Abstract
Enteropathogenic Escherichia coli (EPEC) are diarrhoeagenic E. coli, and are a significant cause of gastrointestinal illness among young children in developing countries. Typical EPEC are identified by the presence of the bundle-forming pilus encoded by a virulence plasmid, which has been linked to an increased severity of illness, while atypical EPEC lack this feature. Comparative genomics of 70 total EPEC from lethal (LI), non-lethal symptomatic (NSI) or asymptomatic (AI) cases of diarrhoeal illness in children enrolled in the Global Enteric Multicenter Study was used to investigate the genomic differences in EPEC isolates obtained from individuals with various clinical outcomes. A comparison of the genomes of isolates from different clinical outcomes identified genes that were significantly more prevalent in EPEC isolates of symptomatic and lethal outcomes than in EPEC isolates of asymptomatic outcomes. These EPEC isolates exhibited previously unappreciated phylogenomic diversity and combinations of virulence factors. These comparative results highlight the diversity of the pathogen, as well as the complexity of the EPEC virulence factor repertoire.
Enteropathogenic E. coli (EPEC) are a cause of moderate to severe diarrhoea in young children, primarily in developing countries1. The Global Enteric Multicenter Study (GEMS), an epidemiological study of children with moderate to severe diarrhoea and children with no diarrhoea, has demonstrated that EPEC is a leading cause of lethality associated with diarrhoea among children that are less than 12 months of age2,3. By definition, EPEC contain the locus of enterocyte effacement (LEE) pathogenicity island, which encodes a type III secretion system (T3SS) involved in the pathogenesis of these organisms4–7. The LEE region is a defining feature of the attaching and effacing E. coli (AEEC), which includes EPEC and the Shiga toxin-producing enterohaemorrhagic E. coli (EHEC), which are associated with severe food-related illness worldwide8–11. EPEC are further categorized by the presence or lack of the plasmid-encoded bundle-forming pilus genes (BFP)8,12, which are commonly found on the EPEC adherence factor (EAF) plasmid and confer localized adherence (LA) to the surface of intestinal epithelial cells13–16. The BFP operon is frequently identified in EPEC associated with diarrhoeal illness, and these isolates are termed typical EPEC (tEPEC)8,17. E. coli that possess the LEE region, but do not contain the BFP or Shiga toxin genes (LEE+/stx−/bfp−), are commonly termed atypical EPEC (aEPEC)17. Previous studies investigating the genetic diversity of aEPEC have demonstrated that LEE+/stx−/bfp− isolates are a diverse group that can include among them isolates that are more related to other E. coli pathovars and commensal isolates18,19. The aEPEC can also include EHEC and EPEC that have lost the Shiga-toxin genes and BFP genes during passage through a host or the environment or after culture in the laboratory18,19.
Investigation of the genetic and virulence factor diversity of tEPEC has focused mainly on isolates within two lineages, EPEC1 and EPEC220, as defined by multi-locus sequence typing (MLST)20. MLST and phylogenetic analysis have also described additional tEPEC lineages, EPEC3 and EPEC420, as well as EPEC5 and EPEC6, which comprise aEPEC isolates19, suggesting that there is probably greater genetic diversity among EPEC isolates than originally anticipated. Until the recent comparative genomic analysis of a collection of diverse AEEC isolates18, which included additional EPEC1, EPEC2 and the first EPEC4 genomes described, the genome sequences available for EPEC isolates were limited to E2348/69, B171, E22 (a rabbit EPEC isolate) and E110019 (an aEPEC isolate)21,22. Even with recent sequencing, the majority of the EPEC genomes sequenced are historical isolates from developed countries, and little is known regarding the genomic diversity of recent EPEC isolates from developing countries, where EPEC has been identified in the recent landmark GEMS analysis as an important pathogen of children, with tEPEC associated with the greatest amount of mortality2.
In the present study we sequenced the genomes and performed comparative genomic analysis of 70 EPEC isolates from children less than 5 years of age enrolled in GEMS2. Phylogenomic analysis of these 70 EPEC isolates highlighted the considerable evolutionary diversity and variability of EPEC virulence mechanisms in more recent EPEC isolates from developing countries. By comparing the genomes of 24 EPEC from lethal cases (LI), 23 EPEC from non-lethal symptomatic cases (NSI) and 23 EPEC from asymptomatic cases (AI), we identified the genes that are more frequently associated with EPEC from different clinical outcomes. Genomic studies such as this provide valuable insight into the diversity and virulence mechanisms of an E. coli pathogen that is associated with increased risk of death among infants in developing countries3. The findings of this study can be used to generate improved methods for molecular diagnostics of EPEC that will provide information regarding the evolutionary history of an isolate as previously described18. The genes that were identified as more frequently associated with lethal or symptomatic EPEC isolate genomes may be further characterized to obtain a deeper understanding of the EPEC pathogenesis and provide additional targets for vaccine and therapeutic development.
Results
Phylogenomic analysis of GEMS site EPEC isolates associated with different clinical outcomes
To investigate the genomic diversity and virulence mechanisms of EPEC isolated from individuals with differing clinical severity we sequenced the genomes of 70 EPEC from multiple geographic sites included in GEMS3. The 70 EPEC isolates were obtained from cases of diarrhoeal illness in children classified as LI or NSI, or as controls with asymptomatic (AI) outcomes. There were a total of 24 EPEC isolates from LI cases, 23 from NSI cases and 23 from AI cases. The 24 EPEC isolates from LI cases were all tEPEC, and 20 of 23 (87%) of the EPEC from NSI cases and 17 of 23 (74%) of the EPEC from AI cases were tEPEC.
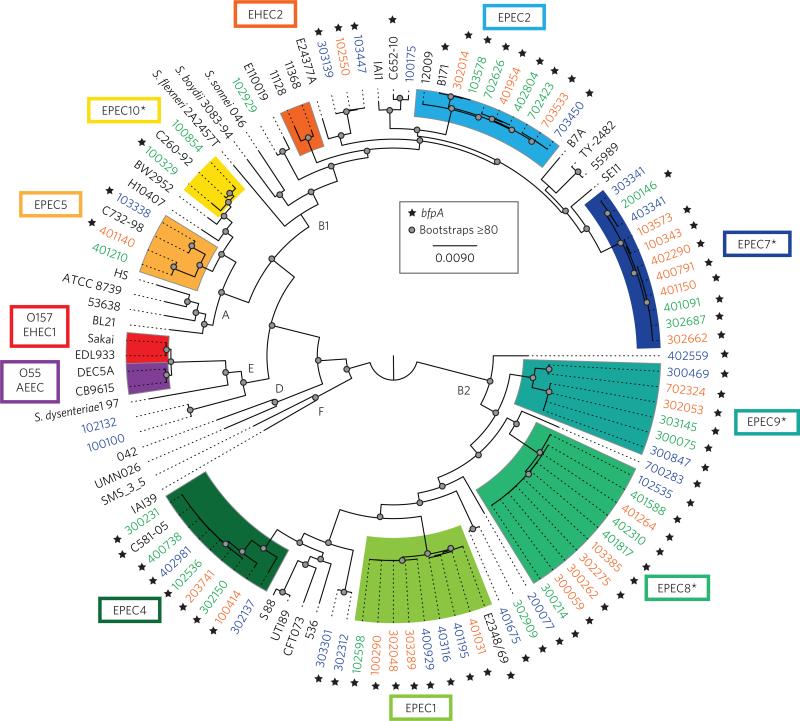
Phylogenomic analysis of the 70 EPEC isolate genomes, together with a collection of previously sequenced AEEC isolates and diverse E. coli and Shigella18, demonstrated that there is greater genomic diversity among recent EPEC isolates from Africa and Asia than in prototype E. coli isolates2,3,23 (Fig. 1). The 70 EPEC isolates were present in E. coli phylogroups A, E, B1 and B218,24, demonstrating considerable genomic diversity for E. coli belonging to a single pathovar (Fig. 1 and Tables 1 and 2). The majority of the isolates were in phylogroups B2 (55.7%, 39/70) and B1 (34.3%, 24/70), each of which included multiple E. coli isolates from various pathovars, as well as laboratory-adapted and commensal E. coli (Fig. 1 and Table 2). Overall, the phylogenomic lineages were not geographically confined, with the exception of the isolates belonging to EPEC lineages in phylogroup A (EPEC5, EPEC10), which were restricted to only two sites (The Gambia and Kenya) (Fig. 1 and Table 2).
