Abstract
Saccharomyces cerevisiae express three isoforms of Snf1 kinase that differ by which β subunit is present, Gal83, Sip1 or Sip2. Here we investigate the abundance, activation, localization and signaling specificity of the three Snf1 isoforms. The relative abundance of these isoforms was assessed by quantitative immunoblotting using two different protein extraction methods and by fluorescence microscopy. The Gal83 containing isoform is the most abundant in all assays while the abundance of the Sip1 and Sip2 isoforms is typically underestimated especially in glass-bead extractions. Earlier studies to assess Snf1 isoform function utilized gene deletions as a means to inactivate specific isoforms. Here we use point mutations in Gal83 and Sip2 and a 17 amino acid C-terminal truncation of Sip1 to inactivate specific isoforms without affecting their abundance or association with the other subunits. The effect of low glucose and alkaline stresses was examined for two Snf1 phosphorylation substrates, the Mig1 and Mig2 proteins. Any of the three isoforms was capable of phosphorylating Mig1 in response to glucose stress. In contrast, the Gal83 isoform of Snf 1 was both necessary and sufficient for the phosphorylation of the Mig2 protein in response to alkaline stress. Alkaline stress led to the activation of all three isoforms yet only the Gal83 isoform translocates to the nucleus and phosphorylates Mig2. Deletion of the SAK1 gene blocked nuclear translocation of Gal83 and signaling to Mig2. These data strongly support the idea that Snf1 signaling specificity is mediated by localization of the different Snf1 isoforms.
Keywords: Saccharomyces cerevisiae, Snf1 kinase, AMP-activated protein kinase, β-subunit
1. Introduction
All cells use signal transduction pathways to sense and respond to changes in nutrient availability and energy status. In eukaryotes, the AMP-activated protein kinase (AMPK) plays a central role in cellular and whole-body energy homeostasis [1]. The AMPK complex is a heterotrimer with a single catalytic subunit, called α in mammalian enzymes and Snf1 in yeast. In addition, the kinase holoenzyme contains two regulatory subunits referred to as the β and γ subunits. The γ subunit binds to high- and low-energy adenylate nucleotides and thereby directly transduces information concerning cellular energy status to the other subunits [2,3]. The β subunit also contributes to the regulation of this kinase complex. The β subunits all contain two highly conserved, signature domains: a C-terminal αγ a domain (AG) that forms an interface for interaction of the α and γ subunits and a carbohydrate binding module (CBM) that can bind glycogen [4]. Two locations for the CBM in structural models of the AMPK holoenzyme have been proposed. In models of the yeast enzyme using the γ subunit and the C-termini of the α and β subunits, the CBM is seen as bound to the γ subunit [5]. In models of the mammalian enzyme using the entire α and γ subunits along with most of the β subunit, the CBM is seen as bound to the N-lobe of the kinase domain of the α subunit [6,7]. These latter structural models seem more likely to be correct with regard to CBM position since they are based on a more complete heterotrimer that includes the kinase domain. Small molecule activators such as A-769662 and salicylate can directly bind to and activate AMPK [8]. The small molecule-binding pocket lies at the interface between the CBM of the β subunit and N-lobe of the α subunit kinase domain [7,9]. Interestingly, this activation occurs much more efficiently with mammalian AMPK complexes containing the β1 subunit compared to those with the β2 subunit [10], supporting the idea that different β subunits play specialized roles in vivo. Whether or not mammals produce ligand(s) that use this small molecule-binding pocket is a subject of great interest.
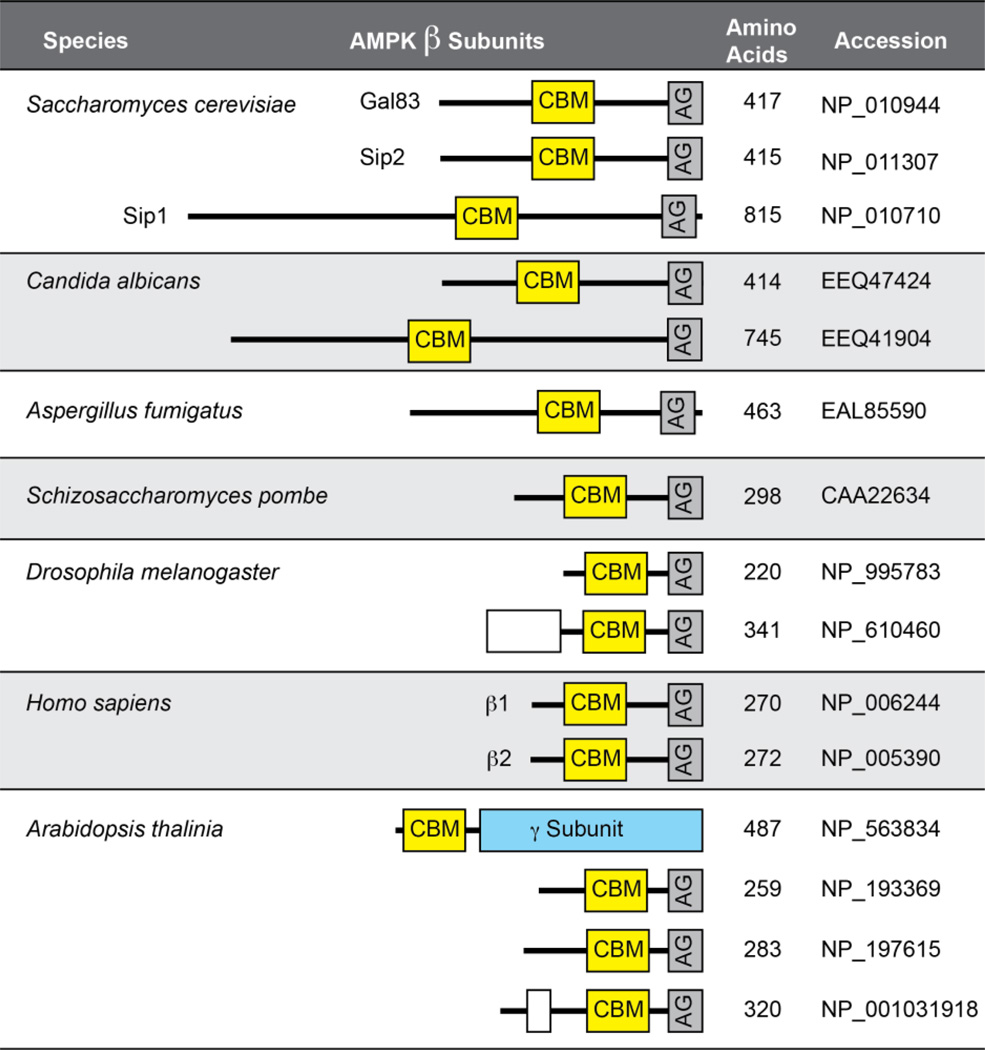
All three subunits of the AMPK enzyme are highly conserved across eukaryotic evolution. The only eukaryotes lacking clear AMPK orthologs are a few intracellular pathogens such as Plasmodium falciparum [11]. Here we focus our attention on the β subunits of the AMPK enzyme of the budding yeast Saccharomyces cerevisiae. Yeast express three isoforms of the AMPK enzyme that differ in subunit composition by having either Gal83, Sip1 or Sip2 as the β subunit in association with Snf1 (α) and Snf4 (γ) [12]. The GAL83 and SIP2 genes are paralogs derived from the whole genome duplication event that occurred 100 million years ago [13]. As such they are very similar in size (417 and 415 residues, respectively) and primary sequence (55% identity and 75% similar over a 360 residue window). In contrast, the Sip1 protein is distinct from Gal83 and Sip2 by being much larger (815 residues) and with greater sequence divergence (19% identity and 44% similarity over a 490 residue window when compared to Gal83). The number and sequence of β subunits in different eukaryotic species is quite variable (Fig. 1). Some fungal species (A. fumigatus and S. pombe) make do with a single β subunit while others have two (C. albicans) that show clear similarity to the smaller Gal83/Sip2 subunits or to the larger Sip1 subunit. In metazoans, the trend has been toward smaller β subunits. Humans and Drosophila express two β subunits that are smaller than their yeast counterparts. Some plants such as A. thaliana express typical β subunits as well as a hybrid AMPK subunit that contains a CBM domain typically found in β subunits fused to the γ subunit. How the diversity of the β subunits contributes to the complexity and specificity of AMPK signaling has not been fully explored.
