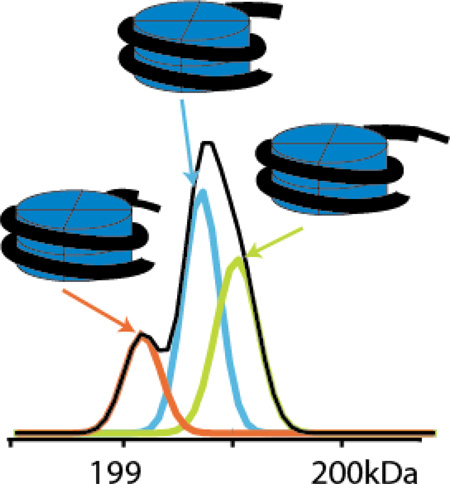
Abstract
Native electrospray-ionization mass spectrometry (native MS) measures biomolecules under conditions that preserve most aspects of protein tertiary and quaternary structure, enabling direct characterization of large intact protein assemblies. However, native spectra derived from these assemblies are often partially obscured by low signal-to-noise as well as broad peak shapes due to residual solvation and adduction after the electrospray process. The wide peak widths together with the fact that sequential charge state series from highly charged ions are closely spaced means that native spectra containing multiple species often suffer from high degrees of peak overlap or else contain highly interleaved charge envelopes. This situation presents a challenge for peak detection, correct charge state and charge envelope assignment, and ultimately extracting the relevant underlying mass values of the non-covalent assemblages being investigated.
In this report we describe a comprehensive algorithm developed for addressing peak detection, peak overlap, and charge state assignment in native mass spectra, called PeakSeeker. Overlapped Peaks are detected by examination of the second derivative of the raw mass spectrum. Charge state distributions of the molecular species are determined by fitting linear combinations of charge envelopes to the overall experimental mass spectrum. This current software is capable of deconvoluting heterogeneous, complex, and noisy native mass spectra of large protein assemblies as demonstrated by analysis of (a) synthetic mononucleosomes containing severely overlapping peaks, (b) an RNA polymerase II/a-amanitin complex with many closely interleaved ion signals, and (c) human TriC complex containing high levels of background noise.
Graphical Abstract
Introduction
Mass spectrometry (MS) now plays an increasingly important role in characterizing large protein assemblies [1–4]. Interacting surfaces between the component proteins of a large complex can be mapped using bottom-up proteomics strategies such as hydrogen-deuterium exchange (HDX) [5], hydroxyl radical protein surface fingerprinting [6], or chemical cross-linking [7, 8]. In native mass spectrometry (Native MS), electrospray is employed to ionize intact non-covalent protein complexes from non-denaturing solutions. Solution interactions between component proteins, ligands, nucleic acids and other biomolecules are preserved allowing determination of the intact mass of an assembly and hence the stoichiometry of the individual subunits [9]. It is therefore complementary to techniques such as cross-linking MS that measure proteolysis products. Through partial disruption of a protein complex either in solution [10] or in the gas phases [11], dissociation pathways can be mapped and the topology of a complex deduced [12–14].
In native MS measurements, intact protein complexes are introduced to the gas phase by nanoelectrospray ionization from aqueous solutions buffered with volatile salts near neutral pH [1]. Gas phase ions of the complexes preserve the major structural and topological features of the complex. The resulting ions are present at higher m/z values and are distributed across a narrower range of charge states than typically observed through denaturing conditions. Observation of ions from native complexes requires mass spectrometers capable of detecting signals beyond 10,000 m/z. This has now been achieved using time-of-flight (TOF) [15], Fourier transform ion cyclotron resonance (FT-ICR) [16–18], and Orbitrap detectors [19]. The mass resolving power of FT-ICR is inversely proportional to m/z while the mass resolving power of TOF and Orbitrap analyzers is inversely proportional to the square root of m/z. In practice however, the instrumental limits of mass resolving power are seldom achieved due to incomplete desolvation of ions from intact protein complexes [2, 15, 20].
Intact protein assemblies are introduced to the vacuum while still partially solvated and require increased collisional energy deposition in either the instrument source region and/or a collision cell to achieve adequately resolved ion signals [21]. However, the ions may still not be fully desolvated at the time of mass measurement. This is demonstrated by the observations that: a) native MS measurements consistently give higher molecular weight values than expected from a given complex, and b) ion signals from native protein complexes are much wider than expected from the calculated isotopic distributions of known primary sequences. The detected signals therefore represent heterogeneous adducts between the protein complex, buffer ions, and water molecules. Collisional or thermal dissociation of these adducts must be balanced against the need to maintain intact assemblies. Since hydrogen bonding, and electrostatic and hydrophobic interactions are major determinants of protein structure [22] and water molecules and metal ions can form intrinsic structural elements, it is unlikely that complete desolvation while simultaneously maintaining secondary, tertiary, and quaternary structure of a native protein ion is an achievable or even desirable outcome.
The broad ion signals of native complexes therefore poses a complication to determining the intact mass of an assembly, particularly in spectra containing multiple species with overlapping and/or highly interleaved ion signals. Overlap resulting from the presence of multiple, unresolved ions distorts the signals in both m/z and intensity and confounds accurate measurement of the underlying species. Highly interleaved charge state distributions, on the other hand, can lead to errors in determining charge envelope membership, assigning charge states, and ultimately, calculating molecular mass. Coupling online ion mobility separation to native MS analysis may reduce the severity of these issues by introducing a dimension of gas-phase separation of different molecular species [23]. However, the availability and performance of suitable peak deconvolution algorithms remains an important problem, as it is likely that improved separation technologies will only prompt the analysis of increasingly complex samples.
Current deconvolution methods can be broadly classified as either working forwards from a theoretical model or backwards from the experimental data. Forward algorithms [24–26] start with a model describing the behavior of proteins in the mass spectrometer. This is used to determine the most probable set of molecular ions that reconstruct the experimental data when convolved by the model. Backwards algorithms [3, 9, 20, 27–31] start by detecting peaks in the experimental spectrum, assigning charge state distributions to sets of peaks, and from these inferring the protein composition. Forward algorithms side-step issues involved in explicit peak detection, but in turn are at risk of returning local minima without robust means of generating initial starting guesses at the protein composition.
Massign [9], developed by the Robinson lab, has been amongst the most effective backwards algorithms. Massign detects peaks and simulates charge envelopes from those peaks using the fact that charge states follow a Gaussian distribution over the charge domain. The modeled envelope that best fits the original spectrum represents the correct charge state. Because this method models both m/z and intensity, it can effectively assign envelope membership in highly interleaved spectra. Furthermore, overlapping signals are handled via a “peeling algorithm” in which envelopes are repeatedly fit to the spectra after subtracting out the previously determined envelopes. The intensity of overlapped peaks is therefore apportioned among the envelopes.
An alternate approach, Automass [30] uses an intensity-independent method which varies the charge state assignment of a set of peaks and examines the standard deviation of the mass for a minimum value as well as the periodicity of this deviation over different charge states. Overlapping signals are not directly assessed as each peak is assumed to belong to exactly one envelope and boundaries between charge envelopes are modeled by a game theory based treatment [31].
