Abstract
Acetylation is a post-translational modification (PTM) that regulates chromatin dynamics and function. Dysregulation of acetylation or acetyltransferase activity has been correlated with several human diseases. Many, if not all histone acetyltransferases (HATs) are regulated in part through tethered domains, association with binding partners or post-translational modification, including predominantly acetylation. This review focuses on what is currently understood at the molecular level of HAT regulation as it occurs via binding partners, associated domains, and autoacetylation.
INTRODUCTION
Acetylation is one of several post-translational modifications (PTMs) that occurs on histones to regulate chromatin dynamics and function in DNA transcription, replication, repair, and other DNA-templated activities. The hypothesis that histone acetylation could regulate gene expression was first proposed by Allfrey and coworkers in 1964.1 However, it was not until over another thirty years that the first histone acetyltransferase HAT1 was identified by Sternglanz and coworkers2 and Gottschling and coworkers.3 In 1996 Allis and coworkers isolated the first acetyltransferase GCN5 from tetrahymena, opening the door to the identification of other histone PTMs, the enzymes that mediated the modifications, and their roles in chromatin regulation.4
Of the multiple histone acetyltransferases (HATs) that have been identified to date, the most studied have been classified into five subfamilies based on sequence homology and substrate acetylation properties: HAT1, GCN5/PCAF, MYST, p300/CBP, and Rtt109. These HATs all share a structurally conserved Acetyl Coenzyme A (AcCoA) binding core, and within these subfamilies proteins share catalytic mechanisms and structurally conserved core flanking regions. Interestingly, despite the structural homology between many of these proteins, sequence homology is limited for HATs, with the exception of GCN5/PCAF, and HAT1 which belong to the GCN5-related N-acetyltransferase (GNAT) superfamily sharing four regions of sequence conservation spanning ~100 amino acids 5 despite the different substrate specificities between GCN5/PCAF and HAT1 as reviewed in6.
Histones are not the only proteins that are acetylated. Through acetylomic studies, we now know that thousands of proteins are acetylated throughout the cell, spanning a wide class spectrum, including transcription factors, kinases, ubiquitin ligases, ribosomal proteins and metabolic enzymes, to mediate a broad range of cellular activities, including cell cycle control, DNA damage check-points, cytoskeleton organization, endocytosis and metabolism.7–10 While some of the non-histone acetyltransferases have been identified, such at the α-tubulin acetyltransferase, αTAT111, 12 and the cohesin acetyltransferase, ESCO1 in humans and Eco1 in yeast,13–15 the biologically relevant enzymes, if any, for many of the other acetylation sites are not known. It should be noted that several HATs such p300/CBP have been shown to also acetylate many non-histone substrates.
Given the pervasiveness of acetylation in the cell, it is not surprising that aberrant protein acetylation or acetyltransferase function has been correlated with several human diseases. For example, the CBP HAT forms translocation products with MLL (mixed lineage leukemia) and the MOZ HAT (monocytic leukemia zinc-finger protein) in a subset of acute myeloid leukemias.16 In addition, the p300 HAT is mutated in a subset of colorectal and gastric cancers making it a bona fide tumor suppressor.17 The leading diabetes drug, Metformin, was recently shown to act through p300/CBP inhibition and heterozygous CBP knockout mice are lean and exhibit increased insulin sensitivity.18
Because of their important biological roles and these links to disease, it is not surprising that HATs, and the HAT domains within proteins in particular, have been extensively studied at the biochemical and structural level. This has lead to a mechanistic understanding of HAT domain architecture, histone substrate binding and enzymology as summarized above and described in more detail in several review articles.6, 19–22 However, many, if not all HATs are regulated themselves through tethered domains, their association with binding partners or through post-translational modification, most notably acetylation. Such HAT regulation appears to be at the heart of HAT structure/function, yet we are only beginning to understand this at the molecular level. In this review we will highlight what is currently known at the molecular level regarding the ways in which HAT proteins are regulated by other HAT-associated domains, their binding partners and by autoacetylation.
REGULATION BY BINDING PARTNERS AND ASSOCIATED DOMAINS
HAT1
Histone acetyltransferase 1 (Hat1) was initially identified in the cytosolic extract of S. cerevisiae and characterized as a type B HAT for its localization and ability to acetylate free histones.2, 3 While Hat1 was subsequently found to be largely localized in the nucleus of cells,23–26 it maintains its classification as a type B HAT for its unique specificity for free histones over nucleosomes. Specifically, Hat1 acetylates Lys5 and Lys12 on nascent histone H4.2, 3 Hat1 was also found to be the main catalytic component of the yeast HAT-B complex, where Hat1p is bound to Hat2p (RbAp46 in humans), which increases its catalytic activity 10-fold, and enhances histone H4 binding capacity.3 Crystal structures of Hat1 in the presence of its cofactor, substrate peptides, and Hat2p binding partner have been reported and help to elucidate the mechanism of enzymatic regulation. These details are reviewed below.
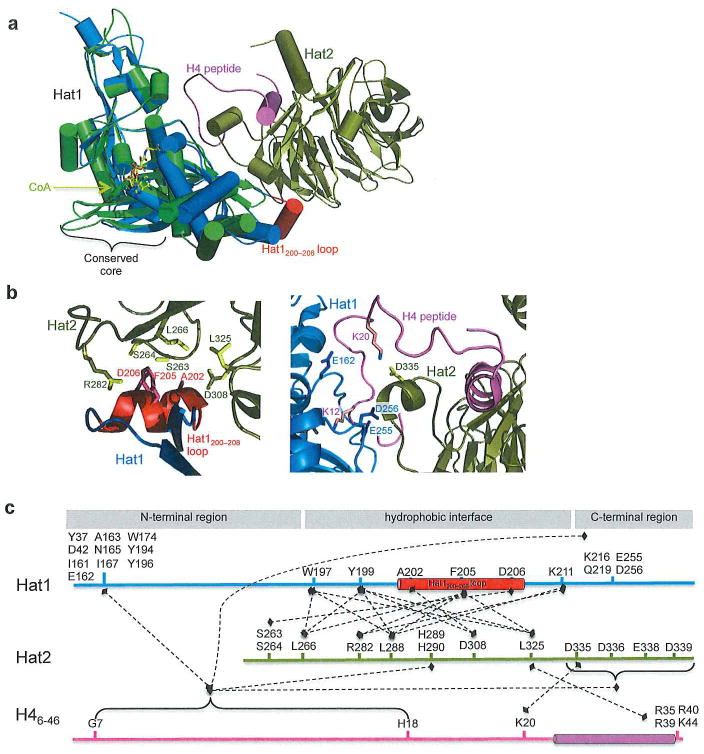
A recent study reports the crystal structure of S. cerevisiae Hat1p in complex with Hat2p bound to Co enzyme A (CoA) and H41-48 peptide at a 1:1:1:1 stoichiometry to 2.0Å (Figure 1A).27 In this structure, the C-terminal region of Hat1 binds the cofactor CoA at the conserved core comprised of three-stranded β-sheet with an α-helix spanning one side. yHat1 in this structure overlays well (RMSD~0.376) with the reported human HAT1 structure bound to AcCoA, implying that binding Hat2p does not induce a conformational change in Hat1p.28 Both the uncomplexed and complexed Hat1 reveal a poorly conserved loop of Hat1 that spans residues 200–208 in yHat1. Surprisingly, this loop is responsible for the bulk of the interactions between Hat1 and Hat2 (Figure 1B, left). Specifically, some key interactions between the Hat1 loop and Hat2 include hydrogen bonding between Ala202 of Hat1 and Asp308 of Hat2, hydrophobic interactions and hydrogen bonds between Phe205 of Hat1 and Leu266 and Leu325 and Arg282 of Hat2, respectively, and side chain hydrogen bonds between Asp206 of Hat1 and Ser263 and Ser264 of Hat2. Furthermore, deletion of this loop significantly compromises the interaction between Hat1 and Hat2.27 Because this loop is not well conserved between Hat1 orthologs of higher eukaryotes, it is hypothesized that another protein could bridge interactions between Hat1 and Hat2 in higher organisms. One report suggests that H4 could potentially fulfill this role as it interacts with both Hat1 and Hat2 in the yeast complex structure (Figure 1b, right).27 Specifically, H4 peptide binds to the Hat1/Hat2 complex in three distinct regions (Figure 1b, right and Figure 1c). The N-terminal portion of H4 binds at the interface between Hat1 and Hat2 making the majority of its contacts with Hat1, confirming what was reported in the HAT1/AcCoA/H4 structure.28 The second portion of H4 contains Lys20, which forms a salt bridge with Asp335 on a long loop of Hat2. The C-terminal portion of the H4 forms a helix and is inserted into a groove on Hat2, in agreement with the reported human structure of Hat2 and H4.29 H4 peptide binding to the Hat1/Hat2 complex reveals that H4K12 is positioned in the active site of Hat1 with CoA on the opposite side. This positioning allows the ε-nitrogen to accept an acetyl moiety in agreement with the catalytic mechanism proposed in the human structure publication, where it was proposed that Glu187 and Glu276 of HAT1 (corresponding to Glu162 and Glu255 in S. cerevisiae) would act as a general base for catalysis with an additional contribution from Asp277 (Asp256 in S. cerevisiae) to deprotonate H4K12 prior to nucleophilic attack on AcCoA. Further biochemical studies also reveal that the Hat1/Hat2 complex is specific for H4K12 in vitro27 while previous reports had shown that Hat1 could acetylate both H4K12 and H4K5 to a lesser extent.3 This data suggests that Hat2 facilitates Hat1 acetylation by increasing the substrate binding specificity of Hat1, and that this specificity appears to occur largely because Hat2 binds a helical portion of the H4 peptide, which helps position H4K12 in the Hat1p active site.27 Furthermore the HAT1/HAT2/CoA/H4 complex was crystalized with H31–15 peptide to 2.5Å27. This structure revealed that H3 could bind to the WD40 domain of Hat2p. Further biochemical studies showed that Hat2 strongly prefers unmodified or monomethylated H3 over trimethylated H3, which may explain how the Hat1/Hat2 complex can select for newly synthesized H3/H4 heterodimers, although more investigation would be needed to fully understand the mechanism of this recognition.
Figure 1.
Hat1/Hat2 complex. (a) Uncomplexed Hat1(green) overlaid with Hat1/Hat2 complex (Hat1 in marine, Hat2 in gold, Hat1200–208 loop in red, CoA in yellow, and histone H41–48 in pink). The region containing the conserved core is indicated. (PDB codes: 2P0W and 4PSW). (b) Zoomed in view of select interactions between Hat1 and Hat2. Left- Interactions between Hat1 loop and Hat2. Right- Interactions between Hat1, Hat2, and histone H4 peptide. (c) Linear representation of select Hat1, Hat2, and H4 peptide interactions. Dashed lines represent interactions. Domain and secondary structure naming correspond to primary texts.
In summary while Hat1 can bind substrate and cofactor independently of a binding partner, the crystal structure of the HAT-B complex reveals that Hat2 likely increases the catalytic efficiency of Hat1 by increasing the overall affinity and specificity for H4K12 acetylation.
MOF
Males absent on the first (MOF) is a histone acetyltransferase member of the MYST family (named for founding members MOZ, Ybf2/Sas3, Sas2 and Tip60). MOF was first identified as the essential acetyltransferase component of the drosophila dosage compensation male specific lethal (MSL) complex, which also contains proteins MSL1, MSL2, MSL3, MLE and noncoding RNAs roX1 and roX2.30, 31 Additional studies of the MSL complex found that MSL1 and MSL3 together in complex with MOF could stimulate acetyltransferase activity by 30-fold and redirect substrate specificity exclusively towards nucleosomal H432. MOF and other members of the MSL complex are conserved throughout higher eukaryotes.33, 34 Four human orthologs have been identified for members of the drosophila MSL complex (MSL1, MSL2, MSL3 and MOF) that target the same substrate as the drosophila complex, lysine 16 on histone H4.
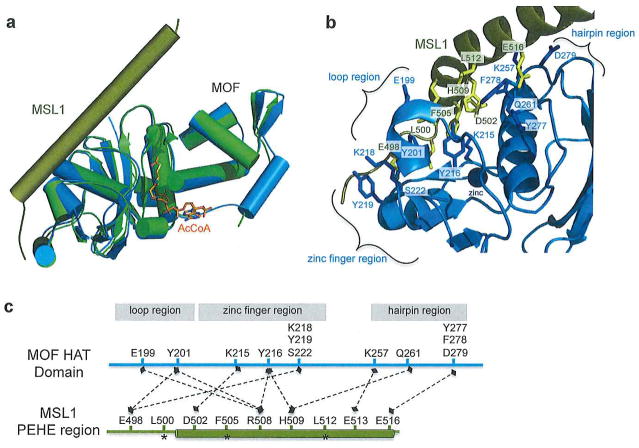
In 2011 the first crystal structures of MOF in complex with MSL1 as well as a structure of MSL1 in complex with MSL3 were reported, providing the first structural insights into how MOF is regulated by its binding partners in the MSL complex.35 The MOF/MSL1 structure reveals the interaction of the MOF HAT domain (residues 174–458) bound to AcCoA and an N-terminal portion of the MSL1 PEHE region (residues 470–540) to 2.8Å (Figure 2a). The MOF portion of this structure overlays with the uncomplexed structure of MOF (PDB code: 2GIV) with an RMSD of 0.510, demonstrating that binding to MSL1 does not induce any major conformational changes in MOF. The MSL1 fragment in this structure forms a loop at the N-terminus followed by a 52Å long helix, which makes numerous, mainly charged, contacts with the N-terminal region of the MOF HAT domain (Figure 2b). MSL1 interacts at three main regions of MOF: the zinc finger (hydrogen bonding occurs between Lys215 of MOF and Asp502 of MSL1; Tyr216 of MOF and Arg508 and His509 of MSL1; and Lys218, Tyr219, and Ser222 of MOF and Glu498 of MSL1), a loop region spanning residues 197–205 (hydrogen bonding occurs between Glu199 of MOF and Arg508 of MSL1; and Tyr201 of MOF and Arg508 and Glu498 of MSL1), and the hairpin region of MOF which spans residues 257–281 (hydrogen bonding occurs between Lys257 of MOF and Glu513 of MSL1; Gln261 of MOF and His509 of MSL1; and Tyr277, Phe278, and Asp279 of MOF and Glu516 of MSL1) (Figure 2c). All of the MSL1 interacting residues are highly conserved between species. Another nearly identical structure of MSL1 in complex with MOF has also been reported where the N-terminal region of MSL1 is resolved and reveals an antiparallel β-strand of MSL1 that interacts with β2 of MOF, forming a β-sheet involving both proteins.36
Figure 2.
MOF/MSL1 complex. (a) Uncomplexed MOF (green) overlaid with MOF/MSL1 complex (MOF in marine, MSL470540 in gold, AcCoA in orange) (PDB codes: 2Y0M and 2GIV). (b) Zoomed in view of select interactions between the MOF HAT domain and MSL1. Zinc atom represented as navy sphere. (c) Linear representation of select interactions between the MOF HAT domain and MSL1 PEHE region. Dashed lines represent interactions. Domain and secondary structure naming correspond to primary texts. Asterisks denote residues that are inserted into hydrophobic pockets of their binding partner.
The structure of an MSL1 fragment (residues 545–597) in complex with the MRG domain of MSL3 (residues 167–517, with loop deletion) to 3Å has also been reported where the MSL1 fragment wraps around the MSL3 protein interacting through a series of conserved residues that are C-terminal to the region of MSL1 that interacts with MOF35. Through biochemical assays it was shown that similar to the drosophila MSL complex, the MSL1 PEHE domain and MSL3 MRG domain were sufficient for enhancing the activity and specificity of human MOF. Specifically, while MOF in the presence or absence of MSL1 and MSL3 is able to acetylate free histones H3 and H4, the complex can only acetylate histone H4 in the context of a nucleosomal substrate 32. Studies of the drosophila MSL complex have shown that MSL1 and MSL3 play a role in recruiting MOF to nucleosomal and free DNA,32 which could perhaps explain how this complex enhances the specificity and efficiency of MOF. This biochemical data, taken together with the two reported structures demonstrates that MSL1 likely acts as a scaffold of the MSL complex to bring together various functional components to enhance MOF activity and specificity. However, to understand the molecular details of this enhancement, further research will need to be done investigating the interaction of different members of the MSL complex with nucleosomes.
The enzymatic activity of the yeast paralog of MOF, Esa1, has been previously dissected. Interestingly, while the isolated Esa1 HAT domain has been shown to employ a ping-pong catalytic mechanism involving residues Cys304 and Glu33837, the Esa1-containing NuA4 complex, shows significant, albeit clearly compromised catalytic activity with the Esa1-C304S mutant,38 arguing against a Cys304-dependent ping-pong catalytic mechanism in the context of the NuA4 complex. This data suggests that Esa1 can use different catalytic mechanism depending on context. Esa1 Cys304 and Glu338, are conserved throughout the MYST family and whether the catalytic mechanism of MOF might also be modulated by its association with binding partners is still an open question that requires further investigation.
p300
p300 was first identified as a protein that bound to the adenovirus early region 1A (E1A) protein.39 Just over 10 years later it was shown that p300, as well as its paralog CBP, were also histone acetyltransferases that could regulate transcriptional activity.40, 41 Since then, studies characterizing p300, its binding partners, substrates, and disease associations have boomed (reviewed in 22). The promiscuity of p300 sets it apart from many of the other HATs. In 2006, p300 was known to interact with more than 312 proteins.42 Today, that number exceeds 411, and further exemplifying the functional reach of p300, approximately 100 proteins have been reported as substrates for p300 HAT activity.22 We can begin to understand the mechanisms of regulation by analyzing the structure of the p300 HAT domain bound to a bisubstrate inhibitor.43 and the structure of the larger multidomain p300 core region.44
The crystal structure of the semi-synthetic p300 HAT domain in complex with the bisubstrate inhibitor Lys-CoA to 1.7Å was reported. 43 This structure revealed that p300 contains a unique ~23-residue substrate-binding loop which encapsulates the Lys-CoA inhibitor and may contribute to the tight cofactor binding demonstrated by p300. Furthermore, p300 contains a highly electronegative patch 10Å away from the substrate lysine-binding site providing insight as to why p300 prefers a proximal basic residue in its substrates. Further biochemical studies revealed that unlike other HATs, p300 does not seem to employ a general base for catalysis. Rather, the two imperative catalytic residues of p300 are Tyr1467 and Trp1436, where Tyr1467 is hypothesized to orient AcCoA and act as a general acid to protonate the leaving group and Trp1436 orients the substrate lysine for nucleophilic attack43.
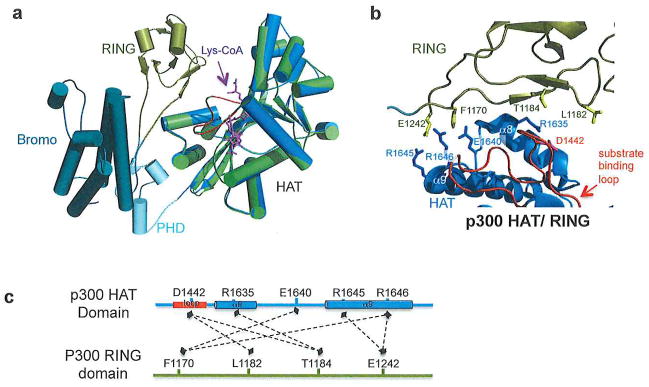
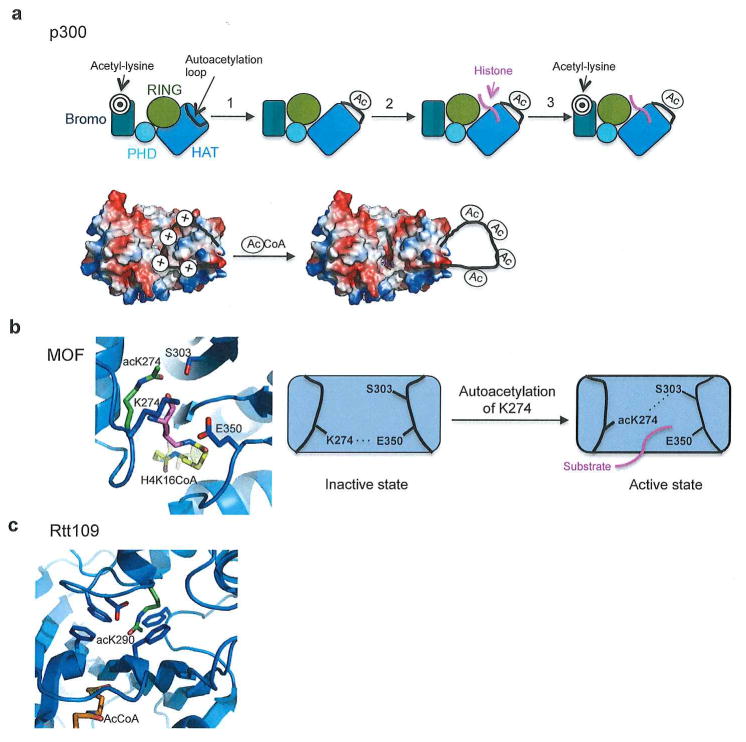
Extending beyond the HAT domain of p300, the crystal structure of the p300 catalytic core, which contains the p300 bromodomain, a discontinuous PHD domain, a RING domain, and CH2 region in addition to the HAT domain in complex with Lys-CoA has been reported (Figure 3a).44 Interestingly, the presence of the PHD and RING domain in this structure came as a surprise, since sequence analysis had failed to predict the presence of these domains. In this structure the domains form a compact module and interact with each other through a series of hydrogen bonds and hydrophobic interactions. The HAT domain of this structure shows little deviation from the isolated p300 HAT domain only structures (RMSD~0.5). The position of the RING domain in this structure implicates a regulatory role as it is flexibly positioned in close proximity to the lysine portion of Lys-CoA and the substrate-binding loop of the HAT domain (Figure 3b). The RING domain electrostatically interacts with the HAT domain through two long flexible linkers including a salt bridge between Glu1242 of the RING domain and Arg1645 and Arg1646 of the HAT domain (Figure 3c). Arg1645 and Glu1640 of the HAT domain also interact with Phe1170 of the RING domain. Additional interactions occur between Asp1442 of the HAT domain and Leu1182 and Thr1184 of the RING domain, and Arg1635 of the HAT domain and Thr1184 of the RING domain. Biochemical studies in which the RING domain was deleted, or the RING-HAT interaction was destabilized, showed an increase in p300 catalytic activity revealing that the RING domain negatively regulates p300 HAT activity.44 Biochemical probing of the bromodomain in this structure revealed that it is the sole mediator for nuclear localization to hyperacetylated chromatin. However, deletion or mutation of the bromodomain had no effect on the HAT activity of p300 implicating that the bromodomain is not essential for substrate acetylation. Using this structure, a model for p300 regulation was proposed (see Figure 5a, top) whereby p300 first becomes autoacetylated (discussed in further detail in the autoacetylation section below) and the ring domain is displaced leading to catalytic activation of p300 thus allowing initial substrate histone acetylation. The bromodomain then targets acetyl-lysine residues on histones to keep p300 bound to histones for subsequent p300-mediated histone acetylation44. We also propose that the bromodomain of p300 could also contribute to the initial histone substrate targeting prior to autoacetylation and subsequent histone acetylation.
Figure 3.
p300 core domain (a) HAT domain of p300 (green) overlaid with p300 catalytic core (HAT domain in marine, RING domain in gold, PHD domain in cyan, Bromodomain in dark teal, substrate binding loop in red, Lys-CoA in purple) (PDB codes: 3BIY, 4BHW). (b) Zoomed in view of select interactions between RING and HAT domains of p300. (c) Linear representation of select interactions between the p300 RING and HAT domains. Dashed lines represent interactions. Secondary structure naming corresponds to primary texts.
Figure 5. HAT Schematics.
(a) Top- Schematic of proposed p300 mechanism. HAT domain is colored marine, RING domain is colored gold, PHD domain is colored cyan, Bromodomain is colored dark teal, and substrate binding loop is colored black, correlating with Figure 1. It is possible that p300 uses its bromodomain initially to recruit target substrates (represented by target symbol). Step 1: p300 becomes acetylated at the autoinhibitory loop that displaces the loop and the RING domain. Step 2: p300 is able to acetylate substrates (shown in pink). Step 3: Bromodomain domain may bind target substrates following initial substrate acetylation to regulate further acetylation. Figure adapted from44. Bottom- Model for p300 activation by autoacetylation. Figure adapted from70. (b) Close up and schematic of autoacetylation of MYST family acetyltransferases (MOF used as a representative member.) When unacetylated K274 forms a hydrogen bond with the general base Glu350. When K274 becomes acetylated (shown in green) it flips 90° out of the active site and forms a hydrogen bond with Ser303, which may permit substrate binding (shown in pink). Overlaying of the H4K16CoA from the ESA1/H4K16CoA structure (PDB code: 3TO6) reveals that the substrate would clash with unacetylated K274. Figure adapted from70. (C) The autoacetylated K290 (shown in green) of Rtt109 in its buried environment. Figure adapted from70.
Rtt109
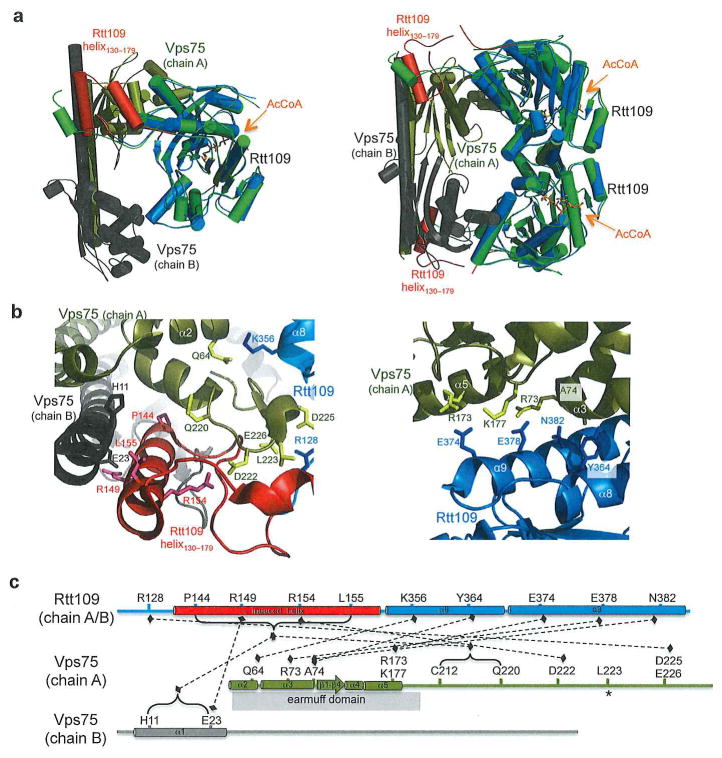
Rtt109 (regulation of Ty1 transposition protein 109) is a fungal-specific HAT that requires one of two binding partners for full activity. Rtt109 transiently associates with histone chaperone Asf1 to target H3K56 for acetylation on nascent histones to mediate nucleosomal assembly.45–47 Rtt109 can also bind with vacuolar protein sorting 75 (Vps75) to form a stable complex with nanomolar binding affinity.48 Although the Rtt109-Vps75 complex has previously been reported to contribute to H3K56 acetylation in vitro 45, 49 more recent studies have demonstrated that this complex mainly targets H3K9 and H3K27 for acetylation within the histone H3 tail.50–53 Structures of Vps75 demonstrate that it forms a symmetrical homodimer.51, 54, 55 Interestingly, biochemical studies on the Rtt109-Vps75 complex have found that it can exist in two different stoichiometries (2Vps75:2Rtt109 and 2Vps75:1Rtt109), possibly suggesting that Vps75 dimers associate with one Rtt109 molecule at a high affinity before binding a second Rtt109 protein at a lower affinity.55 To gain further insights into the mechanism of regulation of Rtt109 by Vps75, two crystal structures at the two different stoichiometries have been reported.53, 56
A crystal structure of full length Rtt109 and the core domain of Vps75 (residues 1–232) to 2.8Å in a 2:2 stoichiometry53 (Figure 4a, right) was reported around the same time as two crystal forms of full length Rtt109 and Vps75 to 2.7Å resolution in a 2:1 stoichiometry50 (Figure 4a, left). Despite their differing stoichiometries, both structures aligned with an RMSD of ~0.7Å for their shared atoms and shared several common features regarding the Rtt109-Vps75 interaction. Both structures reveal that binding of Vps75 to Rtt109 causes the otherwise disordered region of Rtt109 (residues 130–179) to form a helix that is essential for Vps75 association (Figure 4b, left). In the previous structures of Rtt109 in the absence of histone chaperone, this region was either deleted or disordered.55, 57 Confirming this biochemically, both reports show that this region of Rtt109 is sensitive to proteolytic cleavage in the absence of Vps75. With the exception of this region, both the Rtt109 and Vps75 proteins in complex overlay well with their respective nascent structures (RMSD of 0.75 and 1.14 respectively) indicating that association of Vps75 does not induce any conformational changes in Rtt109. Both reports also identified a second interface that was important for the Rtt109-Vps75 interaction that occurs where the Rtt109-α8-α9 associates with the Vps75 earmuff domain at helices α2–α5 (Figure 4b, right). At this region there is a mixture of hydrophobic and electrostatic interactions that drive the association of a mostly negative patch of Rtt109 with a mostly positive patch of Vps75 (Figure 4c). To name a few of the interactions, Lys356 and Glu378 of Rtt109 form hydrogen bonds with Gln64 and Arg73 of Vps75, respectively. Both Arg173 and K177 of Vps75 form salt bridges with Glu374 of Rtt109, and Tyr364 makes hydrophobic interactions with Ala74. Both structures reveal an internal enclosure/reaction core, albeit of different sizes (~30Å diameter for the 2:1 stoichiometry complex versus ~12Å diameter for the 2:2 stoichiometry complex) (Figure 4a). Despite these size differences both studies are led to conclude that this enclosed core acts to properly orient (H3-H4)2 for productive H3 tail acetylation. This is consistent with the finding that the presence of Vps75 results in a 250-fold increase in the catalytic efficiency of Rtt109 with a relatively unchanged Km.51 The Rtt109-Vps75 interaction was further biochemically characterized, finding that the Vps75 dimer is required for full Rtt109 association and acetylation of histones53. Furthermore, while both studies confirm that the helical Vps75 interacting regions of Rtt109 (residues 130–175 and the α8–α9 interface region) are required for optimal Vps75 binding and activation of Rtt109,50, 53 neither of these regions are required for Asf1 stimulation of Rtt109, revealing that the Vps75 and Asfl chaperones activate Rtt109 via distinct mechanisms53.
Figure 4.
Rtt109/Vps75 complex (a) Side by side comparison of the Vps75/Rtt109 in the 2:1 stoichiometry (left) and 2:2 stoichiometry (right). Note that in the 2:2 model residues 129–133 are not modeled. The uncomplexed Rtt109 is shown in green and the complex is shown as follows: Rtt109 in marine, Vps75 chain A in gold, Vps75 chain B in gray, AcCoA in orange. (PDB codes: 2ZFN, 3Q68, and 3Q33). (b) Zoomed in view of select interactions between Rtt109 and Vps75. Left- interactions between Rtt109 and chains A and B of Vps75. Right- Interactions between Rtt109 and the earmuff domain of Vps75 chain A. (c) Linear representation of select interactions between Rtt109 and chains A and B of Vps75 that occur in both the 2:1 and 2:2 model. Domain and secondary structure naming correspond to primary texts. Asterisk denotes residue that is inserted into hydrophobic pocket of binding partner.
Despite the biochemical data supporting the existence of the Rtt109-Vps75 complex in both the 2:2 and 2:1 stoichiometry, more studies will need to be carried out to understand if both stoichiometries are employed in a Rtt109/Vps75-mediated histone H3 tail acetylation pathway or if there are cellular circumstances in which one of the complex stoichiometries would be preferred over the other. Nonetheless, with the information presented in both structures it appears clear that Vps75 regulates Rtt109 activity by delivering substrate to the reactive core of the complex via a pathway independent of Rtt109 activation by Asf1.
AUTOACETYLATION
Another mechanism of regulation for acetyltransferases is autoacetylation. Currently, p300, the MYST family of acetyltransferases, Rtt109, and PCAF are all known to be regulated by acetylation to some degree. Interestingly, despite the prevalence of this modification amongst acetyltransferases, each HAT subfamily appears to be regulated by autoacetylation via unique mechanisms (Table 1). Details are provided below.
Table 1.
Autoacetylation of HATs
| Protein | Relevant autoacetylated lysine(s) | Functions of acetylation |
|---|---|---|
| p300 | K1499, K1549, K1554, K1560 |
|
| MOF | K274 |
|
| Rtt109 | K290 |
|
| PCAF | K416, K428, K430, K431, K442 |
|
Only autoacetylated lysines with a known effect on enzyme biochemistry have been listed. MOF has been listed as a representative member of the MYST family.
p300
Within the p300 HAT domain there exists a ~40 residue proteolytically sensitive lysine-rich loop that can become heavily acetylated to promote catalytic activity. Studies have found that if this loop of p300 (spanning residues 1520–1560) is deleted the enzyme becomes constitutively active decreasing the Km for both AcCoA and substrate peptide rather than increasing the catalytic rate.58 This loop is hypothesized to act as an autoinhibitory loop that regulates p300 activity by blocking substrate binding when unmodified, and exposing the substrate binding region when acetylated.58 This model for autoinhibition of the hypoacetylated loop is consistent with the p300 HAT domain crystal structure, which shows a highly acidic substrate-binding site that could accommodate a basic hypoacetylated loop more avidly than a hyperacetylated loop (Figure 5a, bottom). This mechanism of autoinhibition was further supported when it was shown that deletion of this loop in vitro enhanced doxorubicin induced acetylation of the p300 substrate p7358. Along these lines, the effect of this loop deletion on the ability of p300 to bind the substrate ATF-2 was tested. Not only was the loop unnecessary for substrate binding, but also a hypoacetylated loop blocked substrate binding whereas a hyperacetylated loop enhanced substrate binding providing further evidence that this loop harbors autoinhibitory properties.59
MYST
A number of recent reports have revealed that autoacetylation of a conserved lysine residue promotes catalytic activity in the MYST proteins.60–63 In MOF and Tip60 it was shown that acetylation of K274 and K327, respectively, upregulated catalytic activity.60, 63 Furthermore, arginine and glutamine mutants mimicking lysine and acetylated lysine respectively displayed a >90% loss of catalytic activity for both enzymes, 62, 63 implying that the specific acetyl-lysine modification was required to catalytic activation. Kinetic analysis of deacetylated Tip60 revealed both an increase in Km and a decrease in kcat, implying that autoacetylation in this case affects both substrate binding and catalytic rate.63 Further biochemical assays also showed that hMOF K274 mutants were defective in H4 binding.62 In a separate study, the autoacetylation of hMOF was correlated with decreased recruitment of the protein to chromatin in vivo, and decreased binding to nucleosomes in vitro.61 Furthermore, K274 mutants in dMOF display a decreased overall thermostability profile.35 K274 makes extensive contacts in the acetylated form that may contribute to the maintenance of the loop conformation for productive lysine substrate binding. These include hydrogen bonding of the acetyl carbonyl to Tyr301 and Ser303 and hydrophobic interactions of the acetyl methyl with Phe283 and Phe285.18, 60, 62 The crystal structure of unacetylated hMOF K274, reveals that the lysine is flipped out of the active site by 90° to both block cognate substrate binding and to form a hydrogen bond with the general base E350 (Figure 5b, right), suggesting that that unacetylated lysine residue inhibits cognate substrate lysine acetylation by both preventing substrate binding and by tying up a key catalytic residue (Figure 5b, left). 62
Rtt109
Autoacetylation of Rtt109 at lysine 290 is essential for enzyme activation.64 The acetyl group on this lysine allows it to be inserted into a hydrophobic pocket of conserved residues (Figure 5c).55, 57, 65 Interestingly, while it seemed that this residue would thus be important for the thermostability of Rtt109, it was found that the melting temperature of acetylated Rtt109 did not differ from that of deacetylated Rtt109, demonstrating that AcK290 likely does not contribute to protein stability.64 Rather, it was found that acetylation of K290 activated Rtt109 by increasing the affinity for AcCoA and also increasing the rate of catalytic turnover.64 In contrast to the MYST family of acetyltransferases, acetylation of Rtt109 K290 had no effect on histone substrate binding. The molecular basis by which autoacetylation of Rtt109 facilitates cognate histone lysine acetylation remains unclear.
PCAF
PCAF (p300/CBP-associated factor) was first identified as a p300/CBP binding protein with histone acetyltransferase activity,66 and further studies of PCAF demonstrated that PCAF becomes autoacetylated in vitro.67 Moreover, it was shown that the autoacetylation of PCAF occurs in vivo, and that while PCAF can also be acetylated intermolecularly by p300, it also has the capability to acetylate itself intramolecularly at lysines 416, 428, 430, 441, and 442.68 Following suit with the MYST family, PCAF displayed greater structural stability and a 2–3 fold increase in catalytic activity when autoacetylated.68 Noting that the five lysines targeted for autoacetylation in PCAF occur at the nuclear localization signal (NLS) region of PCAF, the effect of autoacetylation on nuclear localization was investigated. By creating a variety of PCAF constructs and mutants to mimic the unacetylated state, it was shown that acetylated PCAF localized to the nucleus whereas unacetylated PCAF localized to the cytoplasm, indicating that autoacetylation likely plays a role in the regulation of nuclear localization for PCAF.69
CONCLUSIONS
It is clear from the data reviewed here that we are only just beginning to understand how acetyltransferases are regulated and quite likely there is a wealth of information yet to be uncovered. While the majority of the biochemical and structural studies on HATs often focused on the isolated HAT domains of the respective proteins, we are finding that much of their regulation extends to adjacent domains and binding partners. Surprisingly, however, all of the reported structures discussed here reveal that binding partners and other domains are not causing any conformational changes in the HAT domains to alter activity. Rather, the trend seems to be regulation via autoinhibition, substrate delivery and at least in one case, localization. Moreover the structure of the p300 catalytic core, which revealed an unexpected regulatory RING domain, makes a strong case for the importance of structural biology for studying these proteins to fully understand their mechanisms of regulation. In the case of autoacetylation, although this modification appears to be correlated with the enhancement of catalytic activity, the mechanism for this activation is different between the HAT families. Whether such autoacetylation is reversible, and thus regulatory, is still an open question.
References
- 1.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc Natl Acad Sci U S A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleff S, Andrulis ED, Anderson CW, Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem. 1995;270:24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- 3.Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 4.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 5.Neuwald AF, Landsman D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem Sci. 1997;22:154–155. doi: 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- 6.Vetting MW, LPSdC, Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys. 2005;433:212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, Liu CF, Grishin NV, Zhao Y. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Proteomics. 2009;8:215–225. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith KT, Workman JL. Introducing the acetylome. Nat Biotechnol. 2009;27:917–919. doi: 10.1038/nbt1009-917. [DOI] [PubMed] [Google Scholar]
- 10.Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 11.Shida T, Cueva JG, Xu Z, Goodman MB, Nachury MV. The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc Natl Acad Sci U S A. 2010;107:21517–21522. doi: 10.1073/pnas.1013728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akella JS, Wloga D, Kim J, Starostina NG, Lyons-Abbott S, Morrissette NS, Dougan ST, Kipreos ET, Gaertig J. MEC-17 is an alpha-tubulin acetyltransferase. Nature. 2010;467:218–222. doi: 10.1038/nature09324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivanov D, Schleiffer A, Eisenhaber F, Mechtler K, Haering CH, Nasmyth K. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr Biol. 2002;12:323–328. doi: 10.1016/s0960-9822(02)00681-4. [DOI] [PubMed] [Google Scholar]
- 14.Hou F, Zou H. Two human orthologues of Eco1/Ctf7 acetyltransferases are both required for proper sister-chromatid cohesion. Mol Biol Cell. 2005;16:3908–3918. doi: 10.1091/mbc.E04-12-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams BC, Garrett-Engele CM, Li Z, Williams EV, Rosenman ED, Goldberg ML. Two putative acetyltransferases, san and deco, are required for establishing sister chromatid cohesion in Drosophila. Curr Biol. 2003;13:2025–2036. doi: 10.1016/j.cub.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 16.Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26:5395–5407. doi: 10.1038/sj.onc.1210608. [DOI] [PubMed] [Google Scholar]
- 17.Muraoka M, Konishi M, Kikuchi-Yanoshita R, Tanaka K, Shitara N, Chong JM, Iwama T, Miyaki M. p300 gene alterations in colorectal and gastric carcinomas. Oncogene. 1996;12:1565–1569. [PubMed] [Google Scholar]
- 18.He L, Sabet A, Djedjos S, Miller R, Sun X, Hussain MA, Radovick S, Wondisford FE. Metformin and insulin suppress hepatic gluconeogenesis through phosphorylation of CREB binding protein. Cell. 2009;137:635–646. doi: 10.1016/j.cell.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sapountzi V, Cote J. MYST-family histone acetyltransferases: beyond chromatin. Cell Mol Life Sci. 2011;68:1147–1156. doi: 10.1007/s00018-010-0599-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marmorstein R, Zhou MM. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol. 2014;6:a018762. doi: 10.1101/cshperspect.a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parthun MR. Histone acetyltransferase 1: More than just an enzyme? Biochim Biophys Acta. 2012;1819:256–263. doi: 10.1016/j.bbagrm.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dancy BM, Cole PA. Protein lysine acetylation by p300/CBP. Chem Rev. 2015;115:2419–2452. doi: 10.1021/cr500452k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 24.Poveda A, Pamblanco M, Tafrov S, Tordera V, Sternglanz R, Sendra R. Hif1 is a component of yeast histone acetyltransferase B, a complex mainly localized in the nucleus. J Biol Chem. 2004;279:16033–16043. doi: 10.1074/jbc.M314228200. [DOI] [PubMed] [Google Scholar]
- 25.Ruiz-Garcia AB, Sendra R, Galiana M, Pamblanco M, Perez-Ortin JE, Tordera V. HAT1 and HAT2 proteins are components of a yeast nuclear histone acetyltransferase enzyme specific for free histone H4. J Biol Chem. 1998;273:12599–12605. doi: 10.1074/jbc.273.20.12599. [DOI] [PubMed] [Google Scholar]
- 26.Ai X, Parthun MR. The nuclear Hat1p/Hat2p complex: a molecular link between type B histone acetyltransferases and chromatin assembly. Mol Cell. 2004;14:195–205. doi: 10.1016/s1097-2765(04)00184-4. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Zhang L, Liu T, Chai C, Fang Q, Wu H, Agudelo Garcia PA, Han Z, Zong S, Yu Y, Zhang X, Parthun MR, Chai J, Xu RM, Yang M. Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex. Genes Dev. 2014;28:1217–1227. doi: 10.1101/gad.240531.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu H, Moshkina N, Min J, Zeng H, Joshua J, Zhou MM, Plotnikov AN. Structural basis for substrate specificity and catalysis of human histone acetyltransferase 1. Proc Natl Acad Sci U S A. 2012;109:8925–8930. doi: 10.1073/pnas.1114117109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, Laue ED. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bone JR, Lavender J, Richman R, Palmer MJ, Turner BM, Kuroda MI. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev. 1994;8:96–104. doi: 10.1101/gad.8.1.96. [DOI] [PubMed] [Google Scholar]
- 32.Morales V, Straub T, Neumann MF, Mengus G, Akhtar A, Becker PB. Functional integration of the histone acetyltransferase MOF into the dosage compensation complex. EMBO J. 2004;23:2258–2268. doi: 10.1038/sj.emboj.7600235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, Wilm M, Stunnenberg HG, Saumweber H, Akhtar A. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 34.Smith ER, Cayrou C, Huang R, Lane WS, Cote J, Lucchesi JC. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol. 2005;25:9175–9188. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kadlec J, Hallacli E, Lipp M, Holz H, Sanchez-Weatherby J, Cusack S, Akhtar A. Structural basis for MOF and MSL3 recruitment into the dosage compensation complex by MSL1. Nat Struct Mol Biol. 2011;18:142–149. doi: 10.1038/nsmb.1960. [DOI] [PubMed] [Google Scholar]
- 36.Huang J, Wan B, Wu L, Yang Y, Dou Y, Lei M. Structural insight into the regulation of MOF in the male-specific lethal complex and the non-specific lethal complex. Cell Res. 2012;22:1078–1081. doi: 10.1038/cr.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan Y, Barlev NA, Haley RH, Berger SL, Marmorstein R. Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol Cell. 2000;6:1195–1205. doi: 10.1016/s1097-2765(00)00116-7. [DOI] [PubMed] [Google Scholar]
- 38.Berndsen CE, Albaugh BN, Tan S, Denu JM. Catalytic mechanism of a MYST family histone acetyltransferase. Biochemistry. 2007;46:623–629. doi: 10.1021/bi602513x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yee SP, Branton PE. Detection of cellular proteins associated with human adenovirus type 5 early region 1A polypeptides. Virology. 1985;147:142–153. doi: 10.1016/0042-6822(85)90234-x. [DOI] [PubMed] [Google Scholar]
- 40.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 41.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 42.Kasper LH, Fukuyama T, Biesen MA, Boussouar F, Tong C, de Pauw A, Murray PJ, van Deursen JM, Brindle PK. Conditional knockout mice reveal distinct functions for the global transcriptional coactivators CBP and p300 in T-cell development. Mol Cell Biol. 2006;26:789–809. doi: 10.1128/MCB.26.3.789-809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, Cole PA. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 2008;451:846–850. doi: 10.1038/nature06546. [DOI] [PubMed] [Google Scholar]
- 44.Delvecchio M, Gaucher J, Aguilar-Gurrieri C, Ortega E, Panne D. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat Struct Mol Biol. 2013;20:1040–1046. doi: 10.1038/nsmb.2642. [DOI] [PubMed] [Google Scholar]
- 45.Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han J, Zhou H, Li Z, Xu RM, Zhang Z. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J Biol Chem. 2007;282:28587–28596. doi: 10.1074/jbc.M702496200. [DOI] [PubMed] [Google Scholar]
- 48.Albaugh BN, Kolonko EM, Denu JM. Kinetic mechanism of the Rtt109-Vps75 histone acetyltransferase-chaperone complex. Biochemistry. 2010;49:6375–6385. doi: 10.1021/bi100381y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han J, Zhou H, Li Z, Xu RM, Zhang Z. The Rtt109-Vps75 histone acetyltransferase complex acetylates non-nucleosomal histone H3. J Biol Chem. 2007;282:14158–14164. doi: 10.1074/jbc.M700611200. [DOI] [PubMed] [Google Scholar]
- 50.Su D, Hu Q, Zhou H, Thompson JR, Xu RM, Zhang Z, Mer G. Structure and histone binding properties of the Vps75-Rtt109 chaperone-lysine acetyltransferase complex. J Biol Chem. 2011;286:15625–15629. doi: 10.1074/jbc.C111.220715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berndsen CE, Tsubota T, Lindner SE, Lee S, Holton JM, Kaufman PD, Keck JL, Denu JM. Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75. Nat Struct Mol Biol. 2008;15:948–956. doi: 10.1038/nsmb.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh MC, Greenblatt JF. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol Cell Biol. 2008;28:4342–4353. doi: 10.1128/MCB.00182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang Y, Holbert MA, Delgoshaie N, Wurtele H, Guillemette B, Meeth K, Yuan H, Drogaris P, Lee EH, Durette C, Thibault P, Verreault A, Cole PA, Marmorstein R. Structure of the Rtt109-AcCoA/Vps75 complex and implications for chaperone-mediated histone acetylation. Structure. 2011;19:221–231. doi: 10.1016/j.str.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park YJ, Sudhoff KB, Andrews AJ, Stargell LA, Luger K. Histone chaperone specificity in Rtt109 activation. Nat Struct Mol Biol. 2008;15:957–964. doi: 10.1038/nsmb.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang Y, Meeth K, Jiang E, Luo C, Marmorstein R. Structure of Vps75 and implications for histone chaperone function. Proc Natl Acad Sci U S A. 2008;105:12206–12211. doi: 10.1073/pnas.0802393105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ge Z, Wang H, Parthun MR. Nuclear Hat1p complex (NuB4) components participate in DNA repair-linked chromatin reassembly. J Biol Chem. 2011;286:16790–16799. doi: 10.1074/jbc.M110.216846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin C, Yuan YA. Structural insights into histone H3 lysine 56 acetylation by Rtt109. Structure. 2008;16:1503–1510. doi: 10.1016/j.str.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 58.Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, Levrero M, Sartorelli V, Cotter RJ, Cole PA. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol. 2004;11:308–315. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- 59.Karanam B, Wang L, Wang D, Liu X, Marmorstein R, Cotter R, Cole PA. Multiple roles for acetylation in the interaction of p300 HAT with ATF-2. Biochemistry. 2007;46:8207–8216. doi: 10.1021/bi7000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun B, Guo S, Tang Q, Li C, Zeng R, Xiong Z, Zhong C, Ding J. Regulation of the histone acetyltransferase activity of hMOF via autoacetylation of Lys274. Cell Res. 2011;21:1262–1266. doi: 10.1038/cr.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu L, Li L, Lv X, Wu XS, Liu DP, Liang CC. Modulations of hMOF autoacetylation by SIRT1 regulate hMOF recruitment and activities on the chromatin. Cell Res. 2011;21:1182–1195. doi: 10.1038/cr.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan H, Rossetto D, Mellert H, Dang W, Srinivasan M, Johnson J, Hodawadekar S, Ding EC, Speicher K, Abshiru N, Perry R, Wu J, Yang C, Zheng YG, Speicher DW, Thibault P, Verreault A, Johnson FB, Berger SL, Sternglanz R, McMahon SB, Cote J, Marmorstein R. MYST protein acetyltransferase activity requires active site lysine autoacetylation. EMBO J. 2012;31:58–70. doi: 10.1038/emboj.2011.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang C, Wu J, Zheng YG. Function of the active site lysine autoacetylation in Tip60 catalysis. PLoS One. 2012;7:e32886. doi: 10.1371/journal.pone.0032886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Albaugh BN, Arnold KM, Lee S, Denu JM. Autoacetylation of the histone acetyltransferase Rtt109. J Biol Chem. 2011;286:24694–24701. doi: 10.1074/jbc.M111.251579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stavropoulos P, Nagy V, Blobel G, Hoelz A. Molecular basis for the autoregulation of the protein acetyl transferase Rtt109. Proc Natl Acad Sci U S A. 2008;105:12236–12241. doi: 10.1073/pnas.0805813105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 67.Herrera JE, Bergel M, Yang XJ, Nakatani Y, Bustin M. The histone acetyltransferase activity of human GCN5 and PCAF is stabilized by coenzymes. J Biol Chem. 1997;272:27253–27258. doi: 10.1074/jbc.272.43.27253. [DOI] [PubMed] [Google Scholar]
- 68.Santos-Rosa H, Valls E, Kouzarides T, Martinez-Balbas M. Mechanisms of P/CAF auto-acetylation. Nucleic Acids Res. 2003;31:4285–4292. doi: 10.1093/nar/gkg655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blanco-Garcia N, Asensio-Juan E, de la Cruz X, Martinez-Balbas MA. Autoacetylation regulates P/CAF nuclear localization. J Biol Chem. 2009;284:1343–1352. doi: 10.1074/jbc.M806075200. [DOI] [PubMed] [Google Scholar]
- 70.Yuan H, Marmorstein R. Histone acetyltransferases: Rising ancient counterparts to protein kinases. Biopolymers. 2013;99:98–111. [Google Scholar]