Fig. 1.
β subunits of AMPK enzymes in different species. A schematic representation of the β subunits present in 4 fungal species as well as in human, insect and plant species are shown. The number of β subunits in each species, their amino acid length, accession number and the position of the carbohydrate binding module (CBM) and the C-terminal α-γ interaction domain (AG) are shown. Drosophila and Arabidopsis generate β subunit diversity by alternative splicing to include an additional exon (open box).
All β subunits contain two highly conserved, signature domains, the CBM domain and the C-terminal domain that mediates binding to the α and γ subunits. Outside of these domains, β subunits even from closely related species are quite dissimilar. Deletion of the CBM (also called the glycogen-binding domain) activates the Gal83 isoform of the Snf1 enzyme in yeast [14]. In mammals, the CBM is required for the ability of small molecule ligands to activate AMPK [7,9]. The C-terminal AG domain is necessary for heterotrimer assembly and kinase activity [15]. Furthermore, this small domain is by itself sufficient to reconstitute a functional Snf1 kinase [12]. Previous studies have shown that two conserved histidine residues in the αγ interaction domain of the β subunit are in close contact with the kinase domain activation loop and are necessary to stabilize the active conformation of the kinase [2,3,16]. Earlier studies from our lab and others have used gene deletions to probe the isoform specific roles of the Snf1 kinase [12,14,17,18]. However, deletions may disrupt subunit stoichiometry and may result in complementation and mislocalization not observed when all subunits are expressed. Thus, in the complete absence of a given β subunit, an alternative isoform may inappropriately substitute function in a way that would not happen if the catalytic domain were still bound to its rightful β isoform partner. In this study, we have used point mutations in β subunit AG domain as a means to inactivate specific Snf1 isoforms without interfering with isoform assembly. This isoform-specific inactivation of Snf1 allowed us to probe the complexity and specificity of β subunit function in yeast.
2. Materials and methods
2.1 Yeast strains and growth conditions
The yeast strains used in this study were all derived from the S228C lineage. Most yeast strains with specific gene deletions were generated in our laboratory or by the Saccharomyces Genome Deletion project [19] and purchased from Thermo Scientific (Table 1). Cells were grown at 30 °C using standard synthetic complete media lacking nutrients needed for plasmid selection [20]. Plasmids employed in this study are described in Table 2.
Table 1.
Yeast strains
| Strain | Genotype |
|---|---|
| MSY1212 | MATa ura3-52 leu2Δ1 his3Δ200 |
| MSY557 | MATα ura3-52 leu2Δ1 his3Δ200 sip1Δ::HIS3 sip2Δ::HIS3 gal83Δ::HIS3 |
| MSY545 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 sip1Δ::HIS3 sip2Δ::HIS3 |
| MSY552 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 sip1Δ::HIS3 gal83Δ::HIS3 |
| MSY543 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 sip2Δ::HIS3 gal83Δ::HIS3 |
| MSY522 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 gal83Δ::HIS3 |
| MSY528 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 sip1Δ::HIS3 |
| MSY520 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 sip2Δ::HIS3 |
| MSY565 | MATα ura3-52 leu2Δ1 his3Δ200 sip1Δ::HIS3 snf1Δ10 |
| MSY1037 | MATa ura3-52 leu2 his3Δ200 SNF1-3HA sip1Δ::HIS3 sip2Δ::HIS3 gal83Δ::HIS3 |
| MSY955 | MATα ura3-52 leu2Δ1 his3Δ200 trp1Δ63 SNF1-3HA |
| MSY1163 | MATα ura3-52 leu2Δ1 trp1Δ63 gal83Δ::HIS3 SNF1-3HA |
| MSY1165 | MATα ura3-52 leu2Δ1 trp1Δ63 sip2Δ::HIS3 SNF1-3HA |
| MSY1167 | MATα ura3-52 leu2Δ1 trp1Δ63 sip1Δ::HIS3 SNF1-3HA |
| MSY1356 | MATα ura3-52 leu2Δ1 his3Δ200 GAL83-GFP |
| MSY1358 | MATα ura3-52 leu2Δ1 his3Δ200 SIP1-GFP |
| MSY1360 | MATα ura3-52 leu2Δ1 his3Δ200 SIP2-GFP |
| MSY1449 | MATa ura3 leu2 his3 lys2Δ0 GAL83-GFP Δ::KAN |
| MSY1450 | MATa ura3 leu2 his3 lys2Δ0 GAL83-GFP |
Table 2.