Many of the recent developments in deconvolution algorithms have thus focused on improved methods of assigning membership of peaks to charge state distributions. Detection of individual peaks, representing an earlier stage of data analysis, has been somewhat neglected and not tailored to the breadth and heterogeneity of ion signals observed from native complexes. Massign uses a moving average filter as well as a rolling signal threshold to detect shoulder peaks, while Automass uses a Savitzky-Golay filter. However, because these methods depend on local maxima detection, neither of these is able to handle more complex cases of overlap in which peak apexes are shifted by the overlapping signals, and the underlying signals cannot simply be apportioned among the charge envelopes. These cases are particularly serious for native mass spectra, in which the high mass, multiple components, and broadened ion signals can all contribute to overlapping peaks. The distorted peak-shapes can be assigned to the wrong charge envelope, leading to inaccurate mass and abundance determination.
We focus on addressing the problem of peak overlap through improved peak detection and modeling, prior to and during charge envelope assignment. We evaluate multiple peak detection methods to discern true signals from noise. Overlapping signals are deconvoluted through application of a second-derivative based method for peak detection. The second derivative has been extensively used for peak detection in chromatography [32], nuclear magnetic resonance [33], and astronomical spectroscopy [34], but to our knowledge has not been applied to native mass spectrometry data analysis. Similar to the Robinson approach [9], our software simulates charge envelopes in order to best fit the peaks in the mass spectrum. The goodness of fit is determined by a scoring function that combines both mass error and intensity error. Moreover, to further account for overlapping signals, our method fits linear combinations of charge envelopes to the raw data simultaneously, rather than fitting charge state envelopes sequentially as in Massign. Our software is a comprehensive package which can be operated in either automated or manual mode, with processing options at every step to provide great flexibility in addressing spectra with varying amounts of noise and complexity. Furthermore, unlike Massign or Automass, PeakSeeker is freely distributed under an open source license, allowing users the freedom to improve upon the algorithm or adapt it as needed for specific experiments.
We demonstrate the software’s capability to deconvolute native mass spectra of: synthetic nucleosomes containing the core histone octamer surrounded by a tight binding DNA segment, a complex of RNA-polymerase II (pol II) with multiple copies of its inhibitor a-amanitin, and a megaDalton sized human TCP-1 ring complex (TRiC). These spectra have been chosen to demonstrate the three main difficulties of deconvoluting native mass spectra: overlapping peaks, interleaved peaks, and poor signal-to-noise ratio.
Experimental
Sample Preparation and Mass Spectrometry
Mononucleosomes with site-specific methyl lysine analogs (MLA) at Histone 3 Lys 9 (H3K9) were prepared as described [35, 36]. Briefly, individual core histones from Xenopus laevis were expressed recombinantly with a Histone 3 K9C point mutation. The non-native cysteine residue was alkylated by treatment with (2-chloroethyl)-methylammonium chloride to form specific methylated lysine analogs and the core histones were reconstituted to nucleosomes by addition of double stranded DNA with a tight binding sequence of 147 base-pairs prepared by Taq polymerase catalyzed PCR extension. The expected mass value of the MLA nucleosome was calculated from the measured masses of denatured H3K9 Me1 analog and H4, the Uniprot sequences of H2A and H2B with the status of the N-terminal Met residue assigned from MS measurement in proteolytic digests (Glu-C and trypsin), and the calculated mass of the 147 bp Widom 601 sequence [37] including an additional 3’-adenine on both strands from the Taq polymerase reaction.
RNA polymerase II (pol-II) was purified from Saccharomyces cerevisiae as previously described [38]. Pol II /a-amanitin binding samples were prepared by keeping the pol II concentration constant while adding increased concentration of a-amanitin, from 1 mM to 50 mM. The expected mass of pol II was calculated from the primary amino acid sequences contained in the Uniprot database. The presence or absence of N-terminal methionine and acetylation was determined from previous proteomics analysis of these preparations. In addition to the primary sequence, eight zinc ions and the active site magnesium were assumed to remain bound to the complex as in the crystal structure [39].
Human TRiC was purified from HeLa cells in a similar way to bovine TRiC [40] except for cell lysis, which was performed as previously described [41]. To achieve extra purity, TRiC fractions were incubated with 1 mM ATP for 15 min at 37 °C and then reprocessed by Mono-Q HR 16/10 (GE Healthcare, USA) and Superose 6 10/300 GL columns (GE Healthcare, USA) in sequence. TRiC’s folding activity was assessed by luciferase refolding as described [42]. The theoretical mass of TRiC was calculated as described above for pol II, except that metal adducts were not considered.
Native MS measurements were made on an Exactive Plus EMR instrument (Thermo Scientific, Bremen, Germany). The instrument was calibrated in the extended mass range mode (m/z 350–20,000) by reference to an infused CsI solution, which forms well-defined clusters up to 12,000 m/z. Samples were buffer exchanged into 100 mM ammonium acetate buffer at pH 6.8 by four spins on a 10 kDa cutoff centrifugal filter (Millipore). Samples were adjusted to a concentration between 1–5 µM and then introduced to the gas phase by static infusion nanospray ionization using a nanospray source. Spectra were acquired with source collision energy setting generally between 5 and 35 and HCD setting between 150 and 200. Operating pressures in the instrument were typically 1–2 mbar in the S-lens region, 10−4 mbar in the source chamber, and 10−9 mbar in the analyzer chamber. Five to twenty scans were averaged prior to data analysis.
Software Workflow
Algorithms were implemented in Python using the scipy, numpy, and matplotlib libraries. Spectra are read in as text files or from the clipboard exported from the mass spectrometry vendor software, then processed using the workflow illustrated in Supplemental Figure S1. This workflow consists of: 1) spectrum preprocessing, 2) initial peak detection, 3) deconvolution of peak overlap using second derivative, 4) fitting Gaussian functions to the deconvoluted peaks, 5) charge state assignment by fitting to simulated charge envelopes, 6) repeating step 5 for additional masses, and 7) fitting a linear combination of the simulated charge envelopes to the spectrum. In addition to the summary given below, a detailed description of each step is provided in the Supplemental Information while Figure S1 gives an overview of the algorithm. Additionally, Supplemental Table S1 provides a feature comparison between PeakSeeker and several of the other primary deconvolution algorithms.
Spectrum Processing and Peak Detection Methods
Spectra are first aggregated across several scans to enhance the signal-to-noise ratio before being exported to PeakSeeker. After optional smoothing by either a moving average or Savitsky-Golay filter and optional background subtraction, peaks are detected using two levels of processing algorithms. At the first level, one of the following three methods is used to identify the “apparent peaks” with or without overlapping. Method 1 simply detects local maxima above a fixed signal-to-noise ratio threshold. Method 2, adapted from the Massign peak detection algorithm [9], adjusts this threshold based on the intensity of the most recently detected peak to allow for detection of “shoulder” peaks. Method 3 uses the continuous wavelet transform to process noisy spectra. Continuous wavelet transform convolves the spectrum with a Mexican Hat wavelet across a range of peak widths. Narrow and high frequency noise is filtered out while true peaks register as local maxima across multiple widths. This method does not require prior smoothing or background subtraction [43, 44]. At the second level, overlapped peaks are detected using the second derivative across a peak range, defined as the set of consecutive data points whose intensities are above the noise level. The second derivative of a Gaussian shaped peak consists of two zero-crossings surrounding a central minimum. Hence, the number of zero-crossings can be used to derive the number of underlying peaks.
