Abstract
Genome editing in large animals has tremendous practical applications, from more accurate models for medical research through improved animal welfare and production efficiency. Although genetic modification in large animals has a 30 year history, until recently technical issues limited its utility. The original methods – pronuclear injection and integrating viruses – were plagued with problems associated with low efficiency, silencing, poor regulation of gene expression, and variability associated with random integration. With the advent of site specific nucleases such as TALEN and CRISPR/Cas9, precision editing became possible. When used on their own, these can be used to truncate or knockout genes through non-homologous end joining (NHEJ) with relatively high efficiency. When used with a template containing desired gene edits, these can be used to allow insertion of any desired changes to the genome through homologous recombination (HR) with substantially lower efficiency. Consideration must be given to the issues of marker sets and off-target effects. Somatic cell nuclear transfer is most commonly used to create animals from gene edited cells, but direct zygote injection and use of spermatogonial stem cells are alternatives under development. In developing gene editing projects, priority must be given to understanding the potential for off-target or unexpected effects of planned edits, which have been common in the past. Because of the increasing technical sophistication with which it can be accomplished, genome editing is poised to revolutionize large animal genetics, but attention must be paid to the underlying biology in order to maximize benefit.
1. Introduction
Genetic modification in rodents has been routine now for 35 years [1], and first attempts to transfer the technology to large animals began shortly thereafter. Unfortunately, technical and efficiency limitations precluded the practical use of genetic modification in large animals, with few exceptions.
However, genome editing in large animals would be of tremendous utility to medical research, to medicine, and to agriculture. In medical research, the drawbacks of using rodents to model humans are well established [1]. Because of their small size, their short life cycle, their very different diet and dietary priorities, and details of their physiology, mice make poor models for reproductive physiology, pulmonary problems, metabolic regulation, and many other fields of inquiry. They are an improvement on cell culture, but for many important health problems improved preclinical models would be of benefit to research. A clear early example of this is the cystic fibrosis transmembrane conductance regulator (CFTR) knockout pig, which is far more clinically similar to humans than a CFTR knockout in mice [2]. Gene editing in large animals also has the potential to aid human medicine directly, from creation of humanized protein drugs [3] to creation of humanized transplant organs (xenografts) [4, 5].
In addition, there is potential for gene editing in livestock to improve both animal welfare and production efficiency in agricultural applications. From feed conversion and other performance traits, to disease resistance, to improved nutrition, to improving fit to environment, precision gene editing is likely to be an essential tool in improvement of our large animal stock.
2. Historical Methods for Large Animal Engineering
Pronuclear injection, developed in 1980, was the first method of genetically modifying animals, and in the intervening decades has been the most common [6]. In pronuclear injection, DNA containing a desired gene expression construct is injected into a single-cell fertilized egg. Despite somewhat low efficiency it integrates randomly into the DNA of the fertilized egg, which is implanted in a recipient mother, and offspring are checked for presence and expression of the new gene. Because the DNA contained in these constructs was essentially never purely that of the host species, animals produced by this method are termed transgenic.
Pronuclear injection has numerous practical drawbacks. Because integration is random, and because the constructs are usually integrated in multiple copies as concatemers, expression levels were difficult to control. Moreover, because the maximum size of the constructs is somewhat limited, promoter elements, which had to be included, were necessarily abbreviated, and genes were almost always introduced in their fully spliced forms. Regulation of the genes was thus usually rudimentary. Transgenes also had a tendency to be silenced over multiple generations. For research purposes in rodents, the ease of creating transgenics outweighed these concerns.
In large animals, however, the problem was aggravated by efficiency and mechanics of reproduction. In mice, the efficiency of transgene introduction was about 5%–10%. With a gestation period of 3 weeks, and a litter size of 6–10 animals depending on strain, five recipient moms were likely to give you a few founders in just a few weeks. In cattle, however, for unclear reasons, efficiency of transgene introduction was closer to 3%, but with only 18% of blastocysts yielding live calves this dropped the effective rate to a fraction of a percent. With one calf per mother, this meant hundreds of recipients were needed to ensure successful creation of a founder [7, 8]. In some species, for hormonal or other reasons, the method is essentially impossible [9]. The introduction of relatively routine somatic cell nuclear transfer about fifteen years ago reduced the efficiency problem, because cells could be checked for correct integration before creation of animals, but it did not solve all of the other problems with using transgenes [10]. Notwithstanding all of these problems, transgenic sheep, pigs, goats, cattle, and others were created, but most had low practical utility [1].