Plasmids
| Plasmid | Genes | Vector | Source |
|---|---|---|---|
| pF3-Sip1 | Sip1 with triple flag tag at N-terminus | pRS315 | [14] |
| pF3-Sip2 | Sip2 with triple flag tag at N-terminus | pRS315 | [14] |
| pF3-Gal83 | Gal83 with triple flag tag at N-terminus | pRS315 | [14] |
| pF3-Gal83-H379A | Gal83-H379A with triple flag tag at N-terminus | pRS315 | this study |
| pF3-Gal83-H384A | Gal83-H384A with triple flag tag at N-terminus | pRS315 | this study |
| pFG3-H384A-314 | Gal83-H384A with triple flag tag at N-terminus | pRS314 | this study |
| pF3-Sip2-H380A | Sip2-H380A with triple flag tag at N-terminus | pRS315 | this study |
| pFS2-H380A-314 | Sip2-H380A with triple flag tag at N-terminus | pRS314 | this study |
| pF3-Sip1-H772A | Sip1-H772A with triple flag tag at N-terminus | pRS315 | this study |
| pF3-Sip1-Q798 | Sip1 with triple flag tag and stop codon at Q798 | pRS315 | this study |
| pSnf1-3HA | Snf1 with triple HA tag at C-terminus | pRS316 | [22] |
| pMig1-3HA | Mig1 with triple HA tag at C-terminus | pRS316 | [22] |
| pMig2-3HA | Mig2 with triple HA tag at C-terminus | pRS316 | this study |
2.2 Plasmids and epitope tagging
All plasmids used in these studies were low-copy number, centromeric plasmids based on pRS314, pRS315 and pRS316 [21]. Snf1 and Mig1 proteins tagged with 3 copies of the HA epitope have been previously described [22]. The MIG2 gene was subcloned into pRS315 on a 2584 nt genomic StuI-HindIII fragment. Three copies of the HA epitope along with the ADH1 terminator were amplified from pYM2 [23] and appended to the MIG2 gene using gap repair. The resulting plasmid, pMig2-3HA was recovered from yeast and DNA sequence confirmed. The β subunits tagged with three copies of the Flag epitope were described [14]. Amino acid substitutions were generated by oligonucleotide-directed mutagenesis with Pfu polymerase, followed by DpnI digestion of the plasmid template [24] and confirmed by DNA sequencing. The F3-GAL83-H384A and F3-SIP2-H380A alleles were also subcloned into pRS314 (Table 2).
2.3 Mutagenesis of the SIP1 gene
Mutations that compromised SIP1 function were identified by gap repair mutagenesis using pF3-SIP1 plasmid [14] gapped by restriction with NheI and SphI removing the codons for the C-terminal residues 674–815. DNA encoding the C-terminus of Sip1 was amplified with Taq polymerase and used to repair the gapped plasmid. A recipient strain lacking all three β subunit genes allowed assessment of SIP1 function by replica plating Leu+ gap-repaired transformants to raffinose medium. Candidates showing loss of Sip1 function (Raf−) were screened by western blot. None of the sip1− transformants produced full-length Sip1. The candidate producing a Sip1 protein closest to full-length was sequenced and found to contain a stop codon in place of the glutamine codon at position 798. Full-length Sip1 is 815 residues in length. The resulting gene, SIP1-Q798, was used for further studies.
2.4 GFP-tagging the β subunits
Plasmids encoding each of the β subunits were modified such that a 5 residue linker (Ala-Gly-Ala-Gly-Ala) was appended to the C-terminus of each open reading frame followed by the yeast codon-optimized GFP gene derived from pKT127 [25]. Each of the β subunit-GFP genes was excised from the plasmids with sufficient flanking genomic DNA to direct homologous recombination into a yeast strain lacking all three β subunit genes. Integrants were selected by their ability to grow on raffinose media and the correct integration at the endogenous β subunit gene was confirmed by PCR. These strains were then backcrossed to a wild type strain to generate haploid yeast expressing all three β subunits with one bearing a C-terminal GFP tag.
2.5 Protein extract preparations
Glass bead extracts were prepared from cells (approximately 15–20 OD600 units) that were grown to mid-log phase and collected by centrifugation. Cells were washed once in 1 ml and then suspended in 0.2 ml of a lysis buffer consisting of 50 mM Tris pH 8.0, 150 mM NaCl, 0.5% sodium deoxycholate, 50 mM sodium fluoride, 5 mM sodium pyrophosphate supplemented with protease inhibitors (Roche). An equal volume of acid washed glass beads (Sigma; G8772) was added and cells were lysed by vortexing three times for 20 seconds in a FastPrep homogenizer (MP Biomedicals) with 5 minutes recovery between cycles. Sodium dodecyl sulfate (SDS) and Nonidet P40 were added to a final concentration of 0.1% and 1%, respectively and the extract was rotated at 4 °C for 30 minutes. Insoluble material was removed by centrifugation at 15,000 x g for 5 minutes. The supernatant fraction was removed and assayed for protein using the Bradford assay and bovine serum albumin as the standard. Twenty micrograms of total protein was used for each lane in immunoblotting experiments. Alternatively, yeast whole-cell extracts were prepared using a modification of a trichloroacetic acid extraction method [26]. Cells were grown to mid-log phase and absorbance at 600 nm was measured. Cells (2.5 OD600 units) were harvested, washed with cold water, and resuspended in 1 ml of cold water. A 150 µl aliquot of freshly prepared 2 N NaOH, 1 M β-mercaptoethanol was added, mixed and incubated on ice for 15 min. Next, 150 µl of 50% TCA was added, mixed and incubated on ice for 20 min. The sample was then subjected to centrifugation at 15,000 x g for 5 min and the pelleted proteins were resuspended in SDS sample buffer (80 mM Tris-Cl, pH 8.0, 8 mM EDTA, 120 mM dithiothreitol, 3.5% SDS, 0.29% glycerol, 0.08% Tris base, 0.01% bromphenol blue) using 20 µl of SDS sample buffer per OD600 of cells. The extracts were incubated at 37 °C for 30 min. Insoluble material was removed by centrifugation at 10,000 x g for 2 min. The supernatant fraction was removed and 7.5 µl was used for each lane in immunoblotting experiments.
2.6 Immunoblotting and immunoprecipitation
Snf1, Mig1 and Mig2 proteins tagged with the HA epitope were detected with a 1:3000 dilution of mouse anti-HA antibody (Santa Cruz Biotechnology, Santa Cruz, CA). β subunits tagged with the Flag epitope were detected with a 1:1000 dilution of monoclonal mouse anti-Flag antibody (Sigma catalog #F3165). The secondary antibody was goat anti-mouse IgG DyLight 680 (Thermo Scientific, Waltham, MA) diluted 1:10,000. For detection of phosphorylated Snf1, Phospho-AMPKα (Thr172) antibody (Cell Signaling catalog #2535L) diluted 1:1000 was used with goat anti-rabbit IRDye 800CW (Li-Cor) (1:10,000 dilution) as the secondary antibody. Blots were normalized using Sec61 as the control protein detected with affinity-purified anti-Sec61 primary antibody at 1:1000 (gift of Jeffrey Brodsky, Dept of Biological Sciences, Univ. of Pittsburgh) followed by goat anti-rabbit IRDye 800CW (Li-Cor, Lincoln, NE) diluted 1:10,000 as the secondary antibody. Snf1-3HA was immunopreciptated from glass bead extracts using agarose conjugated anti-HA antibody (Santa Cruz). Blots were scanned using an infrared scanner (Odyssey, Li-Cor). Integrated intensity values of bands were quantified by using scanning software supplied by the manufacturer (Li-Cor). Snf1 complexes containing a Flag-tagged Sip1 or Sip2 protein were immunoprecipitated from glass bead extracts (1 mg protein) using 15 µl anti-Flag beads (Sigma catalog #A2220). Beads were washed 3 times in RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 0.1% SDS, 1% Nonidet P40, 0.5% sodium deoxycholate) and bound proteins were eluted 2x SDS sample buffer and analyzed by western blotting with anti-HA, anti-Phospho-AMPKα (Thr172) and anti-Flag antibodies.