Figure 1. Phylogenomic analysis of the 70 EPEC isolates associated with clinical outcomes of differing severity compared with select previously sequenced AEEC genomes and a reference collection of 25 diverse E. coli and Shigella isolate genomes.
The whole-genome assemblies were aligned using Mugsy44 as previously described18. The regions of sequence that aligned in all genomes were concatenated into a single 820,355-bp sequence for each genome, and the concatenated sequences were used to generate a maximum-likelihood phylogeny with 100 bootstrap replications, which was constructed using RAxML v.7.2.845, and visualized using FigTree v.1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/). Bootstrap values of ≥80 are designated on the tree by a filled circle. Genomes examined in this study that were obtained from lethal cases (LI) are indicated in orange, those from non-lethal symptomatic (NSI) cases are indicated in green and isolates from asymptomatic (AI) cases in blue. The presence of bfpA is indicated by a star symbol. The four novel EPEC phylogenomic lineages identified in this study are indicated by an asterisk.
Table 1.
Genome characteristics of the isolates sequenced in this study.
| Specimen ID | Clinical outcome* | Location | BFP† | Intimin type‡ | MLST ST (clonal group) | Phylogenomic lineage§ | Phylogroup§ | Draft genome size (Mb) | No. of contigs | Accession no. |
|---|---|---|---|---|---|---|---|---|---|---|
| 100290 | LI | The Gambia | + | alpha | 236 (6) | EPEC1 | B2 | 5.42 | 115 | JHQV00000000 |
| 100343 | LI | The Gambia | + | beta | 140 (31) | EPEC7 | B1 | 5.07 | 154 | JHQW00000000 |
| 100414 | LI | The Gambia | + | beta2 | ND | EPEC4 | B2 | 5.31 | 116 | JHQX00000000 |
| 102550 | LI | The Gambia | + | epsilon | 555 (NA) | NC | B1 | 5.17 | 209 | JHQY00000000 |
| 103385 | LI | The Gambia | + | mu | ND | EPEC8 | B2 | 5.05 | 149 | JHQZ00000000 |
| 103573 | LI | The Gambia | + | beta | 140 (31) | EPEC7 | B1 | 5.15 | 134 | JHRA00000000 |
| 203741 | LI | Mali | + | beta2 | 23 (7) | EPEC4 | B2 | 5.05 | 446 | JHRB00000000 |
| 300059 | LI | Mozambique | + | mu | ND | EPEC8 | B2 | 4.83 | 122 | JHRC00000000 |
| 300262 | LI | Mozambique | + | mu | ND | EPEC8 | B2 | 4.97 | 111 | JHRD00000000 |
| 302014 | LI | Mozambique | + | beta | 118 (17) | EPEC2 | B2 | 5.23 | 527 | JHRE00000000 |
| 302048 | LI | Mozambique | + | alpha | ND | EPEC1 | B2 | 5.27 | 436 | JHRF00000000 |
| 302053 | LI | Mozambique | + | lambda | ND | EPEC9 | B2 | 4.94 | 420 | JHRG00000000 |
| 302275 | LI | Mozambique | + | mu | ND | EPEC8 | B2 | 5.01 | 394 | JHRH00000000 |
| 302662 | LI | Mozambique | + | beta | 140 (31) | EPEC7 | B1 | 5.13 | 478 | JHRI00000000 |
| 303289 | LI | Mozambique | + | alpha | ND | EPEC1 | B2 | 5.09 | 144 | JHRJ00000000 |
| 400791 | LI | Kenya | + | beta | 140 (31) | EPEC7 | B1 | 5.32 | 170 | JHRK00000000 |
| 401031 | LI | Kenya | + | alpha‡ | 18 (6) | EPEC1 | B2 | 5.04 | 170 | JHRL00000000 |
| 401140 | LI | Kenya | + | epsilon | 433 (23) | EPEC5 | A | 5.00 | 255 | JHRM00000000 |
| 401150 | LI | Kenya | + | beta | 140 (31) | EPEC7 | B1 | 5.08 | 142 | JHRN00000000 |
| 401264 | LI | Kenya | + | mu | ND | EPEC8 | B2 | 5.14 | 228 | JHRO00000000 |
| 401954 | LI | Kenya | + | beta | 118 (17) | EPEC2 | B1 | 5.32 | 287 | JHRP00000000 |
| 402290 | LI | Kenya | + | beta | 140 (31) | EPEC7 | B1 | 5.36 | 109 | JHRQ00000000 |
| 702324 | LI | Pakistan | + | alpha | 228 (5) | EPEC9 | B2 | 5.01 | 96 | JHRR00000000 |
| 703533 | LI | Pakistan | + | beta | ND | EPEC2 | B1 | 5.30 | 549 | JHRS00000000 |
| 100329 | NSI | The Gambia | + | ND‡ | 169 (23) | EPEC10 | A | 4.81 | 390 | JHRT00000000 |
| 100854 | NSI | The Gambia | – | theta | 171 (23) | EPEC10 | A | 4.69 | 101 | JHRU00000000 |
| 102536 | NSI | The Gambia | + | beta2 | 23 (7) | EPEC4 | B2 | 5.30 | 134 | JHRV00000000 |
| 102598 | NSI | The Gambia | + | alpha | 236 (6) | EPEC1 | B2 | 5.36 | 187 | JHRW00000000 |
| 102929 | NSI | The Gambia | – | zeta | 535 (28) | NC | B1 | 5.24 | 147 | JHRX00000000 |
| 103578 | NSI | The Gambia | + | beta‡ | 118 (17) | EPEC2 | B1 | 5.11 | 376 | JHRY00000000 |
| 200146 | NSI | Mali | + | beta | 140 (31) | EPEC7 | B1 | 5.13 | 125 | JHRZ00000000 |
| 300075 | NSI | Mozambique | + | lambda | ND | EPEC9 | B2 | 4.97 | 103 | JHSA00000000 |
| 300214 | NSI | Mozambique | + | mu | ND | EPEC8 | B2 | 5.07 | 104 | JHSB00000000 |
| 300231 | NSI | Mozambique | + | beta2 | 23 (7) | EPEC4 | B2 | 5.13 | 79 | JHSC00000000 |
| 302150 | NSI | Mozambique | + | beta2 | ND | EPEC4 | B2 | 4.94 | 286 | JHSD00000000 |
| 302687 | NSI | Mozambique | + | beta‡ | 140 (31) | EPEC7 | B1 | 5.00 | 394 | JHSE00000000 |
| 302909 | NSI | Mozambique | + | lambda | ND | NC | B2 | 4.79 | 254 | JHSF00000000 |
| 303145 | NSI | Mozambique | + | lambda | ND | EPEC9 | B2 | 4.84 | 102 | JHSG00000000 |
| 400738 | NSI | Kenya | + | beta2 | 23 (7) | EPEC4 | B2 | 5.16 | 102 | JHSH00000000 |
| 401091 | NSI | Kenya | + | beta | 140 (31) | EPEC7 | B1 | 5.09 | 89 | JHSI00000000 |
| 401210 | NSI | Kenya | – | epsilon‡ | 433 (23) | EPEC5 | A | 4.57 | 781 | JHSJ00000000 |
| 401588 | NSI | Kenya | + | mu | ND | EPEC8 | B2 | 5.10 | 100 | JHSK00000000 |
| 401817 | NSI | Kenya | + | mu | ND | EPEC8 | B2 | 4.90 | 96 | JHSL00000000 |
| 402310 | NSI | Kenya | + | mu | ND | EPEC8 | B2 | 5.11 | 94 | JHSM00000000 |
| 402804 | NSI | Kenya | + | beta | 118 (17) | EPEC2 | B1 | 5.48 | 135 | JHSN00000000 |