Using the second derivative derived parameters as a starting guess, we then use a Levenberg-Marquardt algorithm to fit Gaussian functions to the peak range. This method has advantages over other peak detection methods by finding peaks that do not have a local maximum and by better estimating the parameters of adjacent peaks (see Results and Discussion). All fitting and subsequent deconvolution are performed on the original (pre-processed) spectrum.
Charge State Determination and Further Analysis
Possible charge states are iterated to the most intense peak and signals matching the corresponding charge state series are determined. These are modeled by a Gaussian distribution over the charge domain [9, 45]. The quality of the fit is assessed with an automated scoring function that examines both mass error and height error (SI Section 5). The charge state is assigned either automatically, by the scoring function, or optionally by the user, after inspection of the best fitting options.
This is repeated up to four more times using the most intense peak that has not already been assigned envelope membership. Aside from this initial peak, other peaks may still be assigned to the current charge state series even if they already have membership in a different envelope. This accounts for peak overlap that was not detected by the second derivative search. A linear combination of up to five simulated charge envelopes is fit to the original raw spectrum using least squares regression. This method of deconvoluting peak signals is distinct from Massign, which subtracts envelopes from the spectrum one at a time and refits the remaining envelopes. Our method reduces the bias of the order in the subtraction procedure. Finally, the masses and abundances of the molecular species are reported and the residual spectrum is evaluated for the quality of the fit. This process is iterated over the residuals until all peaks are determined.
Results and Discussion
Peak widening due to incomplete desolvation
Peaks in native mass spectra of large complexes are typically much wider than theoretically predicted based on their isotope distributions. This is typically not a limitation of the instrumental mass resolving power. For instance, the Orbitrap Exactive Plus EMR instrument used in these studies was operated at a nominal resolution setting of 8750 determined at 200 m/z. Signals acquired from clusters of CsI ions, which lack isotope peaks, were consistent with these nominal resolution settings (data not shown). Inadequate desolvation and/or additional adduct formation are the primary limiting factors affecting the observed resolution of native mass spectra.
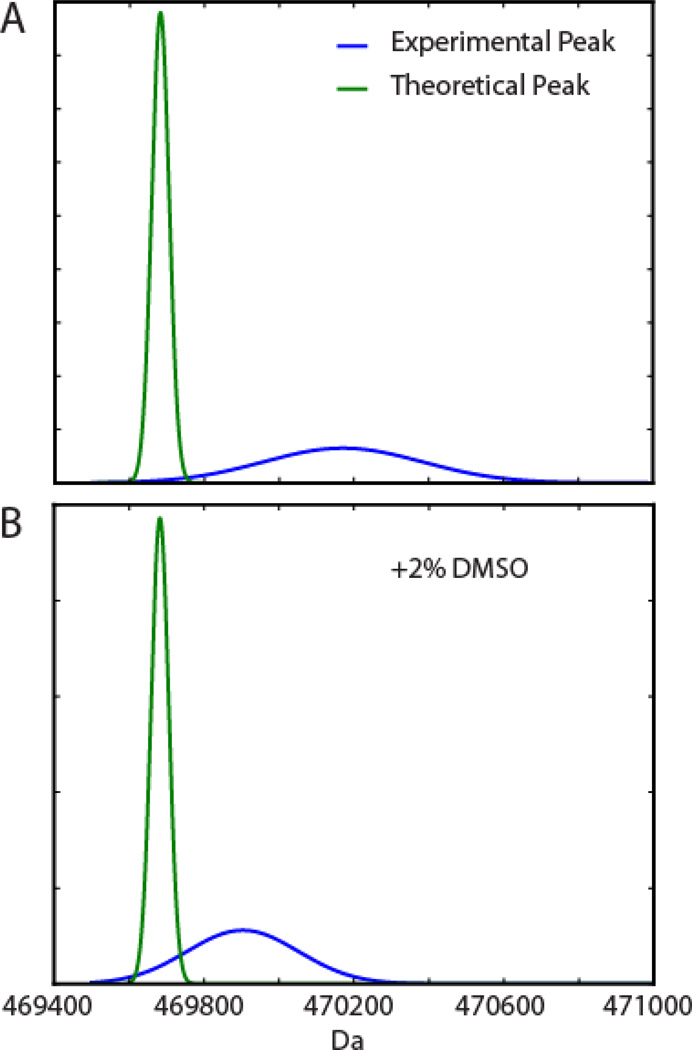
Compared to the expected peak calculated for decameric pol II at this mass resolution (see Experimental), the deconvoluted experimental spectrum is observed at higher mass and wider peak shape (Figure 1A). The experimental peak is shifted by 490 Da and has a width of about 500 Da, whereas the width predicted from the isotope distribution is 60 Da. Assuming the mass increase is purely due to hydration, the complex would retain approximately 27 water molecules. The mass shift is however likely caused by a combination of adducts such as water, sodium, potassium, ammonium, and other anions [1]. The distribution of a variety of adducts clearly dominates the observed peak width.
Figure 1.
Peak broadening by insufficient desolvation. (A): pol II 10-mer theoretical mass peak (green trace, peak width due to isotope distribution) and experimental peak (blue, wider peak due to adducts and hydration) in 100 mM ammonium acetate. (B): pol II 10-mer theoretical peak and experimental peak in 100 mM ammonium acetate and 2% DMSO. Peak areas were normalized for both theoretical and experimental peaks.
In the presence of 2% DMSO (Figure 1B), the width of the peak is decreased to 350 Da, and the mass shift decreased to 220 Da. This observation provides further evidence that adduction determines the effective peak resolution in native MS measurements, as a decreased mass shift represents fewer adducts that are more narrowly distributed. This finding was true across all charge states of pol II. DMSO enhanced the resolution and increased the mass accuracy of native MS and also shifted the observed charge state distribution to higher z while preserving intact decameric and dodecameric pol II complexes (Figure S2). The charge state range was shifted from 43–51+ without DMSO to 55–77+ with DMSO for decameric pol II with similar findings for dodecameric pol II. In pure water, the theoretical upper charge state limit (Zmax=0.078 x √M) for a spherical complex can be estimated from the Rayleigh charging model [46, 47], in which M is the mass of the complex in Da. The Rayleigh limit for the decamer is 54+, near the maximum charge observed in aqueous ammonium acetate (51+). The observation that the maximum charge is shifted to 77+ in the presence of 2% DMSO is consistent with previous findings in which DMSO promotes partial unfolding of proteins in the electrospray droplet inducing a shift to higher charge states [48–50]. Furthermore, these findings suggest that water molecules and other adducts are intrinsic structural elements necessary to maintain the solution-state conformation of the protein complex in the gas-phase.
