Abstract
Therapeutic proteins are powerful next-generation drugs able to effectively treat diverse and devastating diseases, but the development and use of biotherapeutics entails unique challenges and risks. In particular, protein drugs are subject to immune surveillance in the human body, and ensuing antidrug immune responses can cause a wide range of problems including altered pharmacokinetics, loss of efficacy, and even life-threating complications. Here we review recent progress in technologies for engineering deimmunized biotherapeutics, placing particular emphasis on deletion of immunogenic antibody and T cell epitopes via experimentally or computationally guided mutagenesis.
Graphical Abstract
Introduction
Therapeutic proteins represent the cutting edge of modern medicine, and advances in biotechnology are driving growth in the biopharmaceuticals market.[1] As biological entities, however, proteins are subject to immune surveillance in the human body,[2,3] and the induction of antidrug antibody responses can result in a wide range of sequelae including altered pharmacokinetics, loss of efficacy, and more dangerous complications such as hypersensitivity and anaphylactic responses, cross-neutralization of endogenous proteins, and deposition of toxic immune complexes.[4,5] Given the detrimental consequences of anti-biotherapeutic immune responses, there is growing consensus among regulators, physicians, and the biopharma industry that fully exploiting these powerful drugs requires assessment and mitigation of immunogenicity risk.[6-10]
A protein’s immunogenic potential is a complex function of diverse interacting factors.[4,10,11] Thus a variety of different approaches have been pursued to mitigate immunogenicity, including shielding proteins with chemical or biological blocking moieties (e.g., PEGylation,[12,13] XTENylation,[14] PASylation,[15] or reductive methylation[16]), explicitly training the immune system to tolerate proteins,[17] or implicitly rendering proteins tolerable by humanization (with emerging new engineering techniques for antibodies[18-22] as well as non-immunoglobulin proteins[23,24]). In any case, molecular recognition of exogenous proteins by antibodies, antigen presenting cells, and T cells is central to the anti-biotherapeutic immune response, and this review focuses on protein deimmunization by genetic manipulation of immunogenic subsequences, termed “epitopes”.
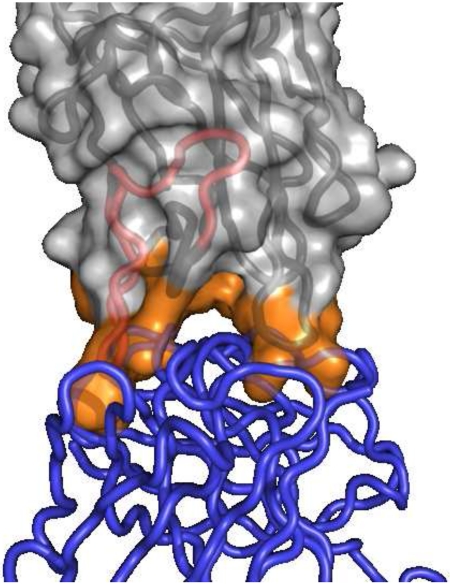
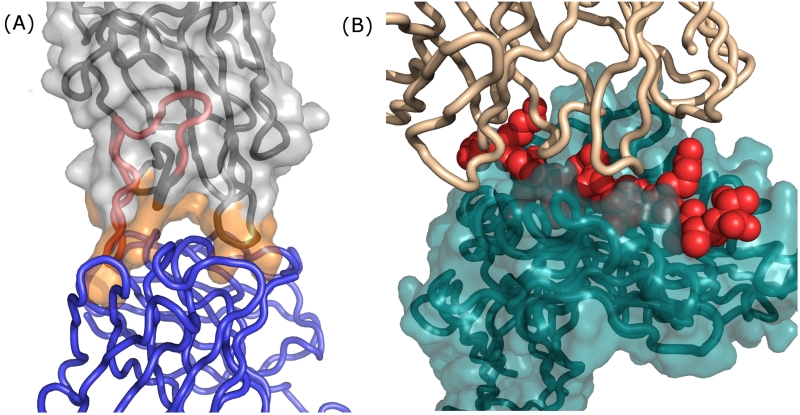
Strategies to deimmunize a protein by mutagenically “deleting” its epitopes are grounded in a detailed understanding of the cellular and molecular mechanisms underlying the antidrug immune response. There exist two very distinct types of epitope. Antibody epitopes are comprised of solvent exposed amino acids on the intact protein, and they constitute the binding sites for B cell receptors and soluble antidrug antibodies (Fig. 1A). T cell epitopes, in contrast, are short peptide fragments proteolytically processed from biotherapeutics (Fig. 1A & B), and they play a key upstream role in the antidrug antibody response. Briefly, a patient’s professional antigen presenting cells can internalize putative protein antigens and degrade them into peptide fragments. Immunogenic peptides, termed T cell epitopes, are then surface displayed via class II major histocompatibility complex molecules (MHC II or HLA in humans), where they can interface with surface receptors on cognate CD4+ helper T cells (Fig. 1B).[25] This ternary molecular recognition event initiates a signaling cascade that activates T cells, drives B cell maturation, and ultimately leads to production of high affinity antidrug antibodies.[4,26] (It bears noting that antidrug antibodies can be generated independent of T cell help, but these T independent responses are generally lower affinity and perhaps less significant than T cell dependent responses.[4,26])
Figure 1.
Recognition and binding of antibody versus T cell epitopes occurs via separate molecular mechanisms. (A) A co-crystal structure of the factor VIII C2 domain in complex with an inhibitory antibody; PDB id 1IQD.[85] The inhibitory antibody is rendered as a blue polypeptide backbone, and the factor VIII C2 domain is rendered as a molecular surface with the underlying polypeptide backbone shown in grey. C2 domain surface residues at the antibody binding interface are colored orange. Independent of the C2 antibody epitope, an experimentally validated C2 T cell epitope (IEDB id 131093)[62] is highlighted as a red segment on the polypeptide backbone. Note that a protein’s T cell epitopes may or may not overlap with its antibody epitopes. (B) Following proteolytic processing from internalized proteins, peptides that represent immunogenic T cell epitopes (red Van der Waals spheres) are bound in the cleft of class II MHC proteins (teal molecular surface with underlying teal peptide backbone); PDB id 1FYT.[86] The CDR regions of a cognate CD4+ T cell receptor are rendered as a tan polypeptide backbone. Formation of this ternary complex represents a key event that drives downstream development of high affinity antidrug antibodies. To reiterate, a T cell epitope need not share any amino acid residues with epitopes of resultant antidrug antibodies, though in this specific example there is at least some overlap. Images rendered with PyMOL (Schrodinger, LLC).
Antibody Epitope Deletion
Antidrug antibodies recognize and bind to sites on the surface of intact biotherapeutic agents, and these epitopes may be composed of either contiguous amino acid sequences (linear epitopes) or discontiguous residues that are close in space (conformational epitopes). Regardless, the protein-protein interaction surface exhibits shape complementarity and finely tuned energetics. Therefore, appropriate amino acid substitutions in a biotherapeutic can disrupt these binding interfaces and effectively delete antibody epitopes.[27•] The target epitopes for deletion are established by a range of strategies including simply focusing on hydrophilic surface residues,[28] random mutagenesis and high throughput screening,[29] more precise mapping using panels of antidrug antibodies,[27•,30•] or in rarer cases structure-guided design via antibody-antigen co-crystals.[31] In virtually all cases, however, substitutions that disrupt antibody binding while retaining biotherapeutic function are selected based on scanning alanine or trial and error mutagenesis.
Due to their importance in anti-cancer therapies, which can be undermined by immunogenicity, toxins have been the focus of substantial reengineering efforts to eliminate antibody epitopes. Diphtheria toxin has been deimmunized by mutagenic substitution of lysine, arginine, glutamine, and glutamic acid residues, amino acids which had been shown previously to be critical contributors to antibody epitopes of Pseudomonas exotoxin A (PE).[32] Inspection of the diphtheria toxin structure identified 24 target residues on the protein surface, and sequential rounds of mutagenesis and testing produced the 7-mutation variant dDTEGF13, which was found to elicit greatly reduced antidrug antibody titers in both BALB/c and C57Bl/6 mice while retaining potent cytotoxic activity both in vitro and in vivo.[28] It was presumed that antibody epitopes were deleted, though T cell epitope deletion (see below) was not definitively ruled out. The PE toxin itself has been the subject of extensive and systematic deimmunization efforts. Murine antibody epitopes of PE were mapped and deleted, yielding immunotoxin HA22-LR-8M that had better than wild-type activity in vitro, near wild-type efficacy in vivo, yet exhibited reduced antibody binding in vitro and reduced immunogenicity in BALB/c mice.[33] These mutations have been leveraged by others to create deimmunized bispecific immunotoxins based on PE.[34] It was subsequently found that human antibodies could bind PE epitopes distinct from those of murine antibodies, prompting a new effort that specifically targeted human antibody epitopes. The resulting variant HA22-LR-LO10 likewise exhibited near wild-type activity in vitro and wild-type efficacy in vivo, yet demonstrated dramatically reduced binding with human anti-serum from patients previously treated with various versions of the wild-type toxin sequence.[30•] These epitope-deleting mutations have been engineered into immunotoxins that exhibit potent anti-cancer activities,[35,36] and in particular the R06927005 immunotoxin variant (formerly RG7787) has entered human trials.
Recent literature describes several therapeutic enzymes and enzyme cofactors engineered for reduced antibody binding. E. coli type II asparaginase, an important therapeutic for acute lymphoblastic leukemia, is one such example. Focusing on a previously identified antibody epitope, the researchers introduced two mutations, K228S and Y176F, that increased cytotoxicity towards leukemic cells, decreased undesirable glutaminase activity, and reduced reactivity with immune serum from both mice and a leukemia patient treated with the wild-type enzyme.[37] Another recent example is deimmunization of factor VIII (FVIII), a life-saving therapy for hemophilia A patients. Guided by a FVIII-antibody co-crystal structure, 43 distinct point mutants within the antibody binding interface were generated, and their binding affinities for known inhibitory monoclonal antibodies (mAb) were measured. Ultimately the F2196K variant, which retained more than 75% wild type activity, was found to evade binding and inhibition by both human and murine mAbs directed against the C2 domain, but it failed to evade a murine mAb directed against the distal A2 domain.[31] This result highlights the challenge inherent to antibody epitope deletion in the context of polyclonal antidrug antibody responses; in the clinic, success will likely require redesign of multiple antigenic surface sites, as described above for the PE toxin. Thus, the F2196K variant represents a proof-of-concept for structure-guided antibody epitope deletion, but additional molecular engineering will be necessary in order to evade the diverse array of inhibitory antibodies that FVIII may elicit.[38]
Other strategies for antibody epitope deletion have leveraged large combinatorial libraries and high throughput screening. For instance, the anti-inflammatory agent chemotaxis inhibitory protein of Staphylococcus aureus (CHIPS) has been engineered by directed evolution so as to minimize binding of human antibodies. Randomly mutated and shuffled CHIPS libraries were displayed on phage, panned for binding to their C5a receptor target, and negatively selected for binding with polyclonal human anti-CHIPS antibodies. Following additional rational design, seven variants bearing five to eight mutations each were characterized and found to maintain reasonable biological activity (6- to 10-fold reduction relative to wild-type), good thermostability (typically better than wild-type), and yet exhibited 40- to 190-fold reductions in binding titer with human anti-CHIPS antibodies.[29]
Clinical complications of antidrug immune responses are the result of antidrug antibodies, and intuition therefore suggests that mutagenic deletion of antibody epitopes is a useful strategy by which to design better biotherapeutics. Importantly, the human immune system (and those of other mammals) is able to generate antibodies against multiple surface epitopes of a given therapeutic protein. See for example studies on the PE toxin, FVIII, and asparaginase [30,32,33,38,39] to name only a few. However, there tends to be substantial binding site overlap among the diverse antibodies that comprise a polyclonal response, and thus a protein’s antibody epitopes can be “grouped” into discrete subsets, each of which might be deleted with one or a few mutations.[27•] Moreover, murine model studies with antibody epitope depleted diphtheria and PE toxins show that the mouse immune system does not readily generate new antibodies against neoepitopes during repeated administration of engineered toxin variants.[28,33] These results, and results from earlier studies,[40-43] suggest that antibody epitope deletion may indeed have clinical utility. At the same time, it should be acknowledged that the human immune system is arguably the world’s most efficient antibody discovery and affinity maturation engine. As a result, there remains the possibility that repeated administration of antibody epitope depleted variants to human subjects might elicit new antibodies directed against alternative epitopes. To mitigate this risk, another compelling deimmunization strategy targets upstream molecular recognition events within the overall antidrug immune response: deletion of T cell epitopes.
Experimentally-driven T Cell Epitope Deletion
Biotherapeutic deimmunization by T cell epitope deletion has a long and well validated history. Indeed, though not explicitly articulated at the time, removal of immunogenic T cell epitopes was an implicit result of early antibody humanization strategies.[44,45] Deimmunization of staphylokinase, to treat myocardial infarctions, is an early seminal study of explicit T cell epitope deletion,[46] and other early examples include deimmunization of Factor VIII domain C1,[47] erythropoietin,[48] interferon beta,[49] and a beta lactamase enzyme.[50] More recently, the general approach has proliferated and become more sophisticated. One case in point is the systematic application of T cell epitope deletion to various FDA approved monoclonal antibodies that suffer from undesirable immunogenicity.[51•] An important conclusion from this work was the fact that even fully human antibodies, such as the #1 selling drug Humira, can be highly immunogenic due to T cell epitopes within hypervariable CDRs, yet epitopes within such functionally critical regions may be amenable to mutagenic deletion.
A prominent example of increasingly sophisticated T cell epitope deletion strategies can be found in work on E. coli type II asparaginase. Guided by bioinformatics prediction of likely T cell epitopes, anchor residues in three putative immunogenic hotspots were subjected to iterative site-directed saturation mutagenesis followed by high-throughput functional screening using a customized flow cytometric assay. Variant enzymes exhibiting high catalytic activity were isolated, and those functional mutations predicted to be disruptive of class II MHC binding were chosen as templates for subsequent rounds of mutagenesis and screening. This neutral drift strategy ultimately produced the 8-mutation variant 3.1.E2 that exhibited high catalytic proficiency (kcat equal to wild-type and only 3-fold higher KM) but was significantly less immunogenic in HLA transgenic mice expressing the human DRB1*0401 MHC II allele (assessed by both ex vivo cellular immunoassays and in vivo anti-drug antibody titers).[52••]
Anti-cancer immunotoxins have proven to be another productive space for T cell epitope deletion. For example, extensive experimental efforts have generated a detailed T cell epitope map for the PE toxin. Using immune cells from both healthy donors and patients previously treated with the immunogenic PE38 variant, eight immunogenic regions were identified and subsequently deleted via a combination of domain truncation and mutagenic substitution of critical epitope anchoring residues. The resulting deimmunized variant LMB-T18 exhibited high in vitro cytotoxic activity, potent in vivo anti-tumor activity, and yet it decreased ex vivo human T-cell activation by 90% compared to the native immunotoxin.[53••] Bouganin is another highly potent toxin with potential utility in cancer therapy, though as a plant protein it runs the risk of eliciting detrimental immune responses. Experimental epitope mapping and trial and error mutagenic T cell epitope deletion yielded the engineered variant de-bouganin, which has demonstrated low immunogenicity and high anti-tumor activity in a large number of preclinical and clinical studies. In human subjects the VB6-845 immunotoxin, a fusion of de-bouganin and a humanized anti-EpCAM Fab antibody, did exhibit undesirable immunogenicity, but the antidrug antibodies were directed almost exclusively against the humanized Fab, as opposed to the de-bouganin toxin.[54••]
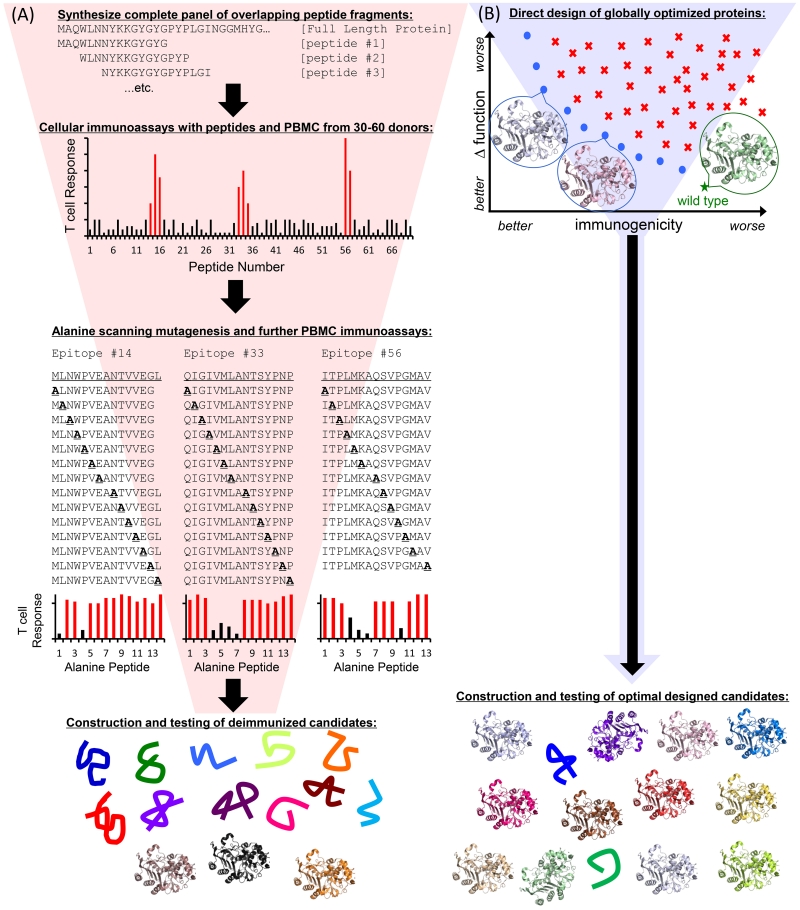
These examples support T cell epitope deletion as an effective strategy by which to suppress antidrug antibodies via upstream disruption of the immune response pathway. However, the above cited successes were derived in whole or in part from time-, labor-, and resource-intensive efforts that included: (i) epitope mapping via cellular immunoassays using large overlapping peptide panels spanning the full length of the protein, (ii) alanine scanning or similar mutagenic deletion of validated epitopes at the peptide level, followed by (iii) introduction of confirmed epitope-deleting mutations into the full length protein to assess structural and functional consequences, which are often unacceptably deleterious (Fig. 2A). Importantly, human class II MHC, composed of α/β heterodimers, are encoded by four different β-chain loci (DRB1, DRB3/4/5, DQB1, DPB1) and corresponding α-chain loci (DRA1, DQA1, and DPA1). Individuals may encode class II MHC from all four loci, and in general MHC II genes are highly polymorphic; nearly 3000 distinct alleles have been identified to date, though many of these alleles appear with low or extremely low frequency in global populations.[55] To better sample the genetic diversity of prospective patients, experimental epitope mapping often employs blood samples from 50 or more donors. Combined with the large number of overlapping peptides to be tested, the scale of such efforts can be both technically and financially imposing. More generally, the hit rate of epitope deleting yet function preserving mutations tends to be low in experimentally driven efforts (i.e., most tested mutations prove unacceptable), and thus more recent strategies have used computational methods to facilitate T cell epitope identification and deletion.
Figure 2.
Schematic diagrams for T cell epitope deletion strategies. (A) Experimentally driven deimmunization is a multistage process, moving top to bottom. A panel of overlapping synthetic peptide fragments spanning the full sequence is synthesized. The peptides are then tested for immune recognition, typically using ex vivo cellular immunoassays with blood cells from large panels of human donors. High responses to overlapping immunogenic peptides are indicated by tall red bars. Identified immunogenic peptides are subjected to alanine scanning mutagenesis and retested with the donor human immune cells. Alanine-substituted peptides that reduce immune cell activation are highlighted as shorter black bars. Confirmed deimmunizing mutations are then engineered back into the full length protein and tested for expression, stability, and activity. Typical low hit rates are indicated by a majority of unfolded variant proteins, with only a few stable and active variants shown as cartoon structures. The process benefits from early identification of bona fide immunogenic peptides, but requires significant time and expense to funnel down to functional deimmunized candidates. (B) Computationally driven deimmunization addresses global protein design as a starting point. The protein design space is shown in two dimensions: predicted immunogenicity (x-axis) and predicted change in function (y-axis). Lower values are better in both objectives. Wild type has good molecular function but high immunogenicity. Sub-optimal designs are shown as red “x”es. The blue circles indicate Pareto optimal designs, or designs that are not simultaneously dominated on both objectives by any other single design. The Pareto frontier spans the full spectrum of optimal tradeoffs between the two objective functions. Representative protein designs are shown as cartoon structures. Individual, globally optimal designs balancing predicted reduction in immunogenicity and maintenance of function are selected for construction and analysis of expression, stability and activity. The high hit rate for folded and functional designs is indicated by a majority of cartoon protein structures. Computational optimization facilitates quick transition to validating candidates predicted to be functionally deimmunized. For both experimentally-driven and computationally-driven deimmunization projects, the final deimmunized candidates must be further tested for immunogenicity using cellular immunoassays and/or humanized murine models.
Computationally-driven T Cell Epitope Deletion
Computational T cell epitope predictors have proven reasonably accurate in benchmark studies.[56,57] While computational analysis enables rapid and facile prediction for large numbers of MHC alleles, extensive experimental studies have revealed a surprising degree of overlap in the peptide binding selectivities of various class II MHC proteins.[58] This in turn has enabled formulation of class II MHC supertypes, or groups of alleles that bind similar peptide repertoires.[59] Thus, the highly polymorphic nature of MHC II alleles can be rendered more tractable by predicting for alleles that are both commonly encoded and broadly representative of key MHC II supertypes.[59,60] Additionally, experimental evidence suggests that immunodominant epitopes are those that bind multiple MHC II alleles, and therefore computational predictions can be further refined by searching for high risk “promiscuous” MHC II binders.[26,61] T cell epitope databases and prediction algorithms are regularly updated and improved,[62] and there exist codified strategies for employing these epitope predictors to guide immunogenicity risk assessment and protein deimmunization.[26,61] The next logical step is to fully integrate epitope predictors with computational protein design methodologies (Fig. 2B).
Conceptually, mutagenic T cell epitope deletion is a dual objective protein design problem: while deimmunizing mutations should disrupt class II MHC recognition and suppress downstream immune responses, each such mutation incurs a risk of compromising a protein’s native fold and therapeutic function. These two objectives, epitope deletion and maintenance of structure and function, are incommensurate and even competing in nature, and the goal of protein design is thus to identify variants that make beneficial trade-offs. Initial deimmunization algorithms employed simple BLOSUM substitution matrices or customized statistical sequence potentials in order to predict the structural and functional impacts of possible deimmunizing mutations.[63,64] More recently, structure-based design algorithms based on OSPREY[65] or Rosetta[66] have been described.[67-69] Pareto optimization approaches knit together both objectives in order to identify designs making optimal trade-offs: a Pareto optimal variant is not simultaneously dominated on both objectives by any other single variant, but instead improvement on one objective comes at the expense of the other.[70]
The P99 beta lactamase protein (P99βL) is a prospective component of Antibody Directed Enzyme Prodrug Therapy (ADEPT), and early work on deimmunizing the enzyme yielded minimally engineered 1- and 2-mutation variants.[50,71] More recent P99βL studies have sought to quantitatively assess the tradeoffs between immunoreactivity, measured by class II MHC binding of peptide fragments, and functionality, measured by thermostability and enzyme kinetics. An analysis of 4-mutation and 5-mutation designs showed that more aggressive sets of mutations were more disruptive of MHC II binding, but the eight engineered enzymes were found to have uniformly high catalytic efficiencies and melting temperatures such that clear tradeoffs were not readily observed.[72] Subsequently, a more expansive analysis was conducted with 18 deimmunized P99βL designs, bearing 1-8 mutations each, that comprised the Pareto frontier, i.e., all undominated designs. This more systematic analysis of the two-dimensional design space showed that the predicted immunoreactivity-functionality trade-offs mapped closely onto experimental observations, concluding that trade-offs are not only predictable but also designable.[73••] In a follow-on study, structure-based molecular modeling replaced the earlier statistical sequence potentials as a predictive measure of P99βL function. Structure-based design again confirmed that more aggressive sets of mutations were more disruptive of MHC II binding, and it yielded seven 8-mutation variants that all retained high level stability and activity (a 100% hit rate in this study).[74] In general, formulating T cell epitope deletion as a dual objective design problem has enabled a systematic and quantitative assessment of the immunoreactivity-functionality tradeoffs that are inherent to the deimmunization process.
In other studies a Rosetta structure-based deimmunization algorithm was used to delete murine MHC II restricted T cell epitopes from GFP. A 9-mutation variant was shown to have wild-type spectral properties, and murine T cells primed with wild-type GFP were significantly less reactive towards a mutant peptide constituting one of three epitope hotspots that had been targeted.[68•] The same algorithm was used to target three known epitopes in the PE toxin, yielding four single point mutants and one 2-mutation design. Three of the point mutants manifested wild-type or better cytotoxicity towards cancer cell lines, and peptide fragments of two were assessed for activation of human T cells and found to have lower overall immunostimulatory potential.[68•] These results further support computational protein design as a useful tool to guide biotherapeutic deimmunization.
Finally, structure-based protein design has enabled functional deimmunization of lysostaphin, a potent anti-MRSA bactericidal enzyme that suffers from undesirable immunogenicity.[75] Using a homology model, structure-guided re-design of lysostaphin’s catalytic domain yielded a large number of 2- to 8-mutation Pareto optimal designs that retained high antibacterial activity, high thermostability, and yet suppressed molecular recognition by human MHC II proteins. Two candidates were shown to have wild-type in vivo efficacy yet significantly decreased in vivo immunogenic potential, as measured by immune cell proliferation in two different humanized mouse models (BLT mice and HLA transgenic mice).[76•] In a separate lysostaphin study, structure-based design algorithms enabled aggressive deletion of putative human HLA DRB1*0401 restricted T cell epitopes. Both individual optimal designs and combinatorial library designs were examined in these experiments, and both methods produced variants that strongly suppressed antidrug antibody responses upon immunization of humanized DR4 mice, which express the human DRB1*0401 HLA allele. Additional humanized mouse experiments demonstrated that the suppressed antidrug antibody response translated into enhanced therapeutic efficacy in a recurrent MRSA infection model.[77••] This latter result represents a milestone for the field: the first systematic and controlled demonstration that deletion of T cell epitopes not only reduces immunogenicity but as a result improves in vivo efficacy.
Discussion and Conclusion
Preclinical assessment of biotherapeutic immunogenicity is itself a complex and multifaceted problem, and the strengths and weaknesses of in silico, in vitro, and in vivo models have been reviewed recently elsewhere.[26,78] For instance, readers interested in the pros and cons of various transgenic and surgically engrafted humanized mouse models are referred to the following articles.[26,78-83] Ultimately, immunogenicity (or lack thereof) must be assessed in human subjects, but appropriate use of preclinical models can yield insights into immunogenic potential, as articulated in these review articles and the primary research articles cited above.
The upstream position of T cell epitope recognition in the immune response pathway suggests that mutagenesis of these subsequences might be particularly useful in designing immunoevasive biologics. In other cases, antibody epitope deletion could be critical for proteins to which patients have experienced prior natural exposures. Moreover, combining T cell and antibody epitope deletion strategies might represent a further step towards comprehensive protein deimmunization. Indeed, published data on the T cell epitope depleted PE immunotoxin LMB-T18 suggests that a subset of T cell epitope deleting mutations fortuitously deleted antibody epitopes as well,[53••] and we have observed similar results with T cell epitope depleted lysostaphin variants (Griswold, unpublished data). More directed efforts to explicitly combine T cell and antibody epitope depleted designs are beginning to emerge, with analysis of the antibody and T cell epitope engineered immunotoxin LMB-T14 having published as the current manuscript was in revision.[84] This 10-mutation PE variant, which retained potent cytotoxic activity towards several cancer cell lines, was broadly evasive of both antibodies and T cells that recognized the wild type toxin. However, peptide fragments of the variant were found to activate T cell populations that predecessor molecules had effectively evaded, and it was subsequently found that mutations designed to delete antibody epitopes inadvertently introduced neo-epitopes recognized by prevalent class II MHC proteins. This work is indicative of the rapid advances being made in the deimmunized biologics space, but it also serves to highlight the remaining challenges and opportunities for further innovation.
With respect to such aggressive molecular engineering, it should be noted that the threshold for achieving “comprehensive” deimmunization remains unclear, as does the necessity of realizing this objective. What fraction of immunogenic epitopes must be deleted to deimmunize a protein? In a murine model study of interferon beta, deletion of one immunodominant T cell epitope did not result in a subsequent response against a subdominant epitope, [49] but in the case of the PE toxin it appears that targeting broadly distributed epitopes with higher mutational loads may be necessary.[53••] What fraction of patients exhibiting an immune response is acceptable? If a deimmunized biologic elicits a response in a significant fraction of patients, yet the response rate is half that of the wild-type predecessor drug, is that not a win? What of the strength of the immune response? Suppose a deimmunized biologic exhibits the same patient response rate as the wild-type protein but induces reduced titers of antidrug antibodies such that more patients may remain on the deimmunized variant longer. Is that an acceptable outcome? Can a highly immunogenic protein be redesigned so as to eliminate the response in all subjects? These questions hinge on a multitude of interdependent technical, clinical and financial factors, and definitive answers may remain elusive far into the future. In the near term, benchmarks for clinically useful deimmunization will likely be assessed on a case-by-case basis for each biotherapeutic and even specific patient populations. As experience with deimmunized biologics grows, perhaps more general rules for what it means to be deimmunized will emerge. In particular, deimmunized biobetters should provide rich data sets by which to probe the above questions in the context of existing clinical data for unmodified originator drugs.
There are a growing number of deimmunized biologics that are either moving towards or are currently in human trials. Long term, we expect that deimmunization technologies will have a profound impact on the biotherapeutics space. Key opportunities include both biobetter versions of FDA approved drugs with known immunogenicity issues as well as innovative new drugs whose therapeutic potential has yet to be tapped due to immunogenicity concerns. Analogous to the manner in which antibody humanization ushered in a wave of engineered monoclonals, one might anticipate that the above deimmunization technologies will yield a revolution for non-immunoglobulin biotherapies.
Highlights.
Immunogenicity as a risk factor for biotherapeutic agents.
Antibody epitope deletion as a strategy to evade antidrug antibodies.
T cell epitope deletion as a strategy to silence the antidrug immune response.
Multi-objective protein design algorithms to facilitate biotherapeutic deimmunization.
Highlights of recent experimental validation for deimmunized biotherapeutics.
Acknowledgements
We wish to generally acknowledge the dedicated community of researchers in the biotherapeutic immunogenicity space. Here, we have specifically highlighted recent work in which deimmunized biotherapeutics were characterized both functionally and immunologically, and we regret any inadvertent omissions and articles not cited due to space and thematic constraints. This work was supported by the National Institute of General Medical Sciences R01-GM-098977 and the National Institute of Allergy and Infectious Diseases 1R21AI098122 and 1R41AI118133. We also gratefully acknowledge computational resources provided by NSF grant CNS-1205521 and the technical support of the Dartmouth Transgenic and Genetic Construct Shared Resource (P30 CA023108) and Immunology COBRE Core C (P20 RR15639).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
Karl E. Griswold and Chris Bailey-Kellogg are Dartmouth faculty and co-members of Stealth Biologics, LLC, a Delaware biotechnology company. These authors acknowledge a potential conflict of interest related to their associations with this company, and they affirm that their above cited works are free of any bias. This article has been reviewed and approved as specified in their Dartmouth conflict of interest management plans.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Aggarwal S. What’s fueling the biotech engine - 2012 to 2013. Nat Biotech. 2014;32:32–39. doi: 10.1038/nbt.2794. [DOI] [PubMed] [Google Scholar]
- 2.Baker MP, Reynolds HM, Lumicisi B, Bryson CJ. Immunogenicity of protein therapeutics: The key causes, consequences and challenges. Self Nonself. 2010;1:314–322. doi: 10.4161/self.1.4.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schellekens H. The Immunogenicity of Therapeutic Proteins. Discovery Medicine. 2010;49:560–564. [PubMed] [Google Scholar]
- 4.Sethu S, Govindappa K, Alhaidari M, Pirmohamed M, Park K, Sathish J. Immunogenicity to Biologics: Mechanisms, Prediction and Reduction. Archivum Immunologiae Et Therapiae Experimentalis. 2012;60:331–344. doi: 10.1007/s00005-012-0189-7. [DOI] [PubMed] [Google Scholar]
- 5.Krishna M, Nadler S. Immunogenicity to Biotherapeutics – the role of Anti-drug Immune complexes. Frontiers in Immunology. 2016;7 doi: 10.3389/fimmu.2016.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CDER: United States Food and Drug Administration, editor. Guidance for Industry Immunogenicity Assessment for Therapeutic Protein Products. 2013. pp. 1–34. vol Draft Guidance. [Google Scholar]
- 7.Buttel IC, Chamberlain P, Chowers Y, Ehmann F, Greinacher A, Jefferis R, Kramer D, Kropshofer H, Lloyd P, Lubiniecki A, et al. Taking immunogenicity assessment of therapeutic proteins to the next level. Biologicals. 2011;39:100–109. doi: 10.1016/j.biologicals.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Kloks C, Berger C, Cortez P, Dean Y, Heinrich J, Bjerring Jensen L, Koppenburg V, Kostense S, Kramer D, Spindeldreher S, et al. A fit-for-purpose strategy for the risk-based immunogenicity testing of biotherapeutics: a European industry perspective. Journal of Immunological Methods. 2015;417:1–9. doi: 10.1016/j.jim.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 9.World Health Organization, editor. WHO: Guidelines on the quality, safety, and efficacy of biotherapeutic protein products prepared by recombinant DNA technology. 2013;814 WHO (Series Editor) [Google Scholar]
- 10.Yin L, Chen X, Vicini P, Rup B, Hickling TP. Therapeutic outcomes, assessments, risk factors and mitigation efforts of immunogenicity of therapeutic protein products. Cell Immunol. 2015;295:118–126. doi: 10.1016/j.cellimm.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Moussa EM, Panchal JP, Moorthy BS, Blum JS, Joubert MK, Narhi LO, Topp EM. Immunogenicity of Therapeutic Protein Aggregates. Journal of Pharmaceutical Sciences. 2016;105:417–430. doi: 10.1016/j.xphs.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Turecek PL, Bossard MJ, Schoetens F, Ivens IA. PEGylation of Biopharmaceuticals: A Review of Chemistry and Nonclinical Safety Information of Approved Drugs. J Pharm Sci. 2016;105:460–475. doi: 10.1016/j.xphs.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 13.Verhoef JJ, Carpenter JF, Anchordoquy TJ, Schellekens H. Potential induction of anti-PEG antibodies and complement activation toward PEGylated therapeutics. Drug Discov Today. 2014;19:1945–1952. doi: 10.1016/j.drudis.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 14.Podust VN, Sim BC, Kothari D, Henthorn L, Gu C, Wang CW, McLaughlin B, Schellenberger V. Extension of in vivo half-life of biologically active peptides via chemical conjugation to XTEN protein polymer. Protein Eng Des Sel. 2013;26:743–753. doi: 10.1093/protein/gzt048. [DOI] [PubMed] [Google Scholar]
- 15.Schlapschy M, Binder U, Borger C, Theobald I, Wachinger K, Kisling S, Haller D, Skerra A. PASylation: a biological alternative to PEGylation for extending the plasma half-life of pharmaceutically active proteins. Protein Eng Des Sel. 2013;26:489–501. doi: 10.1093/protein/gzt023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bachran C, Gupta PK, Bachran S, Leysath CE, Hoover B, Fattah RJ, Leppla SH. Reductive Methylation and Mutation of an Anthrax Toxin Fusion Protein Modulates its Stability and Cytotoxicity. Scientific Reports. 2014;4:8. doi: 10.1038/srep04754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kontos S, Grimm AJ, Hubbell JA. Engineering antigen-specific immunological tolerance. Curr Opin Immunol. 2015;35:80–88. doi: 10.1016/j.coi.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Choi Y, Hua C, Sentman CL, Ackerman ME, Bailey-Kellogg C. Antibody humanization by structure-based computational protein design. mAbs. 2015;7:1045–1057. doi: 10.1080/19420862.2015.1076600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olimpieri PP, Marcatili P, Tramontano A. Tabhu: tools for antibody humanization. Bioinformatics. 2015;31:434–435. doi: 10.1093/bioinformatics/btu667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li T, Pantazes RJ, Maranas CD. OptMAVEn--a new framework for the de novo design of antibody variable region models targeting specific antigen epitopes. PLoS One. 2014;9:e105954. doi: 10.1371/journal.pone.0105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holgate RG, Weldon R, Jones TD, Baker MP. Characterisation of a Novel Anti-CD52 Antibody with Improved Efficacy and Reduced Immunogenicity. PLoS One. 2015;10:e0138123. doi: 10.1371/journal.pone.0138123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi Y, Ndong C, Griswold KE, Bailey-Kellogg C. Computationally-Driven Antibody Engineering Enables Simultaneous Humanization and Thermostabilization. Protein Eng Des Sel. 2016 doi: 10.1093/protein/gzw024. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Jiang N, Wang T, Xie G, Zhang Z, Li H, Yuan J, Sun Z, Chen J. DNA shuffling of uricase gene leads to a more “human like” chimeric uricase with increased uricolytic activity. Int J Biol Macromol. 2016;82:522–529. doi: 10.1016/j.ijbiomac.2015.10.053. [DOI] [PubMed] [Google Scholar]
- 24.Cantor JR, Panayiotou V, Agnello G, Georgiou G, Stone EM. Engineering reduced-immunogenicity enzymes for amino acid depletion therapy in cancer. Methods Enzymol. 2012;502:291–319. doi: 10.1016/B978-0-12-416039-2.00015-X. [DOI] [PubMed] [Google Scholar]
- 25.Trombetta ES, Mellman I. Cell Biology of Antigen Processing in vitro and in vivo. Annual Review of Immunology. 2005;23:975–1028. doi: 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- 26.Jawa V, Cousens LP, Awwad M, Wakshull E, Kropshofer H, De Groot AS. T-cell dependent immunogenicity of protein therapeutics: Preclinical assessment and mitigation. Clinical Immunology. 2013;149:534–555. doi: 10.1016/j.clim.2013.09.006. [DOI] [PubMed] [Google Scholar]
- •27.Nagata S, Pastan I. Removal of B cell epitopes as a practical approach for reducing the immunogenicity of foreign protein-based therapeutics. Advanced Drug Delivery Reviews. 2009;61:977–985. doi: 10.1016/j.addr.2009.07.014. The authors provide an overview of mutagenic antibody epitope deletion, including competition experiments with antibody epitope pairs to map antigenic hotspots. They detail studies in which antibody epitopes were deleted from the PE toxin, and thereby provide a roadmap to pursue similar work on other targets.
- 28.Schmohl JU, Todhunter D, Oh S, Vallera DA. Mutagenic Deimmunization of Diphtheria Toxin for Use in Biologic Drug Development. Toxins (Basel) 2015;7:4067–4082. doi: 10.3390/toxins7104067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gustafsson E, Rosen A, Barchan K, van Kessel KP, Haraldsson K, Lindman S, Forsberg C, Ljung L, Bryder K, Walse B, et al. Directed evolution of chemotaxis inhibitory protein of Staphylococcus aureus generates biologically functional variants with reduced interaction with human antibodies. Protein Eng Des Sel. 2010;23:91–101. doi: 10.1093/protein/gzp062. [DOI] [PubMed] [Google Scholar]
- •30.Liu W, Onda M, Lee B, Kreitman RJ, Hassan R, Xiang L, Pastan I. Recombinant immunotoxin engineered for low immunogenicity and antigenicity by identifying and silencing human B-cell epitopes. Proc Natl Acad Sci U S A. 2012;109:11782–11787. doi: 10.1073/pnas.1209292109. The authors describe the use of phage displayed libraries to isolate monoclonal human antibodies directed against the PE toxin. Competition binding experiments with panels of antidrug antibodies enabled mapping of dominant surface epitopes and subsequent mutagenic deletion. The resulting 7-mutation variant retained potent anti-cancer activity but dramatically reduced binding with human serum from patients that had mounted a response against a predecessor wild-type toxin.
- 31.Lin JC, Ettinger RA, Schuman JT, Zhang A-H, Wamiq-Adhami M, Nguyen P-CT, Nakaya-Fletcher SM, Puranik K, Thompson AR, Pratt KP. Six Amino Acid Residues in a 1200 Å2 Interface Mediate Binding of Factor VIII to an IgG4 Inhibitory Antibody. PLoS ONE. 2015;10:e0116577. doi: 10.1371/journal.pone.0116577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onda M, Nagata S, FitzGerald DJ, Beers R, Fisher RJ, Vincent JJ, Lee B, Nakamura M, Hwang J, Kreitman RJ, et al. Characterization of the B cell epitopes associated with a truncated form of Pseudomonas exotoxin (PE38) used to make immunotoxins for the treatment of cancer patients. J Immunol. 2006;177:8822–8834. doi: 10.4049/jimmunol.177.12.8822. [DOI] [PubMed] [Google Scholar]
- 33.Onda M, Beers R, Xiang L, Lee B, Weldon JE, Kreitman RJ, Pastan I. Recombinant immunotoxin against B-cell malignancies with no immunogenicity in mice by removal of B-cell epitopes. Proc Natl Acad Sci U S A. 2011;108:5742–5747. doi: 10.1073/pnas.1102746108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh S, Tsai AK, Ohlfest JR, Panoskaltsis-Mortari A, Vallera DA. Evaluation of a bispecific biological drug designed to simultaneously target glioblastoma and its neovasculature in the brain. Journal of Neurosurgery. 2011;114:1662–1671. doi: 10.3171/2010.11.JNS101214. [DOI] [PubMed] [Google Scholar]
- 35.Alewine C, Xiang L, Yamori T, Niederfellner G, Bosslet K, Pastan I. Efficacy of RG7787, a next-generation mesothelin-targeted immunotoxin, against triple-negative breast and gastric cancers. Mol Cancer Ther. 2014;13:2653–2661. doi: 10.1158/1535-7163.MCT-14-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hollevoet K, Mason-Osann E, Liu X-f, Imhof-Jung S, Niederfellner G, Pastan I. In Vitro and In Vivo Activity of the Low-Immunogenic Antimesothelin Immunotoxin RG7787 in Pancreatic Cancer. Molecular Cancer Therapeutics. 2014 doi: 10.1158/1535-7163.MCT-14-0089-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehta RK, Verma S, Pati R, Sengupta M, Khatua B, Jena RK, Sethy S, Kar SK, Mandal C, Roehm KH, et al. Mutations in subunit interface and B-cell epitopes improve antileukemic activities of Escherichia coli asparaginase-II: evaluation of immunogenicity in mice. J Biol Chem. 2014;289:3555–3570. doi: 10.1074/jbc.M113.486530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pratt KP. Engineering less immunogenic and antigenic FVIII proteins. Cellular Immunology. 2016;301:12–17. doi: 10.1016/j.cellimm.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Werner A, Rohm KH, Muller HJ. Mapping of B-cell epitopes in E. coli asparaginase II, an enzyme used in leukemia treatment. Biol Chem. 2005;386:535–540. doi: 10.1515/BC.2005.063. [DOI] [PubMed] [Google Scholar]
- 40.Collen D, Bernaerts R, Declerck P, DeCock F, Demarsin E, Jenne S, Laroche Y, Lijnen HR, Silence K, Verstreken M. Recombinant staphylokinase variants with altered immunoreactivity .1. Construction and characterization. Circulation. 1996;94:197–206. doi: 10.1161/01.cir.94.2.197. [DOI] [PubMed] [Google Scholar]
- 41.Collen D, Moreau H, Stockx L, Vanderschueren S. Recombinant staphylokinase variants with altered immunoreactivity .1. Thrombolytic properties and antibody induction. Circulation. 1996;94:207–216. doi: 10.1161/01.cir.94.2.207. [DOI] [PubMed] [Google Scholar]
- 42.Laroche Y, Heymans S, Capaert S, De Cock F, Demarsin E, Collen D. Recombinant staphylokinase variants with reduced antigenicity due to elimination of B-lymphocyte epitopes. Blood. 2000;96:1425–1432. [PubMed] [Google Scholar]
- 43.Mayer A, Sharma SK, Tolner B, Minton NP, Purdy D, Amlot P, Tharakan G, Begent RH, Chester KA. Modifying an immunogenic epitope on a therapeutic protein: a step towards an improved system for antibody-directed enzyme prodrug therapy (ADEPT) Br J Cancer. 2004;90:2402–2410. doi: 10.1038/sj.bjc.6601888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boulianne GL, Hozumi N, Shulman MJ. Production of functional chimaeric mouse/human antibody. Nature. 1984;312:643–646. doi: 10.1038/312643a0. [DOI] [PubMed] [Google Scholar]
- 45.Morrison SL, Johnson MJ, Herzenberg LA, Oi VT. Chimeric human antibody molecules: mouse antigen-binding domains with human constant region domains. Proc Natl Acad Sci U S A. 1984;81:6851–6855. doi: 10.1073/pnas.81.21.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warmerdam PAM, Plaisance S, Vanderlick K, Vandervoort P, Brepoels K, Collen D, De Maeyer M. Elimination of a human T-cell region in staphylokinase by T-cell screening and computer modeling. Thrombosis and Haemostasis. 2002;87:666–673. [PubMed] [Google Scholar]
- 47.Jones TD, Phillips WJ, Smith BJ, Bamford CA, Nayee PD, Baglin TP, Gaston JSH, Baker MP. Identification and removal of a promiscuous CD4+ T cell epitope from the C1 domain of factor VIII. J Thromb Haemost. 2005;3:991–1000. doi: 10.1111/j.1538-7836.2005.01309.x. [DOI] [PubMed] [Google Scholar]
- 48.Tangri S, Mothe BR, Eisenbraun J, Sidney J, Southwood S, Briggs K, Zinckgraf J, Bilsel P, Newman M, Chesnut R, et al. Rationally Engineered Therapeutic Proteins with Reduced Immunogenicity. J Immunol. 2005;174:3187–3196. doi: 10.4049/jimmunol.174.6.3187. [DOI] [PubMed] [Google Scholar]
- 49.Yeung VP, Chang J, Miller J, Barnett C, Stickler M, Harding FA. Elimination of an Immunodominant CD4+ T Cell Epitope in Human IFN-β Does Not Result in an In Vivo Response Directed at the Subdominant Epitope. The Journal of Immunology. 2004;172:6658–6665. doi: 10.4049/jimmunol.172.11.6658. [DOI] [PubMed] [Google Scholar]
- 50.Harding FA, Liu AD, Stickler M, Razo OJ, Chin R, Faravashi N, Viola W, Graycar T, Yeung VP, Aehle W, et al. A Beta-lactamase with reduced immunogenicity for the targeted delivery of chemotherapeutics using antibody-directed enzyme prodrug therapy. Mol Cancer Ther. 2005;4:1791–1800. doi: 10.1158/1535-7163.MCT-05-0189. [DOI] [PubMed] [Google Scholar]
- •51.Harding FA, Stickler MM, Razo J, DuBridge R. The immunogenicity of humanized and fully human antibodies: Residual immunogenicity resides in the CDR regions. mAbs. 2010;2:256–265. doi: 10.4161/mabs.2.3.11641. The authors experimentally mapped T cell epitopes for eight different humanized antibody therapeutics and found that T cell epitopes were located exclusively in the CDR regions. They further showed that appropriate point mutations, identified by trial and error, effectively deleted these T cell epitopes, suppressed recognition by human T cells ex vivo, yet retained high level binding affinity to the target molecule. The article provides an understanding for why humanized and even fully human antibodies can remain immunogenic in human subjects.
- ••52.Cantor JR, Yoo TH, Dixit A, Iverson BL, Forsthuber TG, Georgiou G. Therapeutic enzyme deimmunization by combinatorial T-cell epitope removal using neutral drift. Proceedings of the National Academy of Sciences. 2011;108:1272–1277. doi: 10.1073/pnas.1014739108. The authors describe an innovative “neutral drift” strategy by which to delete T cell epitopes from therapeutic proteins. Asparaginase residues within computationally predicted T cell epitopes were subjected to saturation mutagenesis, and highly functional enzyme variants were isolated using a novel flow cytometric screening assay. Mutations predicted to disrupt MHC II binding were consolidated by directed evolution. The final deimmunized variant retained high catalytic proficiency, yet it was dramatically less immunogenic in a humanized HLA transgenic mouse model.
- ••53.Mazor R, Eberle JA, Hu X, Vassall AN, Onda M, Beers R, Lee EC, Kreitman RJ, Lee B, Baker D, et al. Recombinant immunotoxin for cancer treatment with low immunogenicity by identification and silencing of human T-cell epitopes. Proceedings of the National Academy of Sciences. 2014;111:8571–8576. doi: 10.1073/pnas.1405153111. The authors describe a systematic and expansive effort to map and delete immunodominant T cell epitopes in the PE toxin. Using immune cells from both healthy donors and patients that had previously mounted a response against the wild-type toxin, eight immunodominant T cell epitopes were identified and mutated so as to reduce their immunostimulatory potential. The final engineered variant retained potent anti-cancer activity, reduced stimulatory potential with T cells from diverse human donors, and reduced binding by antibodies directed against predecessor toxin variants.
- ••54.Entwistle J, Kowalski M, Brown J, Cizeau J, MacDonald GC. Antibody-Drug Conjugates and Immunotoxins. Springer; 2013. The Preclinical and Clinical Evaluation of VB6-845: An Immunotoxin with a De-Immunized Payload for the Systemic Treatment of Solid Tumors; pp. 349–367. This article provides an in-depth overview of the engineering, preclinical development, and clinical testing of the VB6-845 immunotoxin based on de-bouganin. De-bouganin is a 4-mutation variant of a plant toxin that was engineered to delete immunodominant human T cell epitopes and thereby reduce immunogenicity. Cumulatively, these results represent the most comprehensive description of preclinical and clinical testing of a T cell epitope engineered biotherapeutic to date.
- 55.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh Steven GE. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Research. 2014 doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang P, Sidney J, Dow C, Mothe B, Sette A, Peters B. A Systematic Assessment of MHC Class II Peptide Binding Predictions and Evaluation of a Consensus Approach. PLoS Comput Biol. 2008;4:e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, Peters B. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11 doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Southwood S, Sidney J, Kondo A, del Guercio M-F, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A. Several Common HLA-DR Types Share Largely Overlapping Peptide Binding Repertoires. J Immunol. 1998;160:3363–3373. [PubMed] [Google Scholar]
- 59.Greenbaum J, Sidney J, Chung J, Brander C, Peters B, Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paul S, Lindestam Arlehamn CS, Scriba TJ, Dillon MB, Oseroff C, Hinz D, McKinney DM, Carrasco Pro S, Sidney J, Peters B, et al. Development and validation of a broad scheme for prediction of HLA class II restricted T cell epitopes. J Immunol Methods. 2015;422:28–34. doi: 10.1016/j.jim.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paul S, Kolla RV, Sidney J, Weiskopf D, Fleri W, Kim Y, Peters B, Sette A. Evaluating the immunogenicity of protein drugs by applying in vitro MHC binding data and the immune epitope database and analysis resource. Clinical & developmental immunology. 2013;2013:467852. doi: 10.1155/2013/467852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, Wheeler DK, Gabbard JL, Hix D, Sette A, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43:D405–412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parker AS, Griswold KE, Bailey-Kellogg C. Optimization of Therapeutic Proteins to Delete T-cell Epitopes while Maintaining Beneficial Residue Interactions. Journal of Bioinformatics and Computational Biology. 2011;9:207–229. doi: 10.1142/s0219720011005471. [DOI] [PubMed] [Google Scholar]
- 64.Parker AS, Zheng W, Griswold KE, Bailey-Kellogg C. Optimization algorithms for functional deimmunization of therapeutic proteins. BMC Bioinformatics. 2010;11:180. doi: 10.1186/1471-2105-11-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gainza P, Roberts KE, Georgiev I, Lilien RH, Keedy DA, Chen CY, Reza F, Anderson AC, Richardson DC, Richardson JS, et al. OSPREY: protein design with ensembles, flexibility, and provable algorithms. Methods Enzymol. 2013;523:87–107. doi: 10.1016/B978-0-12-394292-0.00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang PS, Ban YE, Richter F, Andre I, Vernon R, Schief WR, Baker D. RosettaRemodel: a generalized framework for flexible backbone protein design. PLoS One. 2011;6:e24109. doi: 10.1371/journal.pone.0024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choi Y, Griswold KE, Bailey-Kellogg C. Structure-based redesign of proteins for minimal T-cell epitope content. Journal of Computational Chemistry. 2013;34:879–891. doi: 10.1002/jcc.23213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •68.King C, Garza EN, Mazor R, Linehan JL, Pastan I, Pepper M, Baker D. Removing T-cell epitopes with computational protein design. Proceedings of the National Academy of Sciences. 2014;111:8577–8582. doi: 10.1073/pnas.1321126111. The authors describe development of a customized T cell epitope predictor and its integration with the Rosetta protein design package. The resulting deimmunization algorithm was prospectively tested on both GFP and the PE toxin, and engineered variants were found to retain high molecular function while exhibiting reduced recognition by immune cells (murine for GFP and human for PE).
- 69.Parker AS, Choi Y, Griswold KE, Bailey-Kellogg C. Structure-Guided Deimmunization of Therapeutic Proteins. Journal of Computational Biology. 2013;20:152–165. doi: 10.1089/cmb.2012.0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He L, Friedman AM, Bailey-Kellogg C. A divide-and-conquer approach to determine the Pareto frontier for optimization of protein engineering experiments. Proteins: Structure, Function, and Bioinformatics. 2012;80:790–806. doi: 10.1002/prot.23237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Osipovitch DC, Parker AS, Makokha CD, Desrosiers J, Kett WC, Moise L, Bailey-Kellogg C, Griswold KE. Design and Analysis of Immune-Evading Enzymes for ADEPT Therapy. Protein Eng Des Sel. 2012;25:613–623. doi: 10.1093/protein/gzs044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Salvat RS, Parker AS, Guilliams A, Choi Y, Bailey-Kellogg C, Griswold KE. Computationally driven deletion of broadly distributed T cell epitopes in a biotherapeutic candidate. Cellular and Molecular Life Sciences. 2014;71:4869–4880. doi: 10.1007/s00018-014-1652-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••73.Salvat RS, Parker AS, Guilliams A, Choi Y, Bailey-Kellogg C, Griswold KE. Mapping the Pareto Optimal Design Space for a Functionally Deimmunized Biotherapeutic Candidate. PLoS Comput Biol. 2015;11:e1003988. doi: 10.1371/journal.pcbi.1003988. The authors frame deimmunization as a dual objective optimization problem in which there are inherent tradeoffs between reducing immunogenicity on the one hand and maintaining molecular function on the other hand. They describe coupled computational and experimental mapping of the design space for deimmunized P99βL. A large panel of 15 Pareto optimal deimmunized candidates (i.e., variants not simultaneously dominated on both objectives by any other single variant) was designed, constructed, and tested for immunoreactivity and molecular function. The experimental results mapped closely to the computed design space, thus demonstrating that tradeoffs in the deimmunization process are predictable and designable.
- 74.Salvat RS, Choi Y, Bishop A, Bailey-Kellogg C, Griswold KE. Protein deimmunization via structure-based design enables efficient epitope deletion at high mutational loads. Biotechnology and Bioengineering. 2015;112:1306–1318. doi: 10.1002/bit.25554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kokai-Kun JF. Lysostaphin: a Silver Bullet for Staph. In: Tegos G, Mylonakis E, CABI, editors. Antimicrobial Drug Discovery. edn 1st Vol. 22. Advances in Molecular and Cellular Microbiology; 2012. pp. 147–165. [Google Scholar]
- •76.Blazanovic K, Zhao H, Choi Y, Li W, Salvat RS, Osipovitch DC, Fields J, Moise L, Berwin BL, Fiering SN, et al. Structure-based Redesign of Lysostaphin Yields Potent Anti-Staphylococcal Enzymes that Evade Immune Cell Surveillance. Molecular Therapy - Methods and Clinical Development. 2015;2:15021. doi: 10.1038/mtm.2015.21. The authors describe structured-based computational design to guided T cell epitope deletion in the catalytic domain of the antibacterial protein lysostaphin. Via a staged design process, they generated large panels of 2-8 mutation variants that retained good stability and antibacterial activity. Two candidates were shown to exhibit wild-type in vivo efficacy in a murine lung infection model. In humanized BLT and HLA transgenic mice, the deimmunized variants exhibited significantly reduced immunostimulatory potential, as measured by cellular immunoassays. These studies represent the first comparison of a wild-type and deimmunized biotherapeutic pair in BLT humanized mice, which bear surgically engrafted human immune systems, including human B cells and T cells.
- ••77.Zhao H, Verma D, Li W, Choi Y, Ndong C, Fiering Steven N, Bailey-Kellogg C, Griswold Karl E. Depletion of T Cell Epitopes in Lysostaphin Mitigates Anti-Drug Antibody Response and Enhances Antibacterial Efficacy In Vivo. Chemistry & Biology. 2015;22:629–639. doi: 10.1016/j.chembiol.2015.04.017. The authors used computationally guided Pareto optimal protein design methods to aggressively delete predicted DRB1*0401 restricted T cell epitopes from throughout the lysostaphin antibacterial protein. Both optimal individual variant and optimal combinatorial library design strategies were employed, yielding two variants that each strongly suppressed antidrug antibody development upon immunization of humanized DRB1*0401 HLA transgenic mice. In a first for the field, they then showed that reduced immunogenicity translated into enhanced therapeutic efficacy, compared to wild-type lysostaphin, in treating recurrent MRSA bacteremia in humanized mice.
- 78.Brinks V, Weinbuch D, Baker M, Dean Y, Stas P, Kostense S, Rup B, Jiskoot W. Preclinical Models Used for Immunogenicity Prediction of Therapeutic Proteins. Pharmaceutical Research. 2013;30:1719–1728. doi: 10.1007/s11095-013-1062-z. [DOI] [PubMed] [Google Scholar]
- 79.Brinks V, Jiskoot W, Schellekens H. Immunogenicity of Therapeutic Proteins: The Use of Animal Models. Pharmaceutical Research. 2011;28:2379–2385. doi: 10.1007/s11095-011-0523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiskoot W, Kijanka G, Randolph TW, Carpenter JF, Koulov AV, Mahler H-C, Joubert MK, Jawa V, Narhi LO. Mouse Models for Assessing Protein Immunogenicity: Lessons and Challenges. Journal of Pharmaceutical Sciences. 2016;105:1567–1575. doi: 10.1016/j.xphs.2016.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Scheer N, Snaith M, Wolf CR, Seibler J. Generation and utility of genetically humanized mouse models. Drug Discov Today. 2013;18:1200–1211. doi: 10.1016/j.drudis.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 82.Villaudy J, Schotte R, Legrand N, Spits H. Critical assessment of human antibody generation in humanized mouse models. Journal of Immunological Methods. 2014;410:18–27. doi: 10.1016/j.jim.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 83.Shultz LD, Brehm MA, Garcia-Martinez JV, Greiner DL. Humanized mice for immune system investigation: progress, promise and challenges. Nat Rev Immunol. 2012;12:786–798. doi: 10.1038/nri3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mazor R, Onda M, Park D, Addissie S, Xiang L, Zhang J, Hassan R, Pastan I. Dual B- and T-cell de-immunization of recombinant immunotoxin targeting mesothelin with high cytotoxic activity. Oncotarget. 2016 doi: 10.18632/oncotarget.9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Spiegel PC, Jr., Jacquemin M, Saint-Remy JM, Stoddard BL, Pratt KP. Structure of a factor VIII C2 domain-immunoglobulin G4kappa Fab complex: identification of an inhibitory antibody epitope on the surface of factor VIII. Blood. 2001;98:13–19. doi: 10.1182/blood.v98.1.13. [DOI] [PubMed] [Google Scholar]
- 86.Hennecke J, Carfi A, Wiley DC. Structure of a covalently stabilized complex of a human alphabeta T-cell receptor, influenza HA peptide and MHC class II molecule, HLA-DR1. Embo j. 2000;19:5611–5624. doi: 10.1093/emboj/19.21.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]