2.7 Fluorescence Microscopy
Chromosomally integrated, GFP-tagged β subunits were imaged using a Nikon Ti-Eclipse inverted microscope equipped with a swept-field confocal scan head (Nikon, Chiyoda, Tokyo, Japan; Prairie Instruments, Middleton, WI) or using epifluorescence. An Apo100x objectives (NA 1.49 or 1.45) were used in all imaging and images were captured using an iXon3 EMCCD camera (Andor, Belfast, UK) or an Orca Flash 4.0 cMOS (Hammamatsu, Bridgewater, NJ) and NIS-Elements software (Nikon). Overnight cultures were inoculated into fresh media at an A600 of 0.2 and allowed to grow until they reached mid-logarithmic phase (A600 ~0.5). For imaging of cells shifted from high (2%) to low (0.05%) glucose, cells were pelleted, washed in low glucose media and then resuspended in low glucose media for 10 minutes prior to imaging. For exposure to alkaline stress, cell media was adjusted to pH 8.5 with sodium hydroxide addition and cells were imaged 10–20 minutes after this pH shift. To visualize vacuoles, and aid in subcellular localization assignments, cells were incubated with 250 µM Cell Tracker Blue CMAC dye (Life Technologies, Carlsbad, CA). All images from an experiment were captured with the same imaging parameters and are adjusted equivalently in Adobe Photoshop. For all figures an unsharp mask was applied with a threshold of 5 and a pixel radius of 5. To quantify the total cellular fluorescence for the β subunits, a minimum of 175 cells were outlined manually and the mean pixel intensity was measured in Image J software (NIH, Bethesda, MA).
2.8 Statistical analyses
For all bar plots, mean values using a minimum of three independent measurements are plotted with error bars representing one standard error. Statistical significance was determined using the Student t-test for unpaired variables with equal variance. For the box-and-whisker plot, a one-way ANOVA with Tukey’s post hoc test was performed to assess statistical significance of differences in observed pixel intensities. Statistical significance is indicated as follows: * P < 0.05; ** P < 0.01; *** P < 0.001; ns, P > 0.05.
3. Results
3.1 Abundance of the yeast Snf1 kinase β subunits
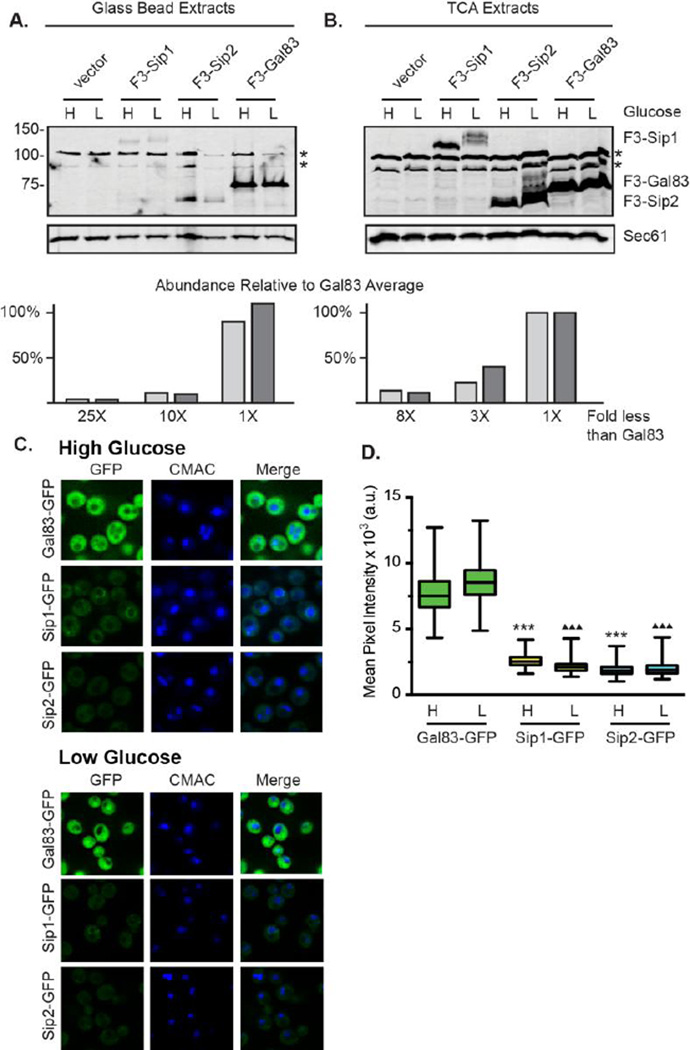
Earlier studies to assess the relative abundance of the yeast proteins globally [27,28] or the Snf1 kinase β subunits specifically [17,29] indicated that the Gal83 protein was by far the most abundant of the β subunits with the Sip1 and Sip2 proteins 5–12 fold less abundant. With the availability of quantitative immunoblotting technologies, we re-examined the relative abundance of the three yeast β subunits expressed from their own cognate promoters on low-copy plasmids and all tagged with three copies of the Flag epitope at their N-termini [30]. Since the β subunits display different subcellular localizations including membrane association [17,31,32], we compared extracts prepared by two different methods: glass bead extraction and TCA extraction, a method that more efficiently extracts membrane-associated proteins [26]. Using Sec61 as a loading control, we found that the Gal83 protein was the most abundant β subunit regardless of the extraction method (Fig. 2A and 2B). With both procedures, Sip2 levels were intermediate and Sip1 was the least abundant. However, we found that the relative abundance of Sip1 and Sip2, the two subunits that have been reported to associate with the vacuolar [17] and plasma membranes [32], respectively, are increased when the TCA extraction method was used. Sip1 was barely detectable and 25-fold less abundant than Gal83 in glass bead extracts but clearly visible and only 8-fold less abundant using the TCA extraction method. Sip2 was 10-fold less abundant than Gal83 in glass bead extracts but only 3 fold less abundant in TCA extracts. Furthermore, the abundance of Sip2 has been reported to be regulated in response to glucose abundance [17,29]. Using TCA whole cell extracts, we found that Sip2 abundance increased by less than a factor of two when cells were shifted to low glucose medium.
Fig. 2.
Abundance of β subunits depends on protein extraction method. Yeast β subunits were each tagged at the N-terminus with 3 copies of Flag epitope, expressed from their own promoters on low-copy number plasmids and analyzed by western blotting. Extracts were prepared from cells grown in high (H) glucose or 1 hour after shifting to low (L) glucose using a glass bead lysis method or a TCA extraction method (see Materials and Methods) as indicated. The gel mobilities of the β subunits are indicated on the right. Relative abundance was determined by comparing the β subunit western signal to that of the Sec61 protein and normalized to the level of Gal83 in high glucose defined as 100%. The fold less than Gal83 for Sip1 and Sip2 is plotted for both extraction methods. Two yeast proteins that cross react with the Flag antibody are indicated by asterisks. (C) Chromosomally integrated, GFP-tagged Gal83, Sip1, and Sip2 were imaged in high glucose media and 30 minutes after shifting to low glucose (0.05%) media. All image acquisition parameters and adjustments were held constant so that direct comparisons of signal intensities between the β subunits can be made. Position of the vacuole is determined by staining with CMAC. (D) Total cellular fluorescence intensity was measured (n, >125 cells for each strain) and the distributions of the pixel intensities were plotted (a.u., arbitrary units). The horizontal midline in each box represents the median; the box is bounded by the upper and lower quartiles; and the whiskers denote the maximal and minimal fluorescence intensities in the populations. A one-way ANOVA with Tukey’s post hoc test was performed to assess statistical significance of differences in observed pixel intensities. (***, P<0.001 relative to Gal83-GFP in high glucose; ♦ ♦ ♦P<0.001 relative to Gal83-GFP in low glucose).
As an alternative approach to monitoring β subunit abundance, we assessed relative levels and localization of β subunits using quantitative fluorescence microscopy (Fig 2C and 2D). We generated chromosomally integrated, GFP-tagged versions of each β subunit, and measured fluorescence in cells grown in high glucose. Gal83 was significantly more abundant than the other β subunits, consistent with the immunoblotting results. Gal83-GFP displayed diffuse cytosolic fluorescence with clear nuclear and vacuolar exclusion under these conditions (Fig. 2C and 7E). Sip1-GFP and Sip2-GFP were 3- and 4-fold less abundant than Gal83 (Figs. 2C–D), respectively, and they too showed diffuse cytoplasmic fluorescence with distinct exclusion from the lumen of the nucleus and vacuole (Fig. 2C, 7C–D). In addition, in some cells Sip1-GFP was localized to the limiting membrane of the vacuole (Fig.2C and 7C). Our findings are consistent with previously reported β subunit localizations, which used plasmid-borne GFP-tagged versions rather than chromosomally integrated versions [17]. This method of quantifying β subunit abundance circumvents the inherit problems of solubility and extraction presented by immunoblotting. Using this approach Sip2 appears to be slightly less abundant than Sip1, which was also observed in proteomic study using mass spectrometry [28] and in a study using qPCR to measure abundance of TAP-tagged proteins [29]. These data suggest that even with the TCA extraction method Sip1 may not be fully solubilized, resulting in a lower relative yield in protein extracts compared to those observed by fluorescence microscopy. Alternatively, the presence of different N-terminal and C-terminal tags might affect the abundance of different β subunits. Taken together, we conclude that the estimation of the relative abundance of the β subunits is greatly influenced by the method employed. Protein extraction and methods that enrich for membrane-associated proteins generate more robust signal for Sip1 and Sip2, and suggest that some earlier studies [17,26] including our own [14] have underestimated the abundance and relative ratios of these isoforms of Snf1 kinase.
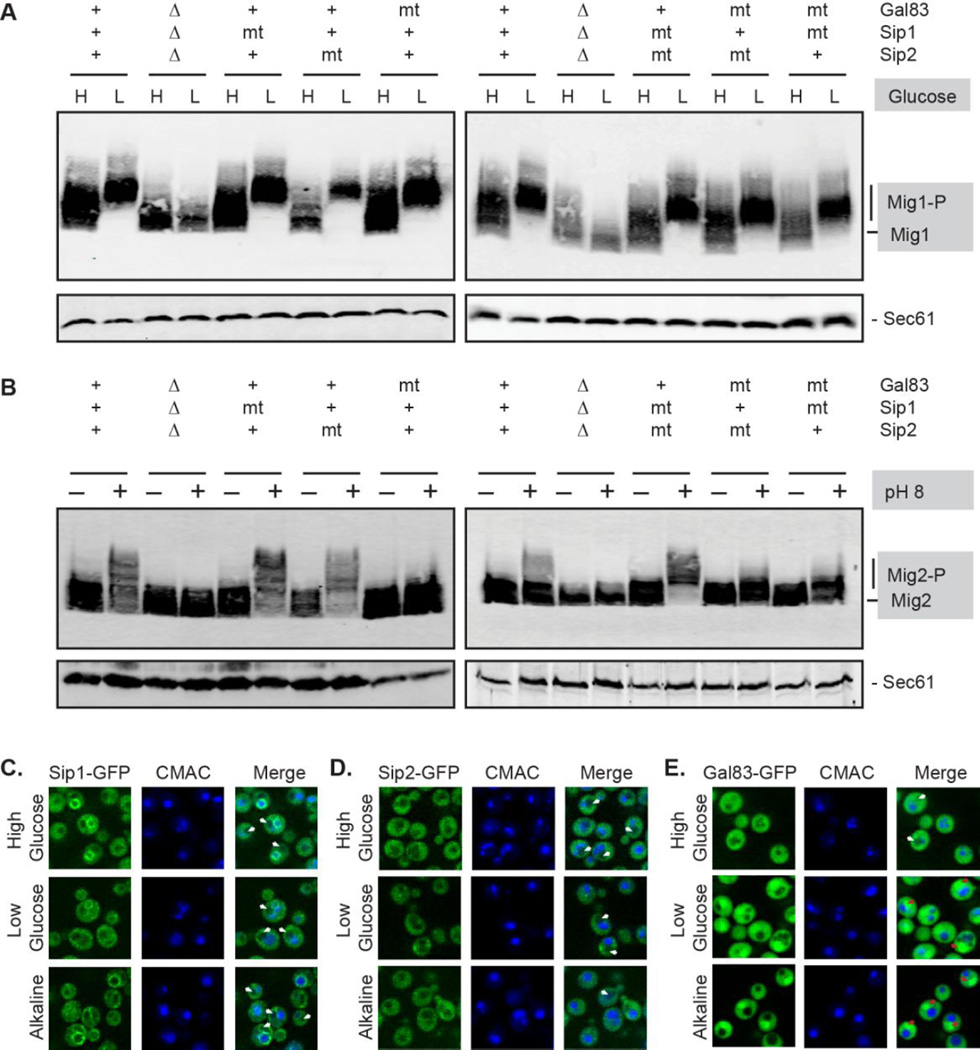
Fig 7.
Snf1 kinase β subunit requirements during glucose and alkaline stress response signaling. Cells expressing wild type β subunits (+), lacking all three β subunits (Δ) or expressing mutant β subunits (mt) with amino acid substitutions (Gal83-H384A, Sip2-H380A) or a truncation mutation (Sip1-Q798) were transformed with plasmids expressing HA-tagged Mig1 protein (A) or HA-tagged Mig2 protein (B). Extracts were prepared from mid-log cells grown in high glucose (H) or 30 minutes after shifting to low glucose (L) and were analyzed by western blotting with antibodies against HA and Sec61. Gel mobility of Mig1 and phosphorylated Mig1 (Mig1-P) are indicated (A). The gel mobility of the Mig2 protein before and 10 minutes after a shift to pH 8 was determined by western blotting. Gel mobility of Mig2 and phospho-Mig2 (Mig2-P) are indicated (B). Extracts were also blotted for Sec61 as a loading control. (C, D, E) Cells expressing the indicated chromosomally integrated, GFP-tagged β subunits were grown in high glucose, and then shifted into low glucose media or alkaline media (pH 8.5) for 10 minutes and imaged using a confocal microscope. CMAC blue stain was used to mark the vacuoles. In merged images, white arrows indicate the fluorescent signal is excluded from the nucleus and red arrows indicate the fluorescent is enriched in the nucleus. The images presented for each β subunit within a panel are adjusted equally; however, the adjustments between the panels are not equivalent so that the localization of each of the β subunits can be optimally visualized.
3.2 Functional role of the conserved histidine residues in the C-terminus of the yeast β subunits
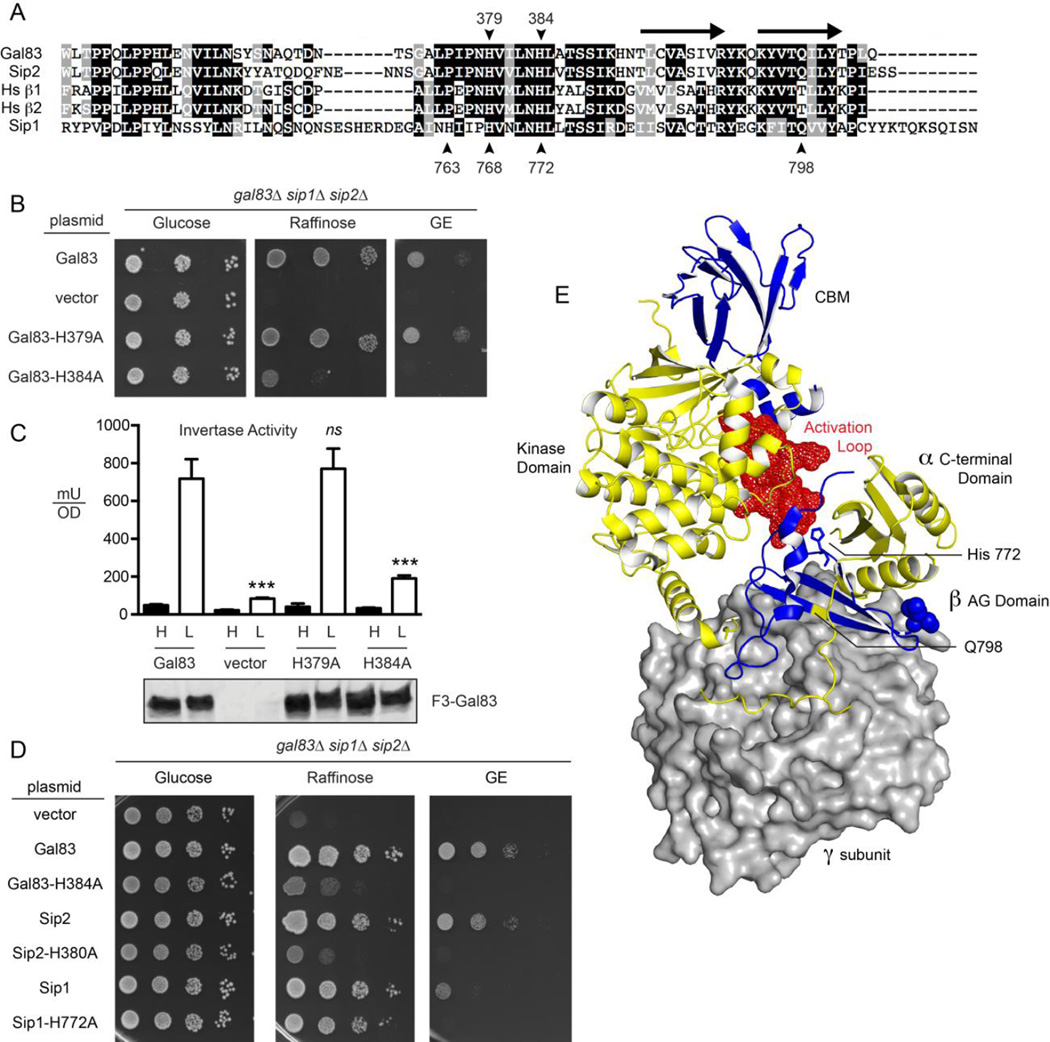
We sought to identify amino acid substitutions in the β subunits that interfered with their ability to activate the Snf1 kinase without affecting their ability to associate with the Snf1 kinase complex. In this way we could assess Snf1 isoform function without creating a complete void as would be the case with deletion of a specific β subunit gene. Gene deletions might allow spurious β subunits to perform biological functions with which they would not otherwise be associated. The β subunits from diverse eukaryotic species share greatest sequence similarity in two domains, the carbohydrate binding module (CBM) and the α-γ interaction domain (AG) at the C-terminus (Fig. 1). The AG domain includes two antiparallel β sheets [3,5,6] that form an interaction interface between the γ subunit and the C-terminal domain of the α subunit (Fig. 3A and 3E). The residues immediately preceding the two β sheets include two highly conserved histidine residues in the sequence context of NHVxNHL that interact with the kinase domain activation loop in the active conformation [33]. Substituting alanine for the first conserved histidine residue (H379) in the Gal83 protein had little effect on Gal83 function while changing the second histidine residue to alanine (Gal83-H384A) caused a severe loss of function when assayed for growth on alternative carbon sources (Fig. 3B) and for invertase induction in response to glucose limitation (Fig. 3C). A similar and severe loss of function is observed when the analogous histidine residue in the Sip2 protein (H380) is changed to alanine (Fig. 3D). Neither the Gal83-H384A nor the Sip2-H380A substitutions affected heterotrimer association as judged by the copurification of the intact Snf1 heterotrimer from yeast and bacteria (data not shown and [2]). In contrast, the Sip1 protein seemed unaffected by this change (Sip1-H772A) when assayed for growth on raffinose (Fig 3D). Previously we showed that cells expressing only the Sip1 isoform of Snf1 kinase grow very poorly on media with glycerol/ethanol as the carbon source [12]. Under those growth conditions, substitution of the Sip1 H772 with alanine did exacerbate the poor growth on this non-fermentable carbon source. However, the lack of a raffinose growth phenotype in cells expressing Sip1-H772A was puzzling. We considered the possibility that other histidine residues in this region (Fig. 3A) might be providing the interaction surface with the activation loop of Snf1 complexes containing Sip1. However, substitutions of Sip1 H763 or H768 as well as the substitution of all three histidine residues had only negligible effect on the Sip1 protein function (data not shown). Therefore, the contacts within the AG domain that stabilize the active form of the Snf1 kinase domain in the complexes containing Gal83 and Sip2 must be different from those used in the Sip1 complex.
Fig 3.
Role of conserved histidine residues in yeast β subunits. (A) Sequence alignment of the α-γ interaction domains of the three β subunits from S. cerevisiae and two β subunits from human (Hs β1 and Hs β2). Positions of the conserved histidine residues are indicated with the numbering on top referring to amino acids in Gal83 and the bottom referring to the Sip1 protein. The position of two β sheets that form the interface between the γ subunit and the C-terminal domain of the α subunit are indicated with arrows above the alignment. The position of Q798 in Sip1 is also indicated. (B) Spot dilution assay to measure β subunit function in yeast lacking all three β subunits but transformed with a low copy plasmid expressing no β subunit (vector), Gal83 or Gal83 with histidine to alanine substitutions at positions 379 or 384. Growth was measured on plates with the glucose, raffinose or a mixture of glycerol and ethanol (GE) as the carbon sources. (C) Invertase enzyme activity was measured in triplicate samples with the mean plotted ±SE. Values statistically different from wild type are indicated. The cells assayed were the same as those used in panel B. Western blot of Flag-tagged Gal83 proteins is shown below. (D) Spot dilution assay to measure β subunit function in yeast lacking all three β subunits but transformed with a low copy plasmid expressing no β subunit (vector), or the β subunit indicated with or without the indicated histidine to alanine substitutions. Growth was measured on plates with the glucose, raffinose or a mixture of glycerol and ethanol (GE) as the carbon sources. (E) Structural model of the Snf1 kinase heterotrimer based on the mammalian AMPK heterotrimer [33]. The γ subunit is shown in surface representation (gray). The α and β subunits are show in the cartoon representation in yellow and blue, respectively. The α subunit kinase domain, C-terminal domain and activation loop (red spheres) are indicated. The β subunit C-terminus and carbohydrate-binding motif (CBM) are indicated. The position of the Sip1 histidine residue 772 is shown in the stick representation while residue glutamine 798 is indicated in yellow. This structural model was created with PyMol (Schrödinger) using the Protein Database file 4CFF.
3.3 Identification of mutations in SIP1 that block Snf1 activation
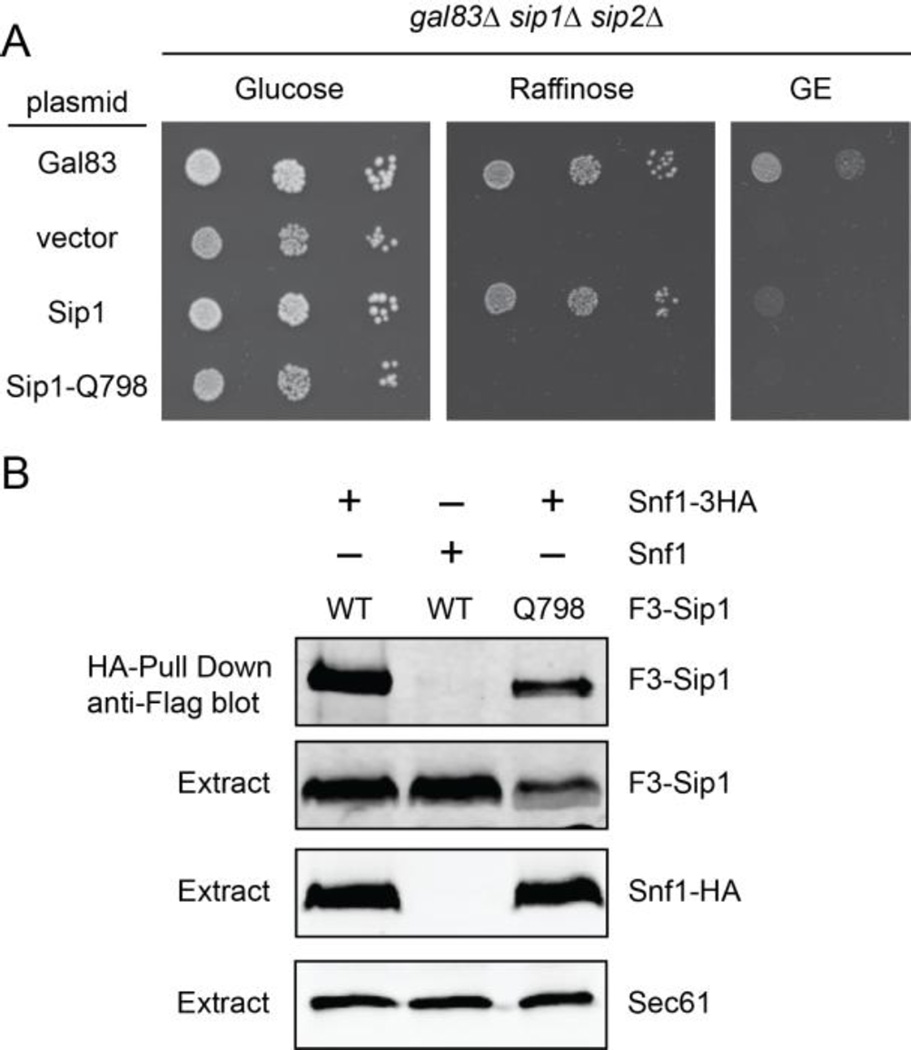
To identify amino acid substitutions in the Sip1 protein that interfered with its ability to activate Snf1 kinase but maintained Snf1 kinase association, we took a random mutagenesis approach. We used polymerase chain reaction to generate random mutations in the DNA encoding the C-terminus of Sip1 and gap repair to introduce those into a Flag-tagged Sip1 plasmid [34]. Transformants were screened for the inability to grow on raffinose and then examined by western blotting. All of the loss-of-function alleles identified made either no Sip1 protein or a truncated Sip1 protein. The clone that made a Sip1 protein closest to full length was sequenced and found to contain a stop codon in place of the glutamine 798 codon. This change causes a truncation of the C-terminal 17 amino acids of the Sip1 protein (Fig. 3A). Cells expressing Sip1-Q798 as the only β subunit are unable to grow on raffinose as well as glycerol ethanol media (Fig. 4A). The Sip1-Q798 protein still assembles into Snf1 kinase complexes as judged by co-immunoprecipitation. HA-tagged Snf1 and associated proteins were isolated from protein extracts using anti-HA beads and the associated Sip1 was detected by immunoblotting with anti-Flag antibodies (Fig 4B). Both wild type Sip1 and Sip1-Q798 associated with HA tagged Snf1. Sip1 was not detected in the pull-down fraction when untagged Snf1 was used. Therefore, the Sip1 protein truncated at Q798 assembles into a complex with Snf1 protein but the kinase complex is not functional.
Fig 4.
Sip1-Q798 is not functional but associates with Snf1. (A) Cells lacking the genes for all three β subunits were transformed with empty plasmid vector or with low-copy plasmids encoding Gal83, Sip1 or Sip1-Q798 (stop codon at 798) as shown. Ten-fold serial dilutions of cells were spotted onto agar media with the indicated carbon sources. (B) Extracts were prepared from snf1Δ sip1Δ cells expressing Snf1 with or without a 3-HA tag and Flag-tagged Sip1 or Sip1-Q798 as shown. Snf1-HA and associated proteins were collected with HA beads and bound F3-Sip1 and F3-Sip1-Q798 were detected with anti-flag antibodies (upper panel). Total Snf1, Sip1 and Sec61 proteins present in extracts were determined by western blotting (lower panels).
3.4 Alkaline and glucose stresses generate distinct Snf1-dependent responses
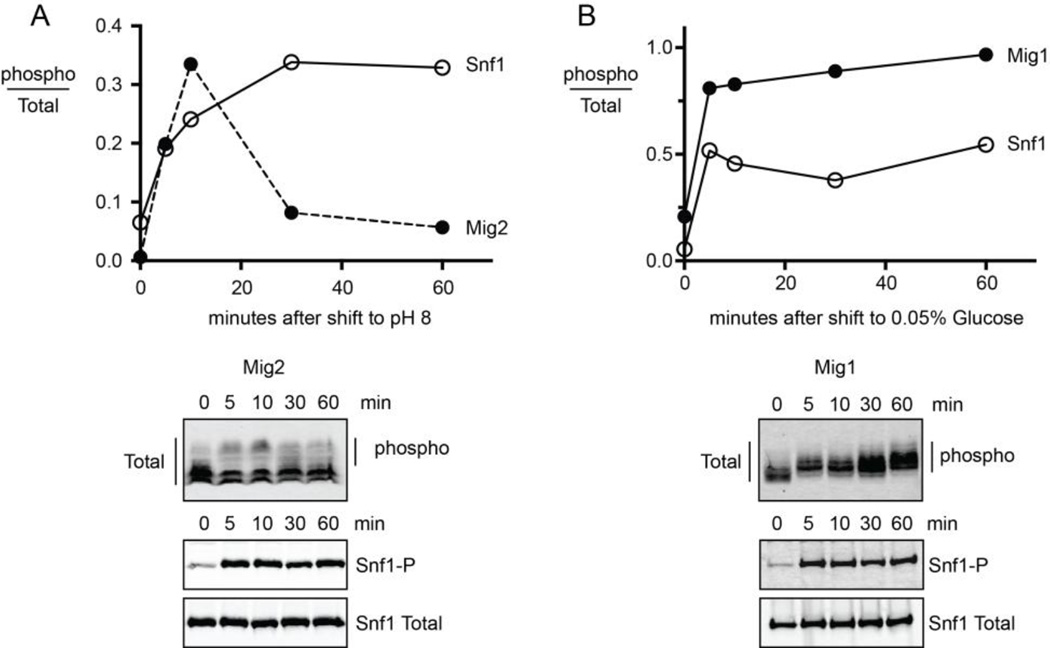
In response to environmental stresses, the Snf1 kinase phosphorylates two zinc-finger transcriptions factors: Mig1 in response to low-glucose stress [35] and Mig2 in response to alkaline stress [36]. Interestingly, the level of phosphorylation, the timing of phosphorylation and their affect on protein abundance is distinct depending on the stress being experienced and the substrate being phosphorylated, making these conditions and substrates ideally suited as initial candidates for assessing β subunit isoform specificity. To analyze the kinetics of Snf1 activation and signaling to Mig1 and Mig2, aliquots were removed from cultures 5, 10, 30 and 60 minutes after shifting cells to media adjusted to pH 8 (Fig. 5A) or to media containing 0.05% glucose (Fig. 5B). Under both stress conditions, Snf1 phosphorylation increased within the first 5 minutes and the increased phosphorylation was sustained throughout the 60 minute time course. The downstream targets of Snf1 displayed very different kinetics of phosphorylation. Mig2 protein showed a transient decrease in gel mobility that peaked at 10 minutes then rapidly declined. The reduced gel mobility was due to phosphorylation since it was eliminated by treatment with calf intestine alkaline phosphatase (not shown). A similar rapid and transient phosphorylation of Mig2 following alkaline stress has been reported previously [36], although this earlier study did not report the sustained activation of Snf1 that we observe following alkaline stress. The Mig1 protein was rapidly phosphorylated following glucose stress and the phosphorylation was sustained over the 60 minutes examined here (Fig. 5B). While both stresses cause rapid and sustained activation of Snf1 kinase, the target proteins display distinct kinetics. The phosphorylated form of Mig2 accumulates in a transient manner despite sustained activation of Snf1.
Fig. 5.
Kinetics of Snf1 response to alkaline stress and glucose limitation. (A) Protein extracts were prepared from cells expressing either HA-tagged Snf1 or HA-tagged Mig2 and analyzed by quantitative western blotting. The phosphorylation status of each protein was measured 0, 5, 10, 30 and 60 minutes after shifting to pH 8. (B) Protein extracts were prepared from cells expressing either HA-tagged Snf1 or HA-tