Abstract
DNA alkylation represents a major type of DNA damage and is generally unavoidable due to ubiquitous exposure to various exogenous and endogenous sources of alkylating agents. Among the alkylated DNA lesions, O2-alkylthymidines (O2-alkyldT) are known to be persistent and poorly repaired in mammalian systems, and have been shown to accumulate in esophagus, lung and liver tissue of rats treated with tobacco-specific N-nitrosamines, i.e. 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and N′-nitrosonornicotine (NNN). In this study, we assessed the replicative bypass of a comprehensive set of O2-alkyldT lesions, with the alkyl group being a Me, Et, nPr, iPr, nBu, iBu or sBu, in template DNA by conducting primer extension assays with the use of major translesion synthesis DNA polymerases. The results showed that human Pol η and, to a lesser degree, human Pol κ, but not human polymerase ι or yeast polymerase ζ, were capable of bypassing all O2-alkyldT lesions and extending the primer to generate full-length replication products. Data from steady-state kinetic measurements showed that human Pol η exhibited high frequencies of misincorporation of dCMP opposite those O2-alkyldT lesions bearing a longer straight-chain alkyl group. However, the nucleotide misincorporation opposite branched-chain lesions was not selective, with dCMP, dGMP, and dTMP being inserted at similar efficiencies, though the total frequencies of nucleotide misincorporation opposite the branched-chain lesions differed and followed the order of O2-iPrdT > O2-iBudT > O2-sBudT. Together, the results from the present study provided important knowledge about the effects of the length and structure of the alkyl group in the O2-alkyldT lesions on the fidelity and efficiency of DNA replication mediated by human Pol η.
Keywords: DNA alkylation, human Pol η, translesion synthesis, O2-alkyldT
Graphical Abstract
Introduction
According to the Center for Disease Control and Prevention, smoking is considered the leading cause of preventable diseases, and it accounts for more than 480,000 premature deaths per year in the United States alone.1 Cigarette smoke contains more than 5,000 chemical species, 73 of which are considered carcinogenic to laboratory animals and/or humans.2 Among these compounds are various polycyclic aromatic hydrocarbons and tobacco-specific N-nitrosamines, which, after metabolic activation, are known to alkylate DNA.2, 3 Depending on the chemical nature of the alkylating agent involved, the size of the alkyl functionality adducted to DNA ranges from simple methyl and ethyl groups, e.g. those arising from ethylating agents present in tobacco,3 to more complex alkyl groups from the tobacco-specific N-nitrosamines. In the latter respect, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and N′-nitrosonornicotine (NNN) can be metabolized by cytochrome P450 enzymes and ultimately result in the addition of a pyridyloxobutyl (POB) or pyridylhydroxybutyl (PHB) group to thymine, cytosine and guanine in DNA.4
Among the various alkylated DNA lesions, O2-alkylthymidines (O2-alkyldT) are known to be resistant to repair and thus persist in mammalian systems.5–7 For instance, O2-POBdT and O2-PHBdT could accumulate in the esophagus, lung and liver tissue of rats treated with NNK and NNN, and were detectable in various human tissues.2, 3, 8–10 Additionally, the levels of O2-ethylthymidine in human leukocyte DNA were found to be significantly higher in smokers than nonsmokers.3 If remained unrepaired, these alkylated DNA adducts may perturb genomic integrity by impeding DNA replication and transcription, and inducing mutations in these processes.2
Unrepaired DNA adducts may stall the replication fork, which results in cell cycle arrest and allows time for the cell to repair the damaged DNA.11–15 Nevertheless, some DNA lesions are more difficult to repair and may result in prolonged stalling of the replication fork, thereby leading to apoptosis. In order to avoid apoptosis, cells are equipped with translesion synthesis (TLS) DNA polymerases which possess more spacious and flexible active sites than replicative DNA polymerases to facilitate lesion bypass.16, 17 The TLS polymerases include polymerases η, ι, κ and Rev1 in the Y-family, and polymerase ζ in the B-family. These polymerases function in the bypass of various DNA lesions, and some are known to bypass specific DNA lesions with similar or better fidelity and efficiency than the corresponding unmodified nucleosides.16–21 In this vein, polymerase η (Pol η) has been shown to be highly efficient and accurate when bypassing the UV-induced thymine-thymine cyclobutane pyrimidine dimers, which is recognized as the major role of Pol η.16, 19, 20, 22, 23 The importance of this polymerase is manifested in patients suffering from the variant form of xeroderma pigmentosum (XPV), and these individuals display elevated UV-induced mutagenesis and susceptibility toward developing skin cancer because they carry inactivating mutations in POLH gene which encodes for Pol η.22–24 Although this is recognized as the main functional role of Pol η, the polymerase is capable of bypassing many other DNA lesions with different efficiency and fidelity.25, 26
Previous studies showed that O2-EtdT is partially blocking to DNA synthesis mediated by T7 DNA polymerase in vitro, and both dAMP and dTMP can be incorporated opposite the lesion.27 Recently, our laboratory showed that polymerase V (an ortholog of human polymerase η) plays a major role in the bypass of various O2-alkylthymidine lesions in Escherichia coli, and the polymerase was indispensable for the lesion-induced T→A and T→G mutations in E. coli cells.28 In addition, Basu and coworkers29 showed that Pol η promotes the replication across the O2-MedT and O2-POBdT lesions in human cells. Thus, in the present study, we chose to focus on human Pol η by characterizing biochemically how the efficiency and fidelity of nucleotide insertion opposite the O2-alkyldT lesions are modulated by the length and structure of the alkyl group.