One approach successfully used by several groups to avoid the problem with low transgene integration efficiency was use of integrating viruses. Integrating viruses retain all of the problems of random integration associated with pronuclear injection, and because of their smaller cargo size, the problems with promoter strength and specificity are usually worse. In addition, because of the viral elements included, progressive silencing over time worsened with viral integration methods [11].
3. Modern Genome Editing Methods
The fundamentally novel technology that has made the impending revolution in gene editing possible is the ability to precisely target specific areas of the genome. This eliminates essentially all of the issues associated with transgenic animals, because native promoter elements and splicing can be used for correct gene regulation, and the variability and gene silencing associated with random integration is eliminated. Instead of a largely random effect, gene editing can now be well controlled.
Gene editing in large animals is primarily different than gene editing in laboratory animals in that the higher expense and longer gestation time in large animals necessitates a lower tolerance for error. In mice, one can tolerate high randomness of results, because litters are large and gestation times are three weeks. In horses or cattle, each embryo must be assured to carry correct edits before gestation is initiated.
There are two relatively new technologies that allow targeting of specific nucleotides: TALEN and CRISPR, each with multiple related technologies. TALEN, and the related technologies of zinc finger nucleases and MegaTAL, use modular protein-based sequence recognition, whereas CRISPR uses RNA-guided sequence recognition. Although variations on these technologies are likely to develop over time, the core technologies are unlikely to change.
Transcription-Activator-Like (TAL) effectors are a class of enzyme first discovered in the plant pathogen Xanothomonas about ten years ago [12], with the code for DNA binding specificity worked out in 2009 [13], and engineered to add a nuclease function for genome editing in 2010 [14]. The combination, TAL effector-like nucleases (TALEN), consist of a modular array of TAL recognition sequences fused to a FokI nuclease [15]. These are inserted in pairs, one for each strand, and work as a dimer to create double-stranded breaks in specific DNA sequences.
There are variants on this; for instance, MegaTAL uses a combination of TAL arrays with a nuclease that has site-specific cleavage, meganuclease, increasing overall specificity of the combination [16]. MegaTAL as a technology is still in development, currently with high cost and complexity; they may be a turnkey solution in a few years, but for now are probably best left to those focusing on method development. Zinc finger nucleases (ZFN) are an older solution, with 20 years of history, and share the use of the FokI nuclease and the need for dimers, but use a different protein-DNA recognition mechanism [17]. In our experience, ZFN are more cumbersome to use, with no compensatory advantages, as compared to TALEN. Both TALEN and ZFN can be created in individual labs, but multiple commercial sources exist for each.
The other major method of making targeted cuts in the genome, CRISPR/Cas9, is also derived from bacteria and archaea in which they are part of a viral defense system [18]. It consists of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) which binds a guide RNA and an associated endonuclease (Cas9). Binding specificity is thus dependent on RNA-DNA interaction strength. The main advantage of the CRISPR/Cas9 over TALEN-based technologies is its speed of production and extremely low cost; its disadvantages, as will be discussed later, are a likely inherently lower specificity, and the legal challenges (for commercial purposes) that are not likely to be resolved for several years [19]. CRISPR/Cas9 are available from both academic and commercial sources.
3.1 Homologous recombination (HR) and non-homologous end joining (NHEJ)
Although all TALEN and CRISPR do, fundamentally, is site-specific cleavage, this allows gene editing through two mechanisms: homologous recombination (HR) and non-homologous end joining (NHEJ) (Figure 1). When TALEN or CRISPR make the site-specific double-stranded break, DNA repair mechanisms are employed to repair the break. If a piece of DNA matching the sites flanking the cleavage site is available, the cell can use HR to repair the damage. If no such DNA is available, the cell will use NHEJ.
