiling (Figure 3B; Table S3; GEO78076). Unsupervised hierarchical clustering segregated out liver, skin, carcinomas and adenomas. Within these clusters, tumours of the same histological type, including metastatic tumours harvested from distal sites, were co-clustered. As a further test of histologic tumour typing, we used our cross-species AGDEX algorithm (Johnson et al., 2010) to compare the transcriptomes of our mouse tumours with those of 154 histologically-matched human primary cancers (Table S4). The transcriptomes of mouse HCC/ICC, skin squamous cell, prostate and uterine carcinomas and colonic adenomas closely matched those of the corresponding human tumour (Figure 3C). Thus, Prom1+ cell populations in some, but not all, mouse organs are susceptible to transformation and generate cancers that recapitulate the histology and transcriptome of the corresponding human tumours.
Prom1+ cell transformation is partly determined by initiating mutation
Organ bias in tumour formation in our mice could be explained in part by tissue-specific susceptibilities to initiating mutations. For example, in keeping with the high rate of WNT pathway mutations in human intestinal tumours (Bienz and Clevers, 2000), 73% (n=16/22) of neonatal and 91% (n=43/47) of adult-induced mice harboring the Ctnnb1flx(ex3) allele developed small intestinal adenomas, compared with only 13% of neonatal (n=12/144) and 3% (n=2/67) of adult-induced mice that lacked the allele (Figure 2B-D, 3A; Table S2; Fisher's exact, P<0.00001). Additionally, consistent with the high rate of PTEN mutations in human uterine cancer (Kong et al., 1997; Risinger et al., 1997), 89% (n=8/9) of female adult-induced mice carrying the Ptenflx/flx allele developed uterine carcinomas versus only 5% (n=1/22) of adult-induced females with wild-type Pten (Fisher's exact, P<0.0001). Different initiating mutations also induced different types of tumours within the same organ. For example, 100% (n=5/5) of liver tumours in neonatal-induced Prom1C-L; RosaZsG; RosaNICD1 mice were Ctnnb1+/Hes1+/GS+/Sall4+ hepatoblastomas, while 100% (n=64/64) of liver tumours arising in the other genotypes were HCC/ICC (Fisher's exact, P<0.00005; Figure 3A; Table S2). Similarly in the stomach, all tumours arising in Prom1C-L; RosaZsG; KrasG12D; Tp53flx/flx mice (n=12/12) and Prom1C-L; RosaZsG; Ptenflx/flx; Tp53flx/flx mice (n=5/5) were gastric adenocarcinomas, while all 37 stomach tumours arising in the other five affected genotypes were gastric adenomas; suggesting loss of Tp53 might be an important determinant of carcinoma formation in the stomach (Fisher's exact, P<0.00005; Figure 3A; Table S2).
But initiating mutations did not fully explain the temporal and topographical bias in tumourigenesis among organs. For example, the liver, small intestine and stomach appeared to show an age specific bias in tumour development in the context of multiple genotypes. Therefore, we used the Mantel-Haenszel-Cochran test to determine if developmental stage and tumour prevalence in these organs were associated even when multiple genotypes were oncogenic. These analyses showed that neonatal-induced mice had a 10-fold average greater incidence of liver tumours than adult-induced mice, which was consistent across genotypes (common odds ratio [COR] P3:P60=9.9; Mantel-Haenstzel-Cochran test [MHC], P<0.0005; Figure 2C,D). Also consistent across genotypes, significantly more stomach tumours arose in adult than neonatal-induced mice (COR P60:P3=2.5, MHC, P<0.05). Furthermore, even those organs that were susceptible to specific initiating mutations demonstrated a temporal bias in cancer formation, including: small intestinal adenomas associated with Ctnnb1 activation that were more common in adult-induced mice (Fishers exact, P<0.05; Figure 2D); Pten-deleted large intestinal and lung adenomas, both of which were more common in neonatal-induced mice (Fishers exact, P<0.05); and perioral and perineal squamous cell carcinomas in Kras-activated/Tp53-deleted mice that were more common in neonatal-induced mice (Fishers exact, P<0.000005). Thus, Prom1+ cell populations in some organs were differentially susceptible to transformation independent of initiating mutation, suggesting that other cell properties might have dictated patterns of cancer development in our mice.
Prom1+ cell generative capacity is a major determinant of organ cancer risk
To begin to understand what cell properties might dictate cancer risk, we used mathematical modeling to explore how Prom1+ cell population size, proliferative capacity, and generative capacity in each organ related to susceptibility to tumourigenesis. All tumorigenesis and Prom1+ cell data from all organs in neonatal and adult-induced mice were incorporated into a generalised linear mixed model with a logit link (GLMM-LL) to assess these relationships since this allowed incorporating multiple tumours within mice; genotype to be treated as a random effect; and mice to be nested within genotype (McCulloch et al., 2008).
In single marker models that incorporated an interaction with age, Prom1+ generative capacity provided the most robust model: the greater the generative capacity of a Prom1+ cell in an organ, the more susceptible it was to tumourigenesis; strongly suggesting that Prom1+ stem cells are especially tumourigenic (Figure 4A; note the smaller the Akaike Information Criterion (AIC) the better the model fit). This notion is further supported by the observation that organs containing the smallest populations of Prom1+ cells were most likely to generate tumours, since stem cells are relatively rare in tissues (Figure 4A; AIC=2255). Interestingly, neonatal and adult Prom1+ cells with equivalent generative capacities displayed markedly different susceptibilities to transformation: those in neonates were significantly more resistant to tumourigenesis (P=5×10−11). Prom1+ proliferative capacity also increased cancer risk, although less dramatically than generative capacity (Figure 4B; AIC=2138).
Figure 4. Relationship of Prom1+ cell characteristics to tumour susceptibility.
Generalised linear mixed models with a logit link (GLMM-LL) of tumour probability vs. Prom1+ cell generative capacity (A), Prom1+ cell population size (B), and Prom1+ cell proliferative capacity (C). Numbers at top of each graph indicate the regression coefficients and p-value of each as well as the Akaike Information Criterion score. D. GLMM-LL iterative multivariable modeling of tumour probability vs. Prom1+ cell generative and proliferative capacities in adult (D) and neonatal (E) tissues. See also Methods and Resources.