Experimental Procedures
Materials
Human Pol η, κ, and yeast Pol ζ were purchased from Enzymax (Lexington, KY), and the recombinant full-length human Pol ι was kindly provided by Prof. Linlin Zhao (Central Michigan University). All other enzymes were obtained from New England BioLabs (Ipswich, MA) and unmodified oligodeoxyribonucleotides (ODNs) were acquired from Integrated DNA Technologies (Coralville, IA). [γ-32P]-ATP was obtained from Perkin-Elmer (Boston, MA), and all other chemicals were from Sigma-Aldrich (St. Louis, MO).
Substrate preparation
20-mer lesion-containing ODNs were generated by ligating 12-mer O2-alkyldT-containing ODNs d(ATGGCGXGCTAT), where ‘X’ designates the O2-alkyldT with the alkyl group being a Me, Et, nPr, nBu, iPr, iBu or sBu (Figure 1)28 to an 8-mer ODN d(GATCCTAG) in the presence of a 27-mer scaffold d(GTAGCTAGGATCATAGCACGCCATTAG), as previously described.25 All ligation products were then purified by polyacrylamide gel electrophoresis (PAGE) and annealed to a 13-mer 32P-labeled primer (10 nM) to yield the primer-template complex for in vitro primer extension and steady-state kinetic measurements (Figure 1).
Figure 1.
(a) The structures of the O2-alkyldT lesions examined in this study and (b) the primer-template complex used for the in vitro primer extension and steady-state kinetic assays, X= dT, O2-MedT, O2-EtdT, O2-nPrdT, O2-iPrdT, O2-nBudT, O2-iBudT or O2-sBudT.
Primer extension assays
Primer extension assays were performed under standing-start conditions by incubating the aforementioned primer-template complex (at a final concentration of 10 nM) at 37°C for 1 hr with various concentrations of human Pol η (Figure 2), κ (Figure S1) and ι (Figure S2), or for 5 hr with yeast Pol ζ (Figure S3), all four dNTPs (250 µM each), MgCl2 (5 mM) and a reaction buffer. The reaction buffer contained 25 mM potassium phosphate (pH 7.0), 5 mM MgCl2, 5 mM DTT, 100 µg/ml BSA and 10% glycerol.30 The reaction was then terminated by adding an equal volume of formamide gel-loading buffer [80% formamide, 10 mM EDTA (pH 8.0), 1 mg/ml xylene cyanol and 1 mg/ml bromophenol blue]. The reaction mixtures were subsequently resolved on a 20% (19:1) denaturing polyacrylamide gel and the gel band intensities analyzed using a Typhoon 9410 Variable Mode Imager (Amersham Biosciences Co.).
Figure 2.
Representative gel images from the primer extension assays under standing-start conditions for primer-template complexes containing an unmodified dT, O2-MedT, O2-EtdT, O2-nPrdT, O2-iPrdT, O2-nBudT, O2-iBudT or O2-sBudT with human Pol η. The final concentration of the primer-template complex was 10 nM, and the final concentrations of human Pol η are indicated.
Steady-state kinetic assay
Standing-start steady-state kinetic assays were performed following previously published procedures.31 The control or lesion-containing primer-template complexes (at a final concentration of 10 nM) were incubated with the above-mentioned reaction buffer, 5 nM human Pol η and various concentrations of individual dNTPs at 37°C for 10 min (Figure 3, S4 and S5). In this vein, the dNTP concentrations were optimized to allow for less than 20% nucleotide incorporation opposite the lesion or the corresponding unmodified nucleoside site. The reaction was again terminated by adding an equal volume of formamide gel-loading buffer. Reaction mixtures were then resolved on a 20% (19:1) denaturing PAGE, and gel-band intensities quantified by phosphorimaging analysis, as described above.
Figure 3.
Representative gel images for steady-state kinetic assays in measuring the individual nucleotide incorporation opposite unmodified dT (a), O2-nPrdT (b) and O2-nBudT (50 nM) (c) with human Pol η. The final concentration of the primer-template complex was 10 nM, and the final concentration of human Pol η was 5 nM. The highest concentrations of individual dNTPs used are indicated in the figure, and the concentration ratio between neighboring lanes was 0.50.