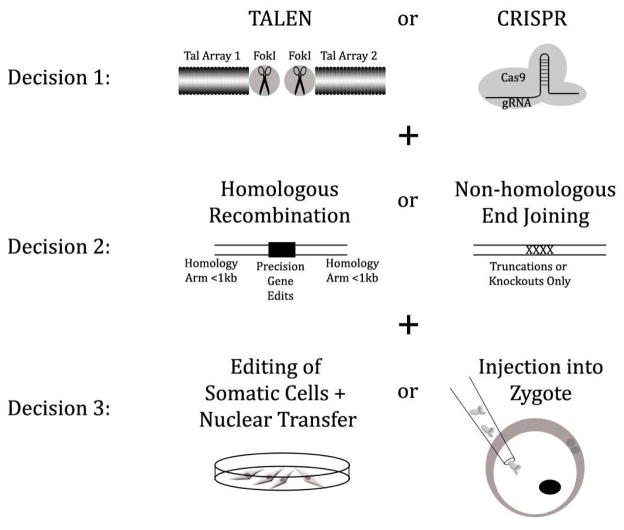
Figure 1.
There are three main decisions to make in a gene editing project. (1) TALEN or CRISPR? CRISPR are lower cost and faster to make, but TALEN appear to have lower error rate and for commercial projects may have fewer legal issues. (2) Homologous Recombination (HR) or Non-Homologous End Joining (NHEJ) ? Including a recombination construct lends tremendous flexibility, but lowers efficiency from over 10–50% to less than 1%. (3) Somatic Cell Nuclear Transfer or Injection into Zygote? Editing somatic cells has a much longer track record, but direct injection into the zygote is faster and likely suitable for NHEJ-based projects because of their higher efficiency.
In NHEJ, although the DNA is rejoined, there is often a deletion or insertion of a small number of nucleotides. If the cleavage site was in the middle of a coding sequence or essential regulatory element, this will serve to destroy gene expression or function. Without addition of a homologous recombination template, then, TALEN and CRISPR are only capable of truncation or knockout, not precision edits. However, the efficiency is relatively high – successfully transfected cells will have DNA insertions or deletions at the target site in from 10% to 60% of cells [20].
Homologous recombination is much more powerful. In HR, the cell is provided with a DNA template to repair the damage caused by TALEN or CRISPR, which precisely matches the surrounding area, aside from the specific gene edits desired. Historically, this technique has been used for creation of precision gene-edited mice for decades. However, without use of TALEN or CRISPR to drive local DNA repair mechanisms, the efficiency was extraordinarily low (less than 1 in 1,000,000 cells) and even then required DNA arms that precisely matched the surrounding sequence with total lengths approaching 10,000 base pairs (bp). This was possible only in specific strains of clonal mice for which libraries of DNA were available. With TALEN and CRISPR, although the theory is the same, the efficiency is dramatically higher (between 1 in 100 and 1 in 1000 cells), and the size of the homology arms can be much smaller (fewer than 1000 bp on each side) [21]. There is evidence that part of the relatively low efficiency of HR is because of its competition with NHEJ, and that inhibition of NHEJ will increase the efficiency substantially [22].
3.2 Marker Sets
Because the efficiency of homologous recombination is so low, it is generally necessary to use a marker set to help find the cells in which it was successful. Antibiotic resistance and fluorescent markers, or both, are used to identify cells with successful integration of the HR construct. The difficulty comes in removing the marker set again.
Removing the markers us generally done with use of site-specific recombinase systems. The most widely used of these is the Cre-loxP system derived from bacteriophage P1, although there are many other less commonly used recombinases. Cre (causes recombination) is a protein that recognizes and mediates site-specific recombination between 34 bp sequences referred to as loxP (locus of crossover (x) in P1 bacteriophage). When Cre protein is introduced into a cell that contains DNA with two loxP sites, everything between the loxP sites is excised, leaving behind one loxP site. However, this remaining site can interfere with gene regulation [23], or cause unexpected translocations and genome reorganizations [24]. This latter problem would increase as additional changes are made to the same genome. Moreover, for regulatory reasons, leaving behind synthetic or bacterial sequences may be problematic. The Cre/loxP system has been in use for many decades, and although many of the reagents are commercially available, they are simple enough to use that commercial vendors beyond expression plasmids are generally not needed.
One solution to this problem of leftover recognition sites is use of transposases, such as Sleeping Beauty and Piggybac. Like recombinases, transposase system consist of a recognition sequence and an enzyme. The base enzyme both excises the sequence, and inserts it semi-randomly in other locations. However, excision-only versions of the enzymes exist. These have the advantage of taking their integration sites with them when they are excised, resulting in the possibility of footprint-free gene editing [25]. The efficiency of excision can be low, but the combination of TALEN with Piggybac has been used for successful footprint-free gene editing in, for instance, correction of cystic fibrosis mutations in human iPS cells [26]. Piggybac reagents are available commercially.
3.3 Off-target effects
To be useful, gene editing tools need to not only be effective, they need to be specific. Causing significant off-target cleavage and NHEJ events would preclude the utility of a technique for either therapies or creation of a line of animals expected to last more than a generation.
Because CRISPR/Cas9 uses RNA-DNA interactions for its specificity, it inherently has a huge problem with off-target effects [27]. A PCR reaction uses the same mechanism to achieve specificity as CRISPR; if done at 37°C it produces a smear rather than a specific band. Numerous methodologies to reduce these off-target effects have been proposed, and reduce these effects in carefully controlled circumstances [28]. Commercial organizations and protocols exist which will screen CRISPR-edited genomes for off-target effects. Nonetheless, in actual practice, there are substantial fidelity and specificity issues, as demonstrated by the detailed analysis done in a recent study in which CRISPR was used to edit human pre-implantation embryos [29].
Although TALEN theoretically could have off-target cleavage, this is much less of an issue for current generation TALEN. Several recent studies using TALEN for gene editing have failed to find any evidence of off-target mutations [30, 31].
3.4 Examples of Use
Because of its relative ease of use, several recent large animals have made use of CRISPR and TALEN, primarily through induction of NHEJ, for creation of animals with specific knock-outs. Fahrenkrug’s group has generated polled cattle and pig models of infertility and colon cancer through these methods [32]. Prather’s group has created pigs with substantial immunity to PRRSv virus by knocking out the PRRSv receptor with random NHEJ [33]. Others have, for instance, knocked out myostatin in pigs [34]. The Roslin institute, the original developers of cloning and one of the great pioneers in this field, have in one of the first examples of use of homologous recombination in large animals, changed a pig allele to a warthog allele to induce resistance to African Swine Fever virus [35]. Thus, for performance, for disease resistance, and for medical research models, large animal genetic engineering is already well on its way.
4. Methods for Creating Large Animals from Edited Genomes
A gene-edited cell is not, of course, an animal. Both somatic cell nuclear transfer (SCNT) from gene-edited somatic cells (for instance, fibroblasts) and direct editing of zygotes have shown success. Based on past successes with older methods of transgenesis, however, editing of spermatogonial stem cells may be a third viable alternative.
4.1 Somatic Cell Nuclear Transfer
In somatic cell nuclear transfer, oocytes generally collected from slaughterhouse-derived ovaries are matured in vitro, enucleated, and fused with the nucleus from the gene-edited cell. Even extended time in culture during the gene editing process does not appear to impair the efficiency of somatic cell nuclear transfer [36]. Although in its infancy 20 years ago, cloning was plagued by extremely low efficiency and health difficulties in first generation clones, as techniques improve, these difficulties have been diminished [37].
4.2 Direct Editing of Zygotes
Injecting TALEN (or presumably other gene editing tools) directly into zygotes has been shown to produce gene-edited progeny [38]. Direct editing of zygotes has the advantages of speed compared to gene editing in culture followed by somatic cell nuclear transfer, and of avoiding potential cloning defects. Unfortunately, the stochastic nature of NHEJ means that the results are quite random, and the process is probably only suitable for animals with large litters from which those usefully edited can be selected.
4.3 Spermatogonial Stem Cells
In 2013, Dobrinski’s group used viral vectors to put GFP into spermatogonial stem cells and transplanted them into boars [39]. Transgene expression persisted in sperm for up to five years, and resulted in the successful creation of transgenic embryos through IVF. Although there are still technical barriers to use of modern gene editing methods instead of viral methods (primarily, ability to expand spermatogonial stem cells in culture), spermatogonial stem cell transfer is a potential alternative method for using modern gene editing techniques to create large animals.
5. A Cautionary Note about Targets
A key point to remember when embarking on a gene editing project is that there is no way to predict what a genetic modification will do without actually seeing the results in a live animal. Results will often translate well from one mammal to another – but as the problem with disease modeling in mice makes clear, even that is not a sure thing.
As a cautionary example, in 1985, only a few years after the first genetic modification techniques were developed, pigs were produced which overexpressed human growth hormone [40]. However, the pigs did not consistently have enhanced growth rate, and health problems included lethargy, lameness, gastric ulcers, and infertility, among others [41]. These problems of unexpected results continue into the modern era. For instance, in the 2012 production of cattle lacking the milk allergen β-lactoglobulin (BLG) by transgenic expression of an inhibitory microRNA, loss of BLG resulted in dramatic changes in most other milk proteins, as well as lack of a tail [42].
This is not to say that even dramatic changes cannot be successful: the most famous example, of course, being Atlantic Salmon transgenic for the growth hormone gene from Chinook Salmon, which are apparently healthy and grow to many times the size of unmodified Atlantic Salmon [43]. Genes taken from related species or breeds are likely to completely retain their function on transfer.
6. Conclusions
We are entering a new era, one in which gene editing will become progressively more straightforward, and more essential to animal welfare and livestock productivity. In the next decades, it may be that every animal brought to state fairs by 4-H youth contains his or her own personal edits, unlocking the creative potential of the next generation in the way that microelectronics or the internet fascinated previous generations of young adults
TALEN and CRISPR/Cas9 enable targeted edits in a way never before possible, and succeeding improved generations of these site-specific nucleases will only increase efficiency and specificity. Right now these are made into live animals through somatic cell nuclear transfer or zygote injection, but one can imagine a not-too distant future in which cells are directly transformed into spermatozoa in a dish [44], making large animal genome editing accessible to a wider population.
Since biology is complex, the effects of any specific gene edit are difficult to predict, unless they have previously been observed in another related animal. The coming years will see an explosion of animals whose genetics have been improved by direct editing. The difficulty is not in making the edits – it is in knowing which edits to make.
Historical genetic modification used random insertion, resulting in low utility.
The new technologies TALEN and CRISPR solve this problem.
Many technical alternatives exist for creating precision gene edits.
Somatic cell nuclear transfer or direct zygote editing are used to introduce edits into animals.
Choice of gene editing targets is of highest importance in producing useful results.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wells DJ. Genetically modified animals and pharmacological research. Handb Exp Pharmacol. 2010:213–226. doi: 10.1007/978-3-642-10324-7_9. [DOI] [PubMed] [Google Scholar]
- 2.Keiser NW, Engelhardt JF. New animal models of cystic fibrosis: what are they teaching us? Curr Opin Pulm Med. 2011;17:478–483. doi: 10.1097/MCP.0b013e32834b14c9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houdebine LM. Production of pharmaceutical proteins by transgenic animals. Comp Immunol Microbiol Infect Dis. 2009;32:107–121. doi: 10.1016/j.cimid.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hata T, Uemoto S, Kobayashi E. Transplantable liver production plan: “Yamaton”--liver project, Japan. Organogenesis. 2013;9:235–238. doi: 10.4161/org.25760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phipatanakul WP, Petersen SA. Porcine small intestine submucosa xenograft augmentation in repair of massive rotator cuff tears. Am J Orthop (Belle Mead NJ) 2009;38:572–575. [PubMed] [Google Scholar]
- 6.Isola LM, Gordon JW. Transgenic animals: a new era in developmental biology and medicine. Biotechnology. 1991;16:3–20. [PubMed] [Google Scholar]
- 7.Pursel VG, Rexroad CE, Jr, Bolt DJ, Miller KF, Wall RJ, Hammer RE, Pinkert CA, Palmiter RD, Brinster RL. Progress on gene transfer in farm animals. Vet Immunol Immunopathol. 1987;17:303–312. doi: 10.1016/0165-2427(87)90149-8. [DOI] [PubMed] [Google Scholar]