Since Prom1+ population size, proliferative capacity, and generative capacity may be interrelated, we used GLMM-LL iterative multivariable modelling to determine the relative association of these factors with cancer risk (Figure 4D,E). Once again, Prom1+ generative capacity was most significantly and positively correlated with tumour incidence (both induction ages, P<2×10−16). Prom1+ proliferative capacity was also related to cancer risk in neonatal-induced mice, but only marginally so in adult-induced mice. Prom1+ population size was not related independently to tumour incidence. Thus, our data strongly suggest that the risk of an organ developing cancer following the introduction of oncogenic mutations into its resident Prom1+ cell population is significantly associated with their life-long generative capacity: this holds true in the presence of multiple genotypes and regardless of developmental stage, supporting the notion that the mutation of stem cells dictates cancer risk.
Hepatic tumourigenesis varies directly with Prom1+ cell generative capacity
To further explore the notion that stem cell phenotype dictates cancer risk, we looked to see if altering stem cell properties in a single organ might concurrently alter tumorigenesis. Similarly sized and located populations of Prom1+ cells exist in neonatal and adult livers; but only those in neonates are generative and tumourigenic (Figures 1-3; Figure S1). Therefore, we hypothesised that adult Prom1+ liver cells might be the quiescent progeny of neonatal Prom1+ liver cells. To test this, we fate mapped Prom1+ liver cells in neonatal Prom1C-L; RosaZsG mice and demonstrated that these cells, or their progeny, constitute 94.6%+0.3SD of Prom1+ cells in adult liver (Figure 5A).
Figure 5. Stem cell activity and tumour susceptibility are functionally related in Prom1+ liver cells.
A. Concurrent β-galactosidase and direct GFP fluorescence of the same section of liver taken from an adult Prom1C-L ; RosaZsG mouse that underwent tamoxifen-recombination as a neonate. Arrows identify β-gal+/GFP+ cells and the percentage of these cells in four separate livers is shown below (scale bar=50μm). B. Prom1+ cell generative capacity in adult mouse livers harbouring two different lineage tracing alleles and damaged by DDC treatment or partial hepatectomy (scale bars=50μm). C. Unsupervised hierarchical clustering of Prom1+ cell transcriptomes isolated normal newborn (NBNT) and adult (ADNT) livers as well as adult livers treated with DDC (ADDDC). D. AGDEX comparison of NBNT and ADDDC Prom1+ cell transcriptomes using the ADNT Prom1+ cell transcriptome as a common comparator. E. Left, survival curves of adult control and DDC treated Prom1C-L ; RosaZsG mice carrying the indicated alleles. P-value=difference in survival. Right, anatomical heatmaps of organ tumour incidence. Below, ratios of tumours in neonatal and adult liver. F=Fishers test of the difference; MHC=Mantel-Haenstzel-Cochran test of the difference. See also Figure S1, Table S2.
Chronic tissue injury plays a critical role in tumourigenesis, particularly in the liver where cancer is rare in the absence of chronic tissue damage (Block et al., 2003; Coussens and Werb, 2002; Pikarsky et al., 2004). Therefore, we next tested if damaging adult livers might reactivate generative and tumourigenic capacity in Prom1+ adult liver cells. Lineage tracing alleles were activated in adult Prom1C-L mice by tamoxifen-recombination and then three days later their livers were damaged by either partial hepatectomy (Michalopoulos and DeFrances, 1997) or administration of diethoxycarbonyl-1,4-dihydrocollidine (DDC) (Preisegger et al., 1999). Both damage mechanisms markedly activated Prom1+ cell proliferation and tissue repair (Figure 5B; Figure S1B). Remarkably, AGDEX and GSEA analyses revealed that while the transcriptomes of Prom1+ cells isolated from damaged adult livers did not resemble those of normal adult liver, they were very similar to those of neonatal Prom1+ liver cells and were enriched with many of the ‘stem cell signatures’ observed in neonatal Prom1+ liver cells (Figure 5C,D; Table S1). Thus, adult Prom1+ liver cells are the quiescent progeny of neonatal Prom1+ liver stem cells: damaging the liver reactivates a ‘neonatal-like’ stem cell program in these cells, promoting their proliferation and liver repair. Since damaged adult liver Prom1+ cells did not upregulate Lgr5 (Figure S1C), they are likely to be distinct from Lgr5+ damage-induced hepatocytes (Huch et al., 2013).
To test if damage-induced generative capacity of adult Prom1+ liver cell increases their susceptibility to tumourigenesis, we repeated our sequential Cre-recombination and DDC-damage protocol in Prom1C-L; RosaZsG mice harboring KrasG12D, KrasG12D; p53flx/flx, Ptenflx/flx; p53flx/flx, or Ptenflx/flx; Cdkn2aflx/flx alleles (n=68). All mice were subjected to tumour surveillance for at least two years and moribund mice underwent full autopsies (DDC treated, n=46; control, n=78; Figure 5E; Table S2). Liver damage dramatically increased the production of liver tumours by mutated Prom1+ cells across all genotypes (COR, DDC:control=38.7, MHC, P<1×10−7; Figure 5E). Although DDC was delivered systemically, the incidence of other cancers remained unchanged; indicating that acceleration of tumourigenesis by DDC was specific to the liver (Table S2). Indeed, consistent rates of tumorigenesis in other organs likely explained why the survival of DDC and non-DDC mice remained relatively similar for most genotypes. For example, non-DDC and DDC treated KrasG12D; p53flx/flx mice succumbed to aggressive gastric adenocarcinomas with similar median survivals (non-DDC=209 days; DDC=193 days), even though all DDC treated animals developed liver tumours. Thus, the reactivation of generative capacity in adult Prom1+ liver cells markedly increases their susceptibility to transformation, providing direct evidence that cell generative capacity can be a major determinant of organ cancer risk.
Cross-species analysis predicts the role of recurrently mutated genes in human cancer
Since Prom1+ stem cells were peculiarly susceptible to transformation, we speculated that gene expression patterns in these cells might inform understanding of cancer, and in particular unmask the role of genes that are recurrently mutated in cancer.