From the gel images, we first determined the observed rate for nucleotide incorporation, Vobs, by dividing the quantified amount of product formed by the incubation time (i.e. 10 min).30, 32, 33 The steady-state kinetic parameters (i.e. Vmax and Km) for nucleotide incorporation were then determined by plotting Vobs versus dNTP concentration and fit to the Michaelis-Menten equation using Origin 6.0 (Origin-Lab, Northampton, MA):31
The kcat values were calculated by dividing Vmax with the concentration of human Pol η employed. The efficiency of nucleotide incorporation was determined by the ratio of kcat/Km, and the frequency of incorrect nucleotide insertion (finc) was calculated from the ratio of kcat/Km obtained for the insertion of incorrect nucleotide over that for the correct nucleotide incorporation:31
It is of note that the 3′ flanking nucleobase of the lesion in the template is a guanine; as a result, we observed, apart from the product with a single dCMP insertion, the product arising from the incorporation of two dCMPs. We included both products for the determination of the steady-state kinetic parameters for dCMP incorporation.
Results
In this study, we aimed to investigate how the structures of the alkyl group in O2-alkyldT lesions influence the replicative bypass mediated by human Pol η, κ, ι and yeast Pol ζ.
Primer extension assay
Primer extension assays were first conducted to assess human Pol η, κ, ι and yeast Pol ζ’s ability to extend a 13-mer primer in the presence of a 20-mer template containing an unmodified dT or site-specifically inserted O2-alkyldT. The results showed that, when all four dNTPs are present, human Pol η was capable of successfully bypassing all seven O2-alkyldT lesions and generating full-length extension products (Figure 2). Quantification of the full-length extension product revealed that human Pol η-mediated primer extension was impeded to a lesser degree by those lesions carrying a branched-chain alkyl group than those with the corresponding straight-chain alkyl group, i.e. O2-iPrdT > O2-nPrdT (53% and 29%, respectively) and O2-iBudT, O2-sBudT > O2-nBudT (22%, 16% and 6.4% respectively) (Table S1). The values represent the percentage of full-length product observed for the lesion-containing substrate relative to that found for the control substrate. Pol κ was also able to bypass all the O2-alkyldT lesions and extend the primer to the end of the DNA template, but the main products formed were 18mer and 19mer products (Figure S1 and Table S1). Pol ι was able to generate shorter 18mer extension products for the templates harboring an O2-MedT or O2-EtdT; this polymerase, however, was incapable of extending further the primer after inserting one nucleotide opposite O2-nPrdT, O2-iPrdT, O2-nBudT, O2-iBudT or O2-sBudT (Figure S2 and Table S1). Yeast Pol ζ, on the other hand, was capable of producing full-length product when bypassing O2-MedT and O2-EtdT (at 50% and 7.4%, respectively), but was blocked by the other O2-alkyldT lesions (Figure S3 and Table S1). Thus, Pol η is the most efficient TLS polymerase involved in bypassing the O2-alkyldT lesions in vitro and was thus chosen for steady-state kinetic analysis.
Steady-state kinetic analysis
We next performed steady-state kinetic assays to assess the efficiency and fidelity of human Pol η in inserting nucleotides opposite the O2-alkyldT lesions (Table 1 and Figures 3, S4, and S5). The results showed that, among the four natural nucleotides, dAMP was inserted opposite the O2-MedT and O2-EtdT at the highest efficiency, though the presence of the two lesions reduces markedly the efficiency of dAMP incorporation, i.e. at 2.09% and 2.15%, respectively, relative to the corresponding insertion for the unmodified control substrate. Additionally, our results showed that human Pol η was more efficient in incorporating the correct nucleotide opposite the O2-alkyldT lesions with a branched-chain alkyl group (i.e. O2-iPrdT, O2-iBudT and O2-sBudT, at 0.42%, 0.34% and 0.18%, respectively, Figure 4) than the corresponding lesions with a straight-chain alkyl group (i.e. O2-nPrdT and O2-nBudT, at 0.08% and 0.07%, respectively, Figure 4).
Table 1.
Steady-state kinetic parameters for human Pol η-mediated incorporation of individual dNTPs opposite the O2-MedT, O2-EtdT, O2-nPrdT, O2-iPrdT, O2-nBudT, O2-iBudT and O2-sBudT and unmodified dT substrates. The results are shown as the mean ± standard deviation of results from at least three independent measurements.
| dNTP | kcat (min−1) | Km (µM) |
kcat/Km (µM−1min−1) |
finca | |
|---|---|---|---|---|---|
| Undamaged dT Substrate | |||||
 |
dTTP | 1.2×10−2 ± 2×10−3 | 12 ± 2 | 9.6×10−4 ± 7×10−5 | 2.0×10−3 |
| dGTP | 5.3×10−2 ± 7×10−3 | 9.2 ± 1 | 5.9×10−3 ± 1×10−3 | 1.2×10−2 | |
| dCTP | 2.5×10−2 ± 4×10−3 | 23 ± 1 | 1.1×10−3 ± 2×10−4 | 2.1×10−3 | |
| dATP | 1.5×10−2 ± 2×10−3 | 3.0×10−2 ± 4×10−4 | 5.1×10−1 ± 8×10−2 | 1.0 | |
| O2-MedT | |||||
 |
dTTP | 5.6×10−2 ± 1×10−2 | 4.8×10+2 ± 8×10+1 | 1.2×10−4 ± 3×10−5 | 1.1×10−2 |
| dGTP | 3.5×10−2 ± 3×10−3 | 89 ± 7 | 3.9×10−4 ± 1×10−5 | 3.7×10−2 | |
| dCTP | 3.7×10−2 ± 3×10−3 | 1.4×10+2 ± 7 | 2.6×10+4 ± 9×10−6 | 2.5×10−2 | |
| dATP | 4.5×10−2 ± 6×10−4 | 4.3×10−1 ± 4×10−2 | 1.1×10+−2 ± 1×10−3 | 1.0 | |
| O2-EtdT | |||||
 |
dTTP | 3.0×10−3 ± 3×10−4 | 22 ± 5 | 1.4×10−4 ± 2×10−5 | 1.3×10−2 |
| dGTP | 2.3×10−2 ± 3×10−3 | 23 ± 4 | 1.0×10−3 ± 1×10−4 | 9.2×10−2 | |
| dCTP | 1.5×10−2 ± 2×10−3 | 48 ± 3 | 3.1×10−4 ± 2×10−5 | 2.9×10−2 | |
| dATP | 8.1×10−3 ± 4×10−4 | 0.77 ± 0.1 | 1.1×10−2 ± 1×10−3 | 1.0 | |
| O2-nPrdT | |||||
 |
dTTP | 2.7×10−2 ± 3×10−3 | 4.4×10+2 ± 6×10+1 | 6.1×10−5 ± 4×10−6 | 1.5×10−1 |
| dGTP | 1.6×10−2 ± 3×10−3 | 1.8×10+2 ± 4 | 8.9×10−5 ± 2×10−5 | 2.2×10−1 | |
| dCTP | 2.2×10−2 ± 3×10−3 | 70 ± 1×10+1 | 3.1×10−4 ± 2×10−5 | 7.6×10−1 | |
| dATP | 3.6×10−2 ± 3×10−3 | 89 ± 1×10+1 | 4.1×10−4 ± 4×10−5 | 1.0 | |
| O2-iPrdT | |||||
 |
dTTP | 4.9×10−3 ± 4×10−4 | 15 ± 2 | 3.4×10−4 ± 2.1×10−5 | 1.6×10−1 |
| dGTP | 2.3×10−2 ± 3×10−2 | 60 ± 7 | 3.9×10−4 ± 2.9×10−5 | 1.9×10−1 | |
| dCTP | 3.4×10−2 ± 1×10−3 | 52 ± 8 | 6.5×10−4 ± 7.9×10−5 | 3.1×10−1 | |
| dATP | 2.4×10−2 ± 3×10−3 | 11 ± 1 | 2.1×10−3 ± 2.2×10−4 | 1.0 | |
| O2-nBudT | |||||
 |
dTTP | 1.7×10−2 ± 2×10−3 | 3.5×10+2 ± 4×10+1 | 4.8×10−5 ± 8×10−6 | 1.4×10−1 |
| dGTP | 2.5×10−2 ± 3×10−3 | 4.4×10+2 ± 6×10+1 | 5.6×10−5 ± 2×10−6 | 1.7×10−1 | |
| dCTP | 1.3×10−2 ± 2×10−3 | 38 ± 6 | 3.4×10−4 ± 4×10−5 | 1.0 | |
| dATP | 2.0×10−2 ± 5×10−3 | 88 ± 7 | 3.4×10−4 ± 4×10−5 | 1.0 | |
| O2-iBudT | |||||
 |
dTTP | 3.9×10−3 ± 6×10−4 | 16 ± 3 | 2.4×10−4 ± 1×10−5 | 1.4×10−1 |
| dGTP | 2.3×10−2 ± 3×10−3 | 69 ± 7 | 3.3×10−4 ± 4×10−5 | 1.9×10−1 | |
| dCTP | 3.1×10−2 ± 5×10−3 | 73 ± 15 | 4.2×10−4 ± 4×10−5 | 2.5×10−1 | |
| dATP | 8.7×10−3 ± 2×10−3 | 5.1± 1 | 1.7×10−3 ± 2×10−4 | 1.0 | |
| O2-sBudT | |||||
 |
dTTP | 2.1×10−2 ± 3×10−3 | 2.2×10+2 ± 2×10+1 | 9.3×10−5 ± 2×10−5 | 1.0×10−1 |
| dGTP | 2.5×10−2 ± 2×10−2 | 3.1×10+3 ± 6×10+1 | 8.1×10−5 ± 1×10−5 | 8.8×10−2 | |
| dCTP | 3.1×10−2 ± 2×10−3 | 90 ± 9 | 1.3×10−4 ± 3×10−5 | 1.4×10−1 | |
| dATP | 8.7×10−2 ± 5×10−3 | 32 ± 3 | 9.2×10−4 ± 1×10−4 | 1.0 | |
Frequency of nucleotide misincorporation = [kcat/Km (incorrect nucleotide)]/[kcat/Km(correct nucleotide)]
Figure 4.
The efficiencies for the human Pol η-catalyzed insertion of the correct nucleotide, dAMP, opposite unmodified dT, O2-MedT, O2-EtdT, O2-nPrdT, O2-iPrdT, O2-nBudT, O2-iBudT or O2-sBudT (relative to dT). The results represent the mean ± standard deviation of results from at least three independent measurements.
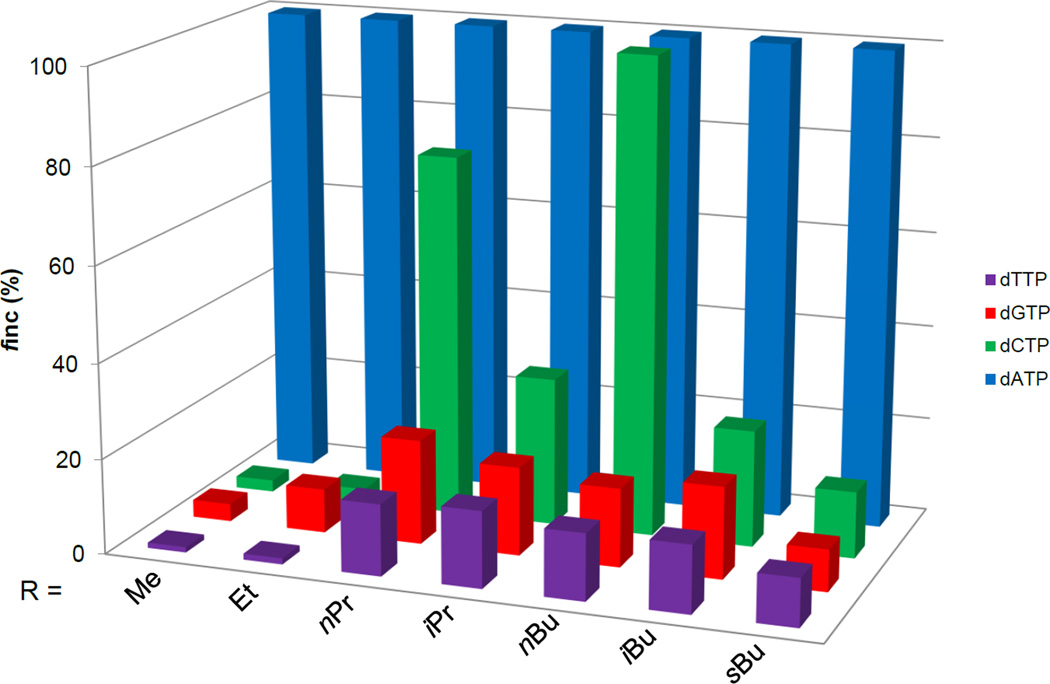
We next analyzed the differences in human Pol η-mediated incorporation of incorrect nucleotides opposite the various O2-alkyldT lesions. Human Pol η displayed high fidelity when bypassing O2-MedT and O2-EtdT. However, human Pol η incorporates the incorrect dCMP opposite O2-nPrdT and O2-nBudT at relatively high frequencies (76% and 100%, respectively, relative to the incorporation of the correct dAMP). In contrast, all the O2-alkyldT lesions with a branched-chain alkyl functionality directed primarily promiscuous nucleotide misincorporation, where no marked preference was found for the misincorporation of dCMP, dGMP or dTMP (Table 1 and Figure 5).
Figure 5.
Relative efficiencies for the Pol η-mediated nucleotide incorporation opposite dT, O2-MedT, O2-EtdT, O2-nPrdT, O2-iPrdT, O2-nBudT, O2-iBudT and O2-sBudT.
Discussion
In the present study, we investigated the replicative bypass of O2-MedT, O2-EtdT, O2-nPrdT, O2-iPrdT, O2-nBudT, O2-iBudT and O2-sBudT by major TLS polymerases, including human Pol η, κ, ι and yeast Pol ζ. We uncovered the different capabilities of the TLS polymerases in bypassing these lesions and generating full-length extension products. We also demonstrated that the structure of the alkyl group attached to the O2 position of thymidine influences the efficiency and fidelity of human Pol η-catalyzed nucleotide incorporation in vitro.
We found that, in the mutual presence of all four dNTPs, human Pol η could bypass all O2-alkyldT lesions and generate full-length replication products (Figure 2). In addition, human Pol η was found to generate more full-length replication product when bypassing those O2-alkyldT lesions bearing a branched-chain alkyl group relative to their straight-chain counterparts, i.e. O2-iPrdT > O2-nPrdT and O2-iBudT, O2-sBudT > O2-nBudT (Table S1). These results suggest that the O2-alkyldT lesions with a branched-chain alkyl group are more readily bypassed by human Pol η than the corresponding straight-chain lesions.
Human Pol κ mainly generated shorter 18mer and 19mer products, though small amounts of full-length extension product could also be detected (Figure S1 and Table S1). Our results about the difficulty of Pol κ in bypassing the O2-alkyldT lesions is in line with previous findings. Pol κ distinguishes itself from the other Y-family polymerases by its capability in bypassing efficiently and accurately dG adducts with various sizes of alkyl groups being attached to the minor-groove N2 position34–36 as well as an N2-N2-guanine interstrand cross-link.37 These abilities are due to the unique structure of Pol κ, whose active site can readily accommodate minor-groove modifications in DNA.34, 38 At first glance, this may suggest that Pol κ is capable of bypassing O2-alkyldT lesions. However, a number of recent studies showed that O2-MedT, O2-EtdT and O2-POBdT are not good substrates for Pol κ or its ortholog in E. coli, i.e. Pol IV.28, 29, 36, 39–41 This may be attributed to the fact that, unlike the N2-modified dG derivatives which still pair favorably with dCTP, the O2-alkyldT lesions do not have strong tendency to form Watson-Crick hydrogen bonding with any of the four canonical nucleotides.28, 29, 36, 39–41
Pol ι, however, was unable to generate full-length replication products, as reflected by the lack of further extension of the primer after insertion of a single nucleotide opposite all the lesions except O2-MedT and O2-EtdT, where Pol ι could generate a shorter 18mer product for templates containing the latter two lesions (Figure S2). Yeast Pol ζ, on the other hand, was capable of yielding full-length replication products as it bypasses O2-MedT and O2-EtdT, with more full-length extension product generated for the former substrate, but was completely blocked by all other O2-alkyldT lesions (Table S1 and Figure S3).
Our steady-state kinetic assay results revealed that the efficiency and fidelity of human Pol η-mediated nucleotide insertion opposite the O2-alkyldT lesions are influenced by the structure of the alkyl group. In particular, we found that human Pol η inserts dAMP opposite O2-MedT and O2-EtdT (relative to dT) more efficiently than the other O2-alkyldT lesions. Interestingly, human Pol η exhibited higher efficiencies in incorporating the correct nucleotide opposite the O2-alkyldT lesions possessing a branched-chain alkyl group, i.e. O2-iPrdT, O2-iBudT and O2-sBudT when compared to their straight-chain counterparts. We also determined the efficiency of human Pol η in incorporating the incorrect nucleotides opposite these minor-groove lesions. We observed that the incorrect nucleotide, dCMP, was incorporated at high frequency opposite the longer straight-chain lesions, i.e. O2-nPrdT and O2-nBudT. In contrast, human Pol η exhibited no strong preference in the incorporation of the incorrect nucleotides opposite the lesions with branched alkyl groups.
These results, in conjunction with previous findings about the major role of Pol V in bypassing the same O2-alkyldT lesions in E. coli, and Pol η’s function in bypassing the O2-MedT and O2-POB-dT in HEK293T human embryonic kidney cells, indicate that human Pol η may be the main TLS polymerase involved in the replicative bypass of the O2-alkyldT lesions.28, 29 Human Pol η is known to accurately bypass thymine-thymine cyclobutane pyrimidine dimers (CPD) with a similar accuracy and efficiency as bypassing the corresponding unmodified nucleosides.16–21 Pol η is unique among the Y-family polymerases due to its spacious active site which has the ability to accommodate the two cross-linked thymine bases in the CPD lesion.36 Additionally, human Pol η is capable of promoting replication through many other DNA lesions including O6-alkyldG, 8-oxodG, O4-alkyldT, O2-MedT and O2-POBdT.30, 36, 41, 42 This may explain why human Pol η is able to incorporate a nucleotide opposite all seven O2-alkyldT lesions and extend past them. Moreover, the high frequency of misincorporation of dCMP opposite the longer straight-chain lesions, i.e. O2-nPrdT and O2-nBudT, may be attributed to Pol η’s recognition of these lesions as cytosine instead of thymine. Nevertheless, the addition of a branched-chain alkyl group to the O2 position of thymine may render it difficult for human Pol η to recognize the hydrogen bonding property of alkylated nucleobase and result in a lack of selectivity in nucleotide misincorporation.
Our laboratory recently investigated the roles of the TLS polymerases in the bypass of the major-groove O4-alkyldT lesions in vitro and in human cells. In keeping with the findings made for the O2-alkyldT lesions in the current study, human Pol η was the only TLS polymerase capable of bypassing all the O4-alkyldT lesions and generating full-length extension product in vitro.30 Additionally, human Pol η and ζ were found to promote the bypass of the O4-alkyldT lesions in HEK293T cells.42 On the other hand, the nature of nucleotide misincorporation mediated by the polymerase is vastly different when bypassing the dT lesions with the same alkyl groups being attached to the O2 and O4 positions. Unlike our current findings where dCMP was the main misincorporation event identified for the minor-groove O2-alkyldT lesions, dGMP was found to be the major nucleotide misincorporated opposite the major-groove O4-alkyldT lesions both in vitro and in cells.30, 42 This study combined with our previous findings demonstrated that the efficiency and fidelity of human Pol η-mediated replicative bypass of the alkylated thymidine lesions are modulated not only by the size and shape of the alkyl group, but also by the position of the alkyl group in duplex DNA.
Together, the results from this study provided important insights into the roles of human Pol η, κ, ι and yeast Pol ζ in bypassing the minor-groove O2-alkyldT lesions, as well as the impact these lesions have on the efficiency and fidelity of human Pol η. Further understanding the impact the structure of the alkyl group on the efficiency and fidelity of human Pol η calls for structural studies in the future. In addition, it will be important to determine the roles of the TLS polymerases in bypassing the O2-alkyldT lesions in human cells as well as the impacts these lesions have on the efficiency and fidelity of cellular DNA replication.
Supplementary Material
Acknowledgments
Funding Source Statement: This work was supported by the National Institutes of Health (R01 ES025121).
Abbreviations
- NOCs
N-nitroso compounds
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- NNN
N′-nitrosonornicotine
- POB
pyridyloxobutyl
- PHB
pyridylhydroxybutyl
- O2-alkyldT
O2-alkylthymidine
- TLS
translesion synthesis
- Pol
polymerase
- ODN
oligodeoxyribonucleotide
- PAGE
polyacrylamide gel electrophoresis
- XPV
xeroderma pigmentosum variant
Footnotes
Supporting Information Available: Standing-start primer extension, steady-state kinetic assay results and quantification of full-length extension product from primer extension assays. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Jamal A, Homa DM, O'Connor E, Babb SD, Caraballo RS, Singh T, Hu SS, King BA. Current cigarette smoking among adults - United States, 2005–2014. Centers for Disease Control and Prevention Morbidity and Mortality Weekly Report. 2015;64:1233–1260. doi: 10.15585/mmwr.mm6444a2. [DOI] [PubMed] [Google Scholar]
- 2.Hecht SS. Lung carcinogenesis by tobacco smoke. Int. J. Cancer. 2012;131:2724–2732. doi: 10.1002/ijc.27816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen H-JC, Wang Y-C, Lin W-P. Analysis of ethylated thymidine adducts in human leukocyte DNA by stable isotope dilution nanoflow liquid chromatography–nanospray ionization tandem mass spectrometry. Anal. Chem. 2012;84:2521–2527. doi: 10.1021/ac203405y. [DOI] [PubMed] [Google Scholar]
- 4.Hecht SS. Progress and challenges in selected areas of tobacco carcinogenesis. Chem. Res. Toxicol. 2007;21:160–171. doi: 10.1021/tx7002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brent T, Dolan M, Fraenkel-Conrat H, Hall J, Karran P, Laval L, Margison GP, Montesano R, Pegg A, Potter P. Repair of O-alkylpyrimidines in mammalian cells: a present consensus. Proc. Natl. Acad. Sci. U. S. A. 1988;85:1759–1762. doi: 10.1073/pnas.85.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bronstein SM, Skopek TR, Swenberg JA. Efficient repair of O6-ethylguanine, but not O4-ethylthymine or O2-ethylthymine, is dependent upon O6-alkylguanine-DNA alkyltransferase and nucleotide excision repair activities in human cells. Cancer Res. 1992;52:2008–2011. [PubMed] [Google Scholar]
- 7.Den Engelse L, De Graaf A, De Brij RJ, Menkveld GJ. O2- and O4-ethylthymine and the ethylphosphotriester dTp(Et)dT are highly persistent DNA modifications in slowly dividing tissues of the ethylnitrosourea-treated rat. Carcinogenesis. 1987;8:751–757. doi: 10.1093/carcin/8.6.751. [DOI] [PubMed] [Google Scholar]
- 8.Lao Y, Yu N, Kassie F, Villalta PW, Hecht SS. Analysis of pyridyloxobutyl DNA adducts in F344 rats chronically treated with (R)- and (S)-N'-nitrosonornicotine. Chem. Res. Toxicol. 2007;20:246–256. doi: 10.1021/tx060208j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lao Y, Yu N, Kassie F, Villalta PW, Hecht SS. Formation and accumulation of pyridyloxobutyl DNA adducts in F344 rats chronically treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol. 2007;20:235–245. doi: 10.1021/tx060207r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Upadhyaya P, Kalscheuer S, Hochalter JB, Villalta PW, Hecht SS. Quantitation of pyridylhydroxybutyl-DNA adducts in liver and lung of F-344 rats treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol. 2008;21:1468–1476. doi: 10.1021/tx8001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houtgraaf JH, Versmissen J, van der Giessen WJ. A concise review of DNA damage checkpoints and repair in mammalian cells. Cardiovasc. Revasc. Med. 2006;7:165–172. doi: 10.1016/j.carrev.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swenberg JA, Lu K, Moeller BC, Gao L, Upton PB, Nakamura J, Starr TB. Endogenous versus exogenous DNA adducts: Their role in carcinogenesis, epidemiology and risk assessment. Toxicol. Sci. 2010;120:S130–S145. doi: 10.1093/toxsci/kfq371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Sancar A, Lindsey-Boltz LA, Ünsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 16.Chang DJ, Cimprich KA. DNA damage tolerance: when it's OK to make mistakes. Nat. Chem. Biol. 2009;5:82–90. doi: 10.1038/nchembio.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA repair. 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 19.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Pol η. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 20.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 21.Yuan B, Cao H, Jiang Y, Hong H, Wang Y. Efficient and accurate bypass of N2-(1-carboxyethyl)-2′-deoxyguanosine by DinB DNA polymerase in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8679–8684. doi: 10.1073/pnas.0711546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biertümpfel C, Zhao Y, Kondo Y, Ramón-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465:1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Limoli CL, Giedzinski E, Morgan WF, Cleaver JE. Polymerase η deficiency in the xeroderma pigmentosum variant uncovers an overlap between the S phase checkpoint and double-strand break repair. Proc. Natl. Acad. Sci. U. S. A. 2000;97:7939–7946. doi: 10.1073/pnas.130182897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat. Rev. Cancer. 2005;5:564–573. doi: 10.1038/nrc1652. [DOI] [PubMed] [Google Scholar]
- 25.Gu C, Wang Y. LC-MS/MS identification and yeast polymerase η bypass of a novel γ-irradiation-induced intrastrand cross-link lesion G[8-5]C. Biochemistry. 2004;43:6745–6750. doi: 10.1021/bi0497749. [DOI] [PubMed] [Google Scholar]
- 26.You C, Swanson AL, Dai X, Yuan B, Wang J, Wang Y. Translesion synthesis of 8,5'-cyclopurine-2'-deoxynucleosides by DNA polymerases η, ɩ and ζ. J. Biol. Chem. 2013;288:28548–28556. doi: 10.1074/jbc.M113.480459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhanot OS, Grevatt PC, Donahue JM, Gabrielides CN, Solomon JJ. In vitro DNA replication implicates O2-ethyldeoxythymidine in transversion mutagenesis by ethylating agents. Nucleic Acids Res. 1992;20:587–594. doi: 10.1093/nar/20.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhai Q, Wang P, Cai Q, Wang Y. Syntheses and characterizations of the in vivo replicative bypass and mutagenic properties of the minor-groove O2-alkylthymidine lesions. Nucleic Acids Res. 2014;42:10529–10537. doi: 10.1093/nar/gku748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weerasooriya S, Jasti VP, Bose A, Spratt TE, Basu AK. Roles of translesion synthesis DNA polymerases in the potent mutagenicity of tobacco-specific nitrosamine-derived O2-alkylthymidines in human cells. DNA Repair (Amst) 2015;35:63–70. doi: 10.1016/j.dnarep.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams NL, Wang P, Wu J, Wang Y. In vitro lesion bypass studies of O4-Alkylthymidines with human DNA polymerase η. Chem. Res. Toxicol. 2016;29:669–675. doi: 10.1021/acs.chemrestox.5b00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodman MF, Creighton S, Bloom LB, Petruska J, Kunkel TA. Biochemical basis of DNA replication fidelity. Crit. Rev. Biochem. Mol. Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- 32.Washington MT, Johnson RE, Prakash S, Prakash L. Accuracy of thymine–thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc. Natl. Acad. Sci. U. S. A. 2000;97:3094–3099. doi: 10.1073/pnas.050491997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase η. J. Biol. Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 34.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol. Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 35.Takeiri A, Wada NA, Motoyama S, Matsuzaki K, Tateishi H, Matsumoto K, Niimi N, Sassa A, Grúz P, Masumura K, Yamada M, Mishima M, Jishage K, Nohmi T. In vivo evidence that DNA polymerase kappa is responsible for error-free bypass across DNA cross-links induced by mitomycin C. DNA Repair (Amst) 2014;24:113–121. doi: 10.1016/j.dnarep.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 36.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 37.Minko IG, Harbut MB, Kozekov ID, Kozekova A, Jakobs PM, Olson SB, Moses RE, Harris TM, Rizzo CJ, Lloyd RS. Role for DNA polymerase kappa in the processing of N2-N2-guanine interstrand cross-links. J. Biol. Chem. 2008;283:17075–17082. doi: 10.1074/jbc.M801238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jha V, Bian C, Xing G, Ling H. Structure and mechanism of error-free replication past the major benzo[a]pyrene adduct by human DNA polymerase κ. Nucleic Acids Res. 2016;44:4957–4967. doi: 10.1093/nar/gkw204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersen N, Wang J, Wang P, Jiang Y, Wang Y. In vitro Replication Studies on O2-Methylthymidine and O4-Methylthymidine. Chem. Res. Toxicol. 2012:2523–2531. doi: 10.1021/tx300325q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andersen N, Wang P, Wang Y. Replication across regioisomeric ethylated thymidine lesions by purified DNA polymerases. Chem. Res. Toxicol. 2013;26:1730–1738. doi: 10.1021/tx4002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gowda AS, Spratt TE. DNA polymerases η and ζ combine to bypass O2-[4-(3-Pyridyl)-4-oxobutyl]thymine, a DNA adduct formed from tobacco carcinogens. Chem. Res. Toxicol. 2016;29:303–316. doi: 10.1021/acs.chemrestox.5b00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu J, Li L, Wang P, You C, Williams NL, Wang Y. Translesion synthesis of O4-alkylthymidine lesions in human cells. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.