Abstract
To determine the association of somatic mutations in acute myeloid leukemia (AML) with risk of relapse after allogeneic hematopoietic stem cell transplantation (alloHSCT), we retrospectively studied pre-transplant genetic profiles obtained from next generation sequencing of 26 genes in 112 adult AML patients who underwent alloHSCT. Univariable and multivariable regression analyses were used to assess the association between the presence of a pathogenic mutation and risk of relapse after alloHSCT. Eighty-six percent (96/112) of patients had at least 1 pathogenic mutation. Mutations in TP53, WT1, and FLT3-ITD were associated with an increased risk of relapse after alloHSCT (adjusted HR [aHR] 2.90, P = .009, aHR 2.51, P = .02, and aHR 1.83, P = .07, respectively). DNMT3A mutation in the absence of FLT3-ITD and NPM1 mutations was associated with a lower relapse risk (aHR 0.22, P = .04). Comparison of pre- and post-alloHSCT genetic profiles showed clonal evolution in 6/6 patients, including acquisition of actionable mutations in 4 patients. In summary, genetic profiling is useful for assessing relapse risk in AML patients undergoing alloHSCT, and may identify patients in need of strategies to reduce this risk. Clonal evolution is present at post-alloHSCT relapse and repeat genetic profiling may uncover acquired actionable mutations.
Keywords: Allogeneic stem cell transplant, acute myeloid leukemia, relapse, next generation sequencing
INTRODUCTION
Disease relapse is the primary reason for treatment failure and death in patients with AML who undergo allogeneic hematopoietic stem-cell transplantation (alloHSCT).1 While the prognostic implications of cytogenetic abnormalities and a growing number of molecular abnormalities are relatively well understood in the context of chemotherapy-treated AML patients,2-11 less is known about their role in determining prognosis after alloHSCT. A cytogenetic scheme has been developed for AML patients undergoing alloHSCT,12 but the prognostic implications of somatic mutations in this setting have only been described for a limited set of common mutations.13 Understanding the prognostic value of an expanded repertoire of somatic mutations, both individually and in combination, is potentially useful for the identification of patients at high risk for relapse after transplant that may benefit from maintenance therapies or other experimental modalities to mitigate this risk. Since the prognostic implications of somatic mutations may differ in patients treated with chemotherapy versus alloHSCT, the significance of recurrent AML mutations needs to be specifically studied in this context.
In addition, the value of repeat molecular profiling at the time of relapse after alloHSCT has not been characterized. Analyzing clonal evolution of AML post-transplant may inform clinical decision-making by identifying previously unrecognized and actionable mutations as well as improve our understanding of the impact of the graft-versus-leukemia (GVL) immune response on molecularly-defined AML clones.
In this study, we describe the utility of hot-spot next generation sequencing (NGS) of genes that are commonly mutated in AML. We report the prognostic value of NGS for the first 112 patients with AML who had genetic profiling prior to alloHSCT at the University of Pennsylvania. We also report the value of repeating NGS at the time of relapse after alloHSCT in order to uncover clonal evolution and potential targets for therapy.
MATERIALS AND METHODS
We retrospectively studied 112 adult (≥ 18 years) AML patients who underwent targeted NGS of leukemia samples and later received related or unrelated alloHSCT from 2005 to 2015 at the Hospital of the University of Pennsylvania. Recipients of umbilical cord blood transplants were excluded. All transplants used T-cell replete grafts and standard calcineurin inhibitor-based graft-versus-host disease prophylaxis. Post-transplant maintenance was not routinely used, but patients with FLT3-ITD-positive AML who received a FLT3-inhibitor pre- and/or post-transplant were included. The Institutional Review Board approved the study and patients provided informed consent for data collection prior to transplant.
NGS was performed on either bone marrow or peripheral blood after confirming the presence of excess blasts. Of the 112 patients, 53 assays were performed retrospectively on samples banked prior to February 14, 2013 in the Tissue Bank of the Stem Cell and Xenograft Core at the University of Pennsylvania and 59 consecutive samples were analyzed as part of standard clinical care in patients diagnosed and treated at Penn on or after February 14, 2013. In 95 (85%) patients, NGS was performed at the time of initial leukemia diagnosis, in 13 (12%) patients at the time of first relapse, and in 4 (4%) patients at the time of persistent disease after induction therapy.
Genomic DNA was extracted from fresh or frozen (banked) bone marrow or peripheral blood samples using the Gentra Puregene cell kit (Qiagen, Venlo, Netherlands). The NGS panel included genes that are frequently mutated in hematologic malignancies, and the current analysis focuses on 26 genes commonly mutated in AML. Target regions of these genes (Supplemental Table 1) were sequenced using the TrueSeq Custom Amplicon (TSCA) (Illumina Inc, San Diego, California, USA). The sequencing libraries were prepared according to manufacturer instructions. Barcoded samples were pooled in equimolar amounts and subjected to 250-nucleotide paired-end sequencing an Illumina MiSeq (San Diego CA) to an average depth of 1 500x and minimal depth of 250x, with a validated calling threshold of 5% allele frequency. Sequence reads were aligned to the human genome (build 37) using a custom bioinformatics algorithm.14 Variant calling was categorized into 5 categories: disease-associated mutations, likely pathogenic mutations, variants of uncertain significance, likely benign, and benign. Disease-associated and likely pathogenic mutations were considered pathogenic for the purposes of this analysis. This categorization was denoted in our prospective internal knowledge database, a laboratory-generated database comparing variants detected to the information from publicly available sources such as PubMed, dbSNP database, COSMIC database, 1000 Genomes, and Exome Variant Server.
Conventional cytogenetic studies were successfully conducted concurrently in 109 of 112 (97%) patients and classified according to Armand et al.12 Additional disease characteristics included the white blood cell (WBC) count at presentation, time from diagnosis to transplant, and disease status immediately prior to transplant classified by percent blasts at pre-transplant marrow assessment (< or ≥ 5% blasts). Patient and transplant variables included donor and recipient age, sex, CMV status, donor type (sibling or unrelated), HLA mismatching, graft source (bone marrow or peripheral blood), and conditioning intensity (myeloablative or reduced intensity).15
Relapse was defined as morphologic evidence of leukemia with similar pre-transplant characteristics. All patients were followed post-transplant at our center where disease re-staging is standard at day 100, at 1 year, and whenever there is clinical suspicion of relapse. Time to relapse was the primary outcome and was determined from date of alloHSCT to date of relapse and analyzed as a cumulative incidence function taking into account non-relapse mortality as a competing risk. The impact of each mutation was analyzed using univariable competing risks regression and each mutation that crossed a threshold of P < .2 was then analyzed in an individual multivariable model. A correction for multiple testing was not performed due to the exploratory nature of this analysis and P ≤ .05 was considered significant. This study was powered to detect approximately a 2-fold or greater increase or reduction in hazard of relapse at the P = 0.05 significance level.
RESULTS
Patients Characteristics and Incidence of Somatic Mutations
Patient characteristics are presented in Table 1. The median follow-up was 23.8 months (range 0.9 – 152.9). Patients were transplanted in first remission (45%), second or later remission (22%), or not in remission (33%). Most patients (80%) had intermediate cytogenetic risk at diagnosis. Figure 1 shows the distribution of genes with pathogenic mutations in individual patients, categorized by cytogenetic risk. The frequency of mutated genes and a list of specific gene mutations in each patient are presented in Supplemental Tables 2 and 3. At least one pathogenic mutation was identified in 86% (96/112) of patients prior to transplant. The most commonly mutated genes were NPM1 (32%), FLT3-ITD (29%), DNMT3A (26%), TET2 (12%), NRAS (12%), IDH1 (12%), and IDH2 (11%). Patients with favorable cytogenetics were less likely to have pathogenic mutations compared to patients with intermediate or adverse cytogenetics (36% vs. 93% or 83%, respectively, P < .001 and P = .04).
Table 1.
Baseline Characteristics (n=112)
| Characteristic | n (%) |
|---|---|
| Disease | |
| Karyotype | |
| Favorable | 11 (10%) |
| Intermediate/unknown | 89 (79%) |
| Unfavorable | 12 (10%) |
| AML type | |
| De novo | 97 (87%) |
| Secondary | 15 (13%) |
| Disease status prior to transplant | |
| < 5% blasts | 75 (67%) |
| ≥ 5% blasts | 37 (33%) |
| WBC count at diagnosis | |
| < 100 ×103 / μL | 89 (82%) |
| ≥ 100 ×103 / μL | 19 (18%) |
| Unknown | 4 |
| Recipient | |
| Age, median (range, in years) | 55.5 (19 – 71) |
| Sex | |
| Male | 61 (54%) |
| Female | 51 (46%) |
| Ethnicity | |
| White | 69 (97%) |
| Black | 2 (3%) |
| Unknown | 41 |
| Donor | |
| Age, median (range, in years) | 40.5 (18 – 70) |
| Sex | |
| Male | 61 (54%) |
| Female | 51 (46%) |
| Transplant | |
| Median time from diagnosis to transplant (range, in months) | 6.2 (2.2 – 95.5) |
| Donor type | |
| Matched unrelated | 59 (53%) |
| Single allele mismatched unrelated | 9 (8%) |
| Matched sibling | 44 (39%) |
| Stem cell source | |
| Peripheral blood | 84 (75%) |
| Bone marrow | 28 (25%) |
| Conditioning intensity | |
| Myeloablative | 77 (69%) |
| Cyclophosphamide + total body irradiation | 44 (39%) |
| Busulfan + cyclophosphamide | 12 (11%) |
| Clofarabine + busulfan | 21 (19%) |
| Reduced intensity | 35 (31%) |
| Fludarabine + busulfan | 35 (31%) |
| CMV Risk | |
| Low risk (Donor and Recipient CMV seronegative) | 57 (51%) |
| High risk (Donor or Recipient CMV seropositive) | 54 (49%) |
| Unknown | 1 |
Abbreviations: WBC, white blood cell; AML, acute myeloid leukemia; CMV, cytomegalovirus
Figure 1. Distribution of somatic mutations in 112 patients with AML.
Each column represents an individual patient sample. The first row indicates grouping by cytogenetic risk (orange, adverse; blue, intermediate/unknown; white, favorable). Patients with mutations in each listed gene are indicated with a blue colored cell.
Impact of Somatic Mutations on Disease Relapse
Entire Cohort
We conducted univariable and multivariable regression analyses of relapse for mutated genes present with greater than 5% frequency in the entire cohort or combinations of mutated genes present with greater than 10% frequency (Table 2). Among the 112 patients, there were 50 relapse events, and 20 deaths of patients in remission. Considering the size of our cohort, we limited the number of additional covariates in each multivariable model to 2 and therefore included only cytogenetic risk and remission status, which are known to affect relapse risk.5, 12, 16-18 The inclusion of conditioning intensity as a third covariate did not alter the results (data not shown), except where noted. Notably, recipient age was not associated with relapse in this cohort and was therefore not included in the multivariable models.
Table 2.
Associations Between Common Mutated Genes and Hazard of Relapse.
| Entire Cohort (n=112) Univariable |
Entire Cohort (n=112) Multivariable* |
Patients in remission (n=75) Univariable |
Patients in remission (n=75) Multivariable* |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | HR Relapse | P | aHR Relapse | P | n | HR Relapse | P | aHR Relapse | P | |
| NPM1 | 36 | 1.06 (95% CI, 0.58 to 1.93) | .86 | 30 | 1.21 (95% CI, 0.59 to 2.47) | .60 | ||||
| FLT3-ITD | 32 | 1.53 (95% CI, 0.86 to 2.74) | .15 | 1.83 (95% CI, 0.95 to 3.51) | .07 | 23 | 1.74 (95% CI, 0.87 to 3.50) | .12 | 1.98 (95% CI, 0.91 to 4.29) | .09 |
| DNMT3A | 29 | 0.63 (95% CI, 0.31 to 1.28) | .20 | 0.60 (95% CI, 0.29 to 1.26) | .18 | 21 | 1.01 (95% CI, 0.46 to 2.21) | .97 | ||
| IDH1 | 13 | 0.93 (95% CI, 0.36 to 2.44) | .89 | 9 | 0.86 (95% CI, 0.25 to 3.01) | .82 | ||||
| NRAS | 13 | 0.81 (95% CI, 0.35 to 1.88) | .63 | 8 | 0.87 (95% CI, 0.30 to 2.51) | .80 | ||||
| TET2 | 13 | 1.19 (95% CI, 0.56 to 2.56) | .65 | 11 | 1.85 (95% CI, 0.82 to 4.16) | .14 | 1.58 (95% CI, 0.63 to 3.95) | .33 | ||
| IDH2 | 12 | 0.59 (95% CI, 0.24 to 1.42) | .24 | 10 | 0.87 (95% CI, 0.36 to 2.13) | .76 | ||||
| TP53 | 9 | 3.05 (95% CI, 1.40 to 6.65) | .005 | 2.90 (95% CI, 1.31 to 6.44) ∞ | .009∞ | 5 | 3.99 (95% CI, 1.17 to 13.58) | .03 | 3.35 (95% CI, 1.10 to 10.22) | .03 |
| ASXL1 | 8 | 0.58 (95% CI, 0.12 to 2.71) | .49 | 5 | 1.79 (95% CI, 0.35 to 9.15) | .48 | ||||
| WT1 | 8 | 2.07 (95% CI, 0.94 to 4.59) | .07 | 2.51(95% CI, 1.14 to 5.49) | .02 | 6 | 1.74 (95% CI, 0.68 to 4.42) | .25 | ||
| FLT3-ITD (+) NPM1 (+) | 18 | 1.22 (95% CI, 0.57 to 2.63) | .61 | 14 | 1.34 (95% CI, 0.56 to 3.20) | .51 | ||||
| FLT3-ITD (+) NPM1 (−) | 14 | 1.68 (95% CI, 0.85 to 3.29) | .13 | 1.84 (95% CI, 0.89 to 3.80) | .10 | 9 | 1.86 (95% CI, 0.87 to 3.99) | .11 | 1.97 (95% CI, 0.89 to 4.39) | .10 |
| FLT3-ITD (−) NPM1 (+) | 18 | 0.88 (95% CI, 0.40 to 1.94) | .76 | 16 | 0.99 (95% CI, 0.40 to 2.42) | .98 | ||||
| FLT3-ITD (+) DNMT3A (−) | 22 | 1.52 (95% CI, 0.82 to 2.81) | .18 | 1.70 (95% CI, 0.87 to 3.34) | .12 | 15 | 1.70 (95% CI, 0.83 to 3.48) | .15 | 1.83 (95% CI, 0.85 to 3.96) | .12 |
| FLT3-ITD (−) DNMT3A (+) | 17 | 0.44 (95% CI, 0.18 to 1.06) | .07 | 0.39 (95% CI, 0.15 to 1.02) | .055 | 13 | 0.82 (95% CI, 0.33 to 2.08) | .68 | ||
| NPM1 (+) DNMT3A (+) | 18 | 1.04 (95% CI, 0.47 to 2.33) | .92 | 16 | 1.13 (95% CI, 0.47 to 2.74) | .78 | ||||
| NPM1 (+) DNMT3A (−) | 18 | 1.05 (95% CI, 0.49 to 2.21) | .91 | 14 | 1.18 (95% CI, 0.50 to 2.81) | .71 | ||||
| FLT3-ITD (−) NPM1 (−) DNMT3A (+) | 11 | 0.28 (95% CI, 0.07 to 1.04) | .06 | 0.22 (95% CI, 0.05 to 0.93) | .04 | 5 | 0.76 (95% CI, 0.22 to 2.66) | .67 | ||
Significant p-values highlighted in bold
Adjusted for cytogenetic risk and remission status
Adjustment for cytogenetic risk not done due to interaction with TP53 mutations
Univariable analysis showed that the presence of a pathogenic mutation in TP53 was associated with an increased hazard of relapse after transplant (HR 3.05, P = .005). This association remained significant in multivariable analysis (adjusted HR (aHR) 2.90, P = .009). Because of an interaction between TP53 status and cytogenetic risk, we only adjusted these results for remission status. The median time to relapse in TP53-mutated AML was 3.3 months (95% CI, 1.6 to 18.6). The cumulative incidence of relapse in TP53-mutated AML patients vs. TP53-wild type AML patients is presented in Figure 2A.
Figure 2. Cumulative incidence plots of relapse in patients carrying mutations or combinations of mutations that impact the relapse risk.
A) TP53 mutated (blue) vs TP53 wt (red), B) WT1 mutated (blue) vs WT1 wt (red), C) DNMT3A mutated, NPM1-wt, FLT3-ITD negative (blue) vs all others (red), and D) FLT3-ITD positive (blue) vs FLT3-ITD negative (red)
Pathogenic mutations in WT1 were also associated with an increased hazard of relapse (HR 2.07, P = .07 in univariable and aHR 2.51, P = .02 in multivariable analysis) with relapse plot shown in Figure 2B. The median time to relapse for WT1-mutated AML patients was 5.3 months (95% CI, 1.45, to not reached).
The presence of a mutation in DNMT3A in the absence of FLT3-ITD and NPM1 mutations was associated with a lower hazard of relapse (aHR 0.22, P = .04, Figure 2C). Interestingly, the protective effect was not associated with the common R882 mutation (aHR 1.31, P = .74), but only with other pathogenic mutations in DNMT3A (aHR .14, P = .048).
FLT3-ITD was associated with an increased risk of relapse (aHR 1.83, P = .07, Figure 2D) when controlled for the cytogenetic risk and remission status. The association was statistically significant when the analysis was also controlled for conditioning regimen intensity (aHR 2.04, P = .047). In addition, we examined the use of FLT3 inhibitors prior to and/or after transplant as maintenance therapy. Of 32 patients with FLT3-ITD, 13 received a FLT3 inhibitor. Controlling for use of a FLT3 inhibitor did not significantly alter the impact of FTL3-ITD on the relapse risk (aHR 1.82, P = .13).
In summary, the 4 genes that were found to have associations with disease relapse (i.e., TP53, WT1, DNTM3A, and FLT3-ITD) provided prognostic information in 54 of 112 patients (48%), the majority of whom were transplanted in remission (Supplemental Table 4).
Patients Transplanted in Remission
We conducted a subgroup analysis in 75 patients who were transplanted with fewer than 5% blasts on a pre-transplant bone marrow aspirate (Table 2). Among these patients, TP53 was again a predictor of relapse (aHR 3.35, P = .03). FLT3-ITD was associated with higher relapse risk but without reaching statistical significance (aHR 1.98, P = .09). Mutated WT1 was not associated with an increased risk of relapse in this subgroup. Interestingly, DNMT3A(+) FLT3-ITD(−) NPM(−) status was not associated with a lower risk of relapse in this subset (aHR 0.76, P = .67), implying that the protective effect of this combination was seen primarily in patients transplanted not in remission. In our cohort, 6 patients with DNMT3A(+) FLT3-ITD(−) NPM(−) status were transplanted with active leukemia (≥5% blasts), and none of them relapsed (median follow-up 9.2 months, range 3.2 - 114.7 months).
Impact of Cytogenetic Risk Group
Subgroup analyses in patients with different cytogenetic risk were underpowered to detect the impact of specific mutations. Among 12 patients with unfavorable cytogenetic risk, 6 had a mutation in TP53. All 6 patients with TP53 mutations relapsed (range, 1.6 to 18.6 months after transplant), whereas of the 6 patients without TP53 mutations, 2 patients relapsed (at 2.0 and 7.5 months), 1 patient was alive at last follow-up (7.2 months), and 3 patients died from non-relapse causes (at 0.9, 9.0, and 14.7 months).
Among patients with intermediate cytogenetic risk, 3 patients had TP53 mutations. Of them, 2 are alive and in remission at last follow-up (5.3 and 22.7 months from transplant) and one had relapsed 6.0 months after transplant.
Clonal Evolution in Post-Transplant Relapse
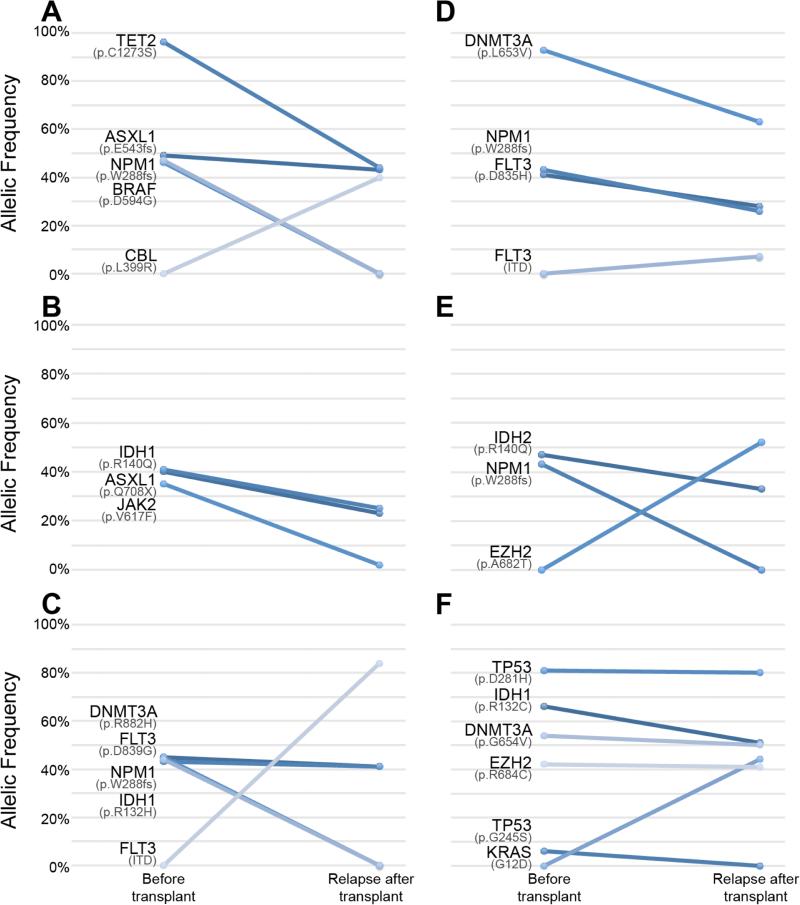
Four patients from our cohort underwent repeat NGS at the time of relapse after alloHSCT; in addition, 2 recipients of umbilical cord blood transplant who were not included in the primary analysis also had NGS conducted pre-transplant and at the time of relapse. In all 6 patients there was evidence for clonal evolution as summarized in Figure 3 (panels A-F each represent an individual patient) and Supplemental Table 5. The pre-transplant and relapse NGS data reveal several interesting and informative aspects about clonal evolution and alloHSCT.
Figure 3. Evolution of the genetic makeup of AML at the time of relapse after alloHSCT.
For each patient, allelic frequencies of mutations in the pre-transplant sample and post-transplant relapse sample are shown.
First, results from all 6 patients indicate that their leukemia was composed of several clones prior to transplant. This can be inferred not only by different allelic frequencies of the mutated genes prior to transplant, but also by changes in the ratios of the frequencies at the time of relapse. For example, patient 1 (Figure 3A) presented with 4 mutations pre-transplant, with 3 of them (ASXL1, BRAF and NPM1) at approximately 50% allele frequency and in addition TET2 at almost 100%, suggesting loss of heterozygosity of TET2 in the dominant clone. These results do not conclusively determine the number of clones. However, at relapse, 2 of the mutations (NPM1 and BRAF) were no longer detected, while the other 2 remained, suggesting that there were at least 2 distinct clones prior to transplant. Similar evidence for multiple clones can be seen in other patients.
In addition, these results indicate selective sensitivity of specific clones to alloHSCT. In 5 of the 6 patients we observe effective eradication or significant reduction of at least one mutated clone, whereas other clones remain dominant with similar allelic frequencies of their mutations at the time of relapse. Whether this clonal selection is induced by the conditioning regimen or the GVL response remains to be investigated.
Finally, new dominant clones have emerged in 5 of the 6 patients. Interestingly, in patient 3, FLT3-ITD appeared at the time of relapse, but was present at 1.51% allelic frequency at the time of initial diagnosis, which was considered below the reporting threshold and was reported as negative (Figure 3C).
With the emergence of new dominant clones, 4 patients acquired potentially actionable mutations (FLT3-ITD, KRAS, and EZH2) and 2 of these patients received FLT3 inhibitors on experimental protocols based on the genetic information available post-transplant.
DISCUSSION
In order to investigate whether certain somatic mutations in AML are associated with post-transplant relapse, we examined 26 frequently mutated genes in 112 pre-transplant samples from AML patients using a NGS panel currently in standard clinical use at our center. We found an association between the presence of a mutation in TP53 or WT1 and an increased risk of relapse, which was independent of other primary drivers of AML relapse after transplant such as remission status and cytogenetic risk. FLT3-ITD was associated with an increased risk of relapse, which was statistically significant after controlling for conditioning intensity. Finally, we identified a protective effect of DNMT3A mutations in the absence of co-occurring FLT3-ITD and NPM1 mutations. These findings suggest that genetic profiling provides important prognostic information for AML patients who undergo alloHSCT.
TP53 was a stronger predictor of relapse than all other recurrently mutated genes. Strikingly, all patients with a mutation in TP53 occurring in the context of adverse cytogenetics relapsed, and being in remission at the time of transplant did not eliminate the increased risk. TP53 mutations have been previously associated with resistance to chemotherapy in AML19 and were recently shown by Middeke et al. to increase risk for relapse after alloHSCT in patients with adverse cytogenetics.20 Our results are consistent with these results and imply that the GVL response does not effectively overcome treatment resistance. Experimental approaches to reduce relapse risk may be appropriate for patients with TP53-mutated AML.
To the best of our knowledge, poor outcome of alloHSCT in patients with WT1 mutations has not been previously described. WT1 is a known mediator of the GVL response and a target for immunotherapies in leukemia.21, 22 The etiology of frequent relapses for WT1-mutated AML is unclear. Mutations may impact the immunogenicity of WT1 and the poor prognosis may also be related to its role in epigenetic regulation in leukemic blasts.23
DNMT3A-mutated AML had favorable outcome with alloHSCT in our cohort, even in the setting of chemo-refractory disease. This finding implies that DNMT3A mutations confer sensitivity to the GVL response. Whether this sensitivity is mediated by expression of leukemia-associated antigens or a different epigenetic mechanism remains to be elucidated. Regardless of mechanism, the favorable outcome for alloHSCT in DNMT3A-mutated AML complements previous reports about a favorable response to high dose induction chemotherapy,9, 24 suggesting that the poor prognosis that was previously associated with this mutation11, 25 could potentially be overcome by using high dose anthracyclines followed by alloHSCT.
The adverse impact of FLT3-ITD in our cohort confirms previous studies.13, 26 Notably, the 2-fold increase in relapse risk for FLT3-ITD vs. non-FLT3-ITD patients was similar to previous reports despite the fact that 13 of 32 patients in our cohort received a FLT3 inhibitor pre- and/or post-transplant. The role of FLT3 inhibitors in patients undergoing alloHSCT will be better defined by ongoing clinical trials.
Our analysis also demonstrated clonal diversity, evolution, and selection in all 6 patients who had NGS performed again in the setting of relapse after alloHSCT. This is the first report describing clonal evolution in the setting of alloHSCT using a NGS assay currently in clinical practice. Similar to the non-transplant setting, where clonal evolution is thought to result from selective pressure induced by cytotoxic chemotherapy,27-29 here we see evidence that the GVL response eradicates certain AML clones but not others, supporting the idea of immunologic pressure leading to clonal selection. In addition to biologic insight, repeat NGS at the time of relapse post-transplant has actionable results. Based on current availability of clinical trials, 4 of these 6 patients acquired potentially actionable mutations (2 FLT3-ITD, 1 EZH2, and 1 KRAS), and two of them in fact received an experimental FLT3 inhibitor. These findings suggest that NGS should be repeated in patients who relapse after transplant in order to uncover potential therapeutic targets. This portion of the analysis was limited to a small subset of our cohort and therefore limits the strength of the conclusions; we hope this descriptive report will inspire further prospective investigation into clonal evolution of AML post-alloHSCT.
The main limitations of our study are its small sample size, heterogeneity of the cohort and retrospective nature; therefore, these results require prospective validation in larger cohorts. Additional work is also needed to better define the number of genes that should be sequenced in AML in order to identify prognostic lesions at diagnosis as well as targetable lesions at both diagnosis and relapse. The optimal coverage of each gene is also not well defined. For example, our panel did not sequence the entire WT1 gene, and as a result we may have under-estimated the impact of WT1 mutations. Finally, the value of NGS for minimal residual disease assessment is rapidly being established for chemotherapy-treated AML,30, 31 and needs to be investigated in alloHSCT recipients but was not possible with the current dataset. With these limitations in mind, this report demonstrates the value of NGS in the management of alloHSCT recipients. This is an important area of unmet need and prospective studies should be encouraged.
Supplementary Material
Highlights.
TP53, WT1, and FLT3-ITD mutations are associated with AML relapse after alloHSCT.
DNMT3A(+) FLT3-ITD(−) NPM1(−) status confers lower risk of post-alloHSCT relapse.
Clonal evolution is present in AML at time of post-alloHSCT relapse.
Repeat genetic profiling at relapse may uncover acquired actionable mutations.
ACKNOWLEDGEMENTS
Financial disclosure: The authors gratefully acknowledge the grant support of T32CA009679 (M.R.L), the National Marrow Donor Program (Amy Strelzer Manasevit Award for the study of post-transplant complications to R.R.), Department of Defense (Career Development Award to R.R.), National Cancer Institute (K23CA178202 to R.R. and P30CA016520 to D.L.P.), Conquer Cancer Foundation (Career Development Award to R.R.), a Hematologic Malignancies Translational Center of Excellence Grant from the Abramson Cancer Center and the Margie and Andy Rooke fund for Leukemia Research (R.R. and D.L.P.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement: There are no conflicts of interest to report.
REFERENCES
- 1.Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 2.Grimwade D, Hills RK, Moorman AV, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2012;116:354–365. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 3.Grimwade D, Walker H, Oliver F, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood. 1998;92:2322–2333. [PubMed] [Google Scholar]
- 4.Byrd JC, Mrozek K, Dodge RK, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood. 2002;100:4325–3236. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 5.Slovak ML, Kopecky KJ, Cassileth PA, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood. 2000;96:4075–4083. [PubMed] [Google Scholar]
- 6.Schlenk RF, Dohner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–1198. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 7.Mrozek K, Marcucci G, Nicolet D, et al. Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. J Clin Oncol. 2012;30:4515–4523. doi: 10.1200/JCO.2012.43.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dohner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 9.Patel JP, Gonen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein K, Kaspers G, Harrison CJ, et al. Clinical impact of additional cytogenetic aberrations, cKIT and RAS mutations, and treatment elements in pediatric t(8;21)-AML: results from an international retrospective study by the International Berlin-Frankfurt-Munster Study Group. J Clin Oncol. 2015;33:4247–4258. doi: 10.1200/JCO.2015.61.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armand P, Kim HT, Zhang MJ, et al. Classifying cytogenetics in patients with acute myelogenous leukemia in complete remission undergoing allogeneic transplantation: a Center for International Blood and Marrow Transplant Research study. Biol Blood Marrow Transplant. 2012;18:280–288. doi: 10.1016/j.bbmt.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmid C, Labopin M, Socie G, et al. Outcome of patients with distinct molecular genotypes and cytogenetically normal AML after allogeneic transplantation. Blood. 2015;126:2062–2069. doi: 10.1182/blood-2015-06-651562. [DOI] [PubMed] [Google Scholar]
- 14.Daber R, Sukhadia S, Morrissette JJ. Understanding the limitations of next generation sequencing informatics, an approach to clinical pipeline validation using artificial data sets. Cancer Genet. 2013;206:441–448. doi: 10.1016/j.cancergen.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Bacigalupo A, Ballen K, Rizzo D, et al. Defining the intensity of conditioning regimens: working definitions. Biol Blood Marrow Transplant. 2009;15:1628–1633. doi: 10.1016/j.bbmt.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlenk RF, Dohner K, Mack S, et al. Prospective evaluation of allogeneic hematopoietic stem-cell transplantation from matched related and matched unrelated donors in younger adults with high-risk acute myeloid leukemia: German-Austrian trial AMLHD98A. J Clin Oncol. 2010;28:4642–4648. doi: 10.1200/JCO.2010.28.6856. [DOI] [PubMed] [Google Scholar]
- 17.Duval M, Klein JP, He W, et al. Hematopoietic stem-cell transplantation for acute leukemia in relapse or primary induction failure. J Clin Oncol. 2010;28:3730–3738. doi: 10.1200/JCO.2010.28.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasquini MC, Zhang MJ, Medeiros BC, et al. Hematopoietic cell transplantation outcomes in monosomal karyotype myeloid malignancies. Biol Blood Marrow Transplant. 2016;22:248–257. doi: 10.1016/j.bbmt.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong TN, Ramsingh G, Young AL, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;518:552–555. doi: 10.1038/nature13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Middeke JM, Herold S, Rucker-Braun E, et al. TP53 mutation in patients with high-risk acute myeloid leukaemia treated with allogeneic haematopoietic stem cell transplantation. Br J Haematol. 2016;172:914–22. doi: 10.1111/bjh.13912. [DOI] [PubMed] [Google Scholar]
- 21.Rezvani K, Yong AS, Savani BN, et al. Graft-versus-leukemia effects associated with detectable Wilms tumor-1 specific T lymphocytes after allogeneic stem-cell transplantation for acute lymphoblastic leukemia. Blood. 2007;110:1924–1932. doi: 10.1182/blood-2007-03-076844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Stasi A, Jimenez AM, Minagawa K, et al. Review of the results of WT1 peptide vaccination strategies for myelodysplastic syndromes and acute myeloid leukemia from nine different studies. Front Immunol. 2015;6:36. doi: 10.3389/fimmu.2015.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rampal R, Alkalin A, Madzo J, et al. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep. 2014;9:1841–1855. doi: 10.1016/j.celrep.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luskin MR, Lee JW, Fernandez HF, et al. Benefit of high dose daunorubicin in AML induction extends across cytogenetic and molecular groups. Blood. 2016;127:1551–1558. doi: 10.1182/blood-2015-07-657403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thol F, Damm F, Ludeking A, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. 2011;29:2889–2896. doi: 10.1200/JCO.2011.35.4894. [DOI] [PubMed] [Google Scholar]
- 26.Brunet S, Labopin M, Esteve J, et al. Impact of FLT3 internal tandem duplication on the outcome of related and unrelated hematopoietic transplantation for adult acute myeloid leukemia in first remission: a retrospective analysis. J Clin Oncol. 2012;30:735–741. doi: 10.1200/JCO.2011.36.9868. [DOI] [PubMed] [Google Scholar]
- 27.Ding L, Ley TJ, Larson DE, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welch JS, Ley TJ, Link DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jan M, Snyder TM, Corces-Zimmerman MR, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med. 2012;4:149ra118. doi: 10.1126/scitranslmed.3004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klco JM, Miller CA, Griffith M, et al. Association between mutation clearance after induction therapy and outcomes in acute myeloid leukemia. JAMA. 2015;314:811–822. doi: 10.1001/jama.2015.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivey A, Hills RK, Simpson MA, et al. Assessment of minimal residual disease in standard-risk AML. N Engl J Med. 2016;374:422–433. doi: 10.1056/NEJMoa1507471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.