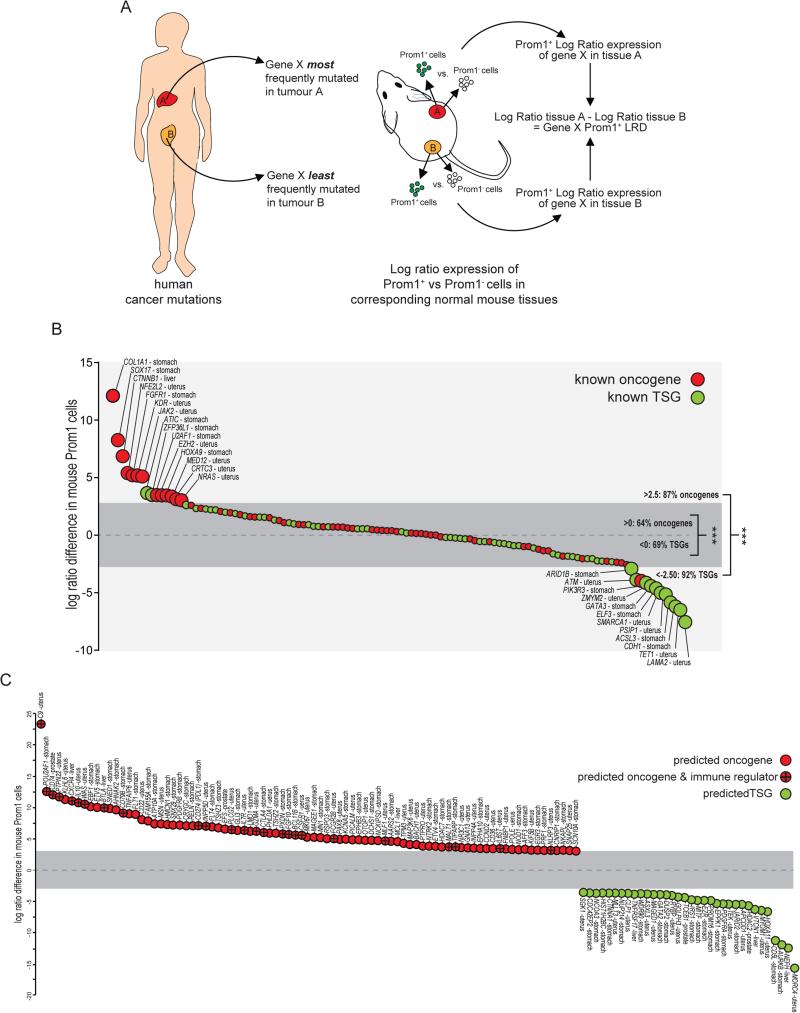
To test this, we first identified 657 genes that The Cancer Genome Atlas reported as recurrently mutated (n>3 samples) in human hepatocellular, gastric, prostatic or uterine carcinomas (www.cbioportal.org). We studied these cancers because the corresponding tumours in our mice arose from Prom1+ stem cells. Only 18% of the 657 mutated genes are validated as either oncogenes (n=58/657) or TSGs (n=60/657); the function in cancer of the remaining 82% (n=539/657) is unknown. Next, we identified the cancers in which each gene was most and least frequently mutated, reasoning that these tissues were most and least likely to be transformed by mutations in the gene (Figure 6A). We then compared the log ratio of expression of each of these genes in Prom1+ vs. Prom1− cells of the corresponding tissues in mice (hereon, Prom1+ log ratio difference [LRD]). Almost twice as many validated oncogenes (n=42/66) than TSGs (n=24/66) had a positive LRD, while more than twice as many TSGs (n=36/52) than oncogenes had a negative LRD (n=16/52; P<0.0005; Figure 6B). Even greater bias was observed among genes with an LRD >3 and <−3, of which almost all are oncogenes (n=13/15) and TSGs (n=10/11; P<0.0005), respectively. Thus, oncogenes and TSGs of specific human cancers are expressed to relatively high and low levels, respectively, in the Prom1+ stem cells that generate these cancers in mice. Therefore, we reasoned that LRD scores might predict which of the 539 unclassified genes are oncogenes or TSGs.
Figure 6. Cross-species comparison of gene mutation and expression in human cancers and tissue-matched Prom1+ stem cells predicts oncogene and TSG function.
A. Overall approach taken to calculate the Log-Ratio Difference (LRD) of each gene (see Supplementary Methods). B. LRDs of known oncogenes and TSGs in liver, stomach, prostate and uterus. Dark grey zone, LRD −3 to +3. C. Predicted function of recurrently mutated genes in liver, stomach, prostate and uterine cancers based on LRD −3 to +3. Note preponderance of immune regulators in predicted oncogenes. See also Methods and Resources.
Fifteen percent (n=85/539) of the unclassified genes had an LRD >3 and are therefore predicted to be oncogenes. Interestingly, these were highly enriched for regulators of immune activation (GO:0006955, FDR = 2.6×10−4), including CD274 (PDL1) and CTLA4 that suppress T cell function and are emerging as a new class of targets of cancer immunotherapy (Pardoll, 2012). While the functional consequences of these mutations remain unclear, PDL1 was recently demonstrated to drive T-ALL malignancies (Casey et al., 2016); therefore, we propose these aberrations enable early transforming stem cells to escape immunosurveillance. A further 6% (n=34/539) of unclassified genes had an LRD <−3 and are therefore predicted to be TSGs (Figure 6C). These included NEFH that is hypermethylated in breast cancer (Calmon et al., 2015) and JARID2, a component of the Polycomb Repressor 2 Complex that negatively regulates haematopoeitic stem cells (Kinkel et al., 2015).
DISCUSSION
Vigorous debate has surrounded two apparently opposing explanations of why cancers are distributed unevenly among organs. One idea proposes that extrinsic carcinogens drive this bias; the other that intrinsic factors e.g., stem cell proliferation, can independently dictate cancer risk. However, since intrinsic and extrinsic factors are likely to cooperate, then correlational analyses of human epidemiological data cannot assess their relative importance. Here, using a series of conditional oncogene and TSG alleles, we selectively introduced mutations into well characterised stem and non-stem cell populations across organs in mice; essentially circumventing the need for mutagenic extrinsic factors and therefore testing the role of intrinsic factors in cancer risk. We show that the risk of an organ developing cancer is significantly associated with the life-long generative capacity of its mutated cells. This relationship held true in the presence of multiple genotypes and regardless of developmental stage, strongly supporting the notion that stem cells can dictate cancer risk among organs.