Limitation of peak deconvolution algorithms
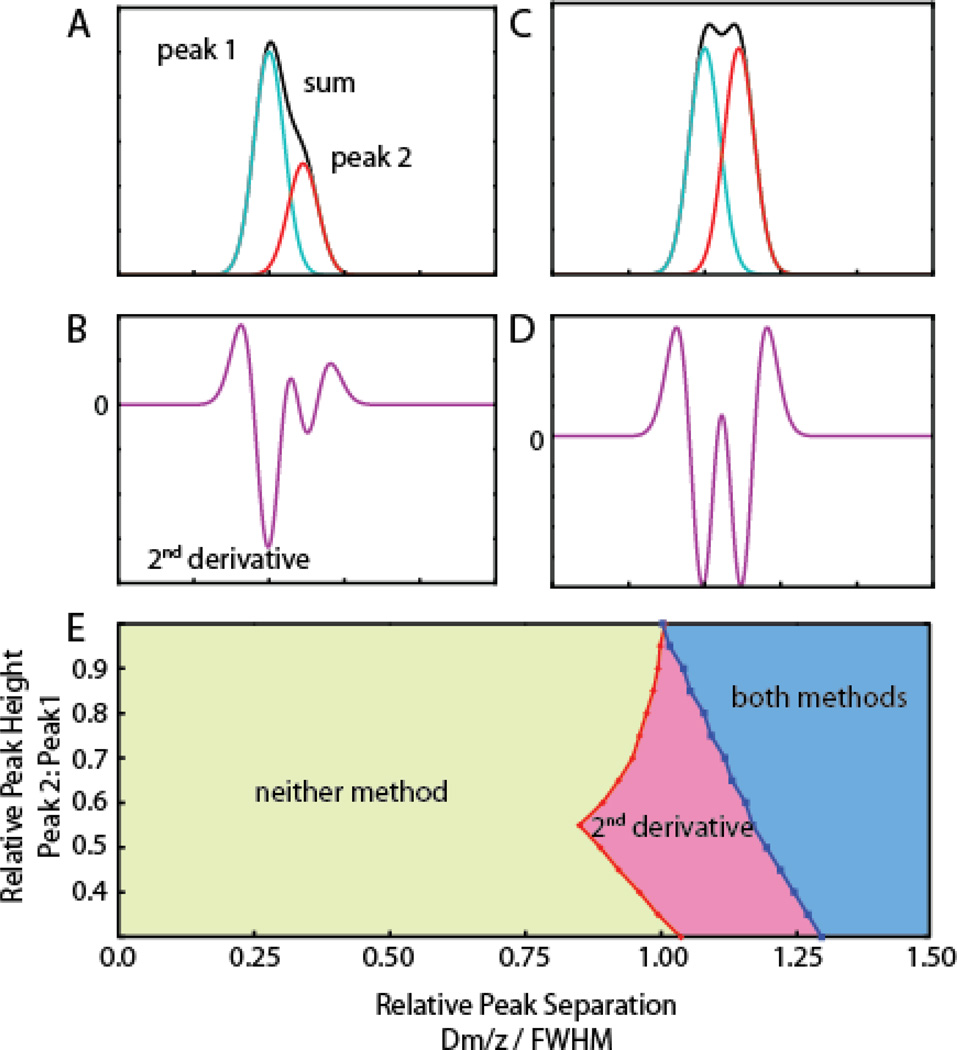
We explored the choice of peak detection methods on deconvolving overlapping ion signals. The theoretical performance of two peak detection algorithms was compared using two simulated, noise-free Gaussian peaks with equal variance but different peak heights and m/z spacing. The combined signal of these two peaks was analyzed using both the second derivative method and a local maxima method. With two closely spaced peaks of different heights (Figure 2A), the second smaller peak appears as a shoulder that is not resolved by local maxima detection. However, the second derivative detects the underlying peak by the presence of multiple minima (Figure 2B).
Figure 2.
Effectiveness of peak detection methods in detecting overlapping ion signals. Peak overlap is modeled by two standardized Gaussian functions of equal variance. The effect of varying the intensity of the 2nd signal (rightmost) relative to the 1st as well as the relative spacing of the signal centroids (plotted as the difference in centroid m/z divided by full width at half maximum) on the performance of peak detection methods was evaluated. A) Peak 2 is half the height of peak 1 with centroids separated by 1 unit of FWHM, and appears as a shoulder in the summed signals. Local maxima detection fails to register the shoulder peak. B) The second derivative shows two local minima separated by zero-crossings indicating that the spectrum contains two underlying peaks. C) When the two signals have equal intensity both local maxima and second derivative detection find the two peaks. D) Second derivative of C). E) State diagram indicating the ranges in which two peaks are successfully deconvoluted by both 2nd derivative and local maxima detection (blue), 2nd derivative only (red), or neither method (green).
Figure 2E shows the range in which the second derivative procedure provides additional deconvoluting power when the relative height and separation (plotted relative to the peak width) of the two simulated peaks is varied. The largest improvement is in finding low intensity shoulder peaks (Figures 2A, 2B, 2E). The second derivative requires ~20% less separation to resolve peak overlap than the local maxima method when the second peak is less than half the height of the first. As the intensity of the two signals approach each other, the advantage of the second derivative declines, and the two methods are comparable for equally sized peaks (Figures 2C, 2D, 2E). In this case, overlapping peaks may also be detected by the unusually large width of a peak compared to its neighbors.
There is a substantial range (Separation < ~1.0) in which neither method can deconvolve the simulated peaks. However, when the two peaks are very closely spaced (separation < 0.48) no distortion is introduced by summing the overlaid signals. Therefore, such overlap does not inhibit charge state assignment. This overlap can be accounted for without explicitly detecting two peaks by apportioning such signals across to multiple charge envelopes in the peak range (see Experimental). Between a separation of 0.48 and approximately 1.0 however, summed peaks will both distort the original peak parameters and escape detection by the second derivative.
Additionally we tested a method in which peaks are initially detected and fit using local maxima detection, followed by examination of the residuals in the subtracted spectrum for any new local maxima. When we tested the residuals procedure by itself, we found it difficult to determine if peaks in the subtracted spectrum are indeed true peaks or simply consequences of an imperfect peak line fit. On the other hand, the second derivative bypasses these confounding factors by searching the original signal for changes in concavity. The first derivative is as sensitive to changes in concavity as the second derivative, but is more difficult to interpret because peaks and valleys represent inflection points in the original spectrum. In contrast, the second derivative shows peaks in the original spectrum clearly as minima separated by zero-crossings (inflection points in the original spectrum). This convenient feature provides a starting guess for the centroid m/z to seed the least squares fitting procedure and simplifies user interpretation.
Native MS Spectrum of Nucleosome
The utility of the second derivative in deconvoluting overlapping ion signals is demonstrated by application to a spectrum obtained from synthetic mononucleosomes. Nucleosomes are the smallest packing units of DNA consisting of approximately two turns of DNA wrapped around an octomeric histone core particle. The posttranslational state of the histone tails and the chromatin structural state of a gene are both fundamental determinants of gene expression, and direct observation of nucleosomes is a first step in developing MS based structural analysis for chromosome architecture.