| 702423 | NSI | Pakistan | + | beta | 140 (31) | EPEC2 | B1 | 5.40 | 181 | JHSO00000000 |
| 702626 | NSI | Pakistan | + | beta | 140 (31) | EPEC2 | B1 | 5.23 | 237 | JHSP00000000 |
| 100100 | AI | The Gambia | – | gamma | 78 (12) | NC | E | 5.27 | 123 | JHSQ00000000 |
| 100175 | AI | The Gambia | – | epsilon2 | 254 (43) | NC | B1 | 4.85 | 89 | JHSR00000000 |
| 102132 | AI | The Gambia | – | gamma | 78 (12) | NC | E | 5.28 | 110 | JHSS00000000 |
| 102535 | AI | The Gambia | + | mu | ND | EPEC8 | B2 | 4.79 | 97 | JHST00000000 |
| 103338 | AI | The Gambia | T | kappa | 933 (NA) | EPEC5 | A | 4.66 | 112 | JHSU00000000 |
| 103447 | AI | The Gambia | + | epsilon | 555 (NA) | NC | B1 | 5.09 | 182 | JHSV00000000 |
| 200077 | AI | Mali | + | eta | 20 (NA) | NC | B2 | 5.09 | 141 | JHSW00000000 |
| 300469 | AI | Mozambique | + | alpha | 228 (5) | EPEC9 | B2 | 4.51 | 64 | JHSX00000000 |
| 300847 | AI | Mozambique | + | lambda | ND | EPEC9 | B2 | 4.85 | 385 | JHSY00000000 |
| 302137 | AI | Mozambique | – | zeta | 378 (7) | EPEC4 | B2 | 5.10 | 44 | JHSZ00000000 |
| 302312 | AI | Mozambique | + | alpha | ND | NC | B2 | 5.15 | 115 | JHTA00000000 |
| 303139 | AI | Mozambique | + | epsilon | 555 (NA) | NC | B1 | 4.93 | 150 | JHTB00000000 |
| 303301 | AI | Mozambique | + | alpha | ND | NC | B2 | 4.91 | 100 | JHTC00000000 |
| 303341 | AI | Mozambique | – | beta | 140 (31) | EPEC7 | B1 | 5.06 | 86 | JHTD00000000 |
| 400929 | AI | Kenya | + | alpha | 236 (6) | EPEC1 | B2 | 5.04 | 139 | JHTE00000000 |
| 401195 | AI | Kenya | + | alpha‡ | 18 (6) | EPEC1 | B2 | 4.89 | 194 | JHTF00000000 |
| 401675 | AI | Kenya | + | lambda | ND | NC | B2 | 4.95 | 125 | JHTG00000000 |
| 402559 | AI | Kenya | + | kappa | 8 (3) | NC | B2 | 4.99 | 288 | JHTH00000000 |
| 402981 | AI | Kenya | + | beta2 | 23 (7) | EPEC4 | B2 | 5.17 | 75 | JHTI00000000 |
| 403116 | AI | Kenya | + | alpha | ND | EPEC1 | B2 | 4.97 | 108 | JHTJ00000000 |
| 403341 | AI | Kenya | – | beta | 140 (31) | EPEC7 | B1 | 5.17 | 111 | JHTK00000000 |
| 700283 | AI | Pakistan | + | kappa | 562 (NA) | NC | B2 | 4.76 | 97 | JHTL00000000 |
| 703450 | AI | Pakistan | + | beta | ND | EPEC2 | B1 | 5.52 | 289 | JHTM00000000 |
ND, not determined; NC, not classified
the clinical outcomes are LI (lethal), NSI (non-lethal symptomatic) and AI (asymptomatic)
a ‘+’ indicates that one or more genes of the BFP operon are present, ‘T’ indicates an isolate that has a truncated bfp operon and ‘–’ indicates that no BFP genes were detected
the intimin types of these isolates are truncated and these are the most related intimin types
the phylogenomic lineage and phylgroup designations are based on the genome-based phylogeny in Fig. 1.
Table 2.
Distribution of the 70 EPEC isolates from different clinical outcomes analysed in this study.
| Phylogroup or lineage* | Total isolates (% of total) | BFP† | Clinical outcomes (% of phylogroup or lineage)‡ |
Location (% of phylogroup or lineage) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LI | NSI | AI | The Gambia | Mali | Mozambique | Kenya | Pakistan | |||
| Total isolates | 70 | 61 (87.1) | 24 (34.2) | 23 (32.9) | 23 (32.9) | 18 (25.7) | 3 (4.3) | 22 (31.4) | 21 (30) | 6 (8.6) |
| Phylogroup B2 | 39 (55.7) | 38 (62.3) | 13 (33.3) | 12 (30.8) | 14 (35.9) | 6 (15.4) | 2 (5.1) | 17 (43.6) | 12 (30.8) | 2 (5.1) |
| EPEC1 | 8 (11.4) | 8 (100) | 4 (50) | 1 (12.5) | 3 (37.5) | 2 (25) | 0 (0) | 2 (25) | 4 (50) | 0 (0) |
| EPEC4 | 8 (11.4) | 7 (87.5) | 2 (25) | 4 (50) | 2 (25) | 2 (25) | 1 (12.5) | 3 (37.5) | 2 (25) | 0 (0) |
| EPEC8 | 10 (14.3) | 10 (100) | 5 (50) | 4 (40) | 1 (10) | 2 (20) | 0 (0) | 4 (40) | 4 (40) | 0 (0) |
| EPEC9 | 6 (8.6) | 6 (100) | 2 (33.3) | 2 (33.3) | 2 (33.3) | 0 (0) | 0 (0) | 5 (83.3) | 0 (0) | 1 (16.7) |
| uAEEC | 7 (10) | 7 (100) | 0 (0) | 1 (14.3) | 6 (85.7) | 0 (0) | 1 (14.3) | 3 (42.8) | 2 (28.6) | 1 (14.3) |
| Phylogroup B1 | 24 (34.3) | 20 (32.8) | 10 (41.7) | 8 (33.3) | 6 (25) | 7 (29.2) | 1 (4.2) | 5 (20.8) | 7 (29.2) | 4 (16.6) |
| EPEC2 | 8 (11.4) | 8 (100) | 3 (37.5) | 4 (50) | 1 (12.5) | 1 (12.5) | 0 (0) | 1 (12.5) | 2 (25) | 4 (50) |
| EPEC7 | 11 (15.7) | 9 (81.8) | 6 (54.5) | 3 (27.3) | 2 (18.2) | 2 (18.2) | 1 (9.1) | 3 (27.3) | 5 (45.4) | 0 (0) |
| uAEEC | 5 (7.1) | 3 (60) | 1 (20) | 1 (20) | 3 (60) | 4 (80) | 0 (0) | 1 (20) | 0 (0) | 0 (0) |
| Phylogroup A | 5 (7.1) | 3 (4.9) | 1 (20) | 3 (60) | 1 (20) | 3 (60) | 0 (0) | 0 (0) | 2 (40) | 0 (0) |
| EPEC5 | 3 (4.3) | 2 (66.7) | 1 (33.3) | 1 (33.3) | 1 (33.3) | 1 (33.3) | 0 (0) | 0 (0) | 2 (66.7) | 0 (0) |
| EPEC10 | 2 (2.9) | 1 (50) | 0 (0) | 2 (100) | 0 (0) | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Phylogroup E | 2 (2.9) | 0 (0) | 0 (0) | 0 (0) | 2 (100) | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| uAEEC | 2 (2.9) | 0 (0) | 0 (0) | 0 (0) | 2 (100) | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Phylogroups B1, B2, E | ||||||||||
| uAEEC Total | 14 (20) | 10 (71.4) | 1 (7.1) | 2 (14.3) | 11 (78.6) | 6 (42.9) | 1 (7.1) | 4 (28.6) | 2 (14.3) | 1 (7.1) |
The phylogenomic lineages are determined based on the genome phylogeny, and uAEEC indicates an ‘uncharacterized’ AEEC that is not in a known lineage
includes isolates that have a truncated BFP operon; the percentages are calculated for each lineage
the clinical outcomes are lethal (LI), non-lethal symptomatic (NSI) and asymptomatic (AI).
An MLST-based phylogeny was also constructed using anchor isolates of the previously described EPEC lineages, EPEC1–EPEC619,20. This allowed the identification of relationships among the 70 EPEC isolates sequenced in the current study to the previous MLST-defined EPEC lineages (Fig. 1 and Supplementary Fig. 1). Remarkably, only 16 (22.9%) of the isolates sequenced were present in the two main previously identified MLST-based lineages of EPEC, EPEC1 and EPEC2, with eight in each lineage (Fig. 1 and Tables 1 and 2). An additional eight genomes (11.4%) were in the EPEC4 lineage (Fig. 1 and Tables 1–2), which has previously been described by MLST and a single genome has been sequenced18,20. Another three genomes of isolates 103338, 401140 and 401210 grouped in the MLST-based phylogeny with an isolate previously designated as EPEC5 using MLST20 (Supplementary Fig. 1). The remaining 43 genomes formed novel EPEC phylogenomic lineages. This finding indicates that there is considerable uncharacterized EPEC genomic diversity identified in this study (Fig. 1). To extend the established MLST-based nomenclature, we are designating four previously undescribed phylogenomic lineages, which each contain five or more genomes, EPEC7–10 (Fig. 1 and Supplementary Table 1). Eleven of these genomes were in the EPEC7 phylogenomic lineage and B1 phylogroup (Table 2). In phylogroup B2 there were ten genomes forming the EPEC8 phylogenomic lineage and six in the EPEC9 phylogenomic lineage (Fig. 1 and Table 2). The remaining two genomes belong to the EPEC10 lineage, which was designated when combined with three previously sequenced LEE+/stx−/bfp−isolates18 (Fig. 1 and Table 2). The four newly described EPEC lineages contain 41.4% (29/70) of the isolates, highlighting the undescribed diversity of global EPEC isolates.
In addition to these novel lineages, there were 14 genomes not assigned to phylogenomic lineages EPEC1–10, which thus represent unclassified EPEC isolates. These isolates were distributed throughout the E. coli phylogeny (Fig. 1 and Table 1). Of these 14 unclassi-fied EPEC isolates, only one was associated with an LI case, two with NSI cases and 11 with AI controls (Fig. 1 and Table 2), and six of these isolates were bfpA− (Fig. 1). Thus, the unclassified EPEC isolates comprised nearly half (11/23, 48%) of the AI isolates, whereas the LI and NSI isolates were primarily associated with phylogenomic lineages that contained one or more tEPEC. These distributions suggest there may be an optimal EPEC genomic content that is required for the greatest virulence.
Distribution of EPEC virulence-associated genes
The expanded genome phylogeny described here identified a previously unrecognized phylogenetic distribution of EPEC isolates; however, it was unclear whether these differences extended to the known EPEC virulence factors. In addition to the T3SS encoded by the LEE pathogenicity island5,25, present in all genomes sequenced in this study, there were additional virulence-associated secretion systems detected in the isolates sequenced in this study (Supplementary Table 1). Among these regions was a type II secretion system (T2SS) and a type VI secretion system (T6SS), both of which exhibited phylogroup- and lineage-specific distributions (Supplementary Table 1). Investigation of the sequence diversity of previously characterized T3SS effectors demonstrated that the effectors exhibited greater similarity by phylogenomic lineage than by clinical outcome (Supplementary Fig. 2).
Phylogenetic analysis of the bfpA nucleotide sequences present in each of the 61 bfpA+ genomes sequenced in this study, with 11 reference bfpA alleles20,26 and 31 bfpA alleles from previously sequenced EPEC genomes18, demonstrated that the majority of the bfpA genes belonged to one of three main phylogenetic groups as defined by Blank and colleagues26 (Supplementary Fig. 3a). Each of the phylogenetic groups of bfpA contains isolates from diverse phylogenomic lineages and clinical outcomes (LI, NSI and AI). This is in contrast to the intimin gene, eae, from the LEE pathogenicity region, which exhibits greater phylogenomic lineage specificity (Supplementary Fig. 3b). This difference suggests that bfpA, and by extension the entire bfp operon and possibly the entire EAF plasmid, have been lost and acquired multiple times by E. coli isolates belonging to diverse EPEC phylogenomic lineages.
Interestingly, all of the LI isolates analysed in this study were found to be bfpA+ by PCR, as previously described18, with the exception of isolate 100414, which was bfpA− (Table 1 and Supplementary Table 1). However, on detailed examination of the genome sequence, EPEC isolate 100414 was determined to encode a bfpA orthologue with 72% nucleotide identity to bfpA of the E2348/69 EAF plasmid, pMAR222. The 100414 bfpA allele exhibited greater phylogenetic similarity to a bfpA-like sequence from the LEE-negative EAEC isolate 101-121 (Supplementary Fig. 3a).
Identification of EPEC genes associated with different clinical outcomes
To identify whether there are genes that are more prevalent among the 70 EPEC from different clinical presentations, we used large-scale BLAST score ratio (LS-BSR) analysis27,28 to analyse the whole genome content. The LS-BSR analysis places predicted homologous genes from each genome into gene clusters that have ≥90% nucleotide identity29. For the 70 genomes analysed in this study, 12,964 gene clusters were identified and 1,080 gene clusters were present in all 70 genomes analysed (LS-BSR ≥ 0.9). These gene clusters represent the conserved EPEC core genome. This is a more conservative approach than was previously used to define the E. coli species core genome and so the absolute number of genes is smaller than the E. coli core genome defined previously21,30.
A comparison of gene cluster prevalence in LI genomes versus AI genomes demonstrated a significant correlation (P < 0.05) of 367 gene clusters (Table 3 and Supplementary Table 2). Among the gene clusters represented in a greater number of LI than AI genomes were genes of the EAF plasmid, flagellin, an allele of the T3SS effector NleG, as well as many hypothetical and phage-associated genes (Supplementary Table 2). There were 111 clusters that were significantly more prevalent in LI genomes or in NSI genomes (Table 3). Among the genes that were more prevalent among the LI genomes were many that encoded hypothetical proteins, putative transcriptional regulators, a putative T3SS effector EspJ and putative phage-associated genes (Supplementary Table 2). Similarly, there were 118 gene clusters that were statistically more prevalent in NSI genomes versus AI genomes (Table 3).
Table 3.
Number of gene clusters identified using LS-BSR that are significantly correlated with one clinical outcome when compared to another clinical outcome.
| Clinical outcomes* | No. of genomes | No. of gene clusters |
|||
|---|---|---|---|---|---|
| Lineage-specific† LS-BSR ≥ 0.9 |
EPEC-specific‡ LS-BSR ≥ 0.8 |
||||
| <0.005 | <0.05 | <0.005 | <0.05 | ||
| All genomes | |||||
| LI vs AI | |||||
| Total | 47 | 20 | 367 | 12 | 198 |
| LI | 24 | 19 | 227 | 11 | 134 |
| AI | 23 | 1 | 140 | 1 | 64 |
| LI vs NSI | |||||
| Total | 47 | 1 | 111 | 0 | 39 |
| LI | 24 | 0 | 31 | 0 | 14 |
| NSI | 23 | 1 | 80 | 0 | 25 |
| NSI vs AI | |||||
| Total | 46 | 7 | 118 | 4 | 67 |
| NSI | 23 | 5 | 63 | 1 | 27 |
| AI | 23 | 2 | 55 | 3 | 40 |
| Typical EPEC genomes only | |||||
| LI vs AI | |||||
| Total | 41 | 11 | 238 | 2 | 134 |
| LI | 24 | 11 | 167 | 2 | 96 |
| AI | 17 | 0 | 71 | 0 | 38 |
| LI vs NSI | |||||
| Total | 44 | 0 | 39 | 0 | 7 |
| LI | 24 | 0 | 9 | 0 | 5 |
| NSI | 20 | 0 | 30 | 0 | 2 |
| NSI vs AI | |||||
| Total | 37 | 7 | 176 | 0 | 87 |
| NSI | 20 | 4 | 89 | 0 | 24 |
| AI | 17 | 3 | 87 | 0 | 63 |
Clinical outcomes are classified as lethal (LI), non-lethal symptomatic (NSI) and asymptomatic (AI)
as part of a lineage-specific gene comparison, genes with ≥90% nucleotide identity were grouped together into gene clusters, and the gene clusters were identified as more prevalent in genomes of one clinical outcome over another by the percentage of genomes of each group that contained the gene cluster with significant similarity (LS-BSR ≥ 0.9)
as part of an EPEC pathovar-specific comparison, genes with ≥80% nucleotide similarity were grouped together into gene clusters and the gene clusters were identified as more prevalent in genomes of one clinical outcome over another by the percentage of genomes of each group that contained the gene cluster with significant similarity (LS-BSR ≥ 0.8) that were not present in three previously characterized E. coli commensal genomes (K-12, SE11, HS).
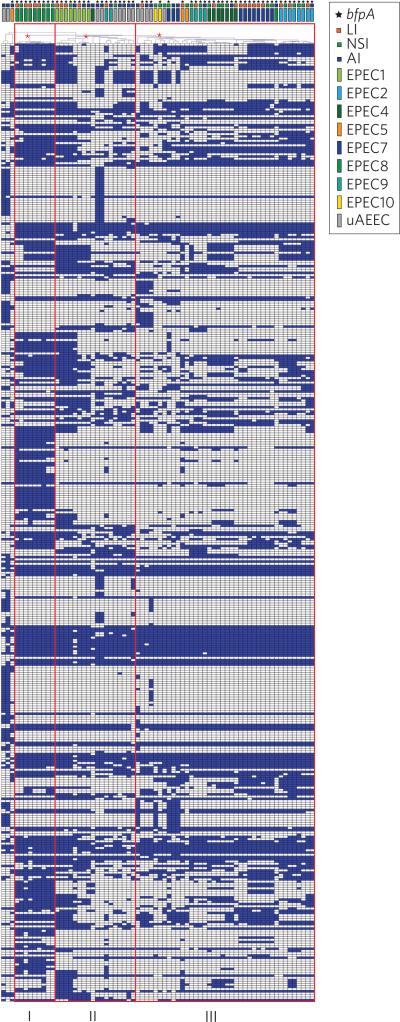
Although we identified gene clusters with a significant correlation with one symptomatic group compared to another symptomatic group (Table 4 and Supplementary Table 3), there were no gene clusters that were detected in all of the LI genomes that were absent from all of the NSI and AI genomes. The absence of universal clinically associated genes may partly be a result of the vast genomic diversity of the isolates associated with each of the clinical outcomes (Fig. 1 and Supplementary Table 3). However, there were 428 gene clusters that were statistically (P < 0.05) more prevalent among the symptomatic (LI and NSI) compared to asymptomatic (AI) genomes, and 40 of these gene clusters had a P value of <0.005 (Table 4 and Supplementary Table 4). These gene clusters that were more prevalent among symptomatic compared to asymptomatic group genomes included numerous hypothetical proteins and phage and plasmid-associated genes (Supplementary Table 4). When the distribution of these 428 gene clusters was compared by hierarchical cluster analysis, the EPEC isolates formed three main groups that included all of the genomes, except three isolates that were outliers (Fig. 2). Group I contained nine of the ten EPEC8 isolates and the only EPEC8 isolate that was not within group I was part of group III and associated with an asymptomatic outcome (Fig. 2). Thus, all of the EPEC isolates of group I were associated with symptomatic outcomes (five LI and four NSI). Meanwhile, group II contained 18 isolates, all belonging to E. coli phylogroup B2. Seven of these isolates (39%) were associated with symptomatic outcomes, while the other 11 (61%) EPEC isolates were from asymptomatic outcomes (Fig. 2). The largest group was group III, which contained 40 isolates, including 31 (78%) from symptomatic outcomes and nine (22%) from asymptomatic outcomes (Fig. 2). The EPEC isolate genomes of group III primarily belonged to phylogroups B1 and A, with the exception of four EPEC9 isolates and seven EPEC4 isolates from phylogroup B2 (Fig. 2).
Table 4.
Number of gene clusters identified using LS-BSR that are significantly correlated with genomes of a particular clinical outcome.
| Clinical outcomes* | No. of genomes | No. of gene clusters |
|||
|---|---|---|---|---|---|
| Lineage-specific† LS-BSR ≥ 0.9 |
EPEC-specific‡ LS-BSR ≥ 0.8 |
||||
| <0.005 | <0.05 | <0.005 | <0.05 | ||
| All genomes | |||||
| Symptomatic vs asymptomatic | |||||
| Total | 70 | 40 | 428 | 24 | 246 |
| LI+NSI | 47 | 25 | 258 | 12 | 109 |
| AI | 23 | 15 | 170 | 12 | 137 |
| Lethal vs non-lethal | |||||
| Total | 70 | 38 | 308 | 7 | 135 |
| LI | 24 | 12 | 170 | 7 | 122 |
| NSI+AI | 46 | 26 | 138 | 0 | 13 |
| Typical EPEC genomes only | |||||
| Symptomatic vs asymptomatic | |||||
| Total | 61 | 31 | 258 | 15 | 141 |
| LI+NSI | 44 | 8 | 151 | 3 | 58 |
| AI | 17 | 23 | 107 | 12 | 83 |
| Lethal vs non-lethal | |||||
| Total | 61 | 11 | 202 | 2 | 86 |
| LI | 24 | 5 | 103 | 2 | 71 |
| NSI+AI | 37 | 6 | 99 | 0 | 15 |
The symptomatic clinical outcomes are classified as lethal (LI) and non-lethal symptomatic (NSI) and non-lethal are the NSI and asymptomatic (AI)
genes with ≥90% nucleotide identity were grouped together into gene clusters, and the gene clusters were identified as more prevalent in genomes of one clinical outcome over another by the percentage of genomes of each group that contained the gene cluster with significant similarity (LS-BSR ≥ 0.9)
genes with ≥80% nucleotide similarity were grouped together into gene clusters, and the gene clusters were identified as more prevalent in genomes of one clinical outcome over another by the percentage of genomes of each group that contained the gene cluster with significant similarity (LS-BSR ≥ 0.8) that were not present in three previously characterized E. coli commensal genomes (K-12, SE11, HS).
Figure 2. Identification of genes associated with symptomatic and asymptomatic EPEC isolates.
The plot is a hierarchical cluster analysis of the 428 LS-BSR gene clusters that were significantly (chi-square test or Fisher's exact test, P < 0.05) more prevalent in genomes of symptomatic (LI and NSI) compared to asymptomatic (AI) cases for all 70 EPEC genomes analysed. The LS-BSR gene clusters, generated using a clustering threshold of 90% nucleotide identity, that were significantly (chi-square test or Fisher's exact test P < 0.05) associated with genomes of symptomatic compared to asymptomatic cases, were compared by hierarchical clustering41. Hierarchical clustering with Pearson correlation and average linkage was performed using MeV42. Each column represents a genome, and each row is an LS-BSR gene cluster. The gene clusters that were present with an LS-BSR value of ≥0.9 are indicated in blue, and the gene clusters that were absent (LS-BSR value of <0.9) in white. Red boxes indicate three groups of genomes, designated I, II and III, and red asterisks identify the nodes that separate the genomes into the three groups. The colour-coded rectangles at the top of the plot denote the phylogenomic lineage, and the colour-coded squares indicate the clinical outcome of each isolate. The colour coding of each symbol is given in the key at the top of the figure. A star symbol denotes the presence of bfpA in each genome.
To investigate whether there were similar trends observed when comparing only the tEPEC isolates, we excluded the nine aEPEC isolates. Comparison of the tEPEC from the three different clinical outcomes (LI versus NSI, LI versus AI and NSI versus AI) identified fewer gene clusters that were significantly (P < 0.05) associated with one clinical outcome over another than were identified when comparing all 70 EPEC genomes (see Table 3 and Supplementary Table 3 for a clinical presentation and Table 4 and Supplementary Table 5 for symptomatic versus asymptomatic comparisons). These findings suggest there is an increased genomic diversity associated with the aEPEC isolates.
Hierarchical cluster analysis of the presence of the 258 gene clusters significantly associated with only tEPEC of symptomatic or asymptomatic outcomes separated the tEPEC isolates into two similarly sized groups (Supplementary Fig. 4). tEPEC group I contained 34 isolates, including 20 (59%) from symptomatic (LI or NSI) outcomes and 14 (41%) from asymptomatic outcomes (Supplementary Fig. 4). Meanwhile, tEPEC group II contained 27 genomes; 24 (89%) from symptomatic outcomes and only three (11%) from asymptomatic outcomes (Supplementary Fig. 4). Within each of these tEPEC groups the isolates were present in subgroups based on phylogenomic lineage. There were 20 gene clusters that were present in all of the genomes of tEPEC group I that were absent from all genomes of tEPEC group II (Supplementary Fig. 4 and Supplementary Table 5) including gene products predicted to be involved in propanediol utilization (Supplementary Table 5), which has been implicated in Salmonella for its role during survival in the host31,32.
EPEC-specific genes associated with different clinical outcomes
To identify genes that were associated with EPEC isolates of different clinical outcomes, while taking into account the considerable underlying genomic diversity of these isolates, we performed LS-BSR analysis using a decreased clustering threshold of 80% nucleotide identity to combine potential alleles. Commensal genomes were included (E. coli HS (NC_009800.1), K-12 (NC_000913.3) and SE11 (NC_011415.1)) in the analysis as a metric for counter selection. This approach provided the opportunity to identify genetic features that were present only in the EPEC, regardless of phylogenomic lineage. For this analysis there were 12,196 total gene clusters. Of those, there were 6,474 gene clusters (53%) that were present in one or more of the EPEC genomes that were absent (LS-BSR < 0.8) from all of the commensal isolates. Using this EPEC-only data set and examining all 70 EPEC isolate genomes, the number of gene clusters that were significantly (P < 0.05) associated with one clinical outcome over another ranged from 39 to 198 (Table 3 and Supplementary Table 6). Similarly, when comparing only the 61 tEPEC genomes, the number of genes associated with genomes of one clinical outcome over another was lower, ranging from 7 to 134 (Table 3 and Supplementary Table 7). Furthermore, the number of genes significantly associated with symptomatic (LI and NSI) compared to asymptomatic (AI), or lethal (LI) compared to non-lethal (NSI and AI) genomes was decreased (Table 4 and Supplementary Table 8). The number of gene clusters associated with symptomatic or asymptomatic genomes was 246 when comparing all 70 EPEC isolates (Table 4, Supplementary Table 8 and Supplementary Fig. 5) and 141 when comparing only the 61 tEPEC isolates (Table 4, Supplementary Table 9 and Supplementary Fig. 6).
Many of the gene clusters that were associated with one clinical outcome were annotated as hypothetical proteins (Supplementary Table 4). To examine the potential function of the predicted peptides, the gene clusters were examined for protein domains identified in membrane-associated or secreted proteins, which would suggest they might be directly involved in surface expression or survival. Of the 39 to 246 gene clusters that were identified as significantly associated with one clinical outcome in the analysis of all 70 EPEC (Tables 3 and 4), the number of gene clusters with protein domains of secreted or surface-associated proteins ranged from 11 to 50 (Supplementary Table 10). Similarly, of the 7 to 141 gene clusters significantly associated with one clinical outcome in the analysis of only the tEPEC genomes (Tables 3 and 4), the number of gene clusters containing membrane-associated or secreted protein domains was low, ranging from 2 to 31 (Supplementary Table 10). Among the gene clusters that were significantly more prevalent in symptomatic compared to asymptomatic genomes were hypothetical proteins, a putative yfdA, an acetyltransferase, a putative pyridoxamine 5-phosphate-dependent dehydrase, a glycosyl transferase family protein, and plasmid conjugal transfer-associated proteins (Supplementary Tables 8 and 9). These analyses provide targets for the functional characterization of these gene products in pathogenesis.
Discussion
The whole-genome sequencing and phylogenomic analysis of 70 EPEC isolates from children enrolled in GEMS2,3 demonstrated that E. coli clinical isolates identified as EPEC based only on their virulence factor content exhibit considerable genomic diversity. Phylogenomic analysis demonstrated that 61% (43/70) of the EPEC isolates examined occupy previously undescribed phylogenomic lineages. This study may have identified newly circulating EPEC in the GEMS sites, but may also highlight the dynamic evolutionary processes that are at work in E. coli pathogens. Of note, a recent study on EPEC demonstrated a shift in the epidemiology from tEPEC to aEPEC isolates1, but this study focused on the tEPEC isolates associated with an adverse outcome. The current study is not meant to be a comprehensive genomic view of all the tEPEC collected with GEMS, but a focused attempt to identify genetic factors associated with the isolates from the most severe outcomes.
The EPEC genome comparisons demonstrated that the degree of genomic difference was greater when comparing the extremes of the clinical presentation, LI to AI genomes, than it was when comparing LI to NSI, or NSI to AI (Table 2). This emphasizes the finding from the phylogenomic analysis that isolates associated with a particular clinical outcome can occur in distantly related EPEC phylogenomic lineages (Fig. 1). Thus, the smaller number of genomic differences identified between the lethal and non-lethal EPEC isolates suggests the differences in the illness severity caused by these isolates may have less to do with the bacterium and more to do with host factors including, but not limited to, co-morbidities, the micro-biome, diet, breast-feeding and access to medical care, among other factors. Overall, these findings suggest that there is not a single gene or genomic region that is responsible for particular EPEC isolates causing more severe clinical outcomes, but it may instead require a collection of genomic regions acting in concert, as well as responding to host factors that will result in more severe infection by EPEC. The gene clusters that are more prevalent in the genomes of EPEC from different clinical outcomes provide a genomic view of what potentially makes certain EPEC isolates more virulent. Among these were many genes with unknown functions, including some that contain predicted protein domains of membrane-associated or secreted proteins that can be investigated for their contribution to the virulence mechanism of EPEC and potentially other pathogenic E. coli. A recent study by Hazen et al.33 describes the comparative transcriptome analysis of four prototype EPEC isolates: E2348/69 (EPEC1), B171 (EPEC2), C581-05 (EPEC4) and E110019 (prototype aEPEC isolate)33. That study identified that there is also transcriptional variation among these prototype isolates33. Further investigation is required to examine the transcriptional variation among the new EPEC lineages described in the current study. The combination of genomics and transcriptomics will provide further insight into the conserved and expressed EPEC features involved in virulence
Large-scale comparative genomic studies that assess the diversity of disease-causing bacteria associated with multiple types of clinical outcome, such as this, provide a framework for understanding the processes that underlie the evolution of pathogenesis. This study describes a number of phylogroup- and lineage-specific differences in the virulence factor and genome content, which suggests that EPEC isolates have continued to acquire genetic changes since their initial acquisition of some of the pathovar-defining features. These studies can also provide insight into the ongoing evolution of the virulence mechanisms of disease-causing bacteria. The emergence of diarrhoea-causing EPEC and the severity of illness attributed to these isolates depend on a suite of genes that includes both lineage-specific virulence factors and genes encoded by plasmid and phage. These regions will provide fertile ground for the examination of EPEC pathogenesis and the development of a possible vaccine against EPEC in the future.
Methods
Bacterial isolates
The bacterial isolates analysed in this study, and the details of each of the genomes sequenced, are listed in Table 1 and also described in a companion study34. The EPEC (LEE+/bfpA+/stx−) isolates analysed in this study were obtained from GEMS as previously described2,23. A total of 24 tEPEC isolates from lethal cases (LI) were obtained, representing all tEPEC isolates associated with a lethal outcome in GEMS2,23. The isolates from lethal outcomes were from only five sites of the seven in GEMS (The Gambia, Mail, Mozambique, Kenya and Pakistan), so there is an over-representation of isolates from Africa. A matching scheme using geography and clinical parameters of the subject was used to select one EPEC isolate from a non-lethal symptomatic case (NSI) and one EPEC isolate from an asymptomatic case (AI) representing controls for each tEPEC from a lethal case as previously described34. One NSI case and one AI case served as controls for two different LI cases, resulting in 23 EPEC from NSI cases and 23 EPEC from AI cases. A tEPEC isolate (bfpA+) was obtained from 20 of the NSI cases and 17 of the AI cases, with the remaining EPEC cases containing an aEPEC (bfpA−). The recent publication by Donnenberg et al.34 describes the case-control aspect of this study and the comparison of the isolates that were directly matched based on patient and clinical parameters. In the current study we delve into the phylogenomic content of the isolates, irrespective of matching criteria and only consider the genotypic presentation of EPEC and the outcome of the infection.
Genome sequencing and assembly
Genomic DNA was isolated from each strain by growing a single colony that was PCR-positive for the LEE-encoded gene escV and/or the EAF plasmid gene bfpA, overnight, in Luria-Bertani (LB) medium at 37 °C with shaking. The genomic DNA was isolated from the overnight culture using the GenElute Genomic kit (Sigma-Aldrich), then sequenced and assembled as previously described18,34.
Phylogenomic analysis
The 70 EPEC genomes sequenced in this study were compared with 37 previously sequenced E. coli and Shigella genomes by whole-genome phylogenomic analysis as previously described18,35.
Gene alignments and phylogenetic analyses
The individual gene phylogenies of eae and bfpA were generated as described previously18. The nucleotide sequences were aligned in MEGA536 using the ClustalW algorithm37. A maximum-likelihood phylogeny was then constructed using the Kimura two-parameter model of distance estimation38 with 1,000 bootstrap replications.
A phylogenetic analysis of seven conserved housekeeping genes that have been used for MLST was generated for the isolates characterized in this study compared to a collection of previously sequenced EPEC and other E. coli isolates as previously described18,20. The EPEC1-4 reference sequences types (STs) included in the phylogeny are those identified by Lacher et al.20 while the EPEC5 and EPEC6 reference sequences were described by Tennant and co-workers.19
BSR analysis
The presence or absence of known virulence-associated genes in the genome sequences generated in this study was determined using BLAST score ratio (BSR) analysis, as described previously18,27,28. The protein-encoding genes that were considered present with significant similarity had BSR values of ≥0.8, while those with BSR values <0.8 but ≥0.4 were considered to be present but divergent.
The level of similarity of protein-encoding genes was compared across genomes in this study using a large-scale BLAST score ratio (LS-BSR) analysis as previously described18,28,29. The gene clusters were assigned using a stringent nucleotide identity threshold of ≥90% (Data Set S1), or using a more inclusive nucleotide identity threshold of ≥80% (Data Set S2). The LS-BSR analysis performed using the more inclusive clustering threshold of ≥80% included the 70 genomes in this study and three commensal genomes: E. coli HS (NC_009800.1), K-12 (NC_000913.3) and SE11 (NC_011415.1). The predicted protein function of each gene cluster was determined using an ergatis-based39 in-house annotation pipeline40.
Hierarchical cluster analysis41 of the LS-BSR gene clusters associated with particular clinical outcomes was performed using Pearson correlation with average linkage using MeV42. The gene clusters compared were considered either present (blue) with an LS-BSR of ≥0.9 (with 90% clustering threshold) or ≥0.8 (with 80% clustering threshold) or absent (white) when <0.9 or <0.8.
Statistical analysis
Statistical significance of the prevalence of predicted gene clusters among genomes associated with different symptomatic groups was determined using the Pearson's chi-square test with Yates’ continuity correction when the number of genomes was five or more, or the Fisher's exact test when the number of genomes in one or both groups being compared was less than five, calculated using R v. 3.1.143. P values of <0.05 were considered statistically significant.
Supplementary Material
Acknowledgements
This project was funded by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services NIH grant no. U19 AI090873.
Footnotes
Author contributions
T.H.H., M.S.D., J.P.N. and D.A.R. conceived and designed the experiments. T.H.H., M.S.D., E.M.B., J.B.K., J.P.N. and D.A.R. performed the experiments. T.H.H., M.S.D., E.M.B., J.B.K., J.P.N. and D.A.R. analysed the data. S.P., M.A., A.H., I.M., J.B.O., T.R., B.T., S.Q., F.Q., A.Z., K.L.K., M.M.L. and J.P.N. contributed materials from the GEMS studies. T.H.H., M.S.D., E.M.B., J.B.K., J.P.N. and D.A.R. co-wrote the paper.
Additional information
Supplementary information is available online.
Competing interests
The authors declare no competing financial interests.
Accession numbers. The genome sequence assemblies generated in this study were deposited in GenBank under the accession numbers listed in Table 1.
References
- 1.Ochoa TJ, Contreras CA. Enteropathogenic Escherichia coli infection in children. Curr. Opin. Infect. Dis. 2011;24:478–483. doi: 10.1097/QCO.0b013e32834a8b8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotloff KL, et al. The global enteric multicenter study (GEMS) of diarrheal disease in infants and young children in developing countries: epidemiologic and clinical methods of the case/control study. Clin. Infect. Dis. 2012;55(Suppl 4):S232–S245. doi: 10.1093/cid/cis753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotloff KL, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 4.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl Acad. Sci. USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliott SJ, et al. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 1998;28:1–4. doi: 10.1046/j.1365-2958.1998.00783.x. [DOI] [PubMed] [Google Scholar]
- 6.Perna NT, et al. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 1998;66:3810–3817. doi: 10.1128/iai.66.8.3810-3817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tauschek M, Strugnell RA, Robins-Browne RM. Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol. Microbiol. 2002;44:1533–1550. doi: 10.1046/j.1365-2958.2002.02968.x. [DOI] [PubMed] [Google Scholar]
- 8.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nature Rev. Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 9.Tarr PI, Gordon CA, Chandler WL. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet. 2005;365:1073–1086. doi: 10.1016/S0140-6736(05)71144-2. [DOI] [PubMed] [Google Scholar]
- 10.Clements A, Young JC, Constantinou N, Frankel G. Infection strategies of enteric pathogenic Escherichia coli. Gut Microbes. 2012;3:71–87. doi: 10.4161/gmic.19182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pennington H. Escherichia coli O157. Lancet. 2010;376:1428–1435. doi: 10.1016/S0140-6736(10)60963-4. [DOI] [PubMed] [Google Scholar]
- 12.Nataro JP, Scaletsky IC, Kaper JB, Levine MM, Trabulsi LR. Plasmid-mediated factors conferring diffuse and localized adherence of enteropathogenic Escherichia coli. Infect. Immun. 1985;48:378–383. doi: 10.1128/iai.48.2.378-383.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bieber D, et al. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 14.Donnenberg MS, Giron JA, Nataro JP, Kaper JB. A plasmid-encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol. Microbiol. 1992;6:3427–3437. doi: 10.1111/j.1365-2958.1992.tb02210.x. [DOI] [PubMed] [Google Scholar]
- 15.Stone KD, Zhang HZ, Carlson LK, Donnenberg MS. A cluster of fourteen genes from enteropathogenic Escherichia coli is sufficient for the biogenesis of a type IV pilus. Mol. Microbiol. 1996;20:325–337. doi: 10.1111/j.1365-2958.1996.tb02620.x. [DOI] [PubMed] [Google Scholar]
- 16.Donnenberg MS, Zhang HZ, Stone KD. Biogenesis of the bundle-forming pilus of enteropathogenic Escherichia coli: reconstitution of fimbriae in recombinant E. coli and role of DsbA in pilin stability—a review. Gene. 1997;192:33–38. doi: 10.1016/s0378-1119(96)00826-8. [DOI] [PubMed] [Google Scholar]
- 17.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hazen TH, et al. Refining the pathovar paradigm via phylogenomics of the attaching and effacing Escherichia coli. Proc. Natl Acad. Sci. USA. 2013;110:12810–12815. doi: 10.1073/pnas.1306836110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tennant SM, et al. Characterisation of atypical enteropathogenic E. coli strains of clinical origin. BMC Microbiol. 2009;9:117. doi: 10.1186/1471-2180-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lacher DW, Steinsland H, Blank TE, Donnenberg MS, Whittam TS. Molecular evolution of typical enteropathogenic Escherichia coli: clonal analysis by multilocus sequence typing and virulence gene allelic profiling. J. Bacteriol. 2007;189:342–350. doi: 10.1128/JB.01472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasko DA, et al. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 2008;190:6881–6893. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iguchi A, et al. Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J. Bacteriol. 2009;191:347–354. doi: 10.1128/JB.01238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panchalingam S, et al. Diagnostic microbiologic methods in the GEMS-1 case/control study. Clin. Infect. Dis. 2012;55(Suppl 4):S294–S302. doi: 10.1093/cid/cis754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nature Rev. Microbiol. 2010;8:207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 25.Deng W, et al. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl Acad. Sci. USA. 2004;101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blank TE, Zhong H, Bell AL, Whittam TS, Donnenberg MS. Molecular variation among type IV pilin (bfpA) genes from diverse enteropathogenic Escherichia coli strains. Infect. Immun. 2000;68:7028–7038. doi: 10.1128/iai.68.12.7028-7038.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasko DA, Myers GS, Ravel J. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinf. 2005;6:2. doi: 10.1186/1471-2105-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sahl JW, Caporaso JG, Rasko DA, Keim P. The large-scale blast score ratio (LS-BSR) pipeline: a method to rapidly compare genetic content between bacterial genomes. PeerJ. 2014;2:e332. doi: 10.7717/peerj.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sahl JW, et al. Evolution of a pathogen: a comparative genomics analysis identifies a genetic pathway to pathogenesis in Acinetobacter. PLoS ONE. 2013;8:e54287. doi: 10.1371/journal.pone.0054287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Touchon M, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conner CP, Heithoff DM, Julio SM, Sinsheimer RL, Mahan MJ. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc. Natl Acad. Sci. USA. 1998;95:4641–4645. doi: 10.1073/pnas.95.8.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heithoff DM, et al. Coordinate intracellular expression of Salmonella genes induced during infection. J. Bacteriol. 1999;181:799–807. doi: 10.1128/jb.181.3.799-807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hazen TH, et al. RNA-Seq analysis of isolate- and growth phase-specific differences in the global transcriptomes of enteropathogenic Escherichia coli prototype isolates. Front. Microbiol. 2015;6:569. doi: 10.3389/fmicb.2015.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donnenberg MS, et al. Bacterial factors associated with lethal outcome of enteropathogenic Escherichia coli infection: genomic case-control studies. PLoS Negl. Trop. Dis. 2015;9:e0003791. doi: 10.1371/journal.pntd.0003791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahl JW, et al. A comparative genomic analysis of diverse clonal types of enterotoxigenic Escherichia coli reveals pathovar-specific conservation. Infect. Immun. 2011;79:950–960. doi: 10.1128/IAI.00932-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 39.Orvis J, et al. Ergatis: a web interface and scalable software system for bioinformatics workflows. Bioinformatics. 2010;26:1488–1492. doi: 10.1093/bioinformatics/btq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galens K, et al. The IGS standard operating procedure for automated prokaryotic annotation. Stand. Genomic Sci. 2011;4:244–251. doi: 10.4056/sigs.1223234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saeed AI, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 43.R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2010. [Google Scholar]
- 44.Angiuoli SV, Salzberg SL. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics. 2011;27:334–342. doi: 10.1093/bioinformatics/btq665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.