But our data by no means undermine the importance of extrinsic factors for cancer risk. The introduction of mutations into Prom1+ cells of the normal adult liver was insufficient to induce tumours; only when extrinsic damage induced these cells to proliferate were they competent to transform. Tissue damage and inflammation are well recognised determinants of cancer risk across a variety of organs; but most studies have suggested that this damage is either mutagenic e.g., through production of reactive oxygen species, or promotes the survival and proliferation of transformed cells (Coussens and Werb, 2002; Shalapour and Karin, 2015). Our data suggest that the ‘carcinogenic’ properties of some extrinsic factors might relate solely to their induction of local tissue damage and activation of stem cell repair, thereby creating an expanded population of cells that are competent for transformation. In this model, organ cancer risk could be determined by a critical combination of factors: the intrinsic proliferative capacity of the resident stem cell population; the presence of local tissue damage that expands the size of this population; the susceptibility of this population to be transformed by acquired mutations; and perhaps the presence of a stem cell transcriptome that allows the evasion of immunosurveillance. This model also allows for the scenario in which damage induces the acquisition of stem cell properties by relatively plastic cell populations (Figure 7).
Figure 7. Proposed model for the role of cell intrinsic and extrinsic factors in determining organ cancer risk.
We propose that extrinsic factors converge specifically on stem cells to induce mutations and/or tissue damage that provokes proliferative repair. Tissue-specific susceptibility of stem cells to induced mutations and their intrinsic, or damage-induced proliferative capacity, create a ‘perfect storm’ that ultimately determines organ cancer risk. The high expression of immune regulators by stem cells might enable early transforming cells to escape immunosurveillance. See also Methods and Resources.
As well as determining the distribution of cancer among different organs, stem cells may also dictate temporal patterns of cancer risk within the same organ. We show on average that neonatal Prom1+ cells are far less likely to undergo transformation than those in adults with equivalent generative capacities; suggesting that neonatal stem cells are intrinsically resistant to transformation. If this biology holds true in humans, then it may explain why cancer rates are many fold lower in children than adults, despite the fact that childhood cancers accrue significant numbers of non-synonymous mutations, and organ growth rates peak in childhood (Jones and Baker, 2014; WHO, 2006). Temporal stem cell context might also dictate cancer type. For example, in our models, hepatoblastoma – a childhood-specific liver tumour – was induced only by aberrant Notch signaling in neonatal Prom1+ liver stem cells. Activation of NOTCH signaling is a recognised feature of hepatoblastoma (Lopez-Terrada et al., 2009).
Our data also suggest that transforming stem cells may do more than merely propagate mutations. Recurrently mutated human cancer genes that were expressed at relatively high levels in Prom1+ stem cells were strikingly enriched for immune regulators, including Ctla4 and Cd274 (Pdl1) that are targets of effective immune checkpoint inhibitors in cancer (Melero et al., 2015). We propose that the expression of these genes enables transforming stem cells to escape immunosurveillance at the earliest stages of tumourigenesis. Further work will be required to determine the precise characteristics that render stem cells vulnerable to transformation; but these may include the tolerance of oncogenic insults that might otherwise induce cellular apoptosis or senescence.
In addition to providing important insights into cancer origins, we identify a number of new Prom1+ stem cell populations in several mouse organs including the neonatal and damaged adult liver, stomach, uterus and prostate. At least two populations of well-characterised stem cells are known to exist in the adult mouse liver: peri-central vein Axin2+ hepatocytes that constantly regenerate undamaged liver (Wang et al., 2015); and damage-induced, peri-portal Lgr5+ cells that repair the liver (Huch et al., 2013). Here, we identify a previously unrecognised population of neonatal Prom1+ liver stem cells with remarkable generative capacity that essentially produce most if not all cells of the early postnatal and juvenile liver. These cells are located within or adjacent to the Krt19+ bile duct epithelium and are therefore distinct from peri-central vein Axin2+ stem cells that perform a similar function in the adult liver. Neonatal Prom1+ liver cells also express low levels of Lgr5+, as do their damage-responsive Prom1+ progeny in adults, suggesting these cells are different from previously described Lgr5+ liver stem cells. Similar to the liver, the number and location of Prom1+ cells in other organs differed little between neonates and adults e.g., lung. Therefore, it will be important to determine if neonatal Prom1+ cells in these organs also give rise to quiescent but reparative and tumourigenic Prom1+ cells in adults.
The various cancer models described here display remarkable histological and transcriptomic similarities to the corresponding human diseases and should therefore add significantly to the existing armamentarium of mouse cancer models available for biology and therapeutic studies. A number of organs in our mice generated few or no cancers e.g., salivary gland, pancreas and kidney, even though these tissues contained large numbers of Prom1+ cells and can be transformed by the same oncogenes and TSGs used in our study (Clark et al., 2011; Hingorani et al., 2005; Raimondi et al., 2006). Our hypothesis proposes that these tissues didn't develop cancer because they lacked Prom1+ stem cells; however, since many of our mice developed multiple tumours, it is possible that cancers with shorter latencies arose and killed mice before they could develop salivary gland, pancreatic or kidney tumours. Although feasible, we believe this is unlikely since the median survival of mice harboring activated Kras and Ctnnb1 alleles in our study, was equal to or greater than the published latency of salivary gland, pancreatic and kidney cancers in mouse models driven by these alleles.
In summary, our data provide direct experimental evidence that the mutation of stem cells is an important determinant of cancer risk. We propose that extrinsic factors converge on these cell populations, inducing mutations and/or proliferation that together create the conditions necessary to drive tumourigenesis.