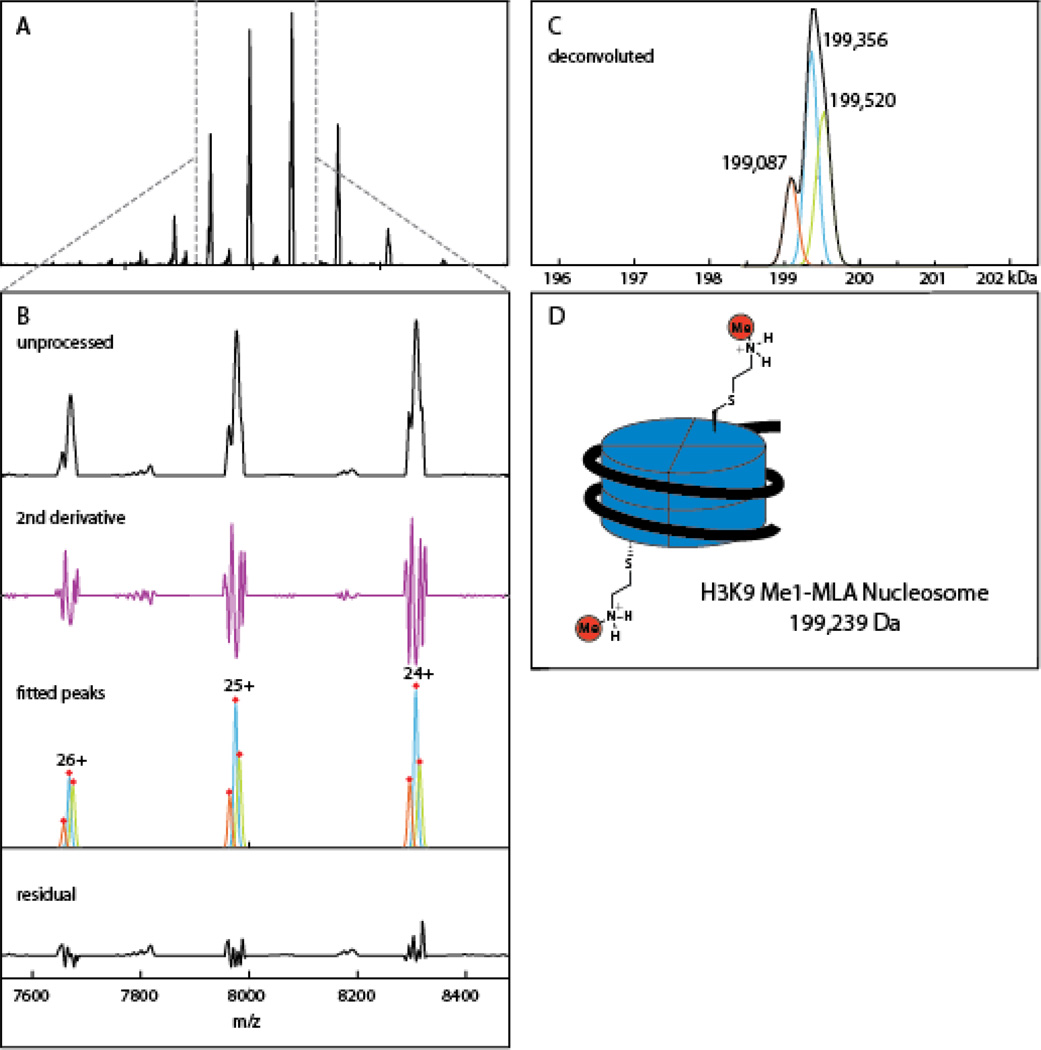
We applied PeakSeeker to a spectrum of a synthetic nucleosome containing a methylysine analog at K9 of Histone 3 (Figure 3). The spectrum contains a charge envelope centered on the 24+ charge state with highly overlapping peaks (Figure 3A). The major ions in charge states of 24–26+ contain shoulder peaks that fail to register as local maxima (Figure 3B). Typical peak detection methods were therefore unable to resolve the underlying signals, whereas the second derivative procedure discerned three species. In the automatic processing mode, our algorithm found three unique masses at 199087, 199356, and 199520 Da with abundances of 19%, 47% and 33% (Figure 3C). The quality of the fit can be manually assessed from inspection of the residual spectrum after subtraction of the fitted envelopes (Figure 3B bottom panel). The spacing of the three species suggests the addition and subtraction of a nucleotide residue mass from the 147 bp segment of DNA reconstituted with the histone octamer. This DNA segment is synthesized by Taq polymerase catalyzed PCR and heterogeneity can result from: the 5’ to 3’ exonuclease activity of Taq, heterogeneity in the original PCR oligonucleotide primers, or incomplete formation of 3’-adenine overhangs left by Taq synthesis. The expected mass of the monomethyl lysine nucleosome is 199239 Da (Figure 3D) which matches the major species to 0.06%. The minor peaks around this then represent the addition nucleotide and the corresponding and loss of a nucleotide residue.
Figure 3.
Deconvolution of methyl lysine analog (MLA) nucleosome spectrum. A) Unprocessed spectrum of MLA nucleosome with charge series centered on the 24+ state. B) A zoomed spectrum shows unresolved peaks in the individual charge signals, which are detected by second derivative examination, modeled with individual Gaussians. The residual spectrum can be manually examined for under- or over-fitting. C) PeakSeeker assigns three deconvoluted mass to the spectrum with spacing around 300 Da, likely due to heterogenous preparations of the DNA component. D) Theoretical mass of the MLA nucleosome based on octameric histone particle and 147 bp strand of DNA.
For comparison, we analyzed the spectrum using MagTran (Amgen Inc.), an implementation of the ZSCORE algorithm [27], Massign (University of Oxford), and Protein Deconvolution (Thermo Scientific) which implements the ReSpect (Positive Probability) maximum entropy algorithm. Magtran finds only the single charge state envelope deconvoluting corresponding to the 199392 Da peak (Supplemental Figure S3). Massign provides more flexible peak fitting parameters for addressing shoulder peaks, and is consequently able to detect either the lower m/z ions or the higher m/z ions but not both (Figure S3). The inability of these algorithms to deconvolute the overlapping signals is predicted from our analysis of the required peak separation and spacing (Figure 2). The spacing of the signal centroids (9–15 m/z units) is approximately equal to the peak widths (FWHM) in the range where second derivative peak processing outperforms detection of local maxima. When the same spectrum is processed with Protein Deconvolution, which performs deconvolution in the forward direction and bypasses explicit peak detection, only charge state signals corresponding to the two higher mass species are detected (Figure S3).
Native MS Spectrum of RNA-Polymerase II/a-amanitin complex
As part of our efforts to measure low affinity ligand binding in the presence of non-specifically bound artifacts (Guan et al, Analytical Chemistry in press) we determined a native MS spectrum of 4.5 µM pol II binding to 20 µM of the fungal inhibitor α-amanitin. α-amanitin is the active component of Amanita phalloides, known as the “death cap” mushroom. Amanitin stalls mRNA synthesis by blocking conformational states of pol II required for translocation along the DNA template [51]. Pol II preparations exist as a mixture of the full 12-subunit enzyme and a 10-subunit form lacking Rpb4 and Rpb7. In the absence of any inhibitor, two species are observed for the 12mer at 514726 Da (theoretical: 514154 Da; 0.1% error) and the 10mer at 470171 Da (theoretical: 469682 Da; 0.1% error) (Figure 4). With the inhibitor added at 20 µM, both charge state series are further split into four peaks resulting from multiple molecules of α-amanitin specifically and non-specifically bound to the complex (Figure 4). Non-specific binding, caused by a high local concentration of ligand in the electrospray droplet, is a common problem confounding native MS binding studies [52, 53]. Furthermore, the many adducts caused by non-specific ligand binding pose a challenge to correct deconvolution and charge state assignment for automated algorithms.
Figure 4.
Deconvolution of native MS spectrum of RNA pol II with high levels of α-amanitin. A) Unprocessed spectrum showing peak splitting due to specific and non-specific inhibitor binding. B) Individual charge state series detected and fitted by PeakSeeker. C) Residual spectrum demonstrating effectiveness of fit. D) Deconvoluted spectrum shows mixture of decameric and dodecameric pol II bound to 1–4 molecules of amanitin. E) theoretical molecular weights for these species.
