Abstract
The development of massively parallel sequencing (or next-generation sequencing) has facilitated a rapid implementation of genomic sequencing in clinical medicine. Genomic sequencing (GS) is now an essential tool for evaluating rare disorders, identifying therapeutic targets in neoplasms, and screening for prenatal aneuploidy. Emerging applications, such as GS for preconception carrier screening and predisposition screening in healthy individuals, are being explored in research settings and utilized by members of the public eager to incorporate genomic information into their health management. The rapid pace of adoption has created challenges for all stakeholders in clinical GS, from standardizing variant interpretation approaches in clinical molecular laboratories to ensuring that nongeneticist clinicians are prepared for new types of clinical information. Clinical GS faces a pivotal moment, as the vast potential of new quantities and types of data enable further clinical innovation and complicated implementation questions continue to be resolved.
Keywords: clinical genomics, exome sequencing, genome sequencing, genomic sequencing, medical genomics,
Abstract
El desarrollo masivo de la secuenciación paralela o de nueva generación ha facilitado una rápida implementación de la secuenciación genómica en la medicina clínica. Hoy en día la secuenciación genómica (SG) es una herramienta esencial para evaluar trastornos raros, identificar blancos terapéuticos en neoplasias y evaluar la aneuploidía prenatal. Las aplicaciones emergentes, como la SG para el tamizaje antes de la concepción de portadores y la evaluación de predisposiciones en sujetos sanos están siendo exploradas en ambientes de investigación y empleadas por miembros de un público ávido de la incorporación de la información genómica para su atención de salud. El rápido ritmo de adopción ha creado desafíos para todos los interesados en la SG clínica, desde los enfoques centrados en la interpretación de variantes estandarizadas en laboratorios moleculares clínicos hasta asegurar que los clínicos no genetistas estén preparados para nuevos tipos de información clínica. La SG clínica se enfrenta a un momento crucial, ya que el gran potencial de nuevas cantidades y tipos de datos posibilitan una mayor innovación clínica mientras continúa la resolución de las preguntas acerca de la complicada implementación.
Abstract
La mise en place rapide du séquençage génomique en médecine clinique a été facilitée par le développement d'un séquençage massivement parallèle (ou séquençage de deuxième génération). Le séquençage génomique (SG) est maintenant un outil essentiel pour l'évaluation des maladies rares, l'identification des cibles thérapeutiques dans les cancers et le dépistage de l'aneuploïdie prénatale. Les applications naissantes comme le SG pour le test de dépistage du statut de porteur sain d'une anomalie avant la conception d'un enfant et le test de prédisposition génétique pour les sujets sains, font l'objet de recherches et sont utilisées par des usagers soucieux d'inclure l'information génomique dans la prise en charge de leur santé. De par cette adoption rapide, tous les acteurs du SG clinique ont été confrontés à des difficultés allant de la standardisation de différentes approches dans l'interprétation des résultats dans les laboratoires de diagnostic moléculaire à l'assurance de la préparation des médecins non généticiens pour recevoir ces nouvelles formes d'information. Le séquençage génomique est à une phase cruciale compte tenu de l'énorme potentiel des nouveautés dans la quantité et le type de données pouvant déboucher sur de futures innovations cliniques ; des problèmes de mise en ?uvre complexe sont encore à résoudre.
Introduction
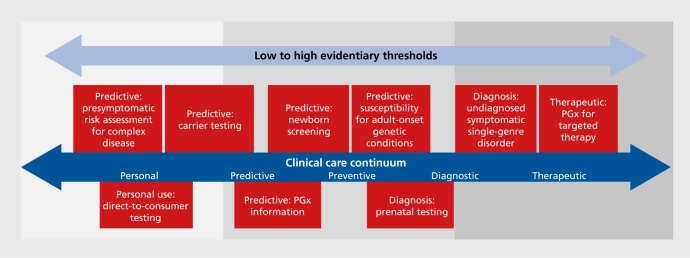
The completion of the Human Genome Project in 2001 initiated a rapid expansion of knowledge about human DNA and genetic variation. However, sequencing technologies remained relatively expensive and time consuming, and clinical applications in subsequent years were mostly limited to the evaluation of rare monogenic Mendelian disorders via the Sanger method of DNA sequencing. The development of massively parallel sequencing (also called next-generation sequencing) has accelerated the implementation of genomic sequencing (GS) in clinical practice, with applications such as exome sequencing and multigene panels ordered regularly in clinical genetics practice and innovative applications steadily emerging. The expanding knowledge base of associations between genetic variation and human disease signifies that the long-anticipated genomic revolution is underway.1-3 Yet the cost of genomic technologies is significant, the interpretation of large-scale genomic data is difficult, and the evidence regarding clinical utility of GS in various settings is scarce (Figure 1) Additionally, many other challenging ethical, financial, and social issues arise in applying genomic knowledge and data to medical practice.
Figure 1. Genomic sequencing use cases and suggested levels of evidence to demonstrate clinical utility. GS, genomic sequencing; PGx, pharmacogenomics. Adapted from ref 59: Delaney SK, Hultner ML, Jacob HJ, et al. Toward clinical genomics in everyday medicine: perspectives and recommendations. Expert Rev Mol Diagn. 2016;16(5):521-532. Copyright © Future Drugs, Ltd, 2016.
This article explores current applications of GS in clinical medicine. We review well-established applications, such as diagnostic sequencing, as well as emerging applications of GS such as predisposition screening for rare variants associated with Mendelian disorders, and disease risk prediction from common genetic variation. Genotyping of neoplasms to individualize cancer treatments and sequencing of cell-free DNA from maternal blood for detection of fetal aneuploidy (eg, trisomy 18 or trisomy 21) and structural rearrangements are also widely used applications of massively parallel sequencing.4-8 However, for the purposes of this review, we will focus on applications to germ line genomics in children and adults because many issues in somatic genomic and prenatal sequencing are unique and these topics have been well explored elsewhere.4-7
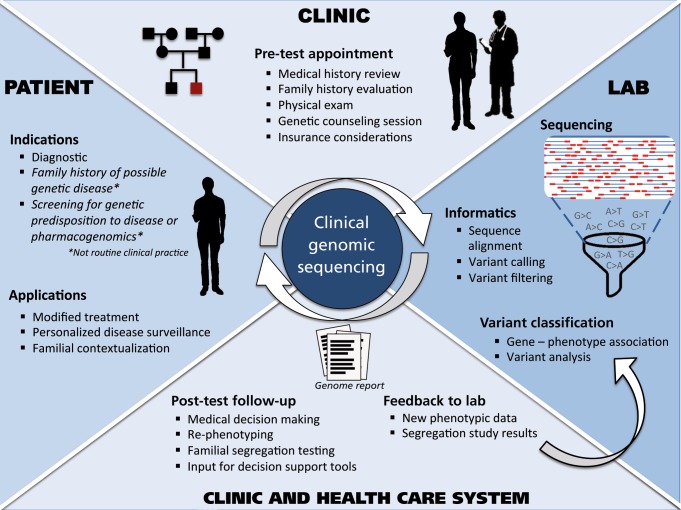
We also review key challenges facing the ongoing implementation of GS, with a focus on preliminary lessons from the MedSeq Project, the first randomized clinical trial to integrate whole-genome sequencing into the practice of medicine for apparently healthy individuals and individuals with disease.9 Throughout this review of applications and challenges, we discuss different components of the clinical GS workflow. A more holistic view of clinical GS is depicted in (Figure 2) Finally, we consider potential future directions, as the newfound availability of genomic and other “omic” data is catalyzing the genesis of “precision medicine” and energizing a trend toward individuals seeking their own genomic data.
Figure 2. A schematic of the workflow of clinical genomic sequencing.
Current and emerging clinical applications of GS
Diagnostic sequencing
To date, the diagnosis of rare Mendelian disease has been the primary clinical application of sequencing the genomes of individual patients. Thousands of pathogenic mutations identified through GS have been reported in recent years, and novel gene-disease associations are proliferating.10-12 Early reports on clinical GS demonstrated that identification of a causative mutation through GS can help to formulate a treatment plan and in other cases offer new opportunities for reproductive planning, as in the first publication reporting a successful diagnosis via GS, which resulted in effective management for a severe autoimmune illness in a young boy.13 Diagnostic GS is indicated for the detection of diagnostic genetic variants in patients with suspected monogenic disorders after known single-gene candidates have been eliminated from consideration or when a multigene testing panel has not yielded a diagnosis. The vast majority of diagnostic GS to date has been performed in children. However, patients can be of any age and presentations of Mendelian disorders in adulthood are probably underrecognized.14
The breadth of possible results from GS requires that thorough counseling and evaluation be performed before ordering GS to ensure proper interpretation of genomic variants, as well as careful clinical contextualization of the results. This process should include gathering detailed family history information, systematically evaluating the patient's and/or family's phenotype, reviewing medical literature and databases for possible overlap with known syndromes or implicated biochemical pathways, and obtaining informed consent. Individuals who consent to clinical GS should be aware that they may learn about disease risks that may also affect their relatives.15
Whereas many clinical molecular laboratories in academic medical centers and commercial laboratories now offer exome sequencing, Baylor College of Medicine and the University of California Los Angeles (UCLA) in the United States have reported on the largest number of clinical sequencing cases and have estimated that they find a causative mutation in 25% to 26% of cases overall,16-18 with lower diagnostic rates for adults than for children.14 Of solved cases, a surprising percentage (4.6%) appear to result from blended phenotypes of two separate Mendelian disorders, each associated with distinct pathogenic variants.18 The combined impact of two distinct Mendelian disease variants often leads to a hybrid phenotype that appears unique and challenging to diagnose.
The application of GS to rare disease has understandably been of intense research interest. In the United States, several National Institutes of Health (NIH) grant programs, including the Clinical Sequencing Exploratory Research (CSER) Consortium, the Centers for Mendelian Genomics, and the Undiagnosed Diseases Program and Network,19-22 have been funded to investigate the application of GS to the diagnosis of rare diseases. The scope of these efforts is broad and includes establishing technical standards for GS and interpretative pipelines (ie, variant filtration algorithms and interpretation protocols), developing and implementing reporting mechanisms, and evaluating the clinical, behavioral, legal, and ethical impacts of GS on clinical practice.
Emerging application: preconception carrier screening
Although targeted carrier screening is well established (eg, focused carrier screening for conditions such as Gaucher, Tay-Sachs, and Canavan disease in individuals of Ashkenazi Jewish descent), genomic technologies offer the opportunity for broader, more comprehensive screening. Preconception screening for carrier variants associated with rare, recessive disorders has been increasingly available in recent years via targeted multiplex genotyping that screens for known mutations in dozens of genes.23 These tests do not necessarily detect extremely rare or novel genetic variants that an unaffected individual may carry, and therefore a “residual risk” of being a carrier remains after negative testing.
Several companies now offer GS for preconception screening. GS affords the opportunity to go beyond a selected subset of recessive disorders to evaluate and report on genes associated with extremely rare recessive conditions.24 Preliminary data from the MedSeq Project,9 which reports results on carrier variants in any gene associated with known autosomal recessive disorders, suggest that approximately 90% of individuals in the general population are carriers for at least one recessive disorder and that most carry two to four carrier variants.25 Due to imperfect coverage of some genes and the low sensitivity of GS for certain types of genetic variation (reviewed below), a negative result on GS does not eliminate the post-test probability of being a carrier, though it generally improves upon the existing residual risk of mutation panel-based approaches.
Discovering that reproductive partners are each carriers for a severe recessive condition enables preimplantation genetic diagnosis (PGD). PGD allows for testing of embryos for a specific genetic variant (or variants, in the case of recessive diseases). Embryos lacking the targeted genetic variants are then implanted, preventing transmission of the genetic disease to offspring. PGD is a complicated and controversial topic both technically and ethically, and has been reviewed thoroughly elsewhere.26
Emerging application: genetic predisposition screening
Several research studies and personal genomics companies have begun to report a broad range of predispositional Mendelian variants to individuals. The general goal of these initiatives is to provide genetically informed predictions of disease risk and medication safety and efficacy, thereby enabling participants to make personalized decisions for disease prevention. Although preliminary data has not demonstrated significant risk of harm, benefits have not been systematically evaluated, and many experts and professional organizations call for caution before adopting GS for generally healthy individuals.27,28 To this end, the PeopleSeq (Personal Genome Sequencing Outcomes) Consortium has been formed as the first systematic large-scale longitudinal study of outcomes of predisposition sequencing and will seek to collect short- and long-term data on participants in GS projects.29
Monogenic variants for Mendelian syndromes that confer a significant risk for a condition, such as the breast cancer susceptibility gene 1 and 2 (BRCA1/2) variants associated with breast and ovarian cancer, may be revealed in GS of persons without a personal or family history. In current clinical practice, these findings are discovered secondary to diagnostic sequencing and are routinely reported for selected genes believed to be clinically actionable. However, in predisposition screening, these variants are a primary finding. Using strict variant-filtering criteria and all genes associated with human disease, the MedSeq Project identified a monogenic variant in 21 out of 100 participants. Identification of these variants has enabled MedSeq physicians to perform deep phenotyping (targeted medical examination and assessment for manifestations of the associated conditions) of asymptomatic individuals with monogenic variants.25
GS can identify common genetic variants that have been associated with risk for complex phenotypes, such as coronary artery disease and type 2 diabetes, in genome-wide association studies (GWAS). Millions of individuals have undergone genotyping for such variants via direct-to-consumer services such as 23andMe, which have utilized chip arrays that identify genotypes at specific single-nucleotide polymorphisms (SNPs). Because many variants identified in GWAS reside outside of exons (protein-coding regions of the genome), such SNPs would not be detectable by exome sequencing. Therefore, with regard to utilizing GS to identify these variants, wholegenome sequencing, instead of exome sequencing, is required. Despite the availability of relevant data from GS and the broad reporting of common disease risks by personal genomic testing companies, there is limited evidence for the clinical validity or utility of risk assessments from common genetic variation. GWAS variants account for a small proportion of variability in the risk of multifactorial phenotypes, known as the “missing heritability” problem (ie, other as yet unidentified genetic factors or interactions between genetic variants must contribute to the heritability of diseases).30 Additionally, risk-assessment methodologies to combine multiple variants remain in flux, and reclassification of individuals from higher risk to average or lower risk is expected to occur in most phenotypes as additional data accrue.31 Nevertheless, some studies have shown that individuals make positive lifestyle changes and become more engaged in their care after receiving such risk predictions.32,33
Utilizing known associations between genetic variants and blood group and antigen subtypes, GS can be used to predict clinically relevant hematological data, such as blood group and platelet antigen types. Antigen subgroup status has potential relevance for individuals who require multiple transfusions secondary to a chronic medical condition, as well as for identifying potential donors who have rare blood group antigens. The analytical algorithms have been developed and validated as part of the MedSeq Project.34
Finally, GS is a powerful tool to screen for multiple pharmacogenomic variants simultaneously, creating the opportunity for personalized medication selection and dosing regimens based on an individual's genotype or haplotype (group of genes inherited together). Pharmacogenomic data offer the opportunity for querying genomic data at the point of care as patients are prescribed medications for the first time and new associations among drugs, genetic variants, and dosing requirements or side effect risks are discovered and validated.35 The topic of pharmacogenomics will be explored more comprehensively in two companion articles in this special issue (p 313 and p 323).
Exploring challenges to the clinical implementation of GS
Standardization of variant classification
Analytic pipelines used to interpret GS after variants have been identified from the raw sequencing data involve two key steps: filtering and analysis. Filtering determines which variants are given further scrutiny for potential reporting. Common metrics used to filter variants include the quality of the variant calls (differences identified between the sequence under study and a reference genome); frequency of the variant in reference population databases, such as the Exome Aggregation Consortium (Ex AC); and its presence or absence in databases of variants believed to be disease causing, such as the Human Gene Mutation Database (HGMD) or ClinVar.36,37 In candidate gene approaches, gene lists are developed before variant filtering and include only those genes known to be associated with the phenotype of interest. Although potentially missing causative variants in genes not previously associated with disease (ie, novel genes), it allows for dedicated analysis of a smaller number of variants identified in those genes of interest.
Variant analysis incorporates a wide range of data, including that from databases of pathogenic variants, allele frequency from population databases, segregation within identified families, evolutionary conservation, and the predicted impact of the genetic variant on the protein.36 The analysis of genetic variants has received considerable attention, in part due to recognized inconsistencies among clinical molecular laboratories.38,39 Recent recommendations from an expert working group integrate multiple lines of supporting evidence into criteria for classifying variants in one of five categories: pathogenic, likely pathogenic, variant of uncertain significance, likely benign, or benign. Although these guidelines represent a significant revision from the previous version, the practice of variant classification remains challenging to standardize.40
Secondary findings
One of the most pressing concerns in clinical genomics has been incidental or secondary findings (SFs), defined as genomic variants of potential medical relevance unrelated to the medical reason for ordering the test.41 This situation is recognized to exist in many specialties; for example, an unexpected adrenal mass discovered in neuroimaging labeled as an incidentaloma. After clinical GS was made available in late 2011, the clinical community recognized a need for guidance on SFs, and recommendations were released by the American College of Medical Genetics and Genomics (ACMG) in 2013 that encouraged the evaluation and reporting of pathogenic or likely pathogenic variants in 56 genes (“ACMG 56”) on all patients for whom GS is ordered, on the basis of presumed clinical utility.42 A working group of medical and laboratory geneticists and genetic counselors decided on principles to guide the selection of genes to recommend reporting as SFs and finalized the list through a process of working group consensus, external review, and final approval by the ACMG Board of Directors.43 In these original recommendations, the ACMG recognized a distinction in the balance of benefit-risk-cost between opportunistic screening after a GS has already been ordered and performed (low cost) versus population screening in which the evidence of benefit would need to be high in order to justify the expense of GS.44,45 The ACMG 56 includes genes and conditions for which there are known interventions that have been proven to improve outcomes and which can be reliably detected using current sequencing technologies. Neuropsychiatric disorders are not currently included on the list as they generally do not have a preventive or curative treatment, and many neurologic disorders are caused by triplet repeat expansions, which are often missed on GS. However, a process for updating the ACMG SF list has been established, and thus the list may evolve as the capabilities of GS technologies advance and as prevention/treatment for neuropsychiatric disorders are developed.
The recommendations to report SFs from the ACMG 56 without seeking a patient's preference to receive them and to report SFs for adult-onset conditions when GS is ordered for minors were controversial and spurred debate among clinical geneticists and medical ethicists. The ACMG reviewed feedback and subsequently modified the original recommendations by calling for an “opt-out” option.46 The recommendation to report the same set of SFs for both children and adults was preserved. Most clinical laboratories now report SFs unless a patient opts out and report variants from genes well beyond the ACMG 56.
Genomics education for nongeneticist clinicians
Many experts have expressed concern over the limited numbers of clinical geneticists and genetic counselors, as well as the lack of genomics knowledge among practicing clinicians. In addition to barriers, such as the high cost of genomic assays and the uncertain utility of some genetic and genomic tests,47 many providers lack confidence in using genomic results in clinical decision making.48,49 One proposed solution, based on the premise that providers who undergo genetic testing themselves will have enhanced engagement and knowledge of genomics, is to incorporate educational programs in which clinicians-in- training receive and interpret their own genomic data.50 However, the impact of initial efforts have been unclear,51-53 and one institution explored but rejected such a project based on safety and ethical concerns.54
The MedSeq Project enrolled both primary care physicians and cardiologists, along with 200 of their current patients, 100 of whom received clinical GS.9,22 A primary goal of the MedSeq Project is to study how nongeneticist physicians interpret results from GS and incorporate these results into clinical management. Before returning GS reports to their patients, physicians received a 6-hour introduction to genomics concepts and types of results being reported. Before receiving genome reports, primary care physicians were worried about their genomic knowledge, and cardiologists were concerned about how to interpret specific types of results and SFs.55
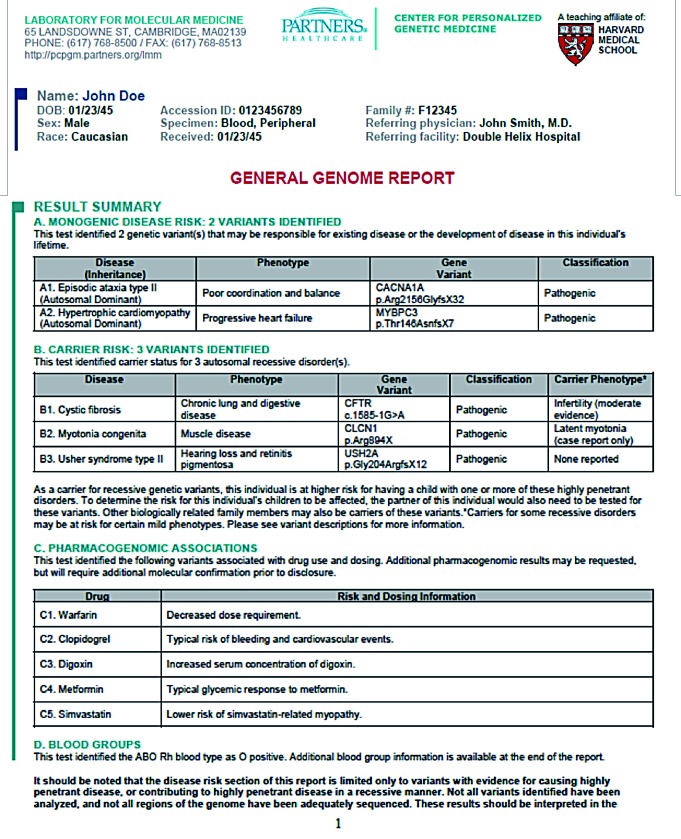
As the intervention of interest, physicians received WGS reports for a sample of their patients. Physicians viewed example reports (Figure 3) during the inperson education sessions, and they learned that they could seek assistance from a Genome Resource Center (GRC), consisting of genetic counselors and medical geneticists. The study team also created an online repository for educational materials.
Figure 3. Example of a MedSeq Genome Report (first page only).
Preliminary results from the MedSeq Project suggest that, with a few exceptions, most GS results were disclosed to patients without safety concerns. No errors deemed “high-risk” to participants, such as an inappropriate recommendation for an invasive procedure or diagnostic test, have yet been identified during the project.56 There have been a small number of errors deemed “moderate risk,” related to insufficient explanation of reproductive risks associated with variants conferring carrier status for relatively common autosomal recessive conditions, such as cystic fibrosis. However, collectively, our experience suggests that nongeneticist physicians quickly adapt to receiving GS results, as physicians have done with other major advancements in clinical care.57 For those who desire additional education about genomic information and its clinical applications, several online resources have been developed (Table I).58
TABLE I. Online genomic information resources. CME, continuing medical education; GWAS, genome-wide association study.
| Category | Organization | Resource name | Resource description | Resource location |
| National Center for Biotechnology Information (NCBI) | ClinVar | A public archive of reports of the relationships among human variations and phenotypes along with supporting evidence. | http://www.ncbi.nlm. nih.gov/clinvar/ | |
| National Institutes of Health (NIH) | Clinical Genome Resource (Clin- Gen) | A resource that defines the clinical relevance of genes and variants. It provides standards, guidance, education resources, and a patient registry. | https://www.clinicalgenome. org/ | |
| National Center for Biotechnology Information (NCBI) | Online Mendelian Inheritance in Man (OMIM) | A compendium of human genes and genetic phenotypes. | http://www.ncbi.nlm. nih.gov/omim | |
| National Center for Biotechnology Information (NCBI) | Genetic Testing Registry | A central location for voluntary submission of genetic test information by laboratories. Includes GeneReviews, a searchable database of expert-authored and peerreviewed disease descriptions. | http://www.ncbi.nlm. nih.gov/gtr/ | |
| Databases/ directories | Pharmacogenomics Knowledgebase (PharmGKB) | PharmGKB database | A searchable database of human genetic variations on drug response, including primary genotype and phenotype data, annotated gene variants, and literature reviews of gene-drug-disease relationships. | http://www.pharmgkb. org |
| Public Health Partners | Links to genomic projects/ information | Overview of resources with links at the federal and international level regarding Public Health Genomics | http://phpartners.org/ public_health_genomics. html | |
| SNPedia | SNPedia | A wiki site that shares information about the effects of variations in DNA, citing peer-reviewed scientific publications. | http://snpedia.com/index. php/SNPedia | |
| Vanderbilt-Ingram Cancer Center | My Cancer Genome | Searchable database of cancer mutations that inform tumor-specific treatment and prognosis. | http://www.mycancergenome. org/ | |
| Professional education | Centers for Disease Control and Prevention (CDC) | Public Health Genomics | The Office of Public Health Genomics hosts and links to a variety of genomic resources for health care providers, including a weekly update, blog, podcasts, A-Z resource guide, and reports and publications. | http://www.cdc.gov/ genomics/ |
| National Human Genome Research Institute | Current Topics In Genome Analysis | A lecture series covering contemporary topics in genomics and bioinformatics. | https://www.genome. gov/12514286/currenttopics- in-genome-analysis- 2016/ | |
| National Society of Genetic Counselors | NSGC 2015 Online Course: Personalized and Precision Medicine | A series of 10 prerecorded presentations by leaders in the personalized and precision medicine community. | http://nsgc.org/p/cm/ld/ fid=378 | |
| Scripps Translational Science Institute | Scripps Genomics Primer | A multifaceted genomics learning tool, including interactive tutorials, a genomics glossary, links to online resources, a recommended reading list, and slide sets covering topics such as sequencing, GWAS, cancer genomics and pharmacogenomics. | http://www.stsiweb. org/index.php/education_ training/primer/ | |
| University of Utah | Genetic Science Learning Center | Educational modules on various genetic topics, including pharmacogenomics, genetic variation, and other basic topics. | http://learn.genetics. utah.edu/ | |
| Counseling and teaching tools | National Coalition for Health Professional Education in Genetics (NCHPEG) | Educational aids and Point-ofcare tools | Educational resources as well as point-ofcare clinical tools for both genetics and nongenetics health professionals (eg, GeneFacts). Includes Medicine's Future: a Genomics Curriculum for Clinicians program that can be hosted for CME credit. | http://www.nchpeg.org/ index.php |
| Wellcome Trust Sanger Institute | yourgenome. org | Online educational tools, including animations, activities, and a glossary | http://www.yourgenome. org/ |
Clinical utility and cost-effectiveness
The limited evidence base to establish clinical utility and cost-effectiveness for most GS applications poses a key challenge to their adoption. The clinical utility of an intervention refers to its benefit to patients' health and its ability to provide information that will guide diagnosis, medical management, and/or disease prevention. The degree of evidence required to establish clinical utility may depend on the context and purpose for which GS is being performed. A stronger evidence base may be required for diagnostic or therapeutic applications than for personal uses (eg, risk assessment for common complex diseases vs recreational applications such as predicting nonmedical traits; see Figure 1).59
Clinical utility as defined by insurance companies is generally limited to predictable, direct medical benefits. However, a position statement from the ACMG asserts that GS may provide value beyond strictly medical implications; for example, GS results that solve a complex diagnostic puzzle may provide immense psychological value to patients and families.60 In diagnostic applications, elucidating a molecular diagnosis may provide clinical utility by predicting prognosis, anticipating future symptoms, enabling early intervention, and identifying treatment possibilities, while avoiding inappropriate interventions. Diagnostic GS on a child may have clinical utility for the child's parents, who may utilize the information for reproductive planning. The clinical utility of GS may extend to at-risk family members who learn of a genetic risk indirectly through the proband. Society may also benefit from a more thorough understanding of genomic contributions to disease risk and penetrance of specific genomic variants. A semiquantitative approach to estimating clinical utility was developed by Berg and colleagues.61 It assigns scores to the severity and likelihood of disease, the efficacy and burden of the intervention, and the knowledge base about the aforementioned components in order to generate a single “actionability” score for each combination of outcome-intervention pair associated with a gene and phenotype (for example, breast cancer surveillance or breast cancer mastectomy for individuals with pathogenic variants in the BRCA1 gene associated with hereditary breast and ovarian cancer).62
Limitations in our current understanding of penetrance, the clinical significance of mild presentations of genetic diseases, interactions between multiple genetic variants, and gene-environment interactions have made it difficult to establish clinical utility. Although certain genetic disorders are associated with a single gene with large effect, most are caused by a combination of genetic and environmental factors, many of which are not fully elucidated. As GS among healthy individuals becomes more widespread, variants once believed to be highly penetrant are identified in asymptomatic individuals,63 raising questions about the degree to which prophylactic measures will prove cost-effective by avoiding the need to treat a disease that could otherwise develop in the future. Thus, the clinical utility of most SFs is limited by our inability to predict the likelihood that an individual with a specific genomic variant will develop the associated disease.
Though few cost-effectiveness studies have been conducted in germ line genomics, early evidence suggests that GS may be cost-effective for diagnosing neurodevelopmental disorders and Lynch syndrome.64-66 SFs from diagnostic GS, and the application of GS as a screening tool among healthy populations, have raised concern that such findings will initiate superfluous tests, exams, procedures, and screening. However, if identification of a pathogenic variant leads to improved preventive measures or more efficacious treatments, there may be long-term cost savings in averting disease, improving symptomatology, or curing the condition, compared with the expense that would have been required to treat the disease over a longer term. Results of initial modeling indicate that return of SFs may be cost-effective in diagnostic settings, but not yet for population screening.67
Preliminary findings from the MedSeq Project provide data on costs accrued in the first 6 months after GS in generally healthy adults and adults with cardiomyopathy. The details of these cost calculations have been described in depth.68 Briefly, among the first 103 cases analyzed 6 months after results disclosure, there was not a significant difference in the median cost of follow-up care among ostensibly healthy participants who received GS, compared with healthy participants who received standard care. Future analyses will incorporate costs generated outside of the health care system (eg, missed work due to health care appointments), quality of life, and longer-term medical followup.69
Insurance discrimination, emotional impact of receiving sequencing results, and ethical/legal/social implications (ELSI)
Concerns about risks to privacy and confidentiality surround many types of health information, particularly genomic information, which may be inherently identifiable.70,71 The fear of discrimination by insurance companies and employers has historically been a barrier to obtaining presymptomatic genetic and genomic testing. US legislation known as the Genetic Information Nondiscrimination Act of 2008 (GINA) prohibits discrimination in health insurance and employment decisions on the basis of genetic information; however, federal protection does not extend to other types of insurance, and laws vary by country. Thus, individuals undergoing GS may wish to secure a life insurance policy (and potentially long-term care and/or disability insurance) before pursuing GS.
The potential for distress caused by learning GS risk information may depend on the specific disease, availability of preventive and/or treatment approaches, and whether or not the information was expected. Starting with the availability of predictive genetic testing for Huntington disease, there have been concerns among the medical genetics and bioethics communities about the emotional impact of learning about one's genetic disease risks, which typically cannot be mitigated by behavioral or medical approaches. The psychological impact of receiving genetic risk information has been assessed in a series of randomized trials that disclosed participants' risk for late-onset Alzheimer disease based on genotype and other factors72,73; other studies have used GWAS associations to provide risk for a variety of common complex diseases.74,75 Participant responses after receiving this information suggest that learning of a heightened risk for these diseases does not have a long-term impact on anxiety or depression.72,73,76-79
The ethical, legal, and social implications (ELSI issues) of GS warrant special consideration for minors. Professional societies, including ACMG and the American Society of Human Genetics (ASHG),80,81 have published position statements recommending against screening minors for genetic predisposition to adult-onset conditions. However, GS presents a different set of considerations; though GS may be performed for a diagnostic indication, SFs may include genetic variants associated with adultonset diseases, and each of those variants is probably inherited from a parent. Such results may be of immediate medical benefit to the parent who, in the absence of a personal or family history of the disease, may have been unaware of his or her risk. The child stands to benefit indirectly if the parent's health and lifespan are extended through knowledge of a genetic disease risk. Thus, although returning SFs on a child removes the child's autonomy to decide whether or not to learn about these risks when she reaches the age of majority, professional guidelines80,81 and physician-researchers in bioethics and medical genetics82 have recommended offering parents actionable SFs for adult-onset conditions on their children who undergo clinical GS.
Future directions and conclusion
Reimagining electronic medical record systems
The potential of DNA to inform medical decisions has been portrayed with a “book of life” analogy, namely that an individual's genomic sequence may be viewed as a life -long guide that clinicians could call upon for information when making currently unanticipated decisions, such as drug selection.3 Now that increasingly large numbers of individuals have undergone genomic sequencing, integration of genomic data with electronic medical records (EMRs) is a largely unsolved issue outside of pilot programs or research consortia, such as the eMERGE (electronic MEdical Records and GeEnomics) Network,83 in which health care organizations, including ours,84 are linking genomic variation to EMR data to assess penetrance and disease expression, as well as to explore medical, behavioral, and economic outcomes of integrating genomic risk variants into the EMR. With the development of relatively nimble and interoperable computing platforms, such as SMART (Substitutable Medical Applications, Reusable Technologies) or FHIR (Fast Healthcare Interoperability Resource), that facilitate integration of external applications and existing EMRs, there is hope and an increasing mandate to develop a medical record system that supports genomic data.
Next “omics”
The application of next-generation sequencing (or massively parallel sequencing) technologies to RNA/expression analysis and the gastrointestinal microbiome, and the emergence of large-scale techniques such as metabolomic profiling and proteomics analysis, have laid the foundation for integrating multiple types of data into a more complete understanding of molecular pathophysiology. Although the potential of such approaches has been illustrated via the deep study of individuals or small cohorts,85 clinical applications of such techniques thus far have been relatively limited. Though exciting, the diversity and scale of these new data have the potential to amplify challenges outlined above, such as physician preparedness and interpretative validity.
Patients lead the way
Through our research initiatives on personal genomic testing and the motivations and outcomes of individuals who pursue it, it is clear that many individuals view genomic data as an opportunity to inform and shape their personal narratives and influence behavioral and health care decisions about their existing medical problems and future well-being.86-88 Although the evidence base for making decisions is currently limited for most types of genomic results, and evidence to date has been collected among cohorts that are not representative of the general population, these data may be informative for the adoption of future technological innovations in the realm of precision medicine.
Conclusion
The development and adoption of massively parallel sequencing has facilitated a dramatic change in the practice of select specialties, including clinical genetics, oncology, and obstetrics, and is increasingly impacting the practices of nongeneticist clinicians. Members of the public are independently pursuing genomic data to inform their own health and well-being decisions. The rapid pace of adoption has created challenges related to both the validity of new clinical tests, as well as for the incorporation of genomic data into clinical decision making, and a need to assess the impacts on health care systems. Clinical GS, therefore, faces a pivotal moment, as the vast potential for access to new quantities and types of data enable further clinical innovation at the same time that complicated questions produced by the first wave of applications continue to be resolved. Thoughtful and rigorous translational investigation remains critically necessary to evaluate the outcomes of GS, as well as to facilitate and achieve the potential of GS to positively impact the health of patients and the public at large.
Acknowledgments
This work was supported by the following National Institutes of Health (NIH) grants: T32GM007748, U01HD006500, U01HG007690, U19HD077671, U41HG006834, and U01 HG008685. Dr Krier has no disclosures. Ms Kalia has received compensation for consulting from Helix and SoundRocket. Dr Green has received compensation for speaking and for advisory services from lllumina. Prudential, Helix, AIA, Invitae, and Roche.
Contributor Information
Joel B. Krier, Genomes2People Research Program, Division of Genetics, Department of Medicine, Brigham and Women's Hospital, Boston, Massachusetts, USA; Harvard Medical School, Boston, Massachusetts, USA.
Robert C. Green, Genomes2People Research Program, Division of Genetics, Department of Medicine, Brigham and Women's Hospital, Boston, Massachusetts, USA; Harvard Medical School, Boston, Massachusetts, USA; Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA.
REFERENCES
- 1.Feero W., Guttmacher A., Collins F. Genomic medicine — an updated primer. N Engl J Med. 2010;362(21):2001–2011. doi: 10.1056/NEJMra0907175. [DOI] [PubMed] [Google Scholar]
- 2.Guttmacher AE., McGuire AL., Ponder B., Stefansson K. Personalized genomic information: preparing for the future of genetic medicine. Nat Rev Genet. 2010;11(2):161–165. doi: 10.1038/nrg2735. [DOI] [PubMed] [Google Scholar]
- 3.Collins FS., Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372(9):793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schweiger MR., Barmeyer C., Timmermann B. Genomics and epigenomics: new promises of personalized medicine for cancer patients. Brief Funct Genomics. 2013;12(5):411–421. doi: 10.1093/bfgp/elt024. [DOI] [PubMed] [Google Scholar]
- 5.Pant S., Weiner R., Marton MJ. Navigating the rapids: the development of regulated next-generation sequencing-based clinical trial assays and companion diagnostics. Front Oncol. 2014;4:78. doi: 10.3389/fonc.2014.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gagan J., Van Allen EM. Next-generation sequencing to guide cancer therapy. Genome Med. 2015;7(1):80. doi: 10.1186/s13073-015-0203-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hui L., Bianchi DW. Recent advances in the prenatal interrogation of the human fetal genome. Trends Genet. 2013;29(2):84–91. doi: 10.1016/j.tig.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Vugt S., Broekhuizen L., Zuithoff N., et al Incidental chest radiographic findings in adult patients with acute cough. Ann Fam Med. 2012;10(6):510–515. doi: 10.1370/afm.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vassy JL., Lautenbach DM., McLaughlin HM., et al The MedSeq Project: a randomized trial of integrating whole genome sequencing into clinical medicine. Trials. 2014;15:85. doi: 10.1186/1745-6215-15-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilissen C., Hoischen A., Brunner HG., Veltman JA. Unlocking mendelian disease using exome sequencing. Genome Biol. 2011;12(9):228. doi: 10.1186/gb-2011-12-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzaga-Jauregui C., Lupski JR., Gibbs RA. Human genome sequencing in health and disease. Annu Rev Med. 2012;63:35–61. doi: 10.1146/annurev-med-051010-162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deciphering Development Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519(7542):223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Worthey EA., Mayer AN., Syverson GD., et al Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13(3):255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 14.Posey JE., Rosenfeld JA., James RA., et al Molecular diagnostic experience of whole-exome sequencing in adult patients. Genet Med. 2016;18(7):678–685. doi: 10.1038/gim.2015.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ACMG Board of Directors Points to consider for informed consent for genome/exome sequencing. Genet Med. 2013;15(9):748–749. doi: 10.1038/gim.2013.94. [DOI] [PubMed] [Google Scholar]
- 16.Lee H., Deignan JL., Dorrani N., et al Clinical exome sequencing for genetic identification of rare mendelian disorders. JAMA. 2014;312(18):1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y., Muzny DM., Reid JG., et al Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369(16):1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y., Muzny DM., Xia F., et al Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312(18):1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bamshad MJ., Shendure JA., Valle D., et al The Centers for Mendelian Genomics: a new large-scale initiative to identify the genes underlying rare mendelian conditions. Am J Med Genet A. 2012;158A(7):1523–1525. doi: 10.1002/ajmg.a.35470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gahl WA., Mulvihill JJ., Toro C., et al The NIH Undiagnosed Diseases Program and Network: applications to modern medicine. Mol Genet Metab. 2016;117(4):393–400. doi: 10.1016/j.ymgme.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gahl WA., Wise AL., Ashley EA. The Undiagnosed Diseases Network of the National Institutes of Health: a national extension. JAMA. 2015;314(17):1797–1798. doi: 10.1001/jama.2015.12249. [DOI] [PubMed] [Google Scholar]
- 22.Green RC., Goddard KA., Jarvik GP., et al Clinical Sequencing Exploratory Research Consortium: accelerating evidence-based practice of genomic medicine. Am J Hum Genet. 2016;98(6):1051–1066. doi: 10.1016/j.ajhg.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nazareth SB., Lazarin GA., Goldberg JD. Changing trends in carrier screening for genetic disease in the United States. Prenat Diagn. 2015;35(10):931–935. doi: 10.1002/pd.4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umbarger MA., Kennedy CJ., Saunders P., et al Next-generation carrier screening. Genet Med. 2014;16(2):132–140. doi: 10.1038/gim.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blout CL., Rehm HL., McGuire AL., et al The MedSeq Project: exploring the integration of whole genome sequencing into the practice of medicine. Paper presented at: Festival of Genomics; June 22-24, 2015 Boston, MA. [Google Scholar]
- 26.Brezina PR., Kutteh WH. Clinical applications of preimplantation genetic testing. BMJ. 2015;350:g7611. doi: 10.1136/bmj.g7611. [DOI] [PubMed] [Google Scholar]
- 27.Zierhut H., McCarthyVeach P., LeRoy b. Canaries inthecoal mine: personal and professional impact of undergoing whole genome sequencing on medical professionals. Am J Med Genet A. 2015;167A(11):2647–2656. doi: 10.1002/ajmg.a.37262. [DOI] [PubMed] [Google Scholar]
- 28.Lewis KL., Hooker GW., Connors PD., et al Participant use and communication of findings from exome sequencing: a mixed-methods study. Genet Med. 2016;18(6):577–583. doi: 10.1038/gim.2015.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linderman MD., Nielsen DE., Green RC. Personal genome sequencing in ostensibly healthy individuals and the PeopleSeq Consortium. J Pers Med. 2016;6(2). doi:10.3390/jpm6020014. doi: 10.3390/jpm6020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456(7218):18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 31.Krier J., Barfield R., Green R., Kraft P. Reclassification of genetic-based predictions as GWAS data accumulate. Genome Med. 2016;8(1):20. doi: 10.1186/s13073-016-0272-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chao S., Roberts JS., Marteau TM., Silliman RA., Green RC. Health behavior changes after genetic risk assessment for Alzheimer disease: the REVEAL Study. Alzheimer Dis Assoc Disord. 2008;22(1):94–97. doi: 10.1097/WAD.0b013e31815a9dcc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kullo IJ., Jouni H., Austin EE., et al Incorporating a genetic risk score into coronary heart disease risk estimates: effect on low-density lipoprotein cholesterol levels (the MI-GENES Clinical Trial). Circulation. 2016;133(12):1181–1188. doi: 10.1161/CIRCULATIONAHA.115.020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lane WJ., Westhoff CM., Uy JM., et al Comprehensive red blood cell and platelet antigen prediction from whole genome sequencing: proof of principle. Transfusion. 2016;56(3):743–754. doi: 10.1111/trf.13416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiita AP., Schrijver I. Clinical application of high throughput molecular screening techniques for pharmacogenomics. Pharmgenomics Pers Med. 2011;4:109–121. doi: 10.2147/PGPM.S15302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duzkale H., Shen J., McLaughlin H., et al A systematic approach to assessing the clinical significance of genetic variants. Clin Genet. 2013;84(5):453–463. doi: 10.1111/cge.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLaughlin HM., Ceyhan-Birsoy O., Christensen KD., et al A systematic approach to the reporting of medically relevant findings from whole genome sequencing. BMC Med Genet. 2014;15:134. doi: 10.1186/s12881-014-0134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amendola LM., Jarvik GP., Leo MC., et al Performance of ACMGAMP variant-interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet. 2016;98(6):1067–1076. doi: 10.1016/j.ajhg.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rehm HL., Berg JS., Brooks LD., et al ClinGen — the Clinical Genome Resource. N Engl J Med. 2015;372(23):2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richards S., Aziz N., Bale SJ., et al Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green RC., Rehm HL., Kohane IS. Clinical genome sequencing. In: Ginsburg GS, Willard HF, eds. Genomic and Personalized Medicine. 2nd ed. San Diego, CA: Academic Press; 2013:102–122. [Google Scholar]
- 42.Green RC., Berg JS., Grody WW., et al ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15(7):565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Green RC., Berg JS., Berry GT., et al Exploring concordance and discordance for return of incidental findings from clinical sequencing. Genet Med. 2012;14(4):405–410. doi: 10.1038/gim.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Green RC., Lupski J., Biesecker LG. Reporting genomic sequencing results to ordering clinicians: incidental, but not exceptional. JAMA. 2013;310(4):365–366. doi: 10.1001/jama.2013.41703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berg JS., Adams M., Nassar N., et al An informatics approach to analyzing the incidentalome. Genet Med. 2013;15(1):36–44. doi: 10.1038/gim.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.ACMG Board of Directors ACMG policy statement: updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet Med. 2015;17(1):68–69. doi: 10.1038/gim.2014.151. [DOI] [PubMed] [Google Scholar]
- 47.Manolio TA., Chisholm RL., Ozenberger B., et al Implementing genomic medicine in the clinic: the future is here. Genet Med. 2013;15(4):258–267. doi: 10.1038/gim.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Powell KP., Cogswell WA., Christiansen CA., et al Primary care physicians' awareness, experience and opinions of direct-to-consumer genetic testing. J Genet Couns. 2012;21(1):113–126. doi: 10.1007/s10897-011-9390-9. [DOI] [PubMed] [Google Scholar]
- 49.Haga SB., Carrig MM., O'Daniel JM., et al Genomic risk profiling: attitudes and use in personal and clinical care of primary care physicians who offer risk profiling. J Gen Intern Med. 2011;26(8):834–840. doi: 10.1007/s11606-011-1651-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Demmer LA., Waggoner DJ. Professional medical education and genomics. Annu Rev Genomics Hum Genet. 2014;15:507–516. doi: 10.1146/annurev-genom-090413-025522. [DOI] [PubMed] [Google Scholar]
- 51.Sharp RR., Goldlust ME., Eng C. Addressing gaps in physician education using personal genomic testing. Genet Med. 2011;13(8):750–751. doi: 10.1097/GIM.0b013e318228821f. [DOI] [PubMed] [Google Scholar]
- 52.Salari K., Karczewski KJ., Hudgins L., Ormond KE. Evidence that personal genome testing enhances student learning in a course on genomics and personalized medicine. PLoS One. 2013;8(7):e68853. doi: 10.1371/journal.pone.0068853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanderson SC., Linderman MD., Kasarskis A., et al Informed decisionmaking among students analyzing their personal genomes on a whole genome sequencing course: a longitudinal cohort study. Genome Med. 2013;5(12):113. doi: 10.1186/gm518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walt DR., Kuhlik A., Epstein SK., et al Lessons learned from the introduction of personalized genotyping into a medical school curriculum. Genet Med. 2011;13(1):63–66. doi: 10.1097/GIM.0b013e3181f872ac. [DOI] [PubMed] [Google Scholar]
- 55.Christensen KD., Vassy JL., Jamal L., et al Are physicians prepared for whole genome sequencing? A qualitative analysis. Clin Genet. 2016;89(2):228–234. doi: 10.1111/cge.12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krier JB., Blout CB., D L., et al Communication and management of genomic sequencing results by non-geneticist physicians. Paper presented at: 65th Annual Meeting of the American Society of Human Genetics; October 6-1 5, 201 5; Baltimore, MD. [Google Scholar]
- 57.Vassy JL., Korf BR., Green RC. How to know when physicians are ready for genomic medicine. So'. Transl Med. 2015;7(287):287fs219. doi: 10.1126/scitranslmed.aaa2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Personalized Medicine Special Interest Group of the National Society of Genetic Counselors Expanding the practice of genomic counseling in personalized medicine: tools for genetic counselors. National Society of Genetic Counselors: Audrey Heimler Special Projects. 2011 . Available at: http:// nsgc.org/p/cm/ld/fid=286. Accessed July 2016. [Google Scholar]
- 59.Delaney SK., Hultner ML., Jacob HJ., et al Toward clinical genomics in everyday medicine: perspectives and recommendations. Expert Rev Mol Diagn. 2016;16(5):521–532. doi: 10.1586/14737159.2016.1146593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.American College of Medical Genetics and Genomics Clinical utility of genetic and genomic services: a position statement of the American College of Medical Genetics and Genomics. Genet Med. 2015;17(6):505–507. doi: 10.1038/gim.2015.41. [DOI] [PubMed] [Google Scholar]
- 61.Berg JS., Foreman AK., O'Daniel JM., et al A semiquantitative metric for evaluating clinical actionability of incidental or secondary findings from genome-scale sequencing. Genet Med. 2016;18(5):467–475. doi: 10.1038/gim.2015.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.ClinGen Actionability Working Group evidence-based summaries. Available at: https://clinicalgenome.org/working-groups/actionability/projects-initiatives/actionability-evidence-based-summaries/ Accessed June 30, 2016. [Google Scholar]
- 63.Chen R., Shi L., Hakenberg J., et al Analysis of 589,306 genomes identifies individuals resilient to severe mendelian childhood diseases. Wat. Biotechnol. 2016;34(5):531–538. doi: 10.1038/nbt.3514. [DOI] [PubMed] [Google Scholar]
- 64.Soden SE., Saunders CJ., Willig LK., et al Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci Transl Med. 2014;6(265):265–168. doi: 10.1126/scitranslmed.3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Snowsill T., Huxley N., Hoyle M., et al A systematic review and economic evaluation of diagnostic strategies for Lynch syndrome. Health Techno! Assess. 2014;18(58):1–406. doi: 10.3310/hta18580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Severin F., Stollenwerk B., Holinski-Feder E., et al Economic evaluation of genetic screening for Lynch syndrome in Germany. Genet Med. 2015;17(10):765–773. doi: 10.1038/gim.2014.190. [DOI] [PubMed] [Google Scholar]
- 67.Bennette CS., Gallego CJ., Burke W., Jarvik GP., Veenstra DL. The costeffectiveness of returning incidental findings from next-generation genomic sequencing. Genet Med. 2015;17(7):587–595. doi: 10.1038/gim.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Christensen KD., Dukhovny D., Siebert U., Green RC. Assessing the costs and cost-effectiveness of genomic sequencing. J Pers Med. 2015;5(4):470–486. doi: 10.3390/jpm5040470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dukhovny D., Christensen KD., Vassy JL., et al Short-term costs of integrating genome sequencing into clinical care: preliminary results from the MedSeq Project. Paper presented at: 65th Annual Meeting of the American Society of Human Genetics; October 6-15, 2015; Baltimore, MD. [Google Scholar]
- 70.Homer N., Szelinger S., Redman M., et al Resolving individuals contributing trace amounts of DNA to highly complex mixtures using highdensity SNP genotyping microarrays. PLoS Genet. 2008;4(8):e1000167. doi: 10.1371/journal.pgen.1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McGuire A., Gibbs R. No longer de-identified. Science. 2006;312(5772):370–371. doi: 10.1126/science.1125339. [DOI] [PubMed] [Google Scholar]
- 72.Roberts J., Cupples L., Relkin N., Whitehouse P., Green RC. Genetic risk assessment for adult children of people with Alzheimer's disease: the Risk Evaluation and Education for Alzheimer's Disease (REVEAL) Study. J Geriatr Psychiatr Neurol. 2005;18(4):250–255. doi: 10.1177/0891988705281883. [DOI] [PubMed] [Google Scholar]
- 73.Green RC., Roberts JS., Cupples LA., et al Disclosure of APOE genotype for risk of Alzheimer's disease. N Engl J Med. 2009;361(3):245–254. doi: 10.1056/NEJMoa0809578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carere DA., Couper MP., Crawford SD., et al Design, methods, and participant characteristics of the Impact of Personal Genomics (PGen) Study, a prospective cohort study of direct-to-consumer personal genomic testing customers. Genome Med. 2014;6(12):96. doi: 10.1186/s13073-014-0096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bloss CS., Ornowski L., Silver E., et al Consumer perceptions of direct-to-consumer personalized genomic risk assessments. Genet Med. 2010;12(9):556–566. doi: 10.1097/GIM.0b013e3181eb51c6. [DOI] [PubMed] [Google Scholar]
- 76.Cassidy MR., Roberts JS., Bird TD., et al Comparing test-specific distress of susceptibility versus deterministic genetic testing for Alzheimer's disease. Alzheimers Dement. 2008;4(6):406–413. doi: 10.1016/j.jalz.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chung WW., Chen CA., Cupples LA., et al A new scale measuring psychologic impact of genetic susceptibility testing for Alzheimer disease. Alzheimer Dis Assoc Disord. 2009;23(1):50–56. doi: 10.1097/wad.0b013e318188429e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ashida S., Koehly LM., Roberts JS., Chen CA., Hiraki S., Green RC. The role of disease perceptions and results sharing in psychological adaptation after genetic susceptibility testing: the REVEAL Study. Eur J Hum Genet. 2010;18(12):1296–1301. doi: 10.1038/ejhg.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bloss CS., Wineinger NE., Darst BF., Schork NJ., Topol EJ. Impact of direct-to-consumer genomic testing at long term follow-up. J Med Genet. 2013;50(6):393–400. doi: 10.1136/jmedgenet-2012-101207. [DOI] [PubMed] [Google Scholar]
- 80.Ross LF., Saal HM., David KL., et al Technical report: ethical and policy issues in genetic testing and screening of children. Genet Med. 2013;15(3):234–245. doi: 10.1038/gim.2012.176. [DOI] [PubMed] [Google Scholar]
- 81.ASHG issues position statement on genetic testing in children and adolescents: statement addresses scientific advances, ethical questions, and implementation [press release]. Bethesda, MD: ASHG Press; July 2, 2015 [Google Scholar]
- 82.Wilfond BS., Fernandez CV., Green RC. Disclosing secondary findings from pediatric sequencing to families: considering the “benefit to families”. J Law Med Ethics. 2015;43(3):552–558. doi: 10.1111/jlme.12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gottesman O., Kuivaniemi H., Tromp G., et al The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15(10):761–771. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smoller JW., Karlson EW., Green R., et al An eMERGE clinical center at Partners Personalized Medicine. J Pers Med. doi: 10.3390/jpm6010005. 2016;6(1) doi: 10.3390/jpm6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen R., Mias Gl., Li-Pook-Than J., et al Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148(6):1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baptista NM., Christensen KD., Carere DA., Broadley SA., Roberts JS., Green RC. Adopting genetics: motivations and outcomes of personal genomic testing in adult adoptees 2016 Jan 28. Epub ahead of print. doi:10.1038/gim. 2015. 192. Genet Med. doi: 10.1038/gim.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meisel SF., Carere DA., Wardle J., et al Explaining, not just predicting, drives interest in personal genomics. Genome Med. 2015;7(1):74. doi: 10.1186/s13073-015-0188-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Van der Wouden CH., Carere DA., Maitland-vander Zee AH., et al Consumer perceptions of interactions with primary care providers after direct-toconsumer personal genomic testing. Ann Intern Med. 2016;164(8):513–522. doi: 10.7326/M15-0995. [DOI] [PubMed] [Google Scholar]