METHODS AND RESOURCES
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Polyclonal anti-phospho-Histone H3 (Ser10) | Millipore | Cat# 06-570; RRID: AB_310177 |
| Rabbit Polyclonal anti-androgen receptor | Sigma-Aldrich | Cat# A9853; RRID: AB_262132 |
| Mouse monoclonal anti-p63 | Santa Cruz | Cat# sc-8431; RRID: AB_628091 |
| Rat monoclonal anti-A6 | gift of N.V. Engelhardt, NIH, Bethesda, MD (Bennoun et al., 1993) | N/A |
| Rat monoclonal anti-Krt19 | gift of J. Friedman, University of Pennsylvania, Philadelphia, PA (Hand et al., 2009) | N/A |
| Rabbit monoclonal anti-Krt19 | Abcam | Cat# ab133496; RRID: AB_11155282 |
| Mouse monoclonal anti-HNF4α | R&D System | Cat# PP-K9218-00; RRID:AB_2116902 |
| Mouse monoclonal anti-β-catenin | BD Biosciences | Cat# 610153; RRID: AB_397554 |
| Rabbit Polyclonal anti-Glutamine Synthetase | Abcam | Cat# ab73593; RRID: AB_2247588 |
| Rabbit Polyclonal anti-Hes-1 | Abcam | Cat# ab71559; RRID: AB_1209570 |
| Rabbit Polyclonal anti-Sall4 | Abcam | Cat# ab29112; RRID: AB_777810 |
| Mouse monoclonal anti-HepPar-1 | Dako | Cat# M 7158; RRID: AB_2335689 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma-Aldrich | Cat# T5648 |
| Rhodamine-labeled DBA | Vector Laboratories | Cat# RL-1032; RRID: AB_2336396 |
| rhodamine-labeled UEA I | Vector Laboratories | Cat# RL-1062; RRID: AB_2336769 |
| GS II, Alexa Fluor® 594 conjugate | Invitrogen | Cat# L-21416 |
| Testosterone-propionate, 2.5 mg/pellet, 21-day release | Innovative Research of America | Cat# A-211 |
| 0.1% DDC rodent chow | Bio-Serv | Diet F4613 |
| Critical Commercial Assays | ||
| RNeasy Mini Kit | Qiagen | Cat# 74104 |
| PicoPure™ RNA extraction kit | Thermo Fisher | Cat# KIT0204 |
| Dead Cell Removal Kit | Miltenyi Biotec | Cat# 130-090-101 |
| MACS with anti-Prominin-1 MicroBeads | Miltenyi Biotec | Cat# 130-092-564 |
| Deposited Data | ||
| Raw and analyzed Affymetrix microarray data | This paper | GEO: GSE78076 |
| Human cancer gene expression profiles | (Engreitz et al., 2010) | GEO: GSE64985 |
| Recurrent mutations in human cancers | cBioPortal for Cancer Genomics | http://www.cbioportal.org |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Mouse: Prom1C-L | Generated in our laboratory (Zhu et al., 2009) | RRID:IMSR_JAX:017743 |
| Mouse: RosaNICD1 | Jackson Laboratory | RRID:IMSR_JAX:008159 |
| Mouse: RosaZsG | Jackson Laboratory | RRID:IMSR_JAX:007906 |
| Mouse: Ptenflx | Jackson Laboratory | RRID:IMSR_JAX:004597 |
| Mouse: Tp53flx | Jackson Laboratory | RRID:IMSR_JAX:008462 |
| Mouse: Cdkn2aflx | Jackson Laboratory | RRID:IMSR_JAX:023323 |
| Mouse: KrasG12D | NCI Mouse Repository | RRID:IMSR_NCIMR:01XJ6 |
| Mouse: Ctnnb1flx(ex3) | from M. Taketo, Kyoto University, Japan (Harada et al., 1999) | N/A |
| Recombinant DNA | ||
| Sequence-Based Reagents | ||
| Software and Algorithms | ||
| AGDEX | (Johnson et al., 2010) | http://www.stjuderesearch.org/site/depts/biostats/agdex |
| GSEA | Broad Institute | http://software.broadinstitute.org/gsea |
| Non-parametric tests (Fisher's Exact, Mantel-Haenszel-Cochran) | Minitab Inc. | Minitab 16.1.0 |
| glmmADMB | (Fournier et al., 2012; Skaug et al., 2014) | http://glmmadmb.r-forge.r-project.org/ |
| Other | ||
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Richard J. Gilbertson (Richard.Gilbertson@cruk.cam.ac.uk).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Mice were Prom1C-L (Zhu et al., 2009), RosaNICD1, RosaZsG, Ptenflx/flx, p53flx/flx, Cdkn2aflx/flx (Jackson Laboratory, Bar Harbor, ME), KrasG12D (NCI Mouse Repository) and Ctnnb1flx(ex3) (from M. Taketo). See “Key Resources Table” for details. All mice were maintained in a specific pathogen-free facility and all animal experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of St. Jude Children's Research Hospital.
METHOD DETAILS
Mouse and Tissue Handling
Cre-recombination in mice was activated with 3mg tamoxifen (TMX)/40g body weight at either neonatal (P1) or adult (typically P60) for two consecutive days by IP injection. Liver damage was induced in mice by partial hepatectomy or chow containing 0.1% DDC (Bio-Serv) commenced 3 days post TMX. Mouse prostate regeneration was induced by orchidectomy and serial testosterone implants. See below for detailed surgical procedures. Full-body necropsy was performed on moribund mice, or the indicated time point, whichever first. Tissue sections of all major mouse organs were subjected to direct GFP microscopy and β-galactosidase staining using 12μm frozen sections or H&E and appropriate diagnostic immunostaining performed on 5μm paraffin sections.
Immunohistochemistry and immunofluorescence
For immunohistochemistry, tissues were fixed in 4% paraformaldehyde, paraffin embedded and sectioned at 5μm. Antibodies for immunohistochemical staining (Figure 3A) include anti-β-catenin, anti-Glutamine Synthetase, anti-Hes-1, anti-Sall4, anti-Krt19 and anti-HepPar-1. For GFP microscopy and immunofluorescence, tissues were perfused and fixed in 2% paraformaldehyde, cryo-protected, frozen in OCT and sectioned at 12μm. Antibodies for immunofluorescence staining (Figure S1 & S2) include anti-phospho Histone H3, anti-androgen receptor, anti-p63, anti-A6, anti-Krt19, and anti-HNF4α. Lectins for gastric mucosa staining (Figure S2) include rhodamine-labeled DBA, rhodamine-labeled UEA I, GS II, Alexa Fluor® 594 conjugate. See “Key Resources Table” for details.
Orchidectomy and testosterone implantation
Two-month-old male Prom1C-L; RosaZsG mice were castrated 3 days post tamoxifen treatment using standard techniques. Briefly, each mouse was anesthetized with 2-3% isoflurane. A small incision was made rostral to the scrotal sac and gentle pressure applied to aid visualizing the testicles. The tunica albuginea was incised to separate and remove the testes from the scrotal ligament. Skin edges were adhered with tissue glue. Mice were allowed to rest for four weeks to ensure involution of the prostate prior to insertion of subcutaneous testosterone pellet implantation. Mice were anesthetized with 2-3% isoflurane and a small skin incision made on the dorsal neck. A testosterone pellet (testosterone-propionate, 2.5 mg/pellet, 21-day release, Innovative Research of America) was inserted and placed 2 cm from the incision by a trochar (Innovative Research of America, Catalog No. MP-182). Tissue glue was used to close the skin edges. The procedure was repeated four times every eight weeks to induce repeated prostatic involution and regeneration that was monitored by ultrasound imaging.
Partial Hepatectomy
Partial hepatectomy in two month old Prom1CreERT2; RosaLacZ mice was performed 3 days post tamoxifen treatment using standard techniques (Boyce and Harrison, 2008). Briefly, each mouse was anesthetized with 2-3% isoflurane and a transverse abdominal incision made to expose the posterior section of the median lobe of the liver. The left and right median lobes of the liver were surgically removed. Sutures used to ligate the hepatic vessels and close the abdominal wall. Wound clips were used to close the skin.
