Abstract
Endocrine disrupting chemicals (EDCs) exert significant effects on health and physiology, many traceable to effects on stem cell programming underlying development. Understanding risk of low-level, chronic EDC exposure will be enhanced by knowledge of effects on stem cells. We exposed rhesus monkey embryonic stem cells to low levels of five EDCs [bisphenol A (BPA), atrazine (ATR), tributyltin (TBT), perfluorooctanoic acid (PFOA), and di-(2-ethylhexyl) phthalate (DEHP)] for 28 days, and evaluated effects on gene expression by RNAseq transcriptome profiling. We observed little effect of BPA, and small numbers of affected genes (≤119) with other EDCs. There was substantial overlap in effects across two, three, or four treatments. Ingenuity Pathway analysis indicated suppression of cell survival genes and genes downstream of several stress response mediators, activation of cell death genes, and modulations in several genes regulating pluripotency, differentiation, and germ layer development. Potential adverse effects of these changes on development are discussed.
Keywords/phrases: endocrine disruptor, atrazine, perfluorooctanoic acid, phthalate, obesogen, tributyltin, bisphenol A, transcriptome, nuclear programming, gene-environment interaction, developmental origins of disease
Introduction
There are nearly 1000 chemicals listed as potential endocrine disruptor chemicals (EDCs) on The Endocrine Disruption Exchange (endocrindisruption.org). These chemicals are known or suspected of disrupting endocrine signaling with a myriad of downstream effects on development, reproduction, immune function, behavior, cognition, diabetes, cancer, and many other diseases and disorders in individuals and their progeny [1]. The broad spectrum of effects, particularly in progeny, reflects the abilities of EDCs to exert significant effects on nuclear programming in the stem cells that contribute to embryogenesis, organogenesis, organ homeostasis, and cancer later in life [2–6].
Understanding the effects of EDCs on stem cells is key for understanding exposure risk and for devising possible strategies for mitigating negative effects of exposures. EDCs are used in a number of industrial processes, including consumer products manufacture and production of herbicides. The widespread industrial use of EDCs is reflected in their presence in serum, urine, amniotic fluid, cord blood, and household dust [1]. Other studies report correlations between these exposures and specific developmental abnormalities and adverse health consequences in humans and other animals [7–20].
One recent study [21] found that species differences in stem cell responses to EDCs can be significant. Consequently, although rodent stem cell and rodent developmental studies provide important information about the potential developmental consequences of EDC exposure, the possibility of species differences needs to be addressed by determining EDC effects in model organisms more closely resembling humans. Some other studies reported that some EDCs affect germ cells and early embryos, but others reported little or no effect [22–32]. Recent studies also illustrate the effects of prolonged low-level exposure on embryonic stem cell (ESC) differentiation [33]. These observations indicate the importance of examining effects on stem cells to better understand potential early developmental consequences of exposure. To explore possible mechanisms underlying EDC effects on early human development, and to establish a foundation for future study in an experimentally tractable animal model closely resembling humans in terms of reproductive physiology, metabolism and developmental mechanisms, we compared effects of long-term culture with each of five EDCs on gene expression patterns in a rhesus monkey embryonic stem cell line. EDCs tested include bisphenol-A (BPA), atrazine (ATR), di-(2-ethylhexyl) phthalate (DEHP), tributyltin (TBT), and perfluorooctanoic acid (PFOA). We applied comparatively low levels of the EDCs, in order to better mimic constant exposures to environmentally relevant concentrations, without complicating influences of acute toxicity, damage to maternal organs, or severe disruptions of maternal endocrine profiles. These compounds were chosen because of their prevalence in the environment and/or in human serum indicating ongoing human exposures, and widespread study. Our results revealed little to no significant effect of BPA with the exposure parameters applied, and significant effects of the other four EDCs. We also observed effects shared between two, three, or four of the latter EDCs, most notably affecting genes related to stress response, cell proliferation, and cell death. Our analysis revealed genes related to specific biological pathways, processes, and functions impacted by these EDCs, and provide novel insight into potential mechanisms whereby early developmental exposure may affect cell reprogramming and thus long-term progeny phenotype.
Methods
Cells and cell culture
A rhesus embryonic stem cell (ESC) line was obtained from the Oregon National Primate Research Center (Beaverton, OR). Oregon Rhesus Macaque Embryonic Stem (ORMES)-6 line cells were maintained on feeder layers of mouse embryonic fibroblast cells that had been mitotically inactivated. This cell line is one of two available (ORMES-6 and -7, 42XX and 42XY, respectively) for which broad differentiation capacity was reported [34]. ORMES-7 proved more difficult to propagate than ORMES-6, and so was not pursued here. Cells were maintained in Dulbecco’s modified Eagle/F-12 medium (DMEM/F12, 11320-033; Invitrogen, Carlsbad, CA) supplemented with 15% fetal bovine serum (Hyclone, Logan, UT), 1% minimum essential medium (MEM) nonessential amino acids, 1 mM L-glutamine, 0.1 mM β-mercaptoethanol, and 0.1% gentamycin in 60-mm cell culture dishes (Mitalipov et al., 2006). Cultures were incubated at 37°C in 5% CO2 and 5% O2. Components for cell culture medium were purchased from Sigma (St. Louis, MO) unless otherwise noted.
For experimental treatments, ESC colonies were cultured in 6-well plates (Falcon 353046), 1 plate for each treatment group. Approximately 40 random colonies per dish were transferred to fresh feeder layers each time ESC colonies were passaged, which occurred every 3 to 4 days. Colonies for passage were taken from a pre-marked, wedge-shaped section of each dish to assure a random sample. Media changes were performed daily (2 mL) and experimental treatments continued for a total of four weeks. A total of six biological replicates were obtained for each treatment group. The cells were harvested and lysed approximately 24 h after the last media change. At harvest, all colonies in each well were cut and lifted, and divided into two microcentrifuge tubes, and centrifuged at 3,000 × g for three min and supernatant removed. Cells were resuspended in 1 mL DPBS with 10% BSA and 25 mM EDTA and centrifuged as above. Supernatant was removed and the remaining cell pellet was lysed in 100 μl PicoPure™ extraction buffer (Thermo Fisher Scientific, Life Technologies, Grand Island, NY).
Chemicals for testing were made as 1000× stocks in methanol and were added to aliquots of ESC maintenance medium up to 48 hours before use and stored at 4°C until use. Chemicals were obtained from Sigma (DEHP: Bis(2-ethylhexyl) phthalate – 47994; PFOA: Perfluorooctanoic acid – 171468; ATR: Atrazine – 45330; TBT: Tributyltin chloride – 442869; BPA: Bisphenol A – 133027). Treatment groups included a vehicle control (methanol) and each of five EDCs at environmentally relevant doses [10 nM BPA (36 nM fetal serum, [35]), 5 μM DEHP (7.7μM pubertal serum, [12]), 30 μM ATR (up to 7 μM drinking water reported, [36]), 100 nM PFOA (average 94 nM ≥12 year old serum, [37]), and 25 nM TBT (0.17 – 534 adult nM serum, [38]). These concentrations are within the range those reported for human serum, for drinking water, or studies reported for mammalian oocytes and preimplantation stage embryos or pluripotent cells in vitro [22, 23, 27, 39–41]. The concentration of ATR used was ~4-fold higher than the maximum concentration reported in one study in drinking water [36], and one study of occupational human serum level (up to 245 nM [42]). The ATR concentration used affects placenta cell gene expression in vitro [43] and is much lower than the doses (200–300 mg/kg) typically applied in rodent studies to test for reproductive effects. Some of these chemicals exert effects on rhesus monkey gonads, embryo or fetal development, and progeny phenotype, or in human reproductive tissues or stem cells. For example, 10–15 nM BPA affects fetal lung development [44], and affects mammary development [45]. DEHP at 25 μM affects monkey Sertoli cell development [46]. TBT (100 nM) negatively affects human embryonal carcinoma cell gene expression and mitochondria [47, 48]. ATR at 200 mg/kg dosing affects gonadogenesis across vertebrate classes [49].
Preparation and sequencing of libraries for RNAseq
RNA was isolated following the PicoPure™ RNA Extraction kit manufacturer protocol, with DNAse digestion to remove any contaminating DNA. To produce libraries for sequencing, 100 ng of each RNA sample (five of the six biological replicates per condition) were processed first using the Ovation RNA-Seq System v2 using Ribo-SPIA™ Technology (NuGen, San Carlos, CA). This was followed by fragmentation to an average of 300 bp using a Covaris-2 sonicator, and then a brief S1 nuclease digestion as described [50]. After purification, the cDNA was processed through the Ovation Ultralow DR Multiplex Systems 1–8 and 9–16 (NuGen) for end repair, barcoding and final library production. Barcoded libraries were pooled, loaded on flowcells and sequenced with Illumina HiSeq 2500 in rapid run mode to generate 50 nt single end reads. To enhance the effectiveness of cluster identification algorithm, the samples were loaded at 65% of optimal loading concentration, along with PhiX Control library (Illumina) – adapter-ligated library obtained from randomly sheared PhiX DNA – added at 10% of loading concentration to increase read sequence complexity. After an initial analysis for differential gene expression, all libraries except the five BPA treatment libraries were submitted for additional sequencing (initial comparison of sequencing data for BPA and Vehicle libraries returned no significantly differentially expressed genes). The total numbers of PF (passed-filter) reads ranged from 27.7M to 68.0M for the re-sequenced libraries and 11.9 to 18.3 for BPA libraries (Table S1). Fraction of Q30 bases ranged from 90.5% to 97.7% and average Q from 36.9 to 38.4. Sequence data will be available in Gene Expression Omnibus (Gene Expression Omnibus accession number GSE86939) and at our Primate Embryo Gene Expression Resource (www.preger.org).
RNAseq data analysis and Ingenuity Pathway Analysis
To eliminate effects of contamination from mouse embryo fibroblast feeders, reads were aligned using TopHat2 [51] to both rhesus monkey genome (MacaM v7, [52]) and mouse genome GRCm38. Reads for which the alignment score to mouse genome was higher than the alignment score to rhesus monkey genome were removed. The fraction of such reads was <20% for all libraries except one DEHP treatment library (61%), which was removed from further analysis. Reads aligned to ribosomal RNA (rRNA) or rRNA-like genes were removed. A total of 7.1M to 17.5M reads were successfully aligned to unique non-rRNA gene transcript sequences (2.5M to 4.8M for BPA treatment libraries). Cuffdiff [53] was used for quantification and differential expression analyses between the group of vehicle libraries and five groups of chemical treatment libraries; genes with q-value (false discovery rate) below 0.05 were considered differentially expressed (DE) genes (DEGs).
The results of Cuffdiff analyses were uploaded into QIAGEN Ingenuity Pathway Analysis® (IPA) and subjected to Core Analysis focusing on DE genes (q≤0.05), in particular Canonical Pathway (CP) analysis, Disease and Functions (DF) analysis, Upstream Regulator (UR) analysis, and Network analysis. IPA calculates overlap p-values for each CP, taking into account the number of DE genes and the number of all molecules in knowledge database that are implicated in that pathway, as well as the number of DE genes and the number of molecules in knowledge database. Similarly, p-values are calculated for DFs based on the number of DE genes implicated in increase or decrease of disease or function, as well as for URs based on the number of DE genes regulated by an UR. In addition to overlap p-values, z-scores are calculated for CPs, DFs, and URs. Z-score is based on how many DE genes’ direction of change (upregulation or downregulation) is consistent with activation (z>0) or inhibition (z<0) of CPs and URs, and with increase (z>0) or decrease (z<0) of DFs. Since P(|z|>1.96)~0.05 for normal N(0,1) distribution, we consider CPs, URs and DFs with z>1.96 to be significantly activated or increased, and those with z<−1.96 to be significantly inhibited or decreased. In addition to these analyses, IPA uses a greedy algorithm to construct networks that incorporate DE genes with some additional genes (or other molecules) where needed, in an attempt to reproduce possible mechanistic networks.
Results
The goal of this study was to assess the impact of long-term exposure to comparatively low levels of five EDCs on nonhuman primate embryonic cells in order to better understand how constant low level exposures affect developing embryos. To do this, rhesus monkey ORMES-6 ESCs were treated for four weeks and then RNA processed for analysis by RNAseq. This treatment period was selected to encompass three passages during treatment, in order to allow time for any DNA replication-dependent epigenetic changes. A long-term exposure of proliferating embryonic stem cells to comparatively low doses of EDCs should reveal potential effects of low-level, constant, environmentally relevant maternal exposure on cells that drive embryogenesis, without the complications of acute toxicity, maternal organ damage, or indirect maternal endocrinological effects that accompany single acute treatments with higher concentrations. A single concentration was selected for each compound due to assay and culture costs. No changes were noted in growth rate or morphology characteristics of ESC colonies during treatment. Hence, the goal of subsequent analysis was to determine effects of low-level chronic exposure on stem cell gene expression profile, and whether those changes include genes that affect stem cell function.
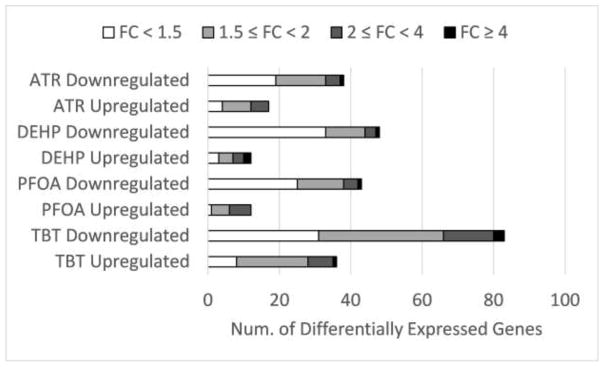
The number of genes with significantly (p ≤ 0.05) affected expression was modest, ranging from zero (BPA) to 119 (TBT) (Fig. 1). Between two and four times as many genes displayed decreased transcript abundance as those showing increased transcript abundance. For BPA, five genes were affected at the confidence level of p < 0.08. Thus, although BPA interacts with nuclear receptors, is an estrogen mimic and anti-androgen, and inhibits PPARγ, thyroid, and glucocorticoid signaling, among other effects [54], there was little effect of treatment detected here, and BPA was not studied further. The maximum fold-change values by treatment ranged from 2.66 (ATR) to 8.50 (DEHP) (Supplemental Tables S2 – S5).
Figure 1.
Summary of differential expression in EDC treated samples. Bar segments illustrate the numbers of genes displaying fold change values in expression indicated.
Atrazine
ATR acts as a G-protein coupled estrogen receptor agonist, induces oxidative stress, and disturbs calcium homeostasis [55, 56]. ATR treatment of ORMES-6 ESCs increased the expression of 17 genes (Table S2); of these 13 were elevated by at least 1.5-fold, and five by at least 2-fold. ATR decreased the expression of 38 genes (Table S2); of these 19 were reduced by at least 1.5-fold, five by 2-fold, and 1 by ≥ 4-fold. The most highly up-regulated mRNAs included FOS (2.66-fold), GSTK1 (2.43-fold), FAIM (2.29-fold), FOSB (2.28-fold), GOS3 (2.03-fold) and JUNB (1.95-fold). Early growth response genes EGR1 and EGR3 were also affected. Most highly down-regulated mRNAs included CCDC184 (4.94-fold), PTX3 (3.7 fold), and CLDN14 (2.86-fold).
The IPA analysis for effects of ATR treatment yielded 111 significantly affected (p ≤ 0.05) biological functions/diseases with four or more affected genes and 20 with ten or more affected genes (Table S6). The analysis returned significant z-scores (|z|>1.96, equivalent to activation p≤.05) for eight biological functions (Table S6) (Table 1). Functions showing activation (z>1.96) included proliferation and growth-related categories (e.g., colony formation, proliferation of connective tissues cells, colony formation of tumor cells, and fibroblast proliferation), and other functions including development of body trunk, cellular homeostasis, and Ca2+ flux. Cell growth and proliferation was also the top-rated affected network appearing in the IPA Network analysis (Table S7) and the UR analysis returned significant activation z-scores for EGF, HMGA1, PDGF and VEGFA, and FOXO3 signaling (Table S8)(Table 2). Significant positive z-scores were also obtained for several cytokines. UR analysis also indicated increased activity for cellular responses to cycloheximide, indicating a possible effect on protein synthesis. The biological functions of tumor cell adhesion, cell movement and necrosis also displayed high activation z-scores, though these fell just below the significance threshold (1.96), indicating possible activation. These processes were repeated amongst similar function categories with lesser activation z-scores. One biological function (secretion) displayed a significant negative z-score, and three other transport-related biological functions yielded strong negative effects falling just below the level of significance (monosaccharide uptake, molecular transport). Apoptosis and glycolysis were among other function categories appearing repeatedly with lesser magnitude negative z-scores. The activation of FOXO3 may contribute to this response. Significantly affected canonical pathways (CPs) with the largest numbers of affected genes included AMPK signaling and NRF2 mediated oxidative stress response (four affected genes each), and GNRH signaling (three affected genes) (Table S9). Two additional stress-related CPs, HIF1A signaling and UVA-induced MAPK signaling, were affected. For comparison, past studies reported that ATR at 60 nM inhibits mouse T-cell proliferation [57], whereas 30 μM ATM does not inhibit proliferation but induces CYP19 activity in human placental cells [43].
Table 1.
Increased and Decreased IPA® Diseases and Functions
| Toxicant | Diseases or Functions | Predicted Activation State | z-score | Num. of DEGs |
|---|---|---|---|---|
| ATR | colony formation of cells | Increased | 2.49 | 7 |
| proliferation of connective tissue cells | Increased | 2.27 | 6 | |
| development of body trunk | Increased | 2.20 | 7 | |
| cellular homeostasis | Increased | 2.13 | 9 | |
| colony formation of tumor cell lines | Increased | 2.13 | 5 | |
| secretion of molecule | Decreased | −2.00 | 5 | |
| cell proliferation of fibroblasts | Increased | 2.00 | 4 | |
| flux of Ca2+ | Increased | 1.98 | 4 | |
| DEHP | damage of lung | Increased | 1.97 | 4 |
| size of tumor | Increased | 1.96 | 4 | |
| PFOA | cellular infiltration by leukocytes | Increased | 2.21 | 5 |
| injury of lung | Increased | 1.98 | 4 | |
| necrosis of liver | Increased | 1.98 | 4 | |
| Hypertrophy | Increased | 1.98 | 5 | |
| TBT | development of body trunk | Increased | 2.35 | 13 |
| development of hematopoietic system | Increased | 2.22 | 7 | |
| development of lymphatic system | Increased | 2.15 | 8 | |
| binding of DNA | Increased | 2.10 | 12 | |
| binding of protein binding site | Increased | 2.08 | 7 | |
| neoplasia of cells | Decreased | −2.07 | 16 |
Table 2.
Predicted Upstream Regulators
| Toxicant | Upstream Regulator | z-score1 | Num. Target DEGs |
|---|---|---|---|
| ATR | cycloheximide | 2.59 | 8 |
| EGF | 2.57 | 7 | |
| HMGA1 | 2.24 | 5 | |
| atorvastatin | 2.24 | 5 | |
| Pdgf (complex) | 2.22 | 5 | |
| fulvestrant | −2.21 | 5 | |
| VEGFA | 2.03 | 7 | |
| Pkg | 2.00 | 4 | |
| dalfampridine | 2.00 | 4 | |
| FOXO3 | 2.00 | 5 | |
| amphetamine | 1.99 | 4 | |
| kainic acid | 1.99 | 4 | |
| Pka | 1.99 | 4 | |
| salirasib | 1.98 | 4 | |
| EPAS1 | −1.98 | 6 | |
| MAPK3 | 1.98 | 4 | |
| CREB1 | 1.98 | 7 | |
| CD38 | −1.98 | 4 | |
| EPO | 1.97 | 5 | |
| CSF3 | 1.97 | 4 | |
| LIF | 1.97 | 4 | |
| Akt | −1.96 | 4 | |
| DEHP | AGT | −2.87 | 10 |
| lipopolysaccharide | −2.73 | 13 | |
| NUPR1 | −2.63 | 7 | |
| HIF1A | −2.53 | 13 | |
| IFNG | −2.48 | 15 | |
| IL5 | −2.45 | 6 | |
| E. coli B5 lipopolysaccharide | −2.43 | 6 | |
| RELA | −2.43 | 6 | |
| SREBF1 | −2.37 | 7 | |
| progesterone | −2.27 | 8 | |
| fluticasone | −2.24 | 5 | |
| fulvestrant | −2.21 | 5 | |
| LEP | −2.21 | 6 | |
| hydrogen peroxide | −2.21 | 6 | |
| CTNNB1 | −2.19 | 5 | |
| Mek | −2.19 | 5 | |
| SB203580 | 2.19 | 5 | |
| CAT | 2.18 | 5 | |
| Ins1 | −2.18 | 5 | |
| N-acetyl-L-cysteine | 2.18 | 5 | |
| PDGF BB | −2.15 | 8 | |
| deferoxamine | −2.05 | 10 | |
| TP53 | −2.05 | 17 | |
| IL1B | −2.04 | 9 | |
| LY294002 | 2.01 | 11 | |
| Akt | −2.00 | 8 | |
| Ifn | −2.00 | 5 | |
| trovafloxacin | −2.00 | 4 | |
| KLF2 | 2.00 | 4 | |
| mifepristone | 1.99 | 5 | |
| FSH | 1.98 | 6 | |
| raloxifene | −1.98 | 4 | |
| TGFBR2 | −1.97 | 4 | |
| P38 MAPK | −1.97 | 5 | |
| LDL | −1.97 | 4 | |
| estrogen | −1.96 | 6 | |
| paclitaxel | −1.96 | 6 | |
| PFOA | HIF1A | −3.19 | 15 |
| salirasib | 2.81 | 8 | |
| Akt | −2.59 | 7 | |
| NUPR1 | −2.50 | 13 | |
| CD38 | −2.43 | 6 | |
| MYC | −2.35 | 9 | |
| fulvestrant | −2.22 | 5 | |
| cobalt chloride | −2.20 | 5 | |
| cycloheximide | 2.00 | 9 | |
| Pkg | 2.00 | 4 | |
| ERBB4 | −2.00 | 4 | |
| resveratrol | 2.00 | 5 | |
| KLF2 | 2.00 | 4 | |
| metribolone | −1.98 | 5 | |
| NOTCH1 | −1.98 | 4 | |
| ARNT | −1.97 | 7 | |
| SREBF1 | −1.96 | 6 | |
| TBT | IL10RA | 2.65 | 7 |
| salirasib | 2.63 | 7 | |
| HIF1A | −2.62 | 21 | |
| CD38 | −2.62 | 7 | |
| cycloheximide | 2.39 | 7 | |
| cobalt chloride | −2.22 | 5 | |
| ESR2 | 2.22 | 9 | |
| STAT4 | −2.21 | 5 | |
| NUPR1 | −2.20 | 13 | |
| lithium chloride | 2.20 | 6 | |
| trichostatin A | 2.20 | 7 | |
| EPO | 2.19 | 6 | |
| Akt | −2.19 | 5 | |
| Growth hormone | 2.18 | 5 | |
| BMP4 | −2.12 | 6 | |
| EPAS1 | −2.01 | 13 | |
| ACVR1C | 2.00 | 4 | |
| Pkg | 2.00 | 4 | |
| COMMD1 | 2.00 | 4 | |
| F3 | 2.00 | 5 | |
| SB-431542 | −1.99 | 5 | |
| di(2-ethylhexyl) phthalate | 1.98 | 4 | |
| NOTCH1 | −1.98 | 5 | |
| CREBBP | 1.98 | 5 | |
| CSF3 | 1.97 | 4 |
Upstream regulators with z-score > 1.96 are predicted to be activated, upstream regulators with z-score < −1.96 are predicted to be inhibited.
DEHP
DEHP can act through multiple mechanisms, with effects as an anti-androgen and inhibitor of steroidogenesis, and may increase expression of DNA methyltransferase genes leading to DNA hypermethylation [58, 59]. DEHP treatment increased the expression of 12 genes (9 ≥ 1.5-fold, 5 ≥ 2-fold, and 2 ≥ 4-fold) and decreased the expression of 48 genes (15 ≥ 1.5-fold, 4 ≥ 2-fold and 1 ≥ 4-fold) (Table S3). The most highly upregulated genes were RL4D (8.5-fold), FAIM (4.61-fold), GSTK1 (3.59-fold), and PARP12 (2.42-fold). The most strongly downregulated genes were MKRN3 (6.03-fold), CDC184 (3.26-fold), SPP1 (2.22-fold), and ADM (2.01-fold).
IPA analysis of biological functions/diseases significantly (p < 0.05) affected by DEHP yielded 172 with four or more affected genes, and 33 with 10 or more affected genes (Table S10). Affected functions/diseases included just two with significant positive z-scores indicating increased activity (lung damage and tumor size) (Table 1), with additional large activation z-scores for bone size, fibrosarcoma cell death and metastasis (Table S10). No significant negative z-scores indicating inhibition were obtained, but functions/diseases with large negative z-scores below the significance threshold included tumor growth, cell spreading, cell migration and carbohydrate metabolism. Cell movement was a component of the two top-rated affected networks (Table S7). These potential negative effects on growth, spreading and migration are somewhat opposite the effects returned for ATR. CP analysis revealed a pathway related to ESC pluripotency that includes two genes downregulated by DEHP treatment (Table S11). UR analysis revealed significant inhibition of stress response mediators (NUPR1, HIF1A, CAT, RLEA, AKT, p38/MAPK, MYC, TP53) and infection/inflammatory response (IFNG, IL5, IL1B, IFN) and inhibition of response to chemical stress-inducing agents (e.g., peroxide) (Table S12)(Table 2). A significant activation z-score was obtained for the anti-inflammatory mediator KLF2.
PFOA
PFOA is a PPARα activator, and modifies genes that function in steroidogenesis, leading to diverse endocrine effects [60, 61], negatively affects thyroid function, and leads to an oxidative stress response and inflammatory response [62]. PFOA treatment increased the expression of 12 genes (11 ≥ 1.5-fold and 6 ≥ 2-fold) and decreased the expression of 43 genes (18 ≥ 1.5-fold, 5 ≥ 2-fold, and 1 ≥ 4-fold) (Table S4). The most highly upregulated genes included GSTK1 (2.87-fold), FOS (2.62-fold), FAIM (2.6-fold), PARP12 (2.05-fold), FOSB (2.03-fold), and TRIB3 (2.02-fold). The most strongly downregulated genes were CDC184 (5.42-fold), NRN1 (3.15-fold), PTX3 (2.63-fold), MKRN3 (2.59-fold), CLDN14 (2.45-fold).
IPA analysis of biological functions/diseases significantly (p ≤ 0.05) affected by PFOA treatment yielded 140 with four or more affected genes, and 30 with ten or more affected genes (Table S13). The affected functions/diseases included four with significant z-scores indicating increased activity (cellular infiltration, injury, necrosis and hypertrophy), with additional high positive z-scores for categories related to cell death and cell movement (Table 1). One function (cell viability) had a high negative z-score value near the threshold of significance and indicating likely inhibition, consistent with the activation of cell death categories. Additional functions with z-scores indicating possible inhibition included cell protrusion, organization of cytoplasm, protein metabolism and synthesis, growth, molecular transport, cell proliferation (largest number of affected genes), and microtubule dynamics. The most significantly affected CP (5 affected members) was NRF2-mediated oxidative stress response (Table S14). Additional indications of changes in genes related to stress response were seen in the UR analysis (Table S15)(Table 2). Highly negative z-scores indicative of inhibition were reported for several key stress response mediators including HIF1A, AKT, NUPR1, MYC, ERBB4, NOTCH1, ARNT, and SREBF1. Additional high inhibition z-scores falling below the significance threshold were obtained for FOS (itself upregulated), SMARCA4, and EPAS1 and for several inflammatory mediators (IL10, NEDD9, IL5). A significant positive z-score was also obtained for the anti-inflammatory mediator KLF2.
A significant positive z-score was obtained for resveratrol, also consistent with activation of oxidative stress response pathway. Positive z-scores for cycloheximide (protein synthesis inhibitor) and the RAS inhibitor Salirasib indicate potential activation of the ER stress response pathway (RAS inhibits ER stress response; [63]), again indicative of a possible inhibitory effect on protein synthesis.
TBT
TBT is an RxR/PPARγ agonist, mitochondrial ATP synthase inhibitor, a possible obesogen, and may cause DNA hypomethylation at some genes [6, 64]. TBT treatment yielded the largest number of affected genes, increasing the expression of 36 mRNAs (28 ≥ 1.5-fold, 8 ≥ 2-fold and 1 ≥ 4-fold) and decreasing expression of 83 mRNAs (52 ≥ 1.5-fold, 17 ≥ 2-fold, 3 ≥ 4-fold) (Table S5). The most highly upregulated mRNAs included HOX2 (4.98-fold), ABCA1 (3.78-fold), CER1 (2.61-fold), FOS (2.44-fold), FOSB (2.43-fold), SLC25A20 (2.17-fold), LEFTY1 (2.03-fold) and RNF219 (2.0-fold). The most highly downregulated mRNAs included CDC184 (8.34-fold), CLDN14 (4.84-fold), PTX3 (4.53-fold), RIMKLA (3.51-fold), and NRN1 (3.36-fold).
IPA analysis yielded 161 biological functions/diseases with significant (p ≤ 0.05) effects containing four or more affected genes (Table S16), and 68 with 10 or more affected genes. Five functions/disease were assigned significant high positive z-scores indicating increased activity: three related to organismal development (including hematopoiesis and lymphopoesis) and two related to DNA and protein binding (Table 1). Networks (Table S7) reiterated this effect on embryonic development of heart, nervous system, and hematological system. Additional biological functions with high positive z-scores included locomotion, differentiation, and transcription. Neoplasia and cancer comprised the top three functions/disease with significant or nearly significant negative z-scores indicating inhibition, followed by cancer, steroid secretion and cell survival. CP analysis revealed effects related to TNFR, BMP and TGFB signaling as well as effects on ES cell transcriptional regulation (Table S17).
UR analysis of TBT effects predicted significantly increased activity for signaling via IL10, ESR2, EPO, growth hormone, CREBBP, and CSF3, and inhibition of signaling via HIF1A, STAT4, CD38, NUPR1, AKT, BMP4, EPAS1, NOTCH1, and possibly NEDD9 (Table 2, Table S18). A number of chemical mediators also emerged from the analysis with significant predicted increases in associated cellular activities, including Salirasib, cycloheximide, lithium, and trichostatin A.
One other notable effect of TBT was to increase expression of genes associated with controlling ESC renewal and germ layer and axis formation (LEFTY1, LEFTY2) [65, 66]. DUSP2, another regulator of cellular potency [67], was also elevated. Also elevated were BMP4, which induces primitive endoderm in monkey ESCs [68], and the hypoblast marker GATA4. Additionally, NANOG emerged as an upstream regulator of six affected genes. Pluripotency also appeared among affected functions for TBT (Table S16) and canonical pathways for DEHP and TBT (Tables S11, S17).
Overlap in effects
There was considerable overlap in differentially expressed gene (DEG) lists between EDCs (Tables 3 and 4). For each gene that was significantly affected by multiple toxicants, those effects had the same directionality, i.e. no gene was found to have expression increased by one toxicant and decreased by another toxicant. Affected genes showing increased expression in treated cells across three treatments included FAIM, GSTK1, EGR1, FOS, and FOSB. Additional genes showing increased expression shared across two treatments included ZGRF1, TRIB3, EGR3, EPB4.1L4A, PARP12, RPUSD3, and ZNF280D. Eight genes showed decreased expression across all four treatments (ANKRD1, CCDC184, EGLN1, HK2, KDM3A, RIMKLA, SLFN5, STC1). Between 8 and 23 genes were affected in common among different combinations of three treatments. All five genes marginally affected (p < 0.08) by BPA (FOS, FOSB, EGR1, HIST1H4C, and HIST1HA2C) were affected in at least one of the other treatments and all but HIST1HA2C were affected in two or more of the other treatments.
Table 3.
Differentially expressed genes affected by two or more toxicant treatments
| Toxicant treatments | Common upregulated genes | Num. of genes1 | |||
|---|---|---|---|---|---|
| ATR | DEHP | PFOA | FAIM, GSTK1 | 2 = 16.7% of 12 | |
| ATR | PFOA | TBT | EGR1, FOS, FOSB | 3 = 25.0% of 12 | |
| ATR | DEHP | FAIM, GSTK1, ZGRF1 | 3 = 25.0% of 12 | ||
| ATR | PFOA | EGR1, FAIM, FOS, FOSB, GSTK1, TRIB3 | 6 = 50.0% of 12 | ||
| ATR | TBT | EGR1, EGR3, EPB41L4A, FOS, FOSB | 5 = 29.4% of 17 | ||
| DEHP | PFOA | FAIM, GSTK1, PARP12, RPUSD3, ZNF280D | 5 = 41.7% of 12 | ||
| PFOA | TBT | EGR1, FOS, FOSB | 3 = 25.0% of 12 | ||
| Toxicant treatments | Common downregulated genes | Num. of genes1 | |||
| ATR | DEHP | PFOA | TBT | ANKRD1, CCDC184, EGLN1, HK2, KDM3A, RIMKLA, SLFN5, STC1 | 8 = 21.1% of 38 |
| ATR | DEHP | PFOA | ANKRD1, CCDC184, EGLN1, HK2, KDM3A, RIMKLA, SLFN5, STC1 | 8 = 21.1% of 38 | |
| ATR | DEHP | TBT | ANKRD1, CCDC184, EGLN1, HIST1H4C, HK2, KDM3A, RIMKLA, SLFN5, STC1 | 9 = 23.7% of 38 | |
| ATR | PFOA | TBT | AK4, ANKRD1, CCDC184, CLDN14, EGLN1, FAM162A, HELZ, HERC3, HK2, KDM3A, NRN1, PFKFB4, PTX3, RAB2B, RIMKLA, RIOK3, SAP30, SLC2A1, SLFN5, STC1, TNIP1, UPRT, ZNF395 | 23 = 60.5% of 38 | |
| DEHP | PFOA | TBT | ANKRD1, BHLHE40, BHLHE41, CCDC184, EGLN1, HK2, HMGCS1, KDM3A, PTPRB, RIMKLA, SLFN5, STC1 | 12 = 27.9% of 43 | |
| ATR | DEHP | ANKRD1, CAV1, CCDC184, EGLN1, GTF3C6, HIST1H2AC, HIST1H4C, HK2, KDM3A, RIMKLA, SLFN5, STC1 | 12 = 31.6% of 38 | ||
| ATR | PFOA | ACTA1, AK4, ANKRD1, CCDC184, CLDN14, EGLN1, FAM162A, HELZ, HERC3, HK2, KDM3A, NRN1, PFKFB4, PTX3, RAB2B, RIMKLA, RIOK3, SAP30, SLC2A1, SLFN5, STC1, TNIP1, UPRT, ZNF395 | 24 = 63.2% of 38 | ||
| ATR | TBT | AK4, ALKBH5, ANKRD1, CCDC184, CLDN14, DDX58, EGLN1, FAM162A, HELZ, HERC3, HIST1H4C, HK2, KDM3A, NRN1, PFKFB4, PTX3, RAB2B, RIMKLA, RIOK3, SAP30, SLC2A1, SLFN5, STC1, TNIP1, UPRT, ZNF395 | 26 = 68.4% of 38 | ||
| DEHP | PFOA | ADM, ANKRD1, BHLHE40, BHLHE41, CCDC184, EGLN1, HK2, HMGCS1, IGFBP5, KDM3A, MKRN3, PTPRB, RIMKLA, SERPINB6, SERPINE1, SLFN5, STC1, TFRC | 18 = 41.9% of 43 | ||
| DEHP | TBT | ANKRD1, BHLHE40, BHLHE41, CCDC184, EGLN1, HIST1H4C, HK2, HMGCS1, KDM3A, NAMPT, PTPRB, RIMKLA, SLFN5, STC1 | 14 = 29.2% of 48 | ||
| PFOA | TBT | ADAMTS20, AK4, ANKRD1, BHLHE40, BHLHE41, CCDC184, CHL1, CLDN14, EGLN1, FAM162A, HELZ, HERC3, HK2, HMGCS1, KCTD12, KDM3A, LRRC58, MAP3K1, NRN1, P4HA1, PFKFB4, PTPRB, PTX3, RAB2B, RIMKLA, RIOK3, SAP30, SLC2A1, SLFN5, STC1, TNIP1, UPRT, ZNF395 | 33 = 76.7% of 43 | ||
Fraction (%) of common genes is relative to the size of the smallest gene set included in the intersection.
Table 4.
Fold-change for genes affected by two or more toxicant treatments
| Upregulated genes | ATR FC |
DEHP FC |
PFOA FC |
TBT FC |
|---|---|---|---|---|
| FAIM | 2.29 | 4.61 | 2.60 | |
| GSTK1 | 2.43 | 3.59 | 2.87 | |
| EGR1 | 1.62 | 1.55 | 1.60 | |
| FOS | 2.66 | 2.62 | 2.44 | |
| FOSB | 2.28 | 2.03 | 2.43 | |
| ZGRF1 | 1.34 | 1.42 | ||
| TRIB3 | 1.89 | 2.02 | ||
| EGR3 | 1.81 | 1.85 | ||
| EPB41L4A | 1.61 | 1.59 | ||
| PARP12 | 2.42 | 2.05 | ||
| RPUSD3 | 1.87 | 1.70 | ||
| ZNF280D | 1.57 | 1.66 |
| Downregulated genes | ATR FC |
DEHP FC |
PFOA FC |
TBT FC |
|---|---|---|---|---|
| ANKRD1 | 1.95 | 1.88 | 1.66 | 1.79 |
| CCDC184 | 4.94 | 3.26 | 5.42 | 8.34 |
| EGLN1 | 1.71 | 1.40 | 1.75 | 2.03 |
| HK2 | 1.51 | 1.35 | 1.44 | 1.66 |
| KDM3A | 1.49 | 1.38 | 1.45 | 1.65 |
| RIMKLA | 2.32 | 1.44 | 1.99 | 3.51 |
| SLFN5 | 1.58 | 1.57 | 1.78 | 1.80 |
| STC1 | 1.86 | 1.62 | 1.91 | 2.97 |
| HIST1H4C | 1.56 | 1.38 | 1.40 | |
| AK4 | 1.45 | 1.34 | 1.44 | |
| CLDN14 | 2.86 | 2.45 | 4.84 | |
| FAM162A | 1.45 | 1.35 | 1.79 | |
| HELZ | 1.37 | 1.38 | 1.55 | |
| HERC3 | 1.54 | 1.44 | 2.23 | |
| NRN1 | 2.76 | 3.15 | 3.36 | |
| PFKFB4 | 1.58 | 1.52 | 2.56 | |
| PTX3 | 3.70 | 2.63 | 4.53 | |
| RAB2B | 1.42 | 1.41 | 1.70 | |
| RIOK3 | 1.43 | 1.39 | 1.66 | |
| SAP30 | 1.61 | 1.45 | 1.71 | |
| SLC2A1 | 1.58 | 1.54 | 1.70 | |
| TNIP1 | 1.43 | 1.34 | 1.85 | |
| UPRT | 1.56 | 1.41 | 1.59 | |
| ZNF395 | 1.44 | 1.40 | 1.61 | |
| BHLHE40 | 1.61 | 1.55 | 1.78 | |
| BHLHE41 | 1.69 | 1.66 | 2.67 | |
| HMGCS1 | 1.49 | 1.44 | 1.42 | |
| PTPRB | 1.52 | 1.59 | 1.56 | |
| CAV1 | 1.45 | 1.42 | ||
| GTF3C6 | 1.33 | 1.35 | ||
| HIST1H2AC | 1.42 | 1.31 | ||
| ACTA1 | 1.39 | 1.38 | ||
| ALKBH5 | 1.33 | 1.39 | ||
| DDX58 | 1.53 | 1.96 | ||
| ADM | 2.01 | 1.95 | ||
| IGFBP5 | 1.81 | 1.55 | ||
| MKRN3 | 6.03 | 2.59 | ||
| SERPINB6 | 1.70 | 1.44 | ||
| SERPINE1 | 1.50 | 1.47 | ||
| TFRC | 1.43 | 1.35 | ||
| NAMPT | 1.38 | 1.55 | ||
| ADAMTS20 | 1.46 | 2.02 | ||
| CHL1 | 1.34 | 1.52 | ||
| KCTD12 | 1.45 | 1.61 | ||
| LRRC58 | 1.34 | 1.41 | ||
| MAP3K1 | 1.36 | 1.45 | ||
| P4HA1 | 1.32 | 1.46 |
Substantial overlap was also seen between treatments examining results of the IPA CP analysis. To identify the most highly shared IPA CPs, we summed the –log10(p) values for the four treatments and ranked the pathways accordingly (Table 5). Top rated CPs included seven affected across three treatments and 22 affected across two treatments. Notable CPs included several stress response pathways, involving, for example, NRF2, AMPK, TNRF1, TNRF2, and HIF1A mediators (affected by two treatments), and UVA response (affected by three treatments).
Table 5.
IPA® Canonical Pathways significantly affected by two or more toxicant treatments
| Canonical Pathways | −log10(p) 1 | ||||
|---|---|---|---|---|---|
| ATR | DEHP | PFOA | TBT | SUM | |
| Circadian Rhythm Signaling | 2.41 | 2.48 | 1.84 | 6.73 | |
|
| |||||
| GNRH Signaling | 2.32 | 2.34 | 1.45 | 6.11 | |
|
| |||||
| Virus Entry via Endocytic Pathways | 1.64 | 2.69 | 1.65 | 5.97 | |
|
| |||||
| PDGF Signaling | 1.75 | 1.70 | 1.77 | 5.22 | |
|
| |||||
| UVA-Induced MAPK Signaling | 1.64 | 1.59 | 1.66 | 4.89 | |
|
| |||||
| CDK5 Signaling | 1.55 | 1.56 | 1.75 | 4.86 | |
|
| |||||
| HIF1α Signaling | 1.53 | 1.47 | 1.54 | 4.54 | |
|
| |||||
| NRF2-mediated Oxidative Stress Response | 2.89 | 3.99 | 6.88 | ||
|
| |||||
| Superpathway of Cholesterol Biosynthesis | 2.55 | 2.62 | 5.17 | ||
|
| |||||
| AMPK Signaling | 2.90 | 1.96 | 4.86 | ||
|
| |||||
| TNFR1 Signaling | 2.14 | 2.59 | 4.73 | ||
|
| |||||
| Mevalonate Pathway I | 3.22 | 1.48 | 4.70 | ||
|
| |||||
| TNFR2 Signaling | 2.59 | 1.95 | 4.54 | ||
|
| |||||
| Superpathway of Geranylgeranyldiphosphate Biosynthesis I (via Mevalonate) | 2.98 | 1.36 | 4.35 | ||
|
| |||||
| April Mediated Signaling | 2.36 | 1.72 | 4.08 | ||
|
| |||||
| Tight Junction Signaling | 2.01 | 2.03 | 4.04 | ||
|
| |||||
| B Cell Activating Factor Signaling | 2.32 | 1.68 | 4.00 | ||
|
| |||||
| Renal Cell Carcinoma Signaling | 1.82 | 1.84 | 3.66 | ||
|
| |||||
| Caveolar-mediated Endocytosis Signaling | 1.82 | 1.76 | 3.58 | ||
|
| |||||
| TGF-β Signaling | 1.67 | 1.90 | 3.57 | ||
|
| |||||
| CD27 Signaling in Lymphocytes | 2.09 | 1.47 | 3.56 | ||
|
| |||||
| RAR Activation | 1.88 | 1.66 | 3.54 | ||
|
| |||||
| EGF Signaling | 2.03 | 1.41 | 3.44 | ||
|
| |||||
| NAD Biosynthesis III | 1.77 | 1.48 | 3.26 | ||
|
| |||||
| Salvage Pathways of Pyrimidine Ribonucleotides | 1.61 | 1.62 | 3.23 | ||
|
| |||||
| LPS/IL-1 Mediated Inhibition of RXR Function | 1.71 | 1.45 | 3.16 | ||
|
| |||||
| Ketogenesis | 1.55 | 1.59 | 3.14 | ||
|
| |||||
| Vitamin-C Transport | 1.41 | 1.42 | 2.83 | ||
|
| |||||
| Glutathione Redox Reactions I | 1.31 | 1.32 | 2.63 | ||
Only values -log10(p)≥1.30 (equivalent to p ≤.05) are shown in the table.
We then examined overlap among affected biological functions/diseases. The largest numbers of shared effects were seen between ATR and PFOA, and between PFOA and TBT (Table S19 and S20). One IPA function (development of body trunk) was significantly increased (z>1.96) for two treatments (ATR and TBT). Other IPA functions displayed trends toward shared increases or decreases across two or three treatments (e.g., |z|≥1.5). Sorting the results by the absolute value of the sum of z-scores yielded highest positive total z-scores (n = number of treatments with |z|≥1.5) for transcription (n=3), and tumor cell death (n=2), and strongest negative z-score sums for tumor growth (n=3), molecular secretion (n=3), molecular transport (n=2), tumor cell proliferation (n=2), and tumor cell viability (n=2) (Table S20).
UR analysis also revealed extensive overlap between treatments (Table S21) with greatest number of shared regulators between PFOA and TBT comparing any two treatments. Three regulators (HIF1A, NUPR1 and AKT) displayed significant (|z|>1.96, p < 0.05) inhibition for DEHP, PFOA, and TBT treatments and a strong negative effect for ATR (Table S22, S23). EPAS1 was inhibited in two treatments (ATR, TBT). Ranking effects according to the absolute value of the sum of z-scores (Table S23) yielded strong activation results (positive sum of z-scores) for chemical mediators (four treatments--cycloheximide, Salirasib, fulvestrant, and Pkg) and signaling ligands (two treatments--PDGF, EPO, KLF2). Strongest inhibition results (negative sum of z-scores) were obtained for stress response regulators: four treatments (HIF1A, NUPR1, AKT), three treatments (CD38), and two treatments (EPAS1, NOTCH1). Additional strong effects (sum z-scores) for two or more treatments but falling below the significance threshold were seen for NEDD9, IL5, MYC, 25-hydroxycholesterol, VEGFA, RELA, PDGF, RELA, and EPO (Table S23). There were a few instances of strong opposite effects amongst treatments; one example was CREB1 with a strong positive activation z-score for ATR but weak negative z-score for DEHP.
Examination of the sum of -log10(p) values (Table S23) confirmed shared significant effects for upstream regulators HIF1A, NUPR1, AKT, EPAS1, NEDD9, EPO, VEGFA, IL5, Pkg, and the chemical regulators Salirisab, cycloheximide, lipopolysaccharide. Additional biological regulators emerged high in that ranking as well, such as MARCH2, ARNT, ADRA1, TNF, PDGFBB, TGFB1, IFNG, NRG1, ELK4, COMMD1, and TP53. Pathways regulated by FOSB and ADM displayed more intermediate p-value sums. Additional strong shared effects were seen for developmental regulators NANOG, GATA4, and BMP4 and weaker shared effects for SOX2, SOX17, POU5F1/OCT4.
Discussion
This is the first analysis of long-term EDC effects in a nonhuman primate embryonic stem cell model. This study reveals four striking observations. Firstly, the number of affected genes is small, ranging as high as just 119. Secondly, while there are some genes affected by just one of the EDCs tested, there is considerable overlap in affected genes among the five EDCs. Thirdly, IPA analysis indicated that long-term treatment with four of the EDCs negatively affects indicators of cell survival, as well as repressing pathways that lie downstream of several stress response mediators. And fourthly, TBT in particular, but also ATR and PFOA treatments modulated genes related to maintaining stem cell potency and cell lineage formation. Canonical pathway analysis for DEHP also indicated possible effects on pluripotency. Thus, all four of these EDCs may alter the balance between proliferation and differentiation, and predisposition for differentiation along certain embryonic lineages.
The small number of genes affected by five different EDCs applied over a four-week period is striking. The modest effect of these EDCs may be a reflection of maintaining the ESCs in a non-differentiated state; further differentiation may be required to develop the endocrine systems that these chemicals disrupt [33, 69, 70]. In studies involving induction of differentiation events from embryos or stem cells, prolonged treatment of up to two weeks in vitro revealed similar small numbers of genes induced or repressed by EDCs [71]. The numbers of affected genes can be much higher for specialized somatic cell lines [72]. In studies of ES or pluripotent cell exposures to higher EDC doses for short periods of 72 h or less, larger numbers of affected genes are seen for some EDCs [73–75], but small numbers for others [75]. The small number of affected genes for all five EDCs seen here may indicate an adaptive response of rhesus monkey ESCs occurring with chronic exposure over a protracted period of four weeks. The ability of ESCs to adapt in this manner, and manifest a limited range of genes with altered expression profiles indicates that ontogenetic effects of chronic exposure to environmental EDCs may be more selective than previously suspected. Consequently, extensive short-term responses to acute exposures affecting thousands of genes may obscure the more selective cellular and developmental consequences of low-level chronic exposures. Focusing attention on the small subsets of genes affected by long-term exposure provides a path to a more specific understanding of the developmental consequences and mechanisms of actions of EDCs present chronically in the environment, relevant to the effects of prevailing environmental exposures, as compared to acute short-term higher concentrations.
The genes affected by chronic exposure to a single compound tested here may provide particular targets of interest for understanding the specific effects of individual compounds (or related families of compounds) on developing embryos. Other studies using mouse ESCs, with or without induction of differentiation to various lineages, but with elevated exposure concentrations for shorter time periods, have indicated gene sets that appear to be characteristic of individual chemicals or chemical families [74], particularly with respect to interference with in vitro differentiation [76–78]. Additionally, analyses of affected biological functions and pathways, as done here, have been suggested to be key in developing biological signatures for exposures to individual chemicals or chemical families. The most highly affected genes observed here do not overlap with published gene lists that emerged as potential biomarkers using mouse ESCs treated with EDCs [74]. Only three genes affected by TBT (KDM4B, CER1, CYP26A1) were reported as affected in mouse ESCs by multiple other EDCs [73, 77]. The difference between the studies employing mouse versus nonhuman primate ESCs in terms of numbers of affected genes could reflect differences in chemical used, duration of treatment (e.g., 10 days), and use of a cell differentiation protocol, concentration (e.g., ID50 1.35 mM for monobutyl phthalate giving 50% inhibition of differentiation, [79]), and gene expression assay and data analysis methods, or could reflect a more fundamental species difference as previously suggested [21]. Further studies would be needed to address these possibilities.
The genes identified here as being affected by two or more separate treatments could reflect common cellular responses to harmful chemicals, or they could represent genes for which embryonic chromatin states are especially susceptible to disruption by a range of environmental factors. A better understanding of the epigenetic regulation of these genes, how their epigenetic states change during normal development, and how these epigenetic states may change in response to stress could provide a new basis for assessing whether certain developmental stages or target embryonic organs are at enhanced risk from exposure to a range of environmental agents, and may provide valuable new targets for biomonitoring.
While there was considerable overlap, there were still differences in effects of the EDCs. These differences may be related to differences in the EDCs’ mechanism of action, or differences in prevalence of downstream mediators of their actions. Understanding how stem cells may modulate downstream pathways responsive to different chemical families will be valuable for understanding effects of chronic low-level exposure.
A deeper understanding of chronic exposure effects and of windows of sensitivity for different developing organs may emerge from identifying the developmental events and stages that may be most highly impacted by the biological functions and processes identified here. The subtle effects seen here on genes related to cellular proliferation and viability, and body trunk development provide potential new connections for assessing potential effects of chronic embryonic and fetal exposure on growth, contribution to congenital malformations, and subtle effects on neurological, immunological, and other functions. The UR analysis revealed potential compensation as part of cellular adaptation to the treatments, which could impact cell proliferation and other functions. For example, the FOS mRNA was upregulated in three of the treatments, but a negative activation z-score was seen for FOS downstream targets for ATR (−1.39) and PFOA (−1.96), indicating inhibition of FOS-regulated downstream functions, and no activation z-score was seen for other treatments.
Two of the EDCs (ATR and PFOA) were previously associated with an activation of stress response. The suppression of stress response pathways here with long term exposure may represent an adaptation of embryonic cells enabling their survival, and may be a general response to a variety of EDCs. The long-term inhibition of the stress response pathways could pose a special risk for developing systems, with far-reaching consequences. Such inhibition could increase the potential for developmental failure following imposition of any additional stressor. Additionally, early embryonic responses to stress are increasingly appreciated for their potential connections to diseases later in life. A variety of stressors (osmotic, nutrient, temperature, hypoxia, hyperoxia, hyperglycemia) lead to long-term effects on gene expression programming in the early embryo, with subsequent effects on progeny phenotype, and even transgenerational effects in subsequent generations [80–84]. An essential benefit of cellular stress responses is to minimize cellular damage to allow cells to recover, and protects them from undergoing necrosis or apoptosis. But even in surviving cells, sub-lethal cytological or genetic damage may occur, leading to changes in mitochondrial characteristics, cellular metabolism, DNA damage, and incorrect epigenetic programming. Suppression of stress response pathways could shift the balance to greater cell death. Indeed, we observed activation of cell death and apoptosis pathways, and reduced cell viability pathways across treatments. Increased stem cell death could reduce the numbers of stem cells available for organogenesis and tissue homeostasis in the embryo, affecting later health. Additionally, inhibition of stress response could lead to a greater degree of sub-lethal cellular damage, leading to propagation in the embryo of cells harboring damaged mitochondria or incorrect epigenetic programming.
Overall, the data presented here illustrate the need for further studies of EDC effects on development using nonhuman primate models. The dynamic pharmacokinetics of EDC metabolism during primate pregnancy [85], effects of EDCs on the uterus [86] and fetal lung [44], and the accessibility for study of nonhuman primate oocytes and embryos as a highly related animal model of human reproduction highlight the value and feasibility of using nonhuman primates to understand exposure risks to human pregnancies. Mechanistic understanding of these effects can be facilitated by determining specific effects of long-term EDC exposures on nonhuman ES and pluripotent cells, and on stem cell commitment and differentiation during successive developmental windows of susceptibility.
Although the study here does not extend to functional effects beyond gene expression changes, the data provide a foundation for further testing of effects of these low-level exposures on stem cell proliferation and developmental potential. Additionally, combinatorial treatments and low-level dose response studies would be valuable for addressing chemical mixture effects and possible additive or synergistic developmental effects of these chemicals in the environment. The effects reported here for cell survival, stress responsiveness, and developmental gene expression warrant further study to assess potential impact of low level chronic EDC exposure on early development in primates.
Supplementary Material
Highlights.
Effects of chronic exposure to low concentrations of 5 EDCs were examined in rhesus monkey embryonic stem cells
There was no apparent gross change in cell morphology or growth in culture
RNAseq revealed small numbers of affected genes
Effects included suppression of genes related to cell survival, increased activity of cell death-related genes, suppression of downstream mediators of several stress response mediators, and modulations of pluripotency and lineage controlling genes
Acknowledgments
The authors thank Dana Hill for her technical assistance. We thank Dr. Almudena Veiga-Lopez for her critical comments on the manuscript. This work was supported by grants from the Office of Research Infrastructure Programs Division of Comparative Medicine Grants R24 OD-012221 (to K.E.L.), and OD011107/RR00169 (California National Primate Research Center) and OD010967/RR025880 (to C.A.V.), by MSU AgBioResearch, and Michigan State University.
Abbreviations
- ATR
Atrazine
- BPA
bisphenol A
- CP
canonical pathway
- DEG
differentially expressed gene
- DEHP
di-(2-ethylhexyl) phthalate
- DF
disease and function
- EDC
endocrine disrupting chemical
- ESC
embryonic stem cell
- IPA
QIAGEN Ingenuity Pathway Analysis®
- PFOA
perfluorooctanoic acid
- RNAseq
RNA sequencing
- TBT
tributyltin
- UR
upstream regulator
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Latham KE, Sapienza C, Engel N. The epigenetic lorax: gene-environment interactions in human health. Epigenomics. 2012;4:383–402. doi: 10.2217/epi.12.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prins GS, Calderon-Gierszal EL, Hu WY. Stem Cells as Hormone Targets That Lead to Increased Cancer Susceptibility. Endocrinology. 2015;156:3451–7. doi: 10.1210/en.2015-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laiosa MD, Tate ER. Fetal Hematopoietic Stem Cells Are the Canaries in the Coal Mine That Portend Later Life Immune Deficiency. Endocrinology. 2015;156:3458–65. doi: 10.1210/en.2015-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strong AL, Shi Z, Strong MJ, Miller DF, Rusch DB, Buechlein AM, Flemington EK, McLachlan JA, Nephew KP, Burow ME, Bunnell BA. Effects of the endocrine-disrupting chemical DDT on self-renewal and differentiation of human mesenchymal stem cells. Environ Health Perspect. 2015;123:42–8. doi: 10.1289/ehp.1408188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X, Xu B, Han X, Mao Z, Chen M, Du G, Talbot P, Wang X, Xia Y. The effects of triclosan on pluripotency factors and development of mouse embryonic stem cells and zebrafish. Arch Toxicol. 2015;89:635–46. doi: 10.1007/s00204-014-1270-2. [DOI] [PubMed] [Google Scholar]
- 6.Kirchner S, Kieu T, Chow C, Casey S, Blumberg B. Prenatal exposure to the environmental obesogen tributyltin predisposes multipotent stem cells to become adipocytes. Mol Endocrinol. 2010;24:526–39. doi: 10.1210/me.2009-0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arbuckle TE, Davis K, Boylan K, Fisher M, Fu J. Bisphenol A, phthalates and lead and learning and behavioral problems in Canadian children 6–11 years of age: CHMS 2007–2009. Neurotoxicology. 2016;54:89–98. doi: 10.1016/j.neuro.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 8.Grote K, Hobler C, Andrade AJ, Grande SW, Gericke C, Talsness CE, Appel KE, Chahoud I. Effects of in utero and lactational exposure to triphenyltin chloride on pregnancy outcome and postnatal development in rat offspring. Toxicology. 2007;238:177–85. doi: 10.1016/j.tox.2007.05.033. [DOI] [PubMed] [Google Scholar]
- 9.Grun F, Watanabe H, Zamanian Z, Maeda L, Arima K, Cubacha R, Gardiner DM, Kanno J, Iguchi T, Blumberg B. Endocrine-disrupting organotin compounds are potent inducers of adipogenesis in vertebrates. Mol Endocrinol. 2006;20:2141–55. doi: 10.1210/me.2005-0367. [DOI] [PubMed] [Google Scholar]
- 10.Fujimoto VY, Kim D, vom Saal FS, Lamb JD, Taylor JA, Bloom MS. Serum unconjugated bisphenol A concentrations in women may adversely influence oocyte quality during in vitro fertilization. Fertil Steril. 2011;95:1816–9. doi: 10.1016/j.fertnstert.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 11.Vandenberg LN, Maffini MV, Wadia PR, Sonnenschein C, Rubin BS, Soto AM. Exposure to environmentally relevant doses of the xenoestrogen bisphenol-A alters development of the fetal mouse mammary gland. Endocrinology. 2007;148:116–27. doi: 10.1210/en.2006-0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durmaz E, Ozmert EN, Erkekoglu P, Giray B, Derman O, Hincal F, Yurdakok K. Plasma phthalate levels in pubertal gynecomastia. Pediatrics. 2010;125:e122–9. doi: 10.1542/peds.2009-0724. [DOI] [PubMed] [Google Scholar]
- 13.Hunt PA, Koehler KE, Susiarjo M, Hodges CA, Ilagan A, Voigt RC, Thomas S, Thomas BF, Hassold TJ. Bisphenol a exposure causes meiotic aneuploidy in the female mouse. Curr Biol. 2003;13:546–53. doi: 10.1016/s0960-9822(03)00189-1. [DOI] [PubMed] [Google Scholar]
- 14.Peden-Adams MM, Keller JM, Eudaly JG, Berger J, Gilkeson GS, Keil DE. Suppression of humoral immunity in mice following exposure to perfluorooctane sulfonate. Toxicol Sci. 2008;104:144–54. doi: 10.1093/toxsci/kfn059. [DOI] [PubMed] [Google Scholar]
- 15.White SS, Calafat AM, Kuklenyik Z, Villanueva L, Zehr RD, Helfant L, Strynar MJ, Lindstrom AB, Thibodeaux JR, Wood C, Fenton SE. Gestational PFOA exposure of mice is associated with altered mammary gland development in dams and female offspring. Toxicol Sci. 2007;96:133–44. doi: 10.1093/toxsci/kfl177. [DOI] [PubMed] [Google Scholar]
- 16.Birnbaum LS, Fenton SE. Cancer and developmental exposure to endocrine disruptors. Environ Health Perspect. 2003;111:389–94. doi: 10.1289/ehp.5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stanko JP, Enoch RR, Rayner JL, Davis CC, Wolf DC, Malarkey DE, Fenton SE. Effects of prenatal exposure to a low dose atrazine metabolite mixture on pubertal timing and prostate development of male Long-Evans rats. Reprod Toxicol. 2010;30:540–9. doi: 10.1016/j.reprotox.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enoch RR, Stanko JP, Greiner SN, Youngblood GL, Rayner JL, Fenton SE. Mammary gland development as a sensitive end point after acute prenatal exposure to an atrazine metabolite mixture in female Long-Evans rats. Environ Health Perspect. 2007;115:541–7. doi: 10.1289/ehp.9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zong T, Lai L, Hu J, Guo M, Li M, Zhang L, Zhong C, Yang B, Wu L, Zhang D, Tang M, Kuang H. Maternal exposure to di-(2-ethylhexyl) phthalate disrupts placental growth and development in pregnant mice. J Hazard Mater. 2015;297:25–33. doi: 10.1016/j.jhazmat.2015.04.065. [DOI] [PubMed] [Google Scholar]
- 20.Midoro-Horiuti T, Tiwari R, Watson CS, Goldblum RM. Maternal bisphenol a exposure promotes the development of experimental asthma in mouse pups. Environ Health Perspect. 2010;118:273–7. doi: 10.1289/ehp.0901259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Habert R, Muczynski V, Grisin T, Moison D, Messiaen S, Frydman R, Benachi A, Delbes G, Lambrot R, Lehraiki A, N’Tumba-Byn T, Guerquin MJ, Levacher C, Rouiller-Fabre V, Livera G. Concerns about the widespread use of rodent models for human risk assessments of endocrine disruptors. Reproduction. 2014;147:R119–29. doi: 10.1530/REP-13-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang XF, Li Y, Gu YH, Liu M, Xu Y, Yuan Y, Sun F, Zhang HQ, Shi HJ. The effects of Di-(2-ethylhexyl)-phthalate exposure on fertilization and embryonic development in vitro and testicular genomic mutation in vivo. PLoS One. 2012;7:e50465. doi: 10.1371/journal.pone.0050465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graves JE, Richardson ME, Bernard RS, Camper ND, Bridges WC. Atrazine effects on in vitro maturation and in vitro fertilization in the bovine oocyte. J Environ Sci Health B. 2002;37:103–12. doi: 10.1081/PFC-120002982. [DOI] [PubMed] [Google Scholar]
- 24.Singh S, Li SS. Epigenetic effects of environmental chemicals bisphenol A and phthalates. Int J Mol Sci. 2012;13:10143–53. doi: 10.3390/ijms130810143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenlee AR, Ellis TM, Berg RL. Low-dose agrochemicals and lawn-care pesticides induce developmental toxicity in murine preimplantation embryos. Environ Health Perspect. 2004;112:703–9. doi: 10.1289/ehp.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cummings AM, Rhodes BE, Cooper RL. Effect of atrazine on implantation and early pregnancy in 4 strains of rats. Toxicol Sci. 2000;58:135–43. doi: 10.1093/toxsci/58.1.135. [DOI] [PubMed] [Google Scholar]
- 27.Casas E, Bonilla E, Ducolomb Y, Betancourt M. Differential effects of herbicides atrazine and fenoxaprop-ethyl, and insecticides diazinon and malathion, on viability and maturation of porcine oocytes in vitro. Toxicol In Vitro. 2010;24:224–30. doi: 10.1016/j.tiv.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Chu DP, Tian S, Qi L, Hao CJ, Xia HF, Ma X. Abnormality of maternal-to-embryonic transition contributes to MEHP-induced mouse 2-cell block. J Cell Physiol. 2013;228:753–63. doi: 10.1002/jcp.24222. [DOI] [PubMed] [Google Scholar]
- 29.Wang T, Han J, Duan X, Xiong B, Cui XS, Kim NH, Liu HL, Sun SC. The toxic effects and possible mechanisms of Bisphenol A on oocyte maturation of porcine in vitro. Oncotarget. 2016;7:32554–65. doi: 10.18632/oncotarget.8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mu X, Liao X, Chen X, Li Y, Wang M, Shen C, Zhang X, Wang Y, Liu X, He J. DEHP exposure impairs mouse oocyte cyst breakdown and primordial follicle assembly through estrogen receptor-dependent and independent mechanisms. J Hazard Mater. 2015;298:232–40. doi: 10.1016/j.jhazmat.2015.05.052. [DOI] [PubMed] [Google Scholar]
- 31.Grossman D, Kalo D, Gendelman M, Roth Z. Effect of di-(2-ethylhexyl) phthalate and mono-(2-ethylhexyl) phthalate on in vitro developmental competence of bovine oocytes. Cell Biol Toxicol. 2012;28:383–96. doi: 10.1007/s10565-012-9230-1. [DOI] [PubMed] [Google Scholar]
- 32.Machtinger R, Combelles CM, Missmer SA, Correia KF, Williams P, Hauser R, Racowsky C. Bisphenol-A and human oocyte maturation in vitro. Hum Reprod. 2013;28:2735–45. doi: 10.1093/humrep/det312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calderon-Gierszal EL, Prins GS. Directed Differentiation of Human Embryonic Stem Cells into Prostate Organoids In Vitro and its Perturbation by Low-Dose Bisphenol A Exposure. PLoS One. 2015;10:e0133238. doi: 10.1371/journal.pone.0133238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitalipov S, Kuo HC, Byrne J, Clepper L, Meisner L, Johnson J, Zeier R, Wolf D. Isolation and characterization of novel rhesus monkey embryonic stem cell lines. Stem Cells. 2006;24:2177–86. doi: 10.1634/stemcells.2006-0125. [DOI] [PubMed] [Google Scholar]
- 35.Vandenberg LN, Hauser R, Marcus M, Olea N, Welshons WV. Human exposure to bisphenol A (BPA) Reprod Toxicol. 2007;24:139–77. doi: 10.1016/j.reprotox.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 36.Fan A, Alexeeff GA, et al. Office of Environmental Health Hazard Assessment California Environmental Protection Agency; Public Health Goal for Atrazine In Drinking Water. 1999. [Google Scholar]
- 37.Calafat AM, Wong LY, Kuklenyik Z, Reidy JA, Needham LL. Polyfluoroalkyl chemicals in the U.S. population: data from the National Health and Nutrition Examination Survey (NHANES) 2003–2004 and comparisons with NHANES 1999–2000. Environ Health Perspect. 2007;115:1596–602. doi: 10.1289/ehp.10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Antizar-Ladislao B. Environmental levels, toxicity and human exposure to tributyltin (TBT)-contaminated marine environment. a review. Environ Int. 2008;34:292–308. doi: 10.1016/j.envint.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 39.Mlynarcikova A, Nagyova E, Fickova M, Scsukova S. Effects of selected endocrine disruptors on meiotic maturation, cumulus expansion, synthesis of hyaluronan and progesterone by porcine oocyte-cumulus complexes. Toxicol In Vitro. 2009;23:371–7. doi: 10.1016/j.tiv.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 40.Ferris J, Mahboubi K, MacLusky N, King WA, Favetta LA. BPA exposure during in vitro oocyte maturation results in dose-dependent alterations to embryo development rates, apoptosis rate, sex ratio and gene expression. Reprod Toxicol. 2016;59:128–38. doi: 10.1016/j.reprotox.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 41.Yamada S, Asanagi M, Hirata N, Itagaki H, Sekino Y, Kanda Y. Tributyltin induces mitochondrial fission through Mfn1 degradation in human induced pluripotent stem cells. Toxicol In Vitro. 2016;34:257–263. doi: 10.1016/j.tiv.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 42.Scutaru B, Cozmei C, Cazuc V, Popa D, Hock B. Immuoenzymatic serum analysis of atrazine exposure among manufacturing workers. THe Journal of Preventive Medicine. 2002;10:31–36. [Google Scholar]
- 43.Thibeault AA, Deroy K, Vaillancourt C, Sanderson JT. A unique co-culture model for fundamental and applied studies of human fetoplacental steroidogenesis and interference by environmental chemicals. Environ Health Perspect. 2014;122:371–7. doi: 10.1289/ehp.1307518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Winkle LS, Murphy SR, Boetticher MV, VandeVoort CA. Fetal exposure of rhesus macaques to bisphenol a alters cellular development of the conducting airway by changing epithelial secretory product expression. Environ Health Perspect. 2013;121:912–8. doi: 10.1289/ehp.1206064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tharp AP, Maffini MV, Hunt PA, VandeVoort CA, Sonnenschein C, Soto AM. Bisphenol A alters the development of the rhesus monkey mammary gland. Proc Natl Acad Sci U S A. 2012;109:8190–5. doi: 10.1073/pnas.1120488109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez-Sosa JR, Bondareva A, Tang L, Avelar GF, Coyle KM, Modelski M, Alpaugh W, Conley A, Wynne-Edwards K, Franca LR, Meyers S, Dobrinski I. Phthalate esters affect maturation and function of primate testis tissue ectopically grafted in mice. Mol Cell Endocrinol. 2014;398:89–100. doi: 10.1016/j.mce.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamada S, Kotake Y, Nakano M, Sekino Y, Kanda Y. Tributyltin induces mitochondrial fission through NAD-IDH dependent mitofusin degradation in human embryonic carcinoma cells. Metallomics. 2015;7:1240–6. doi: 10.1039/c5mt00033e. [DOI] [PubMed] [Google Scholar]
- 48.Yamada S, Kotake Y, Demizu Y, Kurihara M, Sekino Y, Kanda Y. NAD-dependent isocitrate dehydrogenase as a novel target of tributyltin in human embryonic carcinoma cells. Sci Rep. 2014;4:5952. doi: 10.1038/srep05952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayes TB, Anderson LL, Beasley VR, de Solla SR, Iguchi T, Ingraham H, Kestemont P, Kniewald J, Kniewald Z, Langlois VS, Luque EH, McCoy KA, Munoz-de-Toro M, Oka T, Oliveira CA, Orton F, Ruby S, Suzawa M, Tavera-Mendoza LE, Trudeau VL, Victor-Costa AB, Willingham E. Demasculinization and feminization of male gonads by atrazine: consistent effects across vertebrate classes. J Steroid Biochem Mol Biol. 2011;127:64–73. doi: 10.1016/j.jsbmb.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Head SR, Komori HK, Hart GT, Shimashita J, Schaffer L, Salomon DR, Ordoukhanian PT. Method for improved Illumina sequencing library preparation using NuGEN OVation RNA-Seq system. Biotechniques. 2011;50:177–181. doi: 10.2144/000113613. [DOI] [PubMed] [Google Scholar]
- 51.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zimin AV, Cornish AS, Maudhoo MD, Gibbs RM, Zhang X, Pandey S, Meehan DT, Wipfler K, Bosinger SE, Johnson ZP, Tharp GK, Marcais G, Roberts M, Ferguson B, Fox HS, Treangen T, Salzberg SL, Yorke JA, Norgren RB., Jr A new rhesus macaque assembly and annotation for next-generation sequencing analyses. Biol Direct. 2014;9:20. doi: 10.1186/1745-6150-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagel SC, Bromfield JJ. Bisphenol a: a model endocrine disrupting chemical with a new potential mechanism of action. Endocrinology. 2013;154:1962–4. doi: 10.1210/en.2013-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas P, Dong J. Binding and activation of the seven-transmembrane estrogen receptor GPR30 by environmental estrogens: a potential novel mechanism of endocrine disruption. J Steroid Biochem Mol Biol. 2006;102:175–9. doi: 10.1016/j.jsbmb.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 56.Dooley GP, Tjalkens RB, Hanneman WH. The atrazine metabolite diaminochlorotriazine suppresses LH release from murine LbetaT2 cells by suppressing GnRH-induced intracellular calcium transients. Toxicol Res (Camb) 2013;2:180–186. doi: 10.1039/C3TX20088D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thueson LE, Emmons TR, Browning DL, Kreitinger JM, Shepherd DM, Wetzel SA. In vitro exposure to the herbicide atrazine inhibits T cell activation, proliferation, and cytokine production and significantly increases the frequency of Foxp3+ regulatory T cells. Toxicol Sci. 2015;143:418–29. doi: 10.1093/toxsci/kfu242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lai FN, Liu JC, Li L, Ma JY, Liu XL, Liu YP, Zhang XF, Chen H, De Felici M, Dyce PW, Shen W. Di (2-ethylhexyl) phthalate impairs steroidogenesis in ovarian follicular cells of prepuberal mice. Arch Toxicol. 2016 doi: 10.1007/s00204-016-1790-z. (epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 59.Sekaran S, Jagadeesan A. In utero exposure to phthalate downregulates critical genes in Leydig cells of F1 male progeny. J Cell Biochem. 2015;116:1466–77. doi: 10.1002/jcb.25108. [DOI] [PubMed] [Google Scholar]
- 60.Klaunig JE, Hocevar BA, Kamendulis LM. Mode of Action analysis of perfluorooctanoic acid (PFOA) tumorigenicity and Human Relevance. Reprod Toxicol. 2012;33:410–8. doi: 10.1016/j.reprotox.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 61.Kang JS, Choi JS, Park JW. Transcriptional changes in steroidogenesis by perfluoroalkyl acids (PFOA and PFOS) regulate the synthesis of sex hormones in H295R cells. Chemosphere. 2016;155:436–43. doi: 10.1016/j.chemosphere.2016.04.070. [DOI] [PubMed] [Google Scholar]
- 62.Liu C, Yu K, Shi X, Wang J, Lam PK, Wu RS, Zhou B. Induction of oxidative stress and apoptosis by PFOS and PFOA in primary cultured hepatocytes of freshwater tilapia (Oreochromis niloticus) Aquat Toxicol. 2007;82:135–43. doi: 10.1016/j.aquatox.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 63.Yaari-Stark S, Shaked M, Nevo-Caspi Y, Jacob-Hircsh J, Shamir R, Rechavi G, Kloog Y. Ras inhibits endoplasmic reticulum stress in human cancer cells with amplified Myc. Int J Cancer. 2010;126:2268–81. doi: 10.1002/ijc.25102. [DOI] [PubMed] [Google Scholar]
- 64.Kotake Y. Molecular mechanisms of environmental organotin toxicity in mammals. Biol Pharm Bull. 2012;35:1876–80. doi: 10.1248/bpb.b212017. [DOI] [PubMed] [Google Scholar]
- 65.Kim DK, Cha Y, Ahn HJ, Kim G, Park KS. Lefty1 and lefty2 control the balance between self-renewal and pluripotent differentiation of mouse embryonic stem cells. Stem Cells Dev. 2014;23:457–66. doi: 10.1089/scd.2013.0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meno C, Saijoh Y, Fujii H, Ikeda M, Yokoyama T, Yokoyama M, Toyoda Y, Hamada H. Left-right asymmetric expression of the TGF beta-family member lefty in mouse embryos. Nature. 1996;381:151–5. doi: 10.1038/381151a0. [DOI] [PubMed] [Google Scholar]
- 67.Chappell J, Sun Y, Singh A, Dalton S. MYC/MAX control ERK signaling and pluripotency by regulation of dual-specificity phosphatases 2 and 7. Genes Dev. 2013;27:725–33. doi: 10.1101/gad.211300.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kobayashi M, Takada T, Takahashi K, Noda Y, Torii R. BMP4 induces primitive endoderm but not trophectoderm in monkey embryonic stem cells. Cloning Stem Cells. 2008;10:495–502. doi: 10.1089/clo.2008.0030. [DOI] [PubMed] [Google Scholar]
- 69.Yang L, Luo L, Ji W, Gong C, Wu D, Huang H, Liu Q, Xia B, Hu G, Zhang W, Zhang Q, Liu J, Zhang W, Zhuang Z. Effect of low dose bisphenol A on the early differentiation of human embryonic stem cells into mammary epithelial cells. Toxicol Lett. 2013;218:187–93. doi: 10.1016/j.toxlet.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 70.Kim H, Kim YY, Ku SY, Kim SH, Choi YM, Moon SY. The effect of estrogen compounds on human embryoid bodies. Reprod Sci. 2013;20:661–9. doi: 10.1177/1933719112462630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ho SM, Cheong A, Lam HM, Hu WY, Shi GB, Zhu X, Chen J, Zhang X, Medvedovic M, Leung YK, Prins GS. Exposure of Human Prostaspheres to Bisphenol A Epigenetically Regulates SNORD Family Noncoding RNAs via Histone Modification. Endocrinology. 2015;156:3984–95. doi: 10.1210/en.2015-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pereira-Fernandes A, Vanparys C, Vergauwen L, Knapen D, Jorens PG, Blust R. Toxicogenomics in the 3T3-L1 cell line, a new approach for screening of obesogenic compounds. Toxicol Sci. 2014;140:352–63. doi: 10.1093/toxsci/kfu092. [DOI] [PubMed] [Google Scholar]
- 73.van Dartel DA, Pennings JL, de la Fonteyne LJ, Brauers KJ, Claessen S, van Delft JH, Kleinjans JC, Piersma AH. Evaluation of developmental toxicant identification using gene expression profiling in embryonic stem cell differentiation cultures. Toxicol Sci. 2011;119:126–34. doi: 10.1093/toxsci/kfq291. [DOI] [PubMed] [Google Scholar]
- 74.van Dartel DA, Pennings JL, Robinson JF, Kleinjans JC, Piersma AH. Discriminating classes of developmental toxicants using gene expression profiling in the embryonic stem cell test. Toxicol Lett. 2011;201:143–51. doi: 10.1016/j.toxlet.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 75.Theunissen PT, Robinson JF, Pennings JL, van Herwijnen MH, Kleinjans JC, Piersma AH. Compound-specific effects of diverse neurodevelopmental toxicants on global gene expression in the neural embryonic stem cell test (ESTn) Toxicol Appl Pharmacol. 2012;262:330–40. doi: 10.1016/j.taap.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 76.van Dartel DA, Pennings JL, van Schooten FJ, Piersma AH. Transcriptomics-based identification of developmental toxicants through their interference with cardiomyocyte differentiation of embryonic stem cells. Toxicol Appl Pharmacol. 2010;243:420–8. doi: 10.1016/j.taap.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 77.van Dartel DA, Pennings JL, de la Fonteyne LJ, van Herwijnen MH, van Delft JH, van Schooten FJ, Piersma AH. Monitoring developmental toxicity in the embryonic stem cell test using differential gene expression of differentiation-related genes. Toxicol Sci. 2010;116:130–9. doi: 10.1093/toxsci/kfq127. [DOI] [PubMed] [Google Scholar]
- 78.Pennings JL, Theunissen PT, Piersma AH. An optimized gene set for transcriptomics based neurodevelopmental toxicity prediction in the neural embryonic stem cell test. Toxicology. 2012;300:158–67. doi: 10.1016/j.tox.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 79.van Dartel DA, Pennings JL, Hendriksen PJ, van Schooten FJ, Piersma AH. Early gene expression changes during embryonic stem cell differentiation into cardiomyocytes and their modulation by monobutyl phthalate. Reprod Toxicol. 2009;27:93–102. doi: 10.1016/j.reprotox.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 80.Grindler NM, Moley KH. Maternal obesity, infertility and mitochondrial dysfunction: potential mechanisms emerging from mouse model systems. Mol Hum Reprod. 2013;19:486–94. doi: 10.1093/molehr/gat026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Purcell SH, Moley KH. The impact of obesity on egg quality. J Assist Reprod Genet. 2011;28:517–24. doi: 10.1007/s10815-011-9592-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fleming TP, Velazquez MA, Eckert JJ. Embryos, DOHaD and David Barker. J Dev Orig Health Dis. 2015;6:377–83. doi: 10.1017/S2040174415001105. [DOI] [PubMed] [Google Scholar]
- 83.Fernandez-Gonzalez R, Ramirez MA, Bilbao A, De Fonseca FR, Gutierrez-Adan A. Suboptimal in vitro culture conditions: an epigenetic origin of long-term health effects. Mol Reprod Dev. 2007;74:1149–56. doi: 10.1002/mrd.20746. [DOI] [PubMed] [Google Scholar]
- 84.Latham KE. Endoplasmic reticulum stress signaling in mammalian oocytes and embryos: life in balance. Int Rev Cell Mol Biol. 2015;316:227–65. doi: 10.1016/bs.ircmb.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vom Saal FS, VandeVoort CA, Taylor JA, Welshons WV, Toutain PL, Hunt PA. Bisphenol A (BPA) pharmacokinetics with daily oral bolus or continuous exposure via silastic capsules in pregnant rhesus monkeys: Relevance for human exposures. Reprod Toxicol. 2014;45:105–16. doi: 10.1016/j.reprotox.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Calhoun KC, Padilla-Banks E, Jefferson WN, Liu L, Gerrish KE, Young SL, Wood CE, Hunt PA, Vandevoort CA, Williams CJ. Bisphenol A exposure alters developmental gene expression in the fetal rhesus macaque uterus. PLoS One. 2014;9:e85894. doi: 10.1371/journal.pone.0085894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.