- 8.Eyestone WH. Challenges and progress in the production of transgenic cattle. Reprod Fertil Dev. 1994;6:647–652. doi: 10.1071/rd9940647. [DOI] [PubMed] [Google Scholar]
- 9.Hong SG, Kim MK, Jang G, Oh HJ, Park JE, Kang JT, Koo OJ, Kim T, Kwon MS, Koo BC, et al. Generation of red fluorescent protein transgenic dogs. Genesis. 2009;47:314–322. doi: 10.1002/dvg.20504. [DOI] [PubMed] [Google Scholar]
- 10.Su F, Wang Y, Liu G, Ru K, Liu X, Yu Y, Liu J, Wu Y, Quan F, Guo Z, Zhang Y. Generation of transgenic cattle expressing human beta-defensin 3 as an approach to reducing susceptibility to Mycobacterium bovis infection. FEBS J. 2016;283:776–790. doi: 10.1111/febs.13641. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann A, Kessler B, Ewerling S, Kabermann A, Brem G, Wolf E, Pfeifer A. Epigenetic regulation of lentiviral transgene vectors in a large animal model. Mol Ther. 2006;13:59–66. doi: 10.1016/j.ymthe.2005.07.685. [DOI] [PubMed] [Google Scholar]
- 12.Sugio A, Yang B, Zhu T, White FF. Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIAgamma1 and OsTFX1 during bacterial blight of rice. Proc Natl Acad Sci U S A. 2007;104:10720–10725. doi: 10.1073/pnas.0701742104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 14.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Certo MT, Morgan RA. Salient Features of Endonuclease Platforms for Therapeutic Genome Editing. Mol Ther. 2016 doi: 10.1038/mt.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boissel S, Jarjour J, Astrakhan A, Adey A, Gouble A, Duchateau P, Shendure J, Stoddard BL, Certo MT, Baker D, Scharenberg AM. megaTALs: a rarecleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res. 2014;42:2591–2601. doi: 10.1093/nar/gkt1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isalan M, Choo Y, Klug A. Synergy between adjacent zinc fingers in sequence-specific DNA recognition. Proc Natl Acad Sci U S A. 1997;94:5617–5621. doi: 10.1073/pnas.94.11.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rath D, Amlinger L, Rath A, Lundgren M. The CRISPR-Cas immune system: biology, mechanisms and applications. Biochimie. 2015;117:119–128. doi: 10.1016/j.biochi.2015.03.025. [DOI] [PubMed] [Google Scholar]
- 19.Sherkow JS. Law, history and lessons in the CRISPR patent conflict. Nat Biotechnol. 2015;33:256–257. doi: 10.1038/nbt.3160. [DOI] [PubMed] [Google Scholar]
- 20.Nemudryi AA, Valetdinova KR, Medvedev SP, Zakian SM. TALEN and CRISPR/Cas Genome Editing Systems: Tools of Discovery. Acta Naturae. 2014;6:19–40. [PMC free article] [PubMed] [Google Scholar]
- 21.Whitelaw CB, Sheets TP, Lillico SG, Telugu BP. Engineering large animal models of human disease. J Pathol. 2016;238:247–256. doi: 10.1002/path.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meier ID, Bernreuther C, Tilling T, Neidhardt J, Wong YW, Schulze C, Streichert T, Schachner M. Short DNA sequences inserted for gene targeting can accidentally interfere with off-target gene expression. FASEB J. 2010;24:1714–1724. doi: 10.1096/fj.09-140749. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka M, Yamaguchi S, Yamazaki Y, Kinoshita H, Kuwahara K, Nakao K, Jay PY, Noda T, Nakamura T. Somatic chromosomal translocation between Ewsr1 and Fli1 loci leads to dilated cardiomyopathy in a mouse model. Sci Rep. 2015;5:7826. doi: 10.1038/srep07826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Matteo M, Matrai J, Belay E, Firdissa T, Vandendriessche T, Chuah MK. Piggy-Bac toolbox. Methods Mol Biol. 2012;859:241–254. doi: 10.1007/978-1-61779-603-6_14. [DOI] [PubMed] [Google Scholar]
- 26.Camarasa MV, Galvez VM. Robust method for TALEN-edited correction of pF508del in patient-specific induced pluripotent stem cells. Stem Cell Res Ther. 2016;7:26. doi: 10.1186/s13287-016-0275-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting betaglobin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, Lv J, Xie X, Chen Y, Li Y, et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6:363–372. doi: 10.1007/s13238-015-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dreyer AK, Hoffmann D, Lachmann N, Ackermann M, Steinemann D, Timm B, Siler U, Reichenbach J, Grez M, Moritz T, et al. TALEN-mediated functional correction of X-linked chronic granulomatous disease in patient-derived induced pluripotent stem cells. Biomaterials. 2015;69:191–200. doi: 10.1016/j.biomaterials.2015.07.057. [DOI] [PubMed] [Google Scholar]
- 31.Menger L, Gouble A, Marzolini MA, Pachnio A, Bergerhoff K, Henry JY, Smith J, Pule M, Moss P, Riddell SR, et al. TALEN-mediated genetic inactivation of the glucocorticoid receptor in cytomegalovirus-specific T cells. Blood. 2015;126:2781–2789. doi: 10.1182/blood-2015-08-664755. [DOI] [PubMed] [Google Scholar]
- 32.Tan W, Carlson DF, Lancto CA, Garbe JR, Webster DA, Hackett PB, Fahrenkrug SC. Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proc Natl Acad Sci U S A. 2013;110:16526–16531. doi: 10.1073/pnas.1310478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitworth KM, Rowland RR, Ewen CL, Trible BR, Kerrigan MA, Cino-Ozuna AG, Samuel MS, Lightner JE, McLaren DG, Mileham AJ, et al. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat Biotechnol. 2016;34:20–22. doi: 10.1038/nbt.3434. [DOI] [PubMed] [Google Scholar]
- 34.Wang K, Ouyang H, Xie Z, Yao C, Guo N, Li M, Jiao H, Pang D. Efficient Generation of Myostatin Mutations in Pigs Using the CRISPR/Cas9 System. Sci Rep. 2015;5:16623. doi: 10.1038/srep16623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lillico SG, Proudfoot C, King TJ, Tan W, Zhang L, Mardjuki R, Paschon DE, Rebar EJ, Urnov FD, Mileham AJ, et al. Mammalian interspecies substitution of immune modulatory alleles by genome editing. Sci Rep. 2016;6:21645. doi: 10.1038/srep21645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kubota C, Yamakuchi H, Todoroki J, Mizoshita K, Tabara N, Barber M, Yang X. Six cloned calves produced from adult fibroblast cells after long-term culture. Proc Natl Acad Sci U S A. 2000;97:990–995. doi: 10.1073/pnas.97.3.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akagi S, Matsukawa K, Takahashi S. Factors affecting the development of somatic cell nuclear transfer embryos in Cattle. J Reprod Dev. 2014;60:329–335. doi: 10.1262/jrd.2014-057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lillico SG, Proudfoot C, Carlson DF, Stverakova D, Neil C, Blain C, King TJ, Ritchie WA, Tan W, Mileham AJ, et al. Live pigs produced from genome edited zygotes. Sci Rep. 2013;3:2847. doi: 10.1038/srep02847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng W, Tang L, Bondareva A, Honaramooz A, Tanco V, Dores C, Megee S, Modelski M, Rodriguez-Sosa JR, Paczkowski M, et al. Viral transduction of male germline stem cells results in transgene transmission after germ cell transplantation in pigs. Biol Reprod. 2013;88:27. doi: 10.1095/biolreprod.112.104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hammer RE, Pursel VG, Rexroad CE, Jr, Wall RJ, Bolt DJ, Ebert KM, Palmiter RD, Brinster RL. Production of transgenic rabbits, sheep and pigs by microinjection. Nature. 1985;315:680–683. doi: 10.1038/315680a0. [DOI] [PubMed] [Google Scholar]
- 41.Pursel VG, Bolt DJ, Miller KF, Pinkert CA, Hammer RE, Palmiter RD, Brinster RL. Expression and performance in transgenic pigs. J Reprod Fertil Suppl. 1990;40:235–245. [PubMed] [Google Scholar]
- 42.Jabed A, Wagner S, McCracken J, Wells DN, Laible G. Targeted microRNA expression in dairy cattle directs production of beta-lactoglobulin-free, high-casein milk. Proc Natl Acad Sci U S A. 2012;109:16811–16816. doi: 10.1073/pnas.1210057109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Du SJ, Gong ZY, Fletcher GL, Shears MA, King MJ, Idler DR, Hew CL. Growth enhancement in transgenic Atlantic salmon by the use of an “all fish” chimeric growth hormone gene construct. Biotechnology (N Y) 1992;10:176–181. doi: 10.1038/nbt0292-176. [DOI] [PubMed] [Google Scholar]
- 44.Zhou Q, Wang M, Yuan Y, Wang X, Fu R, Wan H, Xie M, Liu M, Guo X, Zheng Y, et al. Complete Meiosis from Embryonic Stem Cell-Derived Germ Cells In Vitro. Cell Stem Cell. doi: 10.1016/j.stem.2016.01.017. [DOI] [PubMed] [Google Scholar]