Ultrasound scans of prostate
Mice were anesthetized with 2-3% isofluorane in O2 (2 liters/min) and their hair removed from the lower ventral abdomen. Animals were then positioned ventrally and secured on the heated platform, allowing physiological monitoring throughout. Warmed Aquasonic 100 (Parker Laboratories, Inc, Fairfield, NJ) coupling gel was applied to the surface of the skin and target organs imaged (40 MHz) using a VEVO-770 High Resolution Ultrasound system (FujiFilm VisualSonics inc., Toronto, Canada). Data were acquired with an in-plane resolution of 40 μm and a step-size of 0.102 mm and processed using the VEVO-770 software to provide volumetric information and rendering.
Two-photon laser-scanning microscopy
Two-photon laser-scanning microscopy was performed using an Ultima imaging system (Prairie Technologies, Middletown WI), a Ti:sapphire Chameleon Ultra femtosecond-pulsed laser (930 nm) (Coherent, Santa Clara, CA), and a 20 × 0.95 NA water-immersion IR objective (Olympus, Center Valley, PA). Stacks of images were reconstructed in 3D using Imaris software (Bitplane, Saint Paul, MN).
Single cells isolation from mouse organs
Liver was dissociated in situ with Liberase-TM (0.1 U/ml; Roche) in Krebs’ Ringer with glucose (3.6 g/L) and calcium (1 mM) as described (Shupe et al., 2009).
Prostates were dissociated with 800μg/ml collagenase type I (Worthington) and 100 μg/ml DNAse I (Roche) in Dulbecco's modified Eagle's medium (Lonza) supplemented with 2mM L-glutamine (Invitrogen), 100 U/ml penicillin, 100 μg/ml streptomycin (Invitrogen) and 10% fetal bovine serum (Atlanta Biologicals) at 37°C for 90 min as described (Xin et al., 2003).
Stomachs were removed from 20 mice, opened and washed extensively in PBS. The mucosa was scraped off the pyloric region and digested for 60 min at 37° in 0.1% Pronase (Roche) and 100 μg/ml DNase I (Roche) in PBS containing 2% BSA and 1mM EDTA as described (Houghton et al., 2004).
Uteruses were dissociated with 0.2% collagenase type I (Worthington) and 100 μg/ml DNAse I (Roche) in Dulbecco's modified Eagle's medium (Lonza) supplemented with 2 mM L-glutamine (Invitrogen), 100 U/ml penicillin, 100 μg/ml streptomycin (Invitrogen) and 10% fetal bovine serum (Atlanta Biologicals) at 37°C for 90 minutes as described (Masuda et al., 2007).
Salivary glands and kidneys were digested with 20 U/ml Papain (Sigma) and 100 μg/ml DNAse I (Roche) in Dulbecco's modified Eagle's medium (Lonza) supplemented with 2 mM L-glutamine (Invitrogen), 100 U/ml penicillin, 100 μg/ml streptomycin (Invitrogen) and 10% fetal bovine serum (Atlanta Biologicals) at 37°C for 30-60 min.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of Prom1+ cell population size, proliferative capacity and generative capacity was performed on multiple tissue sections gathered from 10 separate mice.
Statistical tests used, associated statistical parameters and statistical significance are reported in the Figures and the Figure Legends. Data is judged to be statistically significant when p < 0.05 in applied statistical analysis. In figures, asterisks denote statistical significance (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
Quantification of Prom1+ cell characteristics
Prom1+ cell population size, Prom1+ cell proliferative capacity, Prom1+ cell generative capacity and global organ proliferative capacity were quantified using frozen tissue sections of the appropriate organs taken from Prom1C-L/+; RosaZsG mice that had undergone tamoxifen recombination as described. For each analysis, 12 μm frozen sections were prepared from three separate and representative areas of each organ from a minimum of 10 individual Prom1C-L/+; RosaZsG mice at each time point (‘basal’ and ‘aged’ organs, 30 sections each per organ per time point). For assessments of Prom1+ cell population size, sections were subjected to β-galactosidase staining and counterstained with nuclear fast red (Vector Laboratories). The Prom1+ cell population size was then calculated as the average percentage of β-galactosidase+ nuclei per total nuclei in each organ by counting a minimum of ~15,000 nuclei across the 30 sections of each organ. Note, because different organs display significantly different macro- and microscopic morphologies, including wide variations in cellular density and nuclear size, we did not employ automated imaging software to count cells but performed all counts manually.
We took a very similar approach to calculate Prom1+ cell proliferative capacity, although frozen sections were subjected to pH3 and GFP co-immunofluorescence as described above and counterstained with DAPI (Vector Laboratories). The average percentage of GFP+/pH3+ double-positive nuclei per total GFP+ cells was then calculated for each organ by counting a minimum of 15,000 nuclei across the 30 sections of each organ. To assess the global proliferative capacity of each organ we similarly calculated the average percentage of pH3+ nuclei per total nuclei in each organ. Finally, to calculate the Prom1+ cell generative capacity, we calculated the average percentage of GFP+ nuclei in each ‘basal’ and ‘aged’ organ (one day post tamoxifen treatment, generated as described for the total GFP+ cell count denominator in Prom1+ cell proliferative capacity). Prom1+ cell generative capacity was then calculated by subtracting the average percentage of GFP+ nuclei in ‘basal’ organs from the average percentage of GFP+ nuclei in the corresponding ‘aged’ organs.
Gene expression microarrays, agreement of differential expression (AGDEX) and gene set enrichment analyses (GSEA)
Total RNA of mouse normal and tumor tissues was extract using RNeasy Mini Kit (Qiagen). Total RNA of Prom1+ and Prom1− cells were extracted using PicoPure™ RNA extraction kit (Arcturus). mRNA expression profiles were generated using Affymetrix HT430 (mouse) array. Gene expression data were summarized and normalized using the RMA method as implemented in Partek Genomics Suite 6.6. The data were then imported into Spotfire Decision Site for Unsupervised Gene Hierarchical Clustering. Differences in gene expression between defined groups were calculated using a series of Welch t tests with a Bonferroni threshold at 0.05%. The AGDEX procedure was performed exactly as described previously (Johnson et al., 2010), and GSEA was performed using the online analytical resource from the Broad Institute (http://www.broadinstitute.org/gsea/index.jsp).
Statistical modeling
We used Fisher's exact test or Mantel-Haenszel-Cochran (MHC) Test to capture possible associations between the likelihood of tumor formation and developmental stage across various genotypes within a tissue type. The former was used in tissue types where tumors formed in only 1 genotype for either the adult or the neonatal mice. In tissues where tumors were observed in multiple genotypes both for adult and neonatal mice, we used MHC test to assess this association. MHC is a generalization of the Fisher exact test and we used it to conditionally test the association of Tumor=Y/N vs. adult/neonate in the presence of a third categorical variable, namely genotype. We used the same approach to also study the association between tumor formation and DDC use across genotypes. See supplemental information for details.
DATA AND SOFTWARE AVAILABILITY
Data Resources
All microarray data have been deposited in GEO (GSE78076).
Supplementary Material
HIGHLIGHTS.
Prom1+ cells display different generative and tumourigenic capacities among organs.
Prom1+ cell transformation is determined by life-long generative capacity.
Prom1 marks neonatal liver and damage-induced adult liver stem cells.
Stem cell gene expression patterns predict oncogenes and tumour suppressors.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (R.J.G., P01CA96832, R01 and P30CA021765), the American Lebanese Syrian Associated Charities and Cancer Research UK. We are grateful to the staff of the Hartwell Center for Bioinformatics and Biotechnology, the Cell and Tissue Imaging Shared Resource and the ARC at St Jude Children's Research Hospital for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONTRIBUTIONS
LZ conducted the great majority of experiments. HP, DF, LS, MW, SBP, GN and SU conducted experiments. DLT and DWE provided pathology expertise, KW provided expertise on liver biology. CLG and LS performed AGDEX analyses with guidance from SBP. As co-corresponding authors, A O-T and RJG are responsible for all data, figures and text and confirm that there are no conflicts of interest on behalf of all authors. As Lead Author RJG conceived and oversaw the entire research project and serves as the primary contact for all communication, and reagent and resource sharing.
References
- Arndt K, Grinenko T, Mende N, Reichert D, Portz M, Ripich T, Carmeliet P, Corbeil D, Waskow C. CD133 is a modifier of hematopoietic progenitor frequencies but is dispensable for the maintenance of mouse hematopoietic stem cells. Proceedings of the National Academy of Sciences. 2013;110:5582–5587. doi: 10.1073/pnas.1215438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashford NA, Bauman P, Brown HS, Clapp RW, Finkel AM, Gee D, Hattis DB, Martuzzi M, Sasco AJ, Sass JB. Cancer risk: role of environment. Science. 2015;347:727. doi: 10.1126/science.aaa6246. [DOI] [PubMed] [Google Scholar]
- Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H, van den Born M, et al. Lgr5+ve Stem Cells Drive Self-Renewal in the Stomach and Build Long-Lived Gastric Units In Vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Bennoun M, Rissel M, Engelhardt N, Guillouzo A, Briand P, Weber-Benarous A. Oval cell proliferation in early stages of hepatocarcinogenesis in simian virus 40 large T transgenic mice. The American journal of pathology. 1993;143:1326–1336. [PMC free article] [PubMed] [Google Scholar]
- Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- Block TM, Mehta AS, Fimmel CJ, Jordan R. Molecular viral oncology of hepatocellular carcinoma. Oncogene. 2003;22:5093–5107. doi: 10.1038/sj.onc.1206557. [DOI] [PubMed] [Google Scholar]
- Boyce S, Harrison D. A detailed methodology of partial hepatectomy in the mouse. Lab animal. 2008;37:529–532. doi: 10.1038/laban1108-529. [DOI] [PubMed] [Google Scholar]
- Calmon MF, Jeschke J, Zhang W, Dhir M, Siebenkäs C, Herrera A, Tsai H-C, O'Hagan HM, Pappou EP, Hooker CM, et al. Epigenetic silencing of neurofilament genes promotes an aggressive phenotype in breast cancer. Epigenetics. 2015;10:622–632. doi: 10.1080/15592294.2015.1050173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, Gouw AM, Baylot V, Gutgemann I, Eilers M, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark PE, Polosukhina D, Love H, Correa H, Coffin C, Perlman EJ, de Caestecker M, Moses HL, Zent R. beta-Catenin and K-RAS synergize to form primitive renal epithelial tumors with features of epithelial Wilms’ tumors. The American journal of pathology. 2011;179:3045–3055. doi: 10.1016/j.ajpath.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danaei G, Vander Hoorn S, Lopez AD, Murray CJ, Ezzati M. Causes of cancer in the world: comparative risk assessment of nine behavioural and environmental risk factors. Lancet (London, England) 2005;366:1784–1793. doi: 10.1016/S0140-6736(05)67725-2. [DOI] [PubMed] [Google Scholar]
- Engreitz JM, Daigle BJ, Jr., Marshall JJ, Altman RB. Independent component analysis: mining microarray data for fundamental human gene expression modules. Journal of biomedical informatics. 2010;43:932–944. doi: 10.1016/j.jbi.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier DA, Skaug HJ, Ancheta J, Ianelli J, Magnusson A, Maunder M, Nielsen A, Sibert J. AD Model Builder: using automatic differentiation for statistical inference of highly parameterized complex nonlinear models. Optim Methods Softw. 2012:233–249. [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotay C, Dummer T, Spinelli J. Cancer risk: Prevention is crucial. Science. 2015;347:728. doi: 10.1126/science.aaa6462. [DOI] [PubMed] [Google Scholar]
- Hand NJ, Master ZR, Le Lay J, Friedman JR. Hepatic function is preserved in the absence of mature microRNAs. Hepatology. 2009;49:618–626. doi: 10.1002/hep.22656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, Taketo MM. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. The EMBO journal. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Houghton J, Stoicov C, Nomura S, Rogers AB, Carlson J, Li H, Cai X, Fox JG, Goldenring JR, Wang TC. Gastric cancer originating from bone marrow-derived cells. Science. 2004;306:1568–1571. doi: 10.1126/science.1099513. [DOI] [PubMed] [Google Scholar]
- Howlader N, Noone AM, Krapcho M, Neyman N, Aminou R, Altekruse SF, Kosary CL, Ruhl J, Tatalovich Z, Cho H, et al. SEER Cancer Statistics Review, 1975-2009. National Cancer Institute; Bethesda, MD: 2012. [Google Scholar]