PeakSeeker deconvolutes eight distinct masses from the spectrum of pol II bound α-amanitin (Figure 4). These correspond to states of both the 12mer and the 10mer with 1–4 molecules of amanitin bound. The average difference between the interleaved masses was 983 ± 12 Da, which suggests that α-amanitin (919 Da) forms a complex with additional metal ions or buffer components. The deconvoluted masses are within 0.12% of the theoretical values and showed positive deviations due to incomplete desolvation. The relative abundance of 10mer vs 12 mer was determined to be 57% 10mer to 42% 12mer.
Using Massign we were able to retrieve the same masses and abundances as found with PeakSeeker, indicating that both peak detection methods perform equally well on well-resolved peaks in interleaved envelopes (Figure S4). Magtran, on the other hand, produced no output, due to the complexity of the spectrum (Figure S4). This spectrum poses difficulty for algorithms not designed for native MS. The large number of closely spaced peaks in any given charge state requires discriminating between many combinations of charge state envelopes. PeakSeeker and Massign both extract charge state distributions efficiently from the original data by modeling the intensity of the series as Gaussian distributions. Both methods also allow for a user to select the charge state distribution manually.
Thermo Protein Deconvolution was able to find five of the eight interleaved charge envelopes, but only after extensive tuning of the parameters (Figure S4). For example, the software did not find any of the 12-mer species when the mass range was set from 400 kDa to 600 kDa, but was able to find them when the mass range was extended to 700 kDa. Thus, maximum entropy methods such as Protein Deconvolution can give inconsistent output depending on parameter settings.
Native MS spectrum of Human TRiC complex
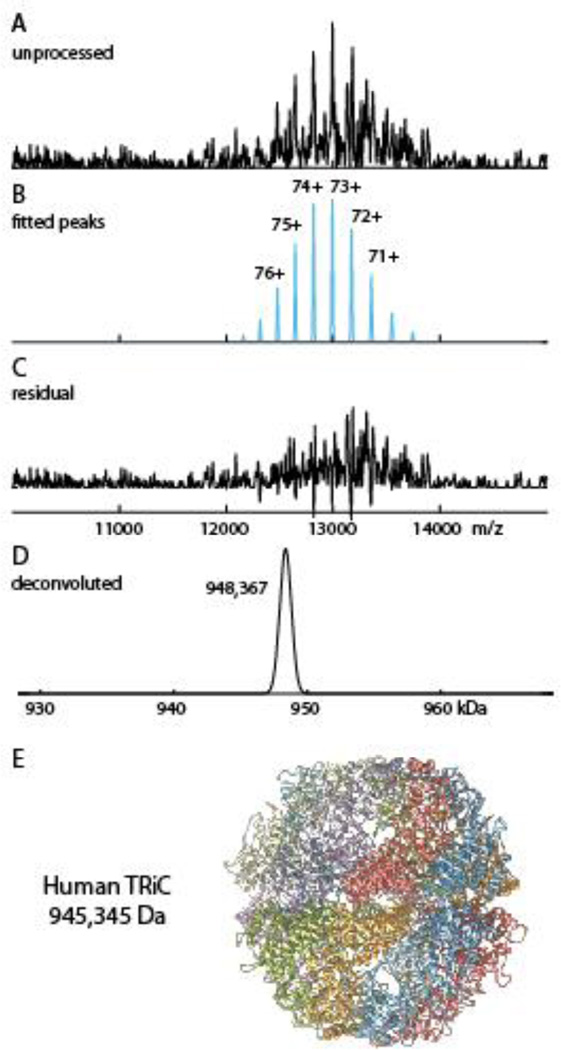
Protein assemblies purified from human cell lines, while clearly more relevant to health related studies, are typically more heterogeneous than complexes reconstituted from individual components (e.g. our nucleosome preparations) or purified from yeast (e.g. our pol II preparation). The heterogeneity can manifest in native spectra as poorly resolved signals amidst high levels of noise and background. We tested our peak detection methods on one such noisy spectrum: a preparation of the human TCP1 ring complex (TRiC), isolated from a HeLa cell line. TRiC is a 1 megaDalton sized chaperonin containing two stacked hetero-octameric rings whose central chambers facilitate folding of client proteins. A broad range of newly synthesized polypeptides from diverse cellular functions are substrates of TRiC [54].
Analysis of the TRiC spectrum presents a unique challenge for deconvolution algorithms due to its poor signal-to-noise ratio. Many peaks detected by Massign without a moving average filter were probably noise peaks. On the other hand, using moving average in Massign, peak parameters may become distorted. PeakSeeker allows testing of multiple processing and detection methods. For instance, we first tested the continuous wavelet transform. This method missed a prominent peak due to its thinness. However smoothing with a Savitzky-Golay filter before peak detection resulted in effective detection. Using manual mode PeakSeeker retrieved the major charge state envelope corresponding to 948300 Da. This deviates from the theoretical MW of 945345 Da by 0.3%. The highest charge state of 76+ agrees well with the Rayleigh limit of 75.9. Using Massign’s manual mode and intensity modeling, we were able to retrieve the same masses and abundances as PeakSeeker (Figure S5). Magtran, on the other hand, produced no output. Finally, Protein Deconvolution retrieves 10 zero charged masses in the 0.9–1 MDa range corresponding to charge envelopes containing subsets of the major ion signals together with different minor peaks and noise signals (Figure S5). The annotations made by Protein Deconvolution for the two most abundant charge envelopes are shown in Figure S5B. The propensity for the maximum entropy method to overfit noisy data demonstrates the pitfalls of omitting explicit peak detection and smoothing/filtering with poor signal-to-noise spectra.
Conclusions
Deconvolution and charge state assignment of electrospray mass spectra from native assemblies presents distinct challenges from those of denatured proteins, including the observation of wide peak widths that are caused by the presence of adducts and incomplete desolvation. We have developed software to specifically address problems we encountered while analyzing native MS spectra from large complexes using other freely and commercially distributed software packages available within our Research Resource. PeakSeeker is capable of mass assignment from native spectra that contain wide ion signals in the context of overlapping and highly interleaved charge distributions and low signal to noise ratio. Multiple methods of peak detection and other signal processing options as well as automated and user-dependent modes provide users with great flexibility in addressing the challenges of a particular analysis. We demonstrate the applicability of this software to the analysis of several large protein complexes with relevance to transcriptional regulation (nucleosomes and pol II) and protein folding (TRiC) and representing leading-edge protein purification techniques. While other software has been developed to address native MS spectra specifically, we present a flexible algorithm that is distributed under the principles of free software, allowing other users to improve upon the algorithms and adapt them for their specific applications.
Supplementary Material
Figure 5.
Native MS spectrum of human TriC complex purified from HeLa cell culture. A) The unprocessed spectrum shows high levels of noise. B) Peaks detected and assigned to charge envelope by PeakSeeker, centered on 73+ ion. C) The residual spectrum demonstrating discrimination between signal and noise. D) Deconvoluted mass of TRiC.
Acknowledgments
This work was supported by the Bio-Organic Biomedical Mass Spectrometry Resource at UCSF (A.L. Burlingame, Director) supported by the Biomedical Technology Research Centers program of the NIH National Institute of General Medical Sciences, NIH NIGMS 8P41GM103481. The ThermoScientific Exactive Plus EMR instrument was supported by NIH NIGMS 8P41GM103481, and UCSF 2013 ETAC Shared Equipment Grant. Initial native MS experiments were also supported by ThermoScientific (Bremen). Additional funding is from NIH P41GM103832, PN2EY016525, GM49985, GM36659, AI21144, AG021601, AG002132, AG010770.
Footnotes
Electronic supplemental material
Additional supporting information may be found in the online version of this article at the publisher’s website.
The software is licensed under the terms of the GNU General Public License as published by the Free Software Foundation. The software is available for download at https://github.com/lujonathanh/PeakSeeker.
References
- 1.Snijder J, Heck AJR. Analytical Approaches for Size and Mass Analysis of Large Protein Assemblies. Annu. Rev. Anal. Chem. 2014;7:43–64. doi: 10.1146/annurev-anchem-071213-020015. [DOI] [PubMed] [Google Scholar]
- 2.Snijder J, Rose RJ, Veesler D, Johnson JE, Heck AJR. Studying 18 MDa Virus Assemblies with Native Mass Spectrometry. Angew. Chem. Int. Ed. 2013;52:4020–4023. doi: 10.1002/anie.201210197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Breukelen B, Barendregt A, Heck AJR, van den Heuvel RHH. Resolving stoichiometries and oligomeric states of glutamate synthase protein complexes with curve fitting and simulation of electrospray mass spectra. Rapid Commun. Mass Spectrom. 2006;20:2490–2496. doi: 10.1002/rcm.2620. [DOI] [PubMed] [Google Scholar]
- 4.Sharon M, Robinson CV. The Role of Mass Spectrometry in Structure Elucidation of Dynamic Protein Complexes. Annu. Rev. Biochem. 2007;76:167–193. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]
- 5.Engen JR, Smith DL. Investigating protein structure and dynamics by hydrogen exchange MS. Anal. Chem. 2001;73:256A–265A. doi: 10.1021/ac012452f. [DOI] [PubMed] [Google Scholar]
- 6.Jain SS, Tullius TD. Footprinting protein-DNA complexes using the hydroxyl radical. Nat. Protoc. 2008;3:1092–1100. doi: 10.1038/nprot.2008.72. [DOI] [PubMed] [Google Scholar]
- 7.Sinz A. Chemical cross-linking and mass spectrometry for mapping three-dimensional structures of proteins and protein complexes. J. Mass Spectrom. 2003;38:1225–1237. doi: 10.1002/jms.559. [DOI] [PubMed] [Google Scholar]
- 8.Walzthoeni T, Leitner A, Stengel F, Aebersold R. Mass spectrometry supported determination of protein complex structure. Curr. Opin. Struct. Biol. 2013;23:252–260. doi: 10.1016/j.sbi.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Morgner N, Robinson CV. Massign: An Assignment Strategy for Maximizing Information from the Mass Spectra of Heterogeneous Protein Assemblies. Anal. Chem. 2012;84:2939–2948. doi: 10.1021/ac300056a. [DOI] [PubMed] [Google Scholar]
- 10.van Duijn E, Barbu IM, Barendregt A, Jore MM, Wiedenheft B, Lundgren M, Westra ER, Brouns SJJ, Doudna JA, van der Oost J, Heck AJR. Native Tandem and Ion Mobility Mass Spectrometry Highlight Structural and Modular Similarities in Clustered-Regularly-Interspaced Shot-Palindromic-Repeats (CRISPR)-associated Protein Complexes From Escherichia coli and Pseudomonas aeruginosa. Mol. Cell. Proteomics. 2012;11:1430–1441. doi: 10.1074/mcp.M112.020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sleno L, Volmer DA. Ion activation methods for tandem mass spectrometry. J. Mass Spectrom. 2004;39:1091–1112. doi: 10.1002/jms.703. [DOI] [PubMed] [Google Scholar]
- 12.Zhou M, Jones CM, Wysocki VH. Dissecting the Large Noncovalent Protein Complex GroEL with Surface-Induced Dissociation and Ion Mobility–Mass Spectrometry. Anal. Chem. 2013;85:8262–8267. doi: 10.1021/ac401497c. [DOI] [PubMed] [Google Scholar]
- 13.Hall Z, Politis A, Robinson CV. Structural Modeling of Heteromeric Protein Complexes from Disassembly Pathways and Ion Mobility-Mass Spectrometry. Structure. 2012;20:1596–1609. doi: 10.1016/j.str.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt C, Zhou M, Marriott H, Morgner N, Politis A, Robinson CV. Comparative cross-linking and mass spectrometry of an intact F-type ATPase suggest a role for phosphorylation. Nat. Commun. 2013;4 doi: 10.1038/ncomms2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sobott F, Robinson CV. Characterising electrosprayed biomolecules using tandem-MS—the noncovalent GroEL chaperonin assembly. Int. J. Mass Spectrom. 2004;236:25–32. [Google Scholar]
- 16.Zhang H, Cui W, Wen J, Blankenship RE, Gross ML. Native electrospray and electron-capture dissociation in FTICR mass spectrometry provide top-down sequencing of a protein component in an intact protein assembly. J. Am. Soc. Mass Spectrom. 2011;21:1966–1968. doi: 10.1016/j.jasms.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Cui W, Wen J, Blankenship RE, Gross ML. Native Electrospray and Electron-Capture Dissociation FTICR Mass Spectrometry for Top-Down Studies of Protein Assemblies. Anal. Chem. 2011;83:5598–5606. doi: 10.1021/ac200695d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin S, Loo JA. Top-Down Mass Spectrometry of Supercharged Native Protein-Ligand Complexes. Int. J. Mass Spectrom. 2011;300:118–122. doi: 10.1016/j.ijms.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose RJ, Damoc E, Denisov E, Makarov A, Heck AJR. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nat. Methods. 2012;9:1084–1086. doi: 10.1038/nmeth.2208. [DOI] [PubMed] [Google Scholar]
- 20.McKay AR, Ruotolo BT, Ilag LL, Robinson CV. Mass Measurements of Increased Accuracy Resolve Heterogeneous Populations of Intact Ribosomes. J. Am. Chem. Soc. 2006;128:11433–11442. doi: 10.1021/ja061468q. [DOI] [PubMed] [Google Scholar]
- 21.Benesch JLP. Collisional activation of protein complexes: Picking up the pieces. J. Am. Soc. Mass Spectrom. 2011;20:341–348. doi: 10.1016/j.jasms.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 22.Pauling L, Corey RB, Branson HR. The Structure of Proteins. Proc. Natl. Acad. Sci. U. S. A. 1951;37:205–211. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanucara F, Holman SW, Gray CJ, Eyers CE. The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat. Chem. 2014;6:281–294. doi: 10.1038/nchem.1889. [DOI] [PubMed] [Google Scholar]
- 24.Ferrige AG, Seddon MJ, Jarvis S, Skilling J, Aplin R. Maximum entropy deconvolution in electrospray mass spectrometry. Rapid Commun. Mass Spectrom. 1991;5:374–377. [Google Scholar]
- 25.Ferrige AG, Seddon MJ, Green BN, Jarvis SA, Skilling J, Staunton J. Disentangling electrospray spectra with maximum entropy. Rapid Commun. Mass Spectrom. 1992;6:707–711. [Google Scholar]
- 26.Marty MT, Baldwin AJ, Marklund EG, Hochberg GKA, Benesch JLP, Robinson CV. Bayesian Deconvolution of Mass and Ion Mobility Spectra: From Binary Interactions to Polydisperse Ensembles. Anal. Chem. 2015;87:4370–4376. doi: 10.1021/acs.analchem.5b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z, Marshall AG. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J. Am. Soc. Mass Spectrom. 1998;9:225–233. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- 28.Sivalingam GN, Yan J, Sahota H, Thalassinos K. Amphitrite: A program for processing travelling wave ion mobility mass spectrometry data. Int. J. Mass Spectrom. 2013:345–347. 54–62. doi: 10.1016/j.ijms.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stengel F, Baldwin AJ, Bush MF, Hilton GR, Lioe H, Basha E, Jaya N, Vierling E, Benesch JLP. Dissecting Heterogeneous Molecular Chaperone Complexes Using a Mass Spectrum Deconvolution Approach. Chem. Biol. 2012;19:599–607. doi: 10.1016/j.chembiol.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tseng Y-H, Uetrecht C, Heck AJR, Peng W-P. Interpreting the Charge State Assignment in Electrospray Mass Spectra of Bioparticles. Anal. Chem. 2011;83:1960–1968. doi: 10.1021/ac102676z. [DOI] [PubMed] [Google Scholar]
- 31.Tseng Y-H, Uetrecht C, Yang S-C, Barendregt A, Heck AJR, Peng W-P. Game-Theory-Based Search Engine to Automate the Mass Assignment in Complex Native Electrospray Mass Spectra. Anal. Chem. 2013;85:11275–11283. doi: 10.1021/ac401940e. [DOI] [PubMed] [Google Scholar]
- 32.Vivó-Truyols G, Torres-Lapasió JR, van Nederkassel AM, Vander Heyden Y, Massart DL. Automatic program for peak detection and deconvolution of multi-overlapped chromatographic signals: Part I. Peak detection. J. Chromatogr. A. 2005;1096:133–145. doi: 10.1016/j.chroma.2005.03.092. [DOI] [PubMed] [Google Scholar]
- 33.Dračínský M, Kaminský J, Bouř P. Relative importance of first and second derivatives of nuclear magnetic resonance chemical shifts and spin-spin coupling constants for vibrational averaging. J. Chem. Phys. 2009;130:094106. doi: 10.1063/1.3081317. [DOI] [PubMed] [Google Scholar]
- 34.Bonoli C. The use of derivative techniques in astronomical spectroscopy. Astrophys. Space Sci. 1983;89:377–385. [Google Scholar]
- 35.Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The Site-Specific Installation of Methyl-Lysine Analogs into Recombinant Histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiau C, Trnka MJ, Bozicevic A, Torres IO, Al-Sady B, Burlingame AL, Narlikar GJ, Fujimori DG. Reconstitution of Nucleosome Demethylation and Catalytic Properties of a Jumonji Histone Demethylase. Chem. Biol. 2013;20:494–499. doi: 10.1016/j.chembiol.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning 1. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 38.Robinson PJJ, Bushnell DA, Trnka MJ, Burlingame AL, Kornberg RD. Structure of the Mediator Head module bound to the carboxy-terminal domain of RNA polymerase II. Proc. Natl. Acad. Sci. 2012;109:17931–17935. doi: 10.1073/pnas.1215241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Armache K-J, Mitterweger S, Meinhart A, Cramer P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J. Biol. Chem. 2005;280:7131–7134. doi: 10.1074/jbc.M413038200. [DOI] [PubMed] [Google Scholar]
- 40.Feldman DE, Spiess C, Howard DE, Frydman J. Tumorigenic Mutations in VHL Disrupt Folding In Vivo by Interfering with Chaperonin Binding. Mol. Cell. 2003;12:1213–1224. doi: 10.1016/s1097-2765(03)00423-4. [DOI] [PubMed] [Google Scholar]
- 41.Knee KM, Sergeeva OA, King JA. Human TRiC complex purified from HeLa cells contains all eight CCT subunits and is active in vitro. Cell Stress Chaperones. 2012;18:137–144. doi: 10.1007/s12192-012-0357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thulasiraman V, Ferreyra RG, Frydman J. Folding assays. Assessing the native conformation of proteins. Methods Mol. Biol. Clifton, NJ. 2000;140:169–177. doi: 10.1385/1-59259-061-6:169. [DOI] [PubMed] [Google Scholar]
- 43.Yang C, He Z, Yu W. Comparison of public peak detection algorithms for MALDI mass spectrometry data analysis. BMC Bioinformatics. 2009;10:4. doi: 10.1186/1471-2105-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du P, Kibbe WA, Lin SM. Improved peak detection in mass spectrum by incorporating continuous wavelet transform-based pattern matching. Bioinformatics. 2006;22:2059–2065. doi: 10.1093/bioinformatics/btl355. [DOI] [PubMed] [Google Scholar]
- 45.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 46.FRS LR. XX. On the equilibrium of liquid conducting masses charged with electricity. Philos. Mag. Ser. 5. 1882;14:184–186. [Google Scholar]
- 47.Heck AJR, van den Heuvel RHH. Investigation of intact protein complexes by mass spectrometry. Mass Spectrom. Rev. 2004;23:368–389. doi: 10.1002/mas.10081. [DOI] [PubMed] [Google Scholar]
- 48.Sterling HJ, Cassou CA, Trnka MJ, Burlingame AL, Krantz BA, Williams ER. The role of conformational flexibility on protein supercharging in native electrospray ionization. Phys. Chem. Chem. Phys. 2011;13:18288–18296. doi: 10.1039/c1cp20277d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sterling HJ, Kintzer AF, Feld GK, Cassou CA, Krantz BA, Williams ER. Supercharging Protein Complexes from Aqueous Solution Disrupts their Native Conformations. J. Am. Soc. Mass Spectrom. 2011;23:191–200. doi: 10.1007/s13361-011-0301-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sterling HJ, Prell JS, Cassou CA, Williams ER. Protein Conformation and Supercharging with DMSO from Aqueous Solution. J. Am. Soc. Mass Spectrom. 2011;22:1178–1186. doi: 10.1007/s13361-011-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bushnell DA, Cramer P, Kornberg RD. Structural basis of transcription: α-Amanitin–RNA polymerase II cocrystal at 2.8 Å resolution. Proc. Natl. Acad. Sci. 2002;99:1218–1222. doi: 10.1073/pnas.251664698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daubenfeld T, Bouin A-P, van der Rest G. A Deconvolution Method for the Separation of Specific Versus Nonspecific Interactions in Noncovalent Protein-Ligand Complexes Analyzed by ESI-FT-ICR Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2006;17:1239–1248. doi: 10.1016/j.jasms.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 53.Dyachenko A, Gruber R, Shimon L, Horovitz A, Sharon M. Allosteric mechanisms can be distinguished using structural mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2013;110:7235–7239. doi: 10.1073/pnas.1302395110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yam AY, Xia Y, Lin H-TJ, Burlingame A, Gerstein M, Frydman J. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat. Struct. Mol. Biol. 2008;15:1255–1262. doi: 10.1038/nsmb.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.