Abstract
Understanding the underlying temporal and mechanistic responses to neurotoxicant exposures during sensitive periods of neuronal development are critical for assessing the impact of these exposures on developmental processes. To investigate the importance of timing of neurotoxicant exposure for perturbation of epigenetic regulation, we exposed human neuronal progenitor cells (hNPCs) to chlorpyrifos (CP) and sodium arsenite (As; positive control) during proliferation and differentiation. CP or As treatment effects on hNPCs morphology, cell viability, and changes in protein expression levels of neural differentiation and cell stress markers, and histone H3 modifications were examined. Cell viability, proliferation/differentiation status, and epigenetic results suggest that hNPCs cultures respond to CP and As treatment with different degrees of sensitivity. Histone modifications, as measured by changes in histone H3 phosphorylation, acetylation and methylation, varied for each toxicant and growth condition, suggesting that differentiation status can influence the epigenetic effects of CP and As exposures.
Keywords: Chlorpyrifos, arsenic, histone 3, differentiation, methylation, acetylation, neurotoxicity, epigenetic
1. Introduction
Neurogenesis is a precisely orchestrated process that occurs in specific structures at specific times during neurodevelopment. It requires temporally and regionally-specific developmental processes that include proliferation, differentiation, synaptogenesis, myelination, and apoptosis (Rice and Barone 2000; Rodier 1995). Maintaining the correct timing and regulation of neurogenesis is critical for nervous system development and function, as disruptions in the process can lead to altered function and disease (Daston et al. 2004; Faustman et al. 2000; Gohlke et al. 2008; Hamby et al. 2008; Moreira et al. 2010; Robinson et al. 2011). Chemical exposure during a key “window of susceptibility” can disrupt brain development and cause lifelong functional impairments (Rodier 1994). For example, epidemiological evidence demonstrates that exposures to developmental neurotoxicants like chlorpyrifos (CP) and arsenic (As) are associated with cognitive effects (Grandjean and Landrigan 2014). In utero exposure to CP in humans is associated with developmental delays in learning, impairments in mental and motor development, and attention problems (Engel et al. 2011; Eskenazi et al. 2010; Eskenazi et al. 2014; Eskenazi et al. 2007; Eskenazi et al. 2008; Horton et al. 2012; Rauh et al. 2006; Rauh et al. 2012). Similarly, prenatal and early postnatal exposures to inorganic arsenic from drinking water are associated with cognitive deficits that are apparent in pre-school children (Grandjean and Landrigan 2014; Hamadani et al. 2011; Wasserman et al. 2007).
The extent of neurotoxicity during development is highly dependent on the timing of exposure and the differentiation status of the exposed tissues. Actively developing regions of the brain tend to be the most vulnerable to toxicant perturbations (Rice and Barone 2000). Proliferating and differentiating neural stem cells have been shown to exhibit differential sensitivity to several toxicants, including the phthalate metabolite mono-(2-ethylhexyl) phthalate (Lim et al. 2009), methylmercury (Theunissen et al. 2010) and valproic acid (Debeb et al. 2010). Therefore, it is important to evaluate toxicant impacts on specific neural developmental processes within various stages of differentiation to evaluate life stage susceptibility.
Epigenetic gene regulation via DNA methylation, histone modification, and noncoding RNA-mediated processes plays an important role in directing the differentiation potential and fate specification of neuronal stem cells (Juliandi et al. 2010b; Sanosaka et al. 2009). Specifically, modulation of chromatin structure by histone tail modifications influences the accessibility of genes for transcription and is thought to be a key regulator of neuronal differentiation (Hsieh and Gage 2004; Juliandi et al. 2010a; Ronan et al. 2013). These regulatory mechanisms are responsive to extracellular signals like cytokines, growth factors, and environmental cues and epigenetic regulation of gene expression has been shown to be responsive to toxicant exposures such as CP and As (Arai et al. 2011; Bailey et al. 2013; Cheng et al. 2012; Cronican et al. 2013; Hou et al. 2012; Juliandi et al. 2010b; Kile et al. 2012; Koestler et al. 2013; Martinez et al. 2011). Toxicant perturbation of chromatin remodeling during neuronal differentiation could mediate neurotoxicity by disrupting normal developmental gene expression patterns. Because chromatin structure changes throughout normal differentiation, toxicant effects on histone modifications are likely to be specific to differentiation state.
hNPCs cultures directed to undergo differentiation provide an opportunity to study early stages of neuronal differentiation processes and have become a valuable tool for neurodevelopmental toxicology (Breier et al. 2008; Radio and Mundy 2008; Shin et al. 2006). ENStem-A , a commercially available hNPCs line derived from human embryonic stem cell line WA09 (Thomson et al. 1998), provides a convenient and accessible model of human neuronal differentiation (Shin et al. 2006). Withdrawal of fibroblast growth factor (bFGF2) leads these multipotent hNPCs cells to undergo a dramatic morphological and functional transition to begin differentiation. Additional morphogenic factors can direct differentiation towards specific regional identities and neuronal subtypes (Young et al. 2011).
We used this model to explore differentiation stage-specific effects of CP exposures along with our positive control As exposures on hNPCs viability, histone modifications and protein expression of differentiation and cell stress markers.
2. Methods
2.1 Cell culture
ENStem-A hNPCs (ArunA Biomedical, Athens, GA) were expanded through passage eight and plated for experiments after reaching 90–100% confluence. Cells were counted by hemocytometer and seeded at 200,000 cells/mL in poly-L-ornithine (20 μg/mL) and laminin (5 μg/mL) double coated tissue culture treated polystyrene 35x10 mm dishes (2 mL) or 96-well black bottom microplates (100 μL) (Becton, Dickinson Co., Franklin Lakes, NJ). Cells were cultured in neural proliferation media (EMD Millipore, Billerica, MA) in a humidifying 37 °C, 5% CO2 incubator. Chemical treatments were administered at the same time as the initiation of differentiation (24 h after plating). Media was replaced either with additional proliferation media containing fibroblast growth factor (FGF) or with HyClone differentiation media without FGF (Thermo Fisher Scientific, Waltham, MA). All results are presented from 6–8 biological replicates and most biological replicates had triple technical replicates.
2.2 Chemical treatment
CP (99.5% purity, ChemService, West Chester, PA) was dissolved in dimethyl sulfoxide (DMSO, Sigma St. Louis, MO) for rapid and complete absorption (Whitney et al. 1995) and the DMSO concentration did not exceed a final volume concentration of 0.1%. The initial CP concentrations tested ranged from 0–570 μM (0, 7, 14, 29, 43, 57, 143, 285, and 570 μM) and working concentrations of 0, 14, 29, 57 μM in 0.1% DMSO were used based on Alamar Blue viability results. Sodium arsenite (As3+), a positive control, was dissolved in DNase/RNase-free distilled water (Invitrogen). Initial As concentrations in media ranged from 0–4 μM (0, 1, 2 and 4 μM) and 1 μM was chosen as the working concentration based on viability results.
2.3 Morphology
All cell culture dishes and plates were monitored with a Nikon inverted microscope equipped with phase-contrast optics (Nikon, Tokyo, Japan) to assess general cellular morphology. Morphological images (not shown) were captured and digitized with a Coolsnap Camera (Roper Scientific, Inc. Duluth, GA).
2.4 Alamar Blue cell viability/proliferation assay
After 24 or 72 h of treatment, concentration-dependent cell viability was measured using the Alamar Blue Assay (Invitrogen). Alamar Blue detects changes in the number of functional cells by measuring mitochondrial reductase activity of live cells by reducing non-fluorescent, blue resazurin dye to fluorescent, red resorufin (O'Brien et al. 2000). 10% Alamar Blue (v/v) was added directly in each well 3 hours in advance of the 24 and 72 h timepoints and incubated at 37 °C in a 5% CO2 incubator for 3 h. After incubation, the 96 well plates were read on a plate reader at 570 nm with reference wavelength 630 nm.
Alamar Blue assay results are presented as average percent of controls (total cells) ± 95% confidence interval. Wells containing media without cells were run to measure background fluorescence every time an assay was performed. The blank media fluorescence measurement was subtracted from fluorescence measurements of cells from the same assay. The percentage normalized by controls was calculated by dividing the adjusted fluorescence levels (FL) of tested cells to the adjusted level of control cells.
2.5 Live, dead and apoptotic cell staining
A three-color fluorescence assay was utilized to determine the number of live, dead and apoptotic cells. Non-fluorescent Calcein AM (Invitrogen, Carlsbad, CA) dye permeates into live cells and is hydrolyzed to green fluorescent Calcein by intracellular esterase. Propidium iodide (PI, Invitrogen) is not permeable to live cells, and selectively stains nuclei of damaged or dead cells with increased membrane permeability with red fluorescence. Hoechst 33342 is used to stain the total nuclei of both living and dead cells with UV fluorescence. The nuclei of live cells are evenly stained, whereas damaged or dead cells have intense and irregular staining. The live/dead assay was performed as previously described (Yu et al. 2005). A Nikon Labophot-2 (Nikon. Tokyo, Japan) was used to visualize the cells and images were captured and digitalized with a Spot Camera at 200x (Diagnostic Instruments, Sterling Heights, MI). MetaMorph software (Molecular Devices LLC. Sunnyvale, CA) was used to process the digitalized images.
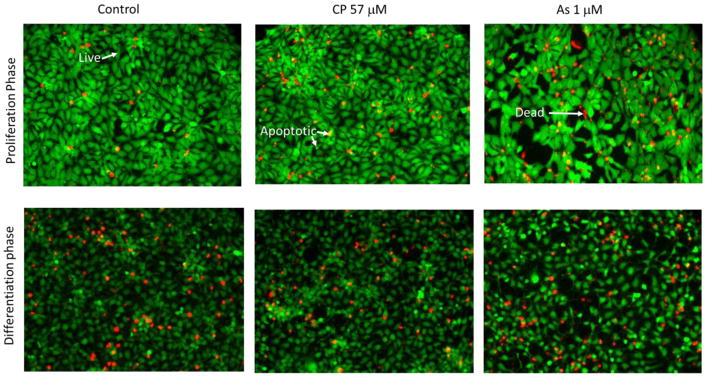
Live/dead cell staining was analyzed quantitatively. The number of Hoechst 33342 stained nuclei were counted manually with the Cell Counting option in the Image J software (National Institute of Health, Maryland, USA). The live cells were Calcein AM positive and PI negative, whereas dead cells were PI positive and Calcein AM negative. Cells were categorized as apoptotic when they either expressed a mixture of Calcein AM and PI, had membrane vesicles and blebs, or Calcein AM was bright and condensed (Figure 2).
Figure 2. Reference staining for live, dead and apoptotic cell quantification.
Examples (arrows) of staining for live, apoptotic, and dead cells stained with Calcein AM (green) and propidium iodide (red) used for cell status enumeration. Apoptotic cells tend to have a mixture of green and red and/or are light green with blebs or are brightly condensed green cells. The top row portrays examples of hNPCs at 72 h under proliferation phase with controls, CP at 57 μM and As at 1 μM. In the row below, control cells under 72 h of differentiation are shown with CP at 57 μM and As at 1 μM.
Total cell counts for each treatment were normalized to 24 h control to assess concentration response. In addition to determining the effect of CP and As treatment on total cell counts, the proportion of live, dead, and apoptotic cells was determined in proliferating and differentiating conditions. Statistical analysis across doses was done comparing to corresponding control (i.e. 24 h control data to 24 h data and 72 h control data to 72 h data). The results of quantitative analysis of the live/dead cell staining assay were presented as the average percent relative to controls (total cells).
2.6 Western blot analysis of cellular neuronal markers
hNPCs in both proliferation and differentiation conditions were harvested 72 h after treatment and lysed in 40 μL cell lysis buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA,1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3Vo4, 1 μg/mL leupeptin (Cell Signaling). The cell lysate was repeatedly frozen and thawed three times and insoluble materials were removed by centrifugation (13,000 rpm for 5 minutes at 4 °C). Protein concentration was measured using the Bio-Rad protein assay kit (Bio-Rad. Hercules, CA). Protein samples were prepared for western blotting by standardizing protein concentrations across samples (by addition of lysis buffer) and adding NuPage reducing agent and sample buffer (Life Technologies). Samples were heated in boiling water for 5 minutes and 10 μg of protein was loaded in each lane of SDS-PAGE gels (Life Technologies). Gels were transferred to polyvinylidene fluoride membrane (Millipore) using a vertical transfer apparatus. Membranes were rinsed briefly in Tris-buffered saline (TBS, pH 7.6) and non-specific binding of the blotted membrane was blocked with 5% non-fat dried milk in TBS with 0.1% Tween-20 (TBBS) for 1 hour. The blocked membranes were incubated with primary antibody (1:1000 – 1:5000) overnight at 4 °C. Following antibody incubation, the membrane was washed three times with TTBS and incubated 2 hours with a secondary antibody. After hybridization with secondary antibodies conjugated to horseradish peroxidase, the immunocomplex was detected with the ECL detection reagent (Amersham Pharmacia Biotech) and exposed to X-ray films. Band intensity was quantified by densitometry using Image J software (National Institute of Health, Maryland, USA). Expression intensity for each protein was first normalized to corresponding β-Actin, then to relevant controls. Statistical analysis was performed as described below. The results from the band densitometric quantification were presented as average fold change to concurrent gel controls ± 95% confidence interval.
The primary antibodies used were nestin (Proteintech), β-tubulin III (Millipore), microtubule-associated protein-2 (MAP-2; Proteintech), p42/44 MAPK (Erk1/2) (Cell Signaling), proliferating cell nuclear antigen (PCNA, Millipore), SOX-2 (Proteintech Group, IL), and caspase 3 (Cell Signaling). β-actin (Sigma) was used as a loading control. A description of the neuronal functional markers can be found in Table 1.
Table 1.
Neuronal functional markers
| Marker | Biological Function | Cells Detected |
|---|---|---|
| SOX-2 | Transcription factor essential for maintaining self-renewal | Embryonic Stem Cells |
| Nestin | Intermediate filament protein involved in radial axon growth | Neural Progenitor Cells |
| β-tubulin III | Component of microtubules found in cytoskeleton and axons | Structural Neuron Specific Marker |
| MAP2 | Microtubule associated protein located primary in dendrites | Structural Neuron Specific Marker |
| p42/44MAPK(Erk1/2) | Ras-Raf-MEK-ERK signal transduction Cascade, stress marker | Ubiquitous |
| PCNA | Proliferation | Proliferating Cells |
| Caspase 3 | Role in apoptosis and in natural brain development | Ubiquitous |
SOX-2 = Sex determining region Y box-2; MAP2 = Microtubule-Associated Protein 2; and PCNA = Proliferating Cell Nuclear Antigen
2.7 Western blot analysis for histone modifications
The methods used for the western blot analysis for analyzing histone modifications were similar to those used above, except for the type of lysis buffer, antibodies, and internal control used. Cells were lysed with 120 μL of 1X SDS buffer (62.5 mM Tris-HCl (pH 6.8 at 25 °C), 2% w/v SDS, 10% glycerol, 50 mM DTT, 0.01% w/v bromophenol blue). Primary antibodies (1:1000) that are specific to Histone H3 with epigenetic modifications at particular amino acid sites (DMH3Lys27, AH3Lys9, pAH3Ser10, DMH3Lys4, DMH3Lys9, DMH3Lys36, DMH3Lys79) and to HDAC4, an enzyme responsible for some epigenetic modifications, were examined. An antibody that detects all histone H3 regardless of site-specific modifications (1:1000–1:3000) was used to normalize the data. All histone-related antibodies were purchased from Cell Signaling Technologies.
Results from densitometric quantification of western bands are presented as average fold change to controls ± 95% confidence interval. Each protein was first normalized to corresponding H3, then to corresponding controls. Statistical analysis was performed as described below.
2.8 Statistical analyses
Mixed effect model analysis was used to investigate significance differences between proliferation and differentiation cell culture conditions and dose response with CP or As exposures. Concentration was used as a continuous fixed effect in the analysis. Random effects for the protein markers and histone H3 were gels and experiment preparation dates.
For the neuronal protein and histone H3 marker endpoints, we used a linear mixed effects model of the change from controls, Y,
where X(i) is the dose of CP or As, A is the intercept and B is the slope for the dose response to CP or As, R(i) is the random effect for gels and preparation date of the ith sample, s is the residual variance after accounting for the fixed effect of dose and the random effects, r is the variance for the random effects.
For Alomar Blue assay, we used a linear model of Y
Statistical analyses were performed using R statistical software (R Core Team, 2016) and the routine “lme4” for mixed effects models (Pinheiro and Bates, 2000; Bates el. Al, 2015).
3. Results
3.1 Cell morphology under proliferation and differentiation conditions
Phase-contrast microscopy demonstrated changes in morphology of untreated cells through time under proliferation and differentiation conditions. Under both culture conditions, cells become increasingly confluent through time. Under differentiation conditions we observed prominent neurite outgrowths after 24 h. By 72 h, cells under differentiation conditions reached full confluence and migrated to form increasingly dense neural networks. This morphology is consistent with what we have observed with our previous in vitro rat embryonic neuronal micromass cultures (Whittaker et al. 1993). CP and As treatments induced a concentration-dependent decrease in cell numbers. Morphological changes were also observed at both 24 and 72 h (data not shown) after the exposure. Fluorescence microscopy images from the live, dead, and apoptotic cell staining assay support these observations and clearly show the changes in morphology in both proliferation and differentiation conditions following toxicant exposure. Under differentiation conditions, exposure to CP (57 μM) or As (1 μM) both resulted in reduced confluence, inhibited neurite outgrowth, and a failure to form clusters.
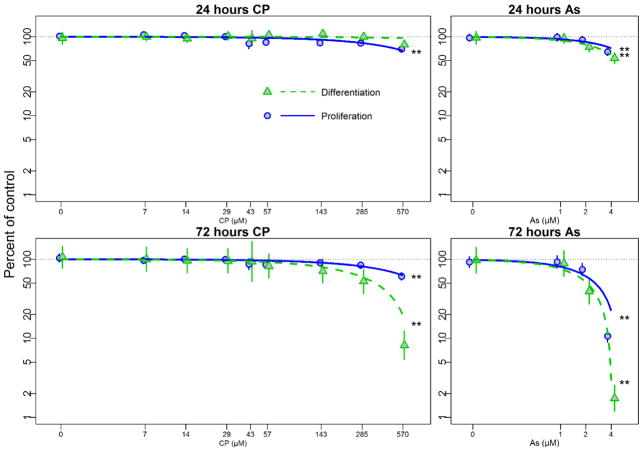
3.2 Cell viability as assessed by the Alamar Blue assay
Alamar Blue cell viability data suggest that differentiating and proliferating hNPCs cultures respond to treatments with different degrees of sensitivity (Figure 1). Concentration-dependent decreases in cell viability of hNPCs are observed in proliferating (p<0.01) cultures with IC10 value of 171 μM after 24 h CP treatment, suggesting that proliferating cells were significantly more sensitive than differentiating cells following 24 h CP exposure. By 72h, CP treatment resulted in a significant concentration-dependent decrease in cell viability of hNPCs under both cell culture conditions (p<0.01). hNPCs under differentiating conditions were significantly more sensitive to CP exposure than those in proliferating conditions (p<0.01). When IC10 were calculated, IC10 in proliferating hNPCs after 72 h of CP exposure was 152 μM while that in differentiating hNPCs was 71 μM.
Figure 1. Alamar Blue cell viability assay with CP and As under proliferation or differentiation conditions.
hNPCs were exposed to chlorpyrifos (CP 0–570 μM) or arsenic (As 0–4 μM) under proliferating (Blue) and differentiating (Green) conditions at 24h and 72h. Plots show means and 95% confidence intervals. Statistical significances, * = p<0.05; ** = p<0.01, are based on a linear mixed effect model indicating significant dose response for each condition. In order to better illustrate the comparison of the data to regression line at low dose, we used a log scale for dose and percent of control, which led to curved trend lines.
Differential susceptibility of cells in proliferating and differentiating conditions was also observed following As treatments. Concentration-dependent decreases in cell viability were observed under both proliferation and differentiation culture conditions following 24 and 72 h of exposure to As. After 24 h of As exposure, proliferating and differentiating hNPCs had IC10 of 1.4 μM and 1.0 μM, respectively. When hNPCs were exposed to As for 72 h, IC10 value dropped to 0.5 μM in proliferation and 0.4 μM in differentiation conditions. These results demonstrate that concentration-dependent decreases in viability in response to As were significantly greater in cells under differentiation cell culture conditions at both time points.
3.3 Cytotoxicity as Assessed by Live, Dead and Apoptotic Cell Staining
Quantitative live/dead cell imaging using Calcein AM (green) and propidium iodide (red) supported our observations regarding differential susceptibility in proliferating and differentiating hNPCs.
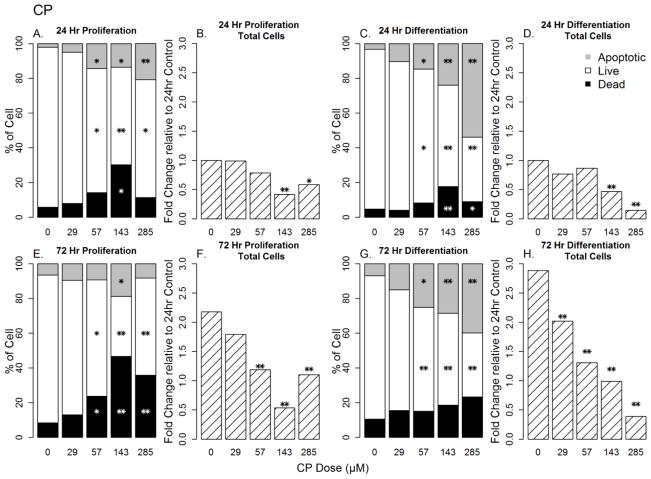
CP exposure resulted in significant concentration-dependent decreases in total cell numbers after 24 h of exposure under both proliferation and differentiation conditions (Figure 3; p<0.01 for both). At CP concentrations of 57–285 μM, there were decreases in the percentage of live cells with concomitant increases in apoptotic and/or dead cells.
Figure 3. Concentration-dependent changes in cell number, cell death, apoptosis, and cell viability in CP-treated hNPCs under proliferation and differentiation conditions.
Sections A, C, E and G represent the distribution of live, dead and apoptotic cells while Sections B, D, F and H represents fold changes in cell count relative to 24 h controls. * = p<0.05; ** = p<0.01
At 72 h exposure of CP exposure, there was a concentration-dependent loss of cell count under both proliferating and differentiating conditions (Figures 3F and H) with proliferating cells having a greater proportion of dead cells than differentiating cells at 57 μM and above (Figure 3E and G).
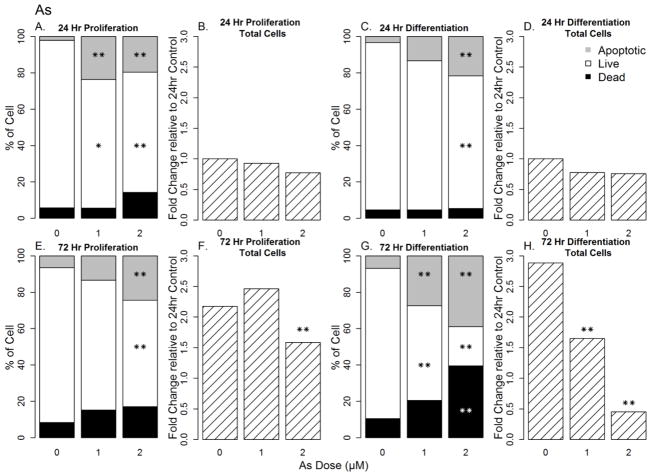
Though exposure to As for 24 h had minimal effect on total cell numbers as assessed by live, dead, and apoptotic cell staining assay in either culture condition (Figure 4B and D, p>0.05), the proportion of live and apoptotic cells were altered under both conditions. This effect was more prominent under proliferating conditions at 24 h (Figure 4A and C). Exposure to As for 72 h significantly reduced the total cell number, increased dead and apoptotic cells and decreased live cells in both proliferation (Figure 4E and F) and differentiation (Figure 4G) conditions (p<0.01). This effect was significantly larger at lower concentrations under differentiation conditions (p<0.01) than in proliferating conditions.
Figure 4. Concentration-dependent changes in cell number, cell death, apoptosis, and cell viability in As-treated hNPCs under proliferation and differentiation conditions.
Sections A, C, E and G represent the distribution of live, dead and apoptotic cells within a culture at a given dose. Significance of the effect is presented relative to controls at the corresponding time point while Sections B, D, F and H represents changes in cell count relative to controls. Significance of the effect is presented relative to the 24 h controls to show the increase in cell number through time. * = p<0.05; ** = p<0.01.
3.4 Neuronal and developmental stage-specific protein marker expression
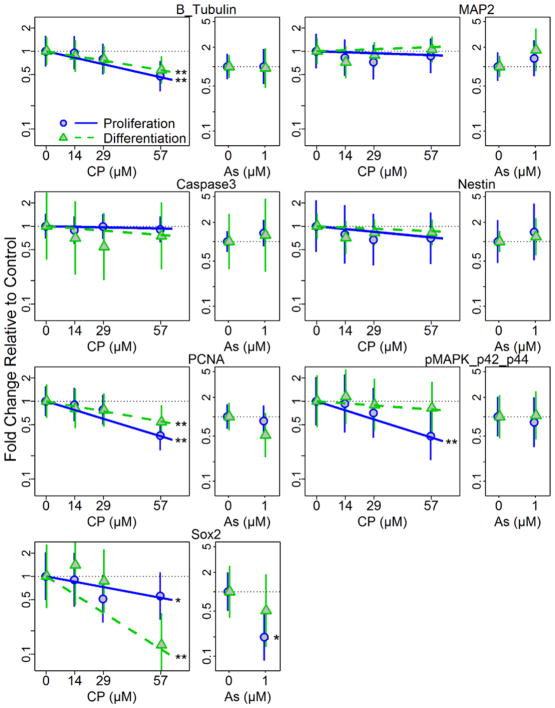
Exposure to CP (0–57 μM) and As (1 μM) resulted in concentration-dependent effects on the protein expression of several differentiation stage-specific markers in hNPCs under both proliferating and differentiating conditions at 72 h (Figure 5, Table 2, Supplement Figure S2–S3). For example, CP exposure resulted in a significant concentration-dependent decrease in the expression of the neuronal marker β-tubulin III (p<0.01), the proliferation marker PCNA (p<0.01), and the stem cell marker SOX-2 (p<0.05) in both proliferating and differentiation conditions (Figure 5, Table 2, Supplement Figure S2) when compared to β-actin. CP exposure in each culture condition had differential effects on expression of p42/44MAPK (Erk1/2), a member of the Ras-Raf-MEK-ERK signal transduction cascade and a mediator of proliferation, differentiation, and stress response processes triggered by extracellular cues. There was a significant concentration-dependent decrease in the expression of p42/44 MAPK (Erk1/2) under proliferation conditions (p<0.01), but no significant effect on cells under differentiating conditions.
Figure 5. Changes in neuronal and stage specific protein marker expressions following treatment of hNPCs cultured under proliferation and differentiation conditions.
Protein expression was quantified from western blotting following 72h treatment with CP or As (positive control) under proliferating or differentiating conditions. Plots show means and 95% confidence intervals. Statistical significances, * = p<0.05; ** = p<0.01, are based on a linear mixed effect model indicating significant dose response for each condition.
Table 2.
Summary of toxicant effects on protein expression
| Marker | CP | AS | ||
|---|---|---|---|---|
| Proliferation | Differentiation | Proliferation | Differentiation | |
| β-TUBULIN III | ↓ ** | ↓ ** | NS | NS |
| MAP2 | NS | NS | NS | NS |
| CASPASE 3 | NS | NS | NS | NS |
| NESTIN | NS | NS | NS | NS |
| PCNA | ↓ ** | ↓ ** | NS | NS |
| MAPK | ↓ ** | NS | NS | NS |
| SOX-2 | ↓ * | ↓ ** | ↓ * | NS |
Note:
= p<0.05;
= p<0.01;
NS = not significant; ↓ = decrease; ↑ = increase
Exposure to As also had differential effects on protein expression (Figure 5, Table 2, Supplement Figure S3). There was a significant decrease in SOX-2 expression unique to proliferation conditions (p<0.05). Expression levels of MAP2, caspase 3 and nestin were not significantly changed at any CP or As concentration in differentiating or proliferating cells.
3.5 Histone Modifications
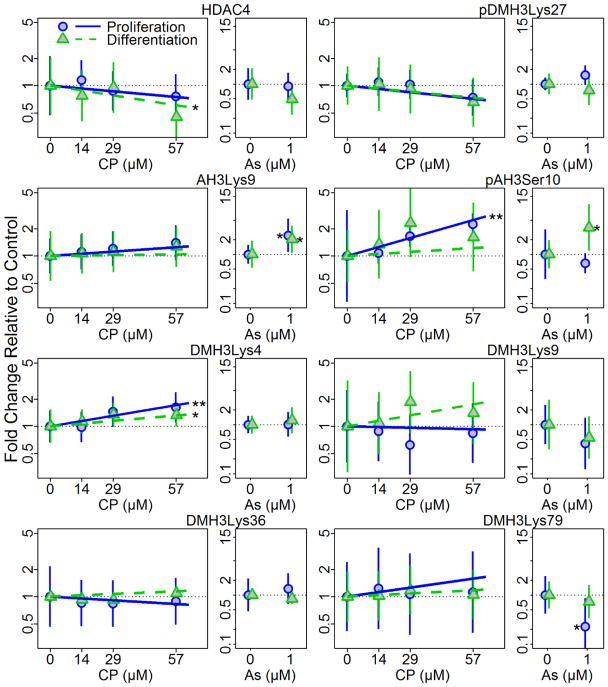
Exposure to CP (0–57 μM) and As (1 μM) differentially changed the relative amounts of several histone modifications in both differentiating and proliferating hNPCs after 72 h (Figure 6, Table 3, Supplement Figure S4–S5). For example, CP induced a significant concentration-dependent increase in pAH3Ser10 in proliferating cells (p<0.01) but not in differentiating cells (Figure 6, Table 3, Supplement Figure S4). Additionally, one of the enzymes responsible for histone modifications, HDAC4, was significantly decreased in a concentration-dependent manner under differentiation conditions (p=0.048) following CP exposure but not under proliferation conditions. CP also induced a significant concentration dependent increase in DMH3Lys4 in both proliferation (p<0.01) and differentiation conditions (p=0.021).
Figure 6. Changes of histone H3 acetylation and methylation following CP and As treatment of hNPCs cultured under proliferation and differentiation conditions at 72 h post treatment.
Histone modifications at specific sites were quantified by western blotting following 72h treatment with CP or As (positive control) under proliferating or differentiating conditions. Plots show means and 95% confidence intervals. Statistical significances, * = p<0.05; ** = p<0.01, are based on a linear mixed effect model indicating significant dose response for each condition.
Table 3.
Summary of toxicant effects on histone modifications
| Marker | CP | AS | ||
|---|---|---|---|---|
| Proliferation | Differentiation | Proliferation | Differentiation | |
| HDAC4 | NS | ↓ * | NS | NS |
| DMH3Lys27 | NS | NS | NS | NS |
| AH3Lys9 | NS | NS | ↑ * | ↑ * |
| pAH3SeR10 | ↑ ** | NS | NS | ↑ * |
| DMH3Lys4 | ↑ ** | ↑ * | NS | NS |
| DMH3Lys9 | NS | NS | NS | NS |
| DMH3Lys36 | NS | NS | NS | NS |
| DMH3Lys79 | NS | NS | ↓ * | NS |
Note:
= p<0.05;
= p<0.01;
NS = not significant; ↓ = decrease; ↑ = increase
Exposure to As significantly increased the expression of AH3Lys9 in both cell growth conditions (p<0.05). However, DHM3Lys79 was decreased only under proliferation conditions (p=0.019) and pAH3Ser10 was significantly increased (p=0.014) only under differentiation conditions following As exposure (Figure 6, Table 3, Supplement Figure S5). These results suggest that differentiation status modifies the effects of CP and As exposure on histone modifications.
4. Discussion
Early life exposures to As or CP can redirect neuronal developmental processes like differentiation (Ivanov and Hei 2013; Visan et al. 2012), neurite outgrowth (Howard et al. 2005; Wang et al. 2010), neuronal subtype fate specification (Jameson et al. 2006), and apoptosis (Lu et al. 2014; Slotkin et al. 2007), and lead to neurological, behavioral and cognitive dysfunctions (Tyler and Allan 2014). Transcriptional control is, in part, influenced by epigenetic gene regulation which has become an important emerging field for understanding the coordination of gene expression patterns during brain development, learning, and memory (Day and Sweatt 2011; Fagiolini et al. 2009; Juliandi et al. 2010b; Rudenko and Tsai 2014; Sanosaka et al. 2009) and is sensitive to perturbation by toxicants (Collotta et al. 2013; Reichard and Puga 2010; Wright and Baccarelli 2007). Previous research has demonstrated that differentiating cells have varying degrees of susceptibility to toxicants that is dependent on the stage of differentiation (Debeb et al. 2010; Lim et al. 2009; Theunissen et al. 2010); however, little work has been done to explore differential toxicant effects on epigenetic endpoints at different developmental stages. In the current research, we demonstrate that hNPCs differentiation state influences the type of histone modifications made in response to the toxicant in addition to modifying the effect of CP or As exposure on viability, apoptosis, morphology, and protein expression. These findings underscore the importance of developmental context and timing of exposure in neurodevelopmental toxicity, and further validate this in vitro hNPCs model as one capable of reflecting stage-specific developmental endpoints.
We characterized the dose-response of hNPCs to a range of concentrations of CP or As (positive control) under proliferating or differentiating conditions. These two toxicants were chosen as they are well-characterized developmental neurotoxicants with the capability of altering the epigenetic landscape in a variety of cell types (Cheng et al. 2012; Cronican et al. 2013; Hou et al. 2012; Salazar-Arredondo et al. 2008; Shin et al. 2014). Results from the Alamar Blue assay revealed no substantial changes in the abundance of functional cells within the working concentration ranges of CP (0–57μM) after 24 h. We focused on concentrations that produced minimal changes in cell viability in the Alamar Blue assay at 24 h to explore protein expression and histone modifications in this study. Results from the quantitative live/dead and apoptotic cell imaging assay for the same time points and concentrations indicated significant decreases in total cell number for proliferating and differentiating cells exposed to 57 μM CP for 72 h. There were also concentration-dependent increases in the proportion of apoptotic cells after CP or As exposures. The image analysis results suggest that the proliferation phase had a more significant concentration-dependent increase in percentage of dead cells compared to the differentiation stage after CP exposures while percentages of apoptotic cells were greater in differentiating hNPCs compared to proliferating conditions. For As, the live/dead cell imaging at 72 h suggests that differentiating cells are more susceptible than proliferating cells based on the results indicated by higher percentages of apoptotic/dead cells and greater reductions in cell numbers with the live, dead, apoptotic cell staining assay.
Protein markers were used to characterize the state of hNPCs differentiation and stress induced by the CP or As exposures. In our previous studies (unpublished), we have shown time dependent changes in markers of proliferation and differentiation where expression of PCNA peaks at day 10 in culture and then decrease while β-tubulin III and MAP-2 expressions increase over 21 days of culture. There were instances where changes in expression levels following treatment occurred for the same proteins in both the proliferation and differentiation conditions; however, the magnitude of this response for some markers was growth condition dependent. For example, CP exposure at 57 μM for 72 h reduced SOX-2 expression, a transcription factor associated with maintaining cellular pluripotency, in differentiating cells approximately sevenfold more than proliferating cells. Expression levels of SOX-2 were also reduced after 72 h of exposure to 1 μM As relative to controls but this effect was more pronounced in the proliferation phase than in differentiating cells. β-tubulin III, a component of microtubules found in the cytoskeleton and axons and a marker of differentiated neurons, was significantly reduced relative to controls following exposure to CP under both proliferation and differentiation conditions. These findings of stage-specific changes in protein expression following toxicant exposure are consistent with previous reports that CP exposure results in distinct transcriptional responses in proliferating and differentiating PC12 cells (Slotkin et al. 2007; Slotkin and Seidler 2012) and that the effects of As on neurite outgrowth in PC12 cells are stage-specific (Frankel et al. 2009). Cell status imaging data suggesting concentration-dependent increases in apoptotic cells with CP and As exposure were not confirmed by concomitant increases of caspase-3, a marker of an activated apoptosis pathway. Indeed, chlorpyrifos (at 29 μM) has been shown to target the genes that drive the apoptotic pathway and cell cycle in differentiated and undifferentiated PC12 neural cells with a higher number of apoptotic genes changed in differentiated cells and a reduction in cell cycle genes from proliferating cells (Slotkin and Seidler 2012).
Histone modification profiles varied between proliferating and differentiating cells in response to exposure to either CP or As which have not been previously demonstrated in vitro. Our histone modification findings are consistent with our previous observation that CP exposure in utero altered the expression of genes associated with chromatin modifications in fetal mouse brains (Moreira et al. 2010). In our hNPC culture system, each toxicant altered the presence of at least one histone modification measured in a differentiation stage-specific manner. Specifically, we found that proliferating hNPCs treated with CP had increased expression of phosphorylated serine 10 in histone H3 (pAH3Ser10) which is functionally linked to apoptosis through PKCδ phosphorylation and Aurora B and VRK1-mediated chromatin condensation during mitosis (Crosio et al. 2002; Kang et al. 2007; Park and Kim 2012).We also found that the enzyme responsible for some of these histone modifications, histone deacetylase 4 (HDAC4), was significantly decreased relative to controls in differentiating but not in proliferating cells exposed to CP. HDAC4 is emerging as an important regulator of brain development (Majdzadeh et al. 2008), plasticity, and learning and memory (Sando et al. 2012).
Methylation of lysine 4 on histone H3 is associated with the activation of gene transcription, in particular, homeotic (HOX) genes which are integral in development and differentiation (Eissenberg and Shilatifard 2010; Ruthenburg et al. 2007). Both proliferating and differentiating hNPCs exposed to CP had increased, relative to controls, di-methylated lysine 4 in histone H3 (DMH3Lys4), suggesting that CP exposure may affect the transcription of genes involved in neurodevelopment and differentiation.
Epigenetic effects associated with As exposure as measured by DNA methylation and histone methylation, acetylation and phosphorylation have been characterized by in vivo and epidemiological studies with a majority of studies reporting global hypomethylation patterns (Arita et al. 2012; Cantone et al. 2011; Chervona et al. 2012; Cronican et al. 2013; Ray et al. 2014). Studies that specifically address the epigenetic effects of As exposure to developing neural tissue or in in vitro systems are limited. Moderate embryonic As exposure in mice resulted in decreased acetylation at lysine 9 in histone H3 (AH3Lys9) in brain tissue which was ultimately associated with behavioral and cognitive impairment in adult mice (Cronican et al. 2013). Changes in histone modification including acetylation of histone H3 lysine 9 were examined in vitro by Pournara et al. (2016), but the study was only done in T-lymphocytes and kidney HEK293 cells. In our hNPCs model, we observed increased acetylation at this same site, relative to controls, in both proliferating and differentiating cells. AH3Lys9 has been found to decline during the first four days of neural differentiation of human embryonic stem cells (hESC) then levels gradually increase during days 4–8 as it is believed to play a dual role in hESC pluripotency maintenance and neural differentiation and is generally associated with an open chromatin state and increased transcriptional competency (Lee and Mahadevan 2009; Peterson and Laniel 2004; Qiao et al. 2015). Similar to proliferating hPNCs exposed to CP, differentiating cells exposed to As had an increase, relative to controls, of pAH3Ser10 that suggests a shift to active gene transcription and apoptosis (Koch et al. 2007; Lo et al. 2000; Park and Kim 2012). Activation of the apoptosis pathway was confirmed with high proportions of apoptotic cells with cell status imaging but not with concomitant increases in caspase 3.
Global increases of di-methylated lysine 79 on histone H3 (DMH3Lys79) have been associated with genome integrity maintenance during mitosis (Fu et al. 2013; Guppy and McManus 2015). Proliferating As-exposed hNPCs had decreased DMH3Lys79, relative to controls, suggesting that genomic stability may be becoming compromised. Indeed, gestational exposure to As in mice has been implicated in increased adult cancer risk and it appears that stem cells are targeted during the process (Tokar et al. 2011).
The consistency of histone modification sites targeted by As and CP across in vivo and in vitro systems demonstrates that our in vitro model is capable of reflecting molecular changes associated with functional outcomes in vivo during sensitive periods of neurodevelopment. It remains to be seen whether the changes in protein expression are a direct result of the histone modifications we observed. The concept of “developmental origins of adult disease” is a phenomenon that is thought to be, in part, mediated via perturbations of the epigenetic program during development (Barker 2007; Skinner 2007). These effects are particularly challenging to study given the long time period between developmental exposures and adult health outcomes. Our ability to detect epigenetic changes in a short-term in vitro assay suggests the exciting potential to rapidly screen for toxicants that have the potential to influence health many years after developmental exposures.
We believe our results of epigenetic changes in hNPCs as a result from As and CP exposure during proliferation and differentiation conditions not only contributes to the greater understanding of the epigenetic processes under the influence of environmental exposures, but also demonstrates the capacity of our in vitro system as a promising model for neurodevelopmental toxicity screening.
Supplementary Material
Highlights.
We evaluated histone modifications in hNPC exposed to chlorpyrifos or arsenic.
Differentiation status influences histone modifications with chlorpyrifos exposure.
Differentiation status influences histone modifications with arsenic exposure.
Protein expression levels of differentiation markers influenced by arsenic.
Protein expression levels of differentiation markers influenced by chlorpyrifos.
Acknowledgments
Funding
Grant sponsor: This publication was made possible by U.S. Environmental Protection Agency (US EPA) grants RD83451401 and RD83170901 and National Institute for Environmental Health Sciences (NIEHS) grants 5P01ES009601 and 5P30ES007033. Its contents are solely the responsibility of the grantee and do not necessarily represent the official views of the US EPA. Further, the US EPA does not endorse the purchase of any commercial products or services mentioned in the publication.
Abbreviations
- AH3Lys9
acetylated histone H3 lysine 9
- As
sodium arsenite
- CP
chlorpyrifos
- DMH3Lys4
dimethylated histone H3 at lysine 4
- DMH3Lys9
dimethylated histone H3 at lysine 9
- DMH3Lys27
dimethylated histone H3 at lysine 27
- DMH3Lys36
dimethylated histone H3 at lysine 36
- DMH3Lys79
dimethylated histone H3 at lysine 79
- FGF
fibroblast growth factor
- HDAC4
histone deacetylase 4
- HDAC7
histone deacetylase 7
- hNPCs
human neuronal progenitor cells
- MAP2
Microtubule-Associated Protein 2
- pAH3Ser10
phosphorylated histone 3 at serine 10
- PCNA
Proliferating Cell Nuclear Antigen
- SOX2
Sex determining region Y box-2
Footnotes
Conflict of interest statement
The author has no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arai Y, Ohgane J, Yagi S, Ito R, Iwasaki Y, Saito K, Akutsu K, Takatori S, Ishii R, Hayashi R, et al. Epigenetic Assessment of Environmental Chemicals Detected in Maternal Peripheral and Cord Blood Samples. Journal of Reproduction and Development. 2011;57(4):507–517. doi: 10.1262/jrd.11-034a. [DOI] [PubMed] [Google Scholar]
- Arita A, Shamy MY, Chervona Y, Clancy HA, Sun H, Hail MN, Qu Q, Gamble MV, Costa M. The effect of exposure to carcinogenic metals on histone tail modifications and gene expression in human subjects. J Trace Elem Med Biol. 2012;26(2–3):174–8. doi: 10.1016/j.jtemb.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey KA, Wu MC, Ward WO, Smeester L, Rager JE, Garcia-Vargas G, Del Razo LM, Drobna Z, Styblo M, Fry RC. Arsenic and the Epigenome: Interindividual Differences in Arsenic Metabolism Related to Distinct Patterns of DNA Methylation. Journal of Biochemical and Molecular Toxicology. 2013;27(2):106–15. doi: 10.1002/jbt.21462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Machler M, Bolker B, Walker S. Fitting Linear Mixed-Effects Models Using lme4. Journal of Statistical Software. 2015;67:1, 1–48. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- Barker DJ. The origins of the developmental origins theory. J Intern Med. 2007;261(5):412–7. doi: 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- Breier JM, Radio NM, Mundy WR, Shafer TJ. Development of a high-throughput screening assay for chemical effects on proliferation and viability of immortalized human neural progenitor cells. Toxicological sciences : an official journal of the Society of Toxicology. 2008;105(1):119–33. doi: 10.1093/toxsci/kfn115. [DOI] [PubMed] [Google Scholar]
- Cantone L, Nordio F, Hou L, Apostoli P, Bonzini M, Tarantini L, Angelici L, Bollati V, Zanobetti A, Schwarz J, Bertazzi PA, Baccarelli A. Inhalable metal-rich air particles and histone H3K4 dimethylation and H3K9 acetylation in a cross sectional study of steel workers. Environ Health Perspect. 2011;119(7):964–9. doi: 10.1289/ehp.1002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng TF, Choudhuri S, Muldoon-Jacobs K. Epigenetic targets of some toxicologically relevant metals: a review of the literature. Journal of Applied Toxicology. 2012;32(9):643–653. doi: 10.1002/jat.2717. [DOI] [PubMed] [Google Scholar]
- Chervona Y, Hall MN, Arita A, Wu F, Sun H, Tseng HC, Ali E, Uddin MN, Liu X, Zoroddu MA, Gamble MV, Costa M. Associations between arsenic exposure and global posttranslational histone modifications among adults in Bangladesh. Cancer Epidemiol Biomarkers Prev. 2012;21(12):2252–60. doi: 10.1158/1055-9965.EPI-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collotta M, Bertazzi PA, Bollati V. Epigenetics and pesticides. Toxicology. 2013;307:35–41. doi: 10.1016/j.tox.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Cronican AA, Fitz NF, Carter A, Saleem M, Shiva S, Barchowsky A, Koldamova R, Schug J, Lefterov I. Genome-wide alteration of histone H3K9 acetylation pattern in mouse offspring prenatally exposed to arsenic. PLoS One. 2013;8(2):e53478. doi: 10.1371/journal.pone.0053478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daston G, Faustman E, Ginsberg G, Fenner-Crisp P, Olin S, Sonawane B, Bruckner J, Breslin W, McLaughlin TJ. A framework for assessing risks to children from exposure to environmental agents. Environ Health Perspect. 2004;112(2):238–56. doi: 10.1289/ehp.6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. Epigenetic mechanisms in cognition. Neuron. 2011;70(5):813–29. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeb BG, Xu W, Mok H, Li L, Robertson F, Ueno NT, Reuben J, Lucci A, Cristofanilli M, Woodward WA. Differential radiosensitizing effect of valproic acid in differentiation versus self-renewal promoting culture conditions. Int J Radiat Oncol Biol Phys. 2010;76(3):889–95. doi: 10.1016/j.ijrobp.2009.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2010;339(2):240–9. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel SM, Wetmur J, Chen J, Zhu C, Barr DB, Canfield RL, Wolff MS. Prenatal exposure to organophosphates, paraoxonase 1, and cognitive development in childhood. Environ Health Perspect. 2011;119(8):1182–8. doi: 10.1289/ehp.1003183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskenazi B, Huen K, Marks A, Harley KG, Bradman A, Barr DB, Holland N. PON1 and neurodevelopment in children from the CHAMACOS study exposed to organophosphate pesticides in utero. Environ Health Perspect. 2010;118(12):1775–81. doi: 10.1289/ehp.1002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskenazi B, Kogut K, Huen K, Harley KG, Bouchard M, Bradman A, Boyd-Barr D, Johnson C, Holland N. Organophosphate pesticide exposure, PON1, and neurodevelopment in school-age children from the CHAMACOS study. Environ Res. 2014;134:149–57. doi: 10.1016/j.envres.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskenazi B, Marks AR, Bradman A, Harley K, Barr DB, Johnson C, Morga N, Jewell NP. Organophosphate pesticide exposure and neurodevelopment in young Mexican-American children. Environ Health Perspect. 2007;115(5):792–8. doi: 10.1289/ehp.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskenazi B, Rosas LG, Marks AR, Bradman A, Harley K, Holland N, Johnson C, Fenster L, Barr DB. Pesticide toxicity and the developing brain. Basic Clin Pharmacol Toxicol. 2008;102(2):228–36. doi: 10.1111/j.1742-7843.2007.00171.x. [DOI] [PubMed] [Google Scholar]
- Fagiolini M, Jensen CL, Champagne FA. Epigenetic influences on brain development and plasticity. Curr Opin Neurobiol. 2009;19(2):207–12. doi: 10.1016/j.conb.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustman EM, Silbernagel SM, Fenske RA, Burbacher TM, Ponce RA. Mechanisms underlying Children's susceptibility to environmental toxicants. Environ Health Perspect. 2000;108(Suppl 1):13–21. doi: 10.1289/ehp.00108s113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel S, Concannon J, Brusky K, Pietrowicz E, Giorgianni S, Thompson WD, Currie DA. Arsenic exposure disrupts neurite growth and complexity in vitro. Neurotoxicology. 2009;30(4):529–37. doi: 10.1016/j.neuro.2009.02.015. [DOI] [PubMed] [Google Scholar]
- Fu H, Maunakea AK, Martin MM, Huang L, Zhang Y, Ryan M, Kim R, Lin CM, Zhao K, Aladjem MI. Methylation of histone H3 on lysine 79 associates with a group of replication origins and helps limit DNA replication once per cell cycle. PLoS Genet. 2013;9(6):e1003542. doi: 10.1371/journal.pgen.1003542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohlke JM, Hiller-Sturmhofel S, Faustman EM. A systems-based computational model of alcohol's toxic effects on brain development. Alcohol Research & Health. 2008;31(1):76–83. [PMC free article] [PubMed] [Google Scholar]
- Grandjean P, Landrigan PJ. Neurobehavioural effects of developmental toxicity. Lancet Neurol. 2014;13(3):330–8. doi: 10.1016/S1474-4422(13)70278-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guppy BJ, McManus KJ. Mitotic accumulation of dimethylated lysine 79 of histone H3 is important for maintaining genome integrity during mitosis in human cells. Genetics. 2015;199(2):423–33. doi: 10.1534/genetics.114.172874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamadani JD, Tofail F, Nermell B, Gardner R, Shiraji S, Bottai M, Arifeen SE, Huda SN, Vahter M. Critical windows of exposure for arsenic-associated impairment of cognitive function in pre-school girls and boys: a population-based cohort study. Int J Epidemiol. 2011;40(6):1593–604. doi: 10.1093/ije/dyr176. [DOI] [PubMed] [Google Scholar]
- Hamby ME, Coskun V, Sun YE. Transcriptional regulation of neuronal differentiation: the epigenetic layer of complexity. Biochim Biophys Acta. 2008;1779(8):432–7. doi: 10.1016/j.bbagrm.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton MK, Kahn LG, Perera F, Barr DB, Rauh V. Does the home environment and the sex of the child modify the adverse effects of prenatal exposure to chlorpyrifos on child working memory? Neurotoxicol Teratol. 2012;34(5):534–41. doi: 10.1016/j.ntt.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Zhang X, Wang D, Baccarelli A. Environmental chemical exposures and human epigenetics. Int J Epidemiol. 2012;41(1):79–105. doi: 10.1093/ije/dyr154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard AS, Bucelli R, Jett DA, Bruun D, Yang D, Lein PJ. Chlorpyrifos exerts opposing effects on axonal and dendritic growth in primary neuronal cultures. Toxicol Appl Pharmacol. 2005;207(2):112–24. doi: 10.1016/j.taap.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Gage FH. Epigenetic control of neural stem cell fate. Curr Opin Genet Dev. 2004;14(5):461–9. doi: 10.1016/j.gde.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Ivanov VN, Hei TK. Induction of apoptotic death and retardation of neuronal differentiation of human neural stem cells by sodium arsenite treatment. Exp Cell Res. 2013;319(6):875–87. doi: 10.1016/j.yexcr.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson RR, Seidler FJ, Qiao D, Slotkin TA. Chlorpyrifos affects phenotypic outcomes in a model of mammalian neurodevelopment: critical stages targeting differentiation in PC12 cells. Environ Health Perspect. 2006;114(5):667–72. doi: 10.1289/ehp.8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliandi B, Abematsu M, Nakashima K. Chromatin remodeling in neural stem cell differentiation. Curr Opin Neurobiol. 2010a;20(4):408–15. doi: 10.1016/j.conb.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Juliandi B, Abematsu M, Nakashima K. Epigenetic regulation in neural stem cell differentiation. Dev Growth Differ. 2010b;52(6):493–504. doi: 10.1111/j.1440-169X.2010.01175.x. [DOI] [PubMed] [Google Scholar]
- Kile ML, Baccarelli A, Hoffman E, Tarantini L, Quamruzzaman Q, Rahman M, Mahiuddin G, Mostofa G, Hsueh YM, Wright RO, Christiani DC. Prenatal Arsenic Exposure and DNA Methylation in Maternal and Umbilical Cord Blood Leukocytes. Environ Health Perspect. 2012;120(7):1061–6. doi: 10.1289/ehp.1104173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17(6):691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestler DC, Avissar-Whiting M, Houseman EA, Karagas MR, Marsit CJ. Differential DNA Methylation in Umbilical Cord Blood of Infants Exposed to Low Levels of Arsenic In Utero. Environ Health Perspect. 2013;121(8):971–7. doi: 10.1289/ehp.1205925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BM, Mahadevan LC. Stability of histone modifications across mammalian genomes: implications for 'epigenetic' marking. J Cell Biochem. 2009;108(1):22–34. doi: 10.1002/jcb.22250. [DOI] [PubMed] [Google Scholar]
- Lim CK, Kim SK, Ko DS, Cho JW, Jun JH, An SY, Han JH, Kim JH, Yoon YD. Differential cytotoxic effects of mono-(2-ethylhexyl) phthalate on blastomere-derived embryonic stem cells and differentiating neurons. Toxicology. 2009;264(3):145–54. doi: 10.1016/j.tox.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R, Berger SL. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell. 2000;5(6):917–26. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- Lu TH, Tseng TJ, Su CC, Tang FC, Yen CC, Liu YY, Yang CY, Wu CC, Chen KL, Hung DZ, et al. Arsenic induces reactive oxygen species-caused neuronal cell apoptosis through JNK/ERK-mediated mitochondria-dependent and GRP 78/CHOP-regulated pathways. Toxicol Lett. 2014;224(1):130–40. doi: 10.1016/j.toxlet.2013.10.013. [DOI] [PubMed] [Google Scholar]
- Majdzadeh N, Wang L, Morrison BE, Bassel-Duby R, Olson EN, D'Mello SR. HDAC4 inhibits cell-cycle progression and protects neurons from cell death. Dev Neurobiol. 2008;68(8):1076–92. doi: 10.1002/dneu.20637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez L, Jimenez V, Garcia-Sepulveda C, Ceballos F, Delgado JM, Nino-Moreno P, Doniz L, Saavedra-Alanis V, Castillo CG, Santoyo ME, Gonzalez-Amaro R, Jimenez-Capdeville ME. Impact of Early Developmental Arsenic Exposure on PromotorCpG-island Methylation of Genes Involved in Neuronal Plasticity. Neurochem Int. 2011;58(5):574–81. doi: 10.1016/j.neuint.2011.01.020. [DOI] [PubMed] [Google Scholar]
- Moreira EG, Yu X, Robinson JF, Griffith W, Hong SW, Beyer RP, Bammler TK, Faustman EM. Toxicogenomic profiling in maternal and fetal rodent brains following gestational exposure to chlorpyrifos. Toxicol Appl Pharmacol. 2010;245(3):310–25. doi: 10.1016/j.taap.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien J, Wilson I, Orton T, Pognan F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur J Biochem. 2000;267(17):5421–6. doi: 10.1046/j.1432-1327.2000.01606.x. [DOI] [PubMed] [Google Scholar]
- Park CH, Kim KT. Apoptotic phosphorylation of histone H3 on Ser-10 by protein kinase Cdelta. PLoS One. 2012;7(9):e44307. doi: 10.1371/journal.pone.0044307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14(14):R546–51. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Pinheiro JC, Bates DM. Mixed effects models in S and S-Plus. New York: Springer; 2000. [Google Scholar]
- Pournara A, Kippler M, Holmlund T, Ceder R, Grafström R, Vahter M, Broberg K, Wallberg AE. Arsenic alters global histone modifications in lymphocytes in vitro and in vivo. Cell Biol Toxicol. 2016;32(4):275–84. doi: 10.1007/s10565-016-9334-0. [DOI] [PubMed] [Google Scholar]
- Qiao Y, Wang R, Yang X, Tang K, Jing N. Dual roles of histone H3 lysine 9 acetylation in human embryonic stem cell pluripotency and neural differentiation. J Biol Chem. 2015;290(4):2508–20. doi: 10.1074/jbc.M114.603761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. URL https://www.R-project.org/ [Google Scholar]
- Radio NM, Mundy WR. Developmental neurotoxicity testing in vitro: models for assessing chemical effects on neurite outgrowth. Neurotoxicology. 2008;29(3):361–76. doi: 10.1016/j.neuro.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Rauh VA, Garfinkel R, Perera FP, Andrews HF, Hoepner L, Barr DB, Whitehead R, Tang D, Whyatt RW. Impact of prenatal chlorpyrifos exposure on neurodevelopment in the first 3 years of life among inner-city children. Pediatrics. 2006;118(6):e1845–59. doi: 10.1542/peds.2006-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauh VA, Perera FP, Horton MK, Whyatt RM, Bansal R, Hao X, Liu J, Barr DB, Slotkin TA, Peterson BS. Brain anomalies in children exposed prenatally to a common organophosphate pesticide. Proc Natl Acad Sci U S A. 2012;109(20):7871–6. doi: 10.1073/pnas.1203396109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PD, Yosim A, Fry RC. Incorporating epigenetic data into the risk assessment process for the toxic metals arsenic, cadmium, chromium, lead, and mercury: strategies and challenges. Front Genet. 2014;5:201. doi: 10.3389/fgene.2014.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard JF, Puga A. Effects of arsenic exposure on DNA methylation and epigenetic gene regulation. Epigenomics. 2010;2(1):87–104. doi: 10.2217/epi.09.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice D, Barone S., Jr Critical periods of vulnerability for the developing nervous system: evidence from humans and animal models. Environ Health Perspect. 2000;108(Suppl 3):511–33. doi: 10.1289/ehp.00108s3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JF, Yu X, Moreira EG, Hong S, Faustman EM. Arsenic- and cadmium-induced toxicogenomic response in mouse embryos undergoing neurulation. Toxicol Appl Pharmacol. 2011;250(2):117–29. doi: 10.1016/j.taap.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier PM. Vulnerable periods and processes during central nervous system development. Environ Health Perspect. 1994;102(Suppl 2):121–4. doi: 10.1289/ehp.94102121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier PM. Developing brain as a target of toxicity. Environ Health Perspect. 1995;103(Suppl 6):73–6. doi: 10.1289/ehp.95103s673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14(5):347–59. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko A, Tsai LH. Epigenetic regulation in memory and cognitive disorders. Neuroscience. 2014;264:51–63. doi: 10.1016/j.neuroscience.2012.12.034. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25(1):15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Salazar-Arredondo E, de Jesus Solis-Heredia M, Rojas-Garcia E, Hernandez-Ochoa I, Quintanilla-Vega B. Sperm chromatin alteration and DNA damage by methyl-parathion, chlorpyrifos and diazinon and their oxon metabolites in human spermatozoa. Reprod Toxicol. 2008;25(4):455–60. doi: 10.1016/j.reprotox.2008.05.055. [DOI] [PubMed] [Google Scholar]
- Sando R, 3rd, Gounko N, Pieraut S, Liao L, Yates J, 3rd, Maximov A. HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell. 2012;151(4):821–34. doi: 10.1016/j.cell.2012.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanosaka T, Namihira M, Nakashima K. Epigenetic mechanisms in sequential differentiation of neural stem cells. Epigenetics. 2009;4(2):89–92. doi: 10.4161/epi.4.2.8233. [DOI] [PubMed] [Google Scholar]
- Shin HS, Seo JH, Jeong SH, Park SW, Park Y, Son SW, Kim JS, Kang HG. Exposure of pregnant mice to chlorpyrifos-methyl alters embryonic H19 gene methylation patterns. Environ Toxicol. 2014;29(8):926–35. doi: 10.1002/tox.21820. [DOI] [PubMed] [Google Scholar]
- Shin S, Mitalipova M, Noggle S, Tibbitts D, Venable A, Rao R, Stice SL. Long-term proliferation of human embryonic stem cell-derived neuroepithelial cells using defined adherent culture conditions. Stem Cells. 2006;24(1):125–38. doi: 10.1634/stemcells.2004-0150. [DOI] [PubMed] [Google Scholar]
- Skinner MK. Endocrine disruptors and epigenetic transgenerational disease etiology. Pediatr Res. 2007;61(5 Pt 2):48R–50R. doi: 10.1203/pdr.0b013e3180457671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, MacKillop EA, Ryde IT, Seidler FJ. Ameliorating the developmental neurotoxicity of chlorpyrifos: a mechanisms-based approach in PC12 cells. Environ Health Perspect. 2007;115(9):1306–13. doi: 10.1289/ehp.10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Seidler FJ. Developmental neurotoxicity of organophosphates targets cell cycle and apoptosis, revealed by transcriptional profiles in vivo and in vitro. Neurotoxicol Teratol. 2012;34(2):232–41. doi: 10.1016/j.ntt.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen PT, Schulpen SH, van Dartel DA, Hermsen SA, van Schooten FJ, Piersma AH. An abbreviated protocol for multilineage neural differentiation of murine embryonic stem cells and its perturbation by methyl mercury. Reprod Toxicol. 2010;29(4):383–92. doi: 10.1016/j.reprotox.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tokar EJ, Qu W, Waalkes MP. Arsenic, stem cells, and the developmental basis of adult cancer. Toxicol Sci. 2011;120(Suppl 1):S192–203. doi: 10.1093/toxsci/kfq342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler CR, Allan AM. The Effects of Arsenic Exposure on Neurological and Cognitive Dysfunction in Human and Rodent Studies: A Review. Curr Environ Health Rep. 2014;1:132–147. doi: 10.1007/s40572-014-0012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visan A, Hayess K, Sittner D, Pohl EE, Riebeling C, Slawik B, Gulich K, Oelgeschlager M, Luch A, Seiler AE. Neural differentiation of mouse embryonic stem cells as a tool to assess developmental neurotoxicity in vitro. Neurotoxicology. 2012;33(5):1135–46. doi: 10.1016/j.neuro.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Wang X, Meng D, Chang Q, Pan J, Zhang Z, Chen G, Ke Z, Luo J, Shi X. Arsenic inhibits neurite outgrowth by inhibiting the LKB1-AMPK signaling pathway. Environ Health Perspect. 2010;118(5):627–34. doi: 10.1289/ehp.0901510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman GA, Liu X, Parvez F, Ahsan H, Factor-Litvak P, Kline J, van Geen A, Slavkovich V, Loiacono NJ, Levy D, et al. Water arsenic exposure and intellectual function in 6-year-old children in Araihazar, Bangladesh. Environ Health Perspect. 2007;115(2):285–9. doi: 10.1289/ehp.9501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney KD, Seidler FJ, Slotkin TA. Developmental neurotoxicity of chlorpyrifos: cellular mechanisms. Toxicol Appl Pharmacol. 1995;134(1):53–62. doi: 10.1006/taap.1995.1168. [DOI] [PubMed] [Google Scholar]
- Whittaker SG, Wroble JT, Silbernagel SM, Faustman EM. Characterization of cytoskeletal and neuronal markers in micromass cultures of rat embryonic midbrain cells. Cell Biol Toxicol. 1993;9(4):359–75. doi: 10.1007/BF00754465. [DOI] [PubMed] [Google Scholar]
- Wright RO, Baccarelli A. Metals and neurotoxicology. J Nutr. 2007;137(12):2809–13. doi: 10.1093/jn/137.12.2809. [DOI] [PubMed] [Google Scholar]
- Young A, Machacek DW, Dhara SK, Macleish PR, Benveniste M, Dodla MC, Sturkie CD, Stice SL. Ion channels and ionotropic receptors in human embryonic stem cell derived neural progenitors. Neuroscience. 2011;192:793–805. doi: 10.1016/j.neuroscience.2011.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Sidhu JS, Hong S, Faustman EM. Essential role of extracellular matrix (ECM) overlay in establishing the functional integrity of primary neonatal rat Sertoli cell/gonocyte co-cultures: an improved in vitro model for assessment of male reproductive toxicity. Toxicol Sci. 2005;84(2):378–93. doi: 10.1093/toxsci/kfi085. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.