- Huch M, Dorrell C, Boj SF, van Es JH, Li VSW, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RA, Wright KD, Poppleton H, Mohankumar KM, Finkelstein D, Pounds SB, Rand V, Leary SE, White E, Eden C, et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466:632–636. doi: 10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C, Baker SJ. Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat Rev Cancer. 2014 doi: 10.1038/nrc3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkel SA, Galeev R, Flensburg C, Keniry A, Breslin K, Gilan O, Lee S, Liu J, Chen K, Gearing LJ, et al. Jarid2 regulates hematopoietic stem cell function by acting with polycomb repressive complex 2. Blood. 2015;125:1890–1900. doi: 10.1182/blood-2014-10-603969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D, Suzuki A, Zou TT, Sakurada A, Kemp LW, Wakatsuki S, Yokoyama T, Yamakawa H, Furukawa T, Sato M, et al. PTEN1 is frequently mutated in primary endometrial carcinomas. Nature genetics. 1997;17:143–144. doi: 10.1038/ng1097-143. [DOI] [PubMed] [Google Scholar]
- Lee A, Kessler JD, Read T-A, Kaiser C, Corbeil D, Huttner WB, Johnson JE, Wechsler-Reya RJ. Isolation of neural stem cells from the postnatal cerebellum. Nat Neurosci. 2005;8:723–729. doi: 10.1038/nn1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Terrada D, Gunaratne PH, Adesina AM, Pulliam J, Hoang DM, Nguyen Y, Mistretta TA, Margolin J, Finegold MJ. Histologic subtypes of hepatoblastoma are characterized by differential canonical Wnt and Notch pathway activation in DLK+ precursors. Human pathology. 2009;40:783–794. doi: 10.1016/j.humpath.2008.07.022. [DOI] [PubMed] [Google Scholar]
- Masuda H, Maruyama T, Hiratsu E, Yamane J, Iwanami A, Nagashima T, Ono M, Miyoshi H, Okano HJ, Ito M, et al. Noninvasive and real-time assessment of reconstructed functional human endometrium in NOD/SCID/gamma c(null) immunodeficient mice. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1925–1930. doi: 10.1073/pnas.0604310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch CE, Searle SR, Neuhaus JM. Generalized, Linear and Mixed Models. 2nd ed Wiley; 2008. [Google Scholar]
- Melero I, Berman DM, Aznar MA, Korman AJ, Gracia JLP, Haanen J. Evolving synergistic combinations of targeted immunotherapies to combat cancer. Nat Rev Cancer. 2015;15:457–472. doi: 10.1038/nrc3973. [DOI] [PubMed] [Google Scholar]
- Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- O'Callaghan M. Cancer risk: Accuracy of literature. Science. 2015;347:729. doi: 10.1126/science.aaa6212. [DOI] [PubMed] [Google Scholar]
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, Gutkovich-Pyest E, Urieli-Shoval S, Galun E, Ben-Neriah Y. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–466. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- Potter JD, Prentice RL. Cancer risk: Tumors excluded. Science. 2015;347:727. doi: 10.1126/science.aaa6507. [DOI] [PubMed] [Google Scholar]
- Preisegger KH, Factor VM, Fuchsbichler A, Stumptner C, Denk H, Thorgeirsson SS. Atypical ductular proliferation and its inhibition by transforming growth factor beta1 in the 3,5-diethoxycarbonyl-1,4-dihydrocollidine mouse model for chronic alcoholic liver disease. Laboratory investigation; a journal of technical methods and pathology. 1999;79:103–109. [PubMed] [Google Scholar]
- Raimondi AR, Vitale-Cross L, Amornphimoltham P, Gutkind JS, Molinolo A. Rapid development of salivary gland carcinomas upon conditional expression of K-ras driven by the cytokeratin 5 promoter. The American journal of pathology. 2006;168:1654–1665. doi: 10.2353/ajpath.2006.050847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- Risinger JI, Hayes AK, Berchuck A, Barrett JC. PTEN/MMAC1 mutations in endometrial cancers. Cancer research. 1997;57:4736–4738. [PubMed] [Google Scholar]
- Shalapour S, Karin M. Immunity, inflammation, and cancer: an eternal fight between good and evil. The Journal of Clinical Investigation. 2015;125:3347–3355. doi: 10.1172/JCI80007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shupe TD, Piscaglia AC, Oh SH, Gasbarrini A, Petersen BE. Isolation and characterization of hepatic stem cells, or “oval cells,” from rat livers. Methods in molecular biology (Clifton, NJ) 2009;482:387–405. doi: 10.1007/978-1-59745-060-7_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Skaug H, Fournier D, Bolker B, Magnusson A, Nielsen A. Generalized Linear Mixed Models using AD Model Builder. R package version 080. 2014 [Google Scholar]
- Song M, Giovannucci EL. Cancer risk: Many factors contribute. Science. 2015;347:728–729. doi: 10.1126/science.aaa6094. [DOI] [PubMed] [Google Scholar]
- Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhao L, Fish M, Logan CY, Nusse R. Self-renewing diploid Axin2+ cells fuel homeostatic renewal of the liver. Nature. 2015;524:180–185. doi: 10.1038/nature14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO WHO Child Growth Standards based on length/height, weight and age. Acta Paediatr Suppl. 2006;450:76–85. doi: 10.1111/j.1651-2227.2006.tb02378.x. [DOI] [PubMed] [Google Scholar]
- Wild C, Brennan P, Plummer M, Bray F, Straif K, Zavadil J. Cancer risk: Role of chance overstated. Science. 2015;347:728. doi: 10.1126/science.aaa6799. [DOI] [PubMed] [Google Scholar]
- Wu S, Powers S, Zhu W, Hannun YA. Substantial contribution of extrinsic risk factors to cancer development. Nature. 2015 doi: 10.1038/nature16166. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L, Ide H, Kim Y, Dubey P, Witte ON. In vivo regeneration of murine prostate from dissociated cell populations of postnatal epithelia and urogenital sinus mesenchyme. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(Suppl 1):11896–11903. doi: 10.1073/pnas.1734139100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin AH, Miraglia S, Zanjani ED, Almeida-Porada G, Ogawa M, Leary AG, Olweus J, Kearney J, Buck DW. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90:5002–5012. [PubMed] [Google Scholar]
- Zhu L, Gibson P, Currle DS, Tong Y, Richardson RJ, Bayazitov IT, Poppleton H, Zakharenko S, Ellison DW, Gilbertson RJ. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature. 2009;457:603–607. doi: 10.1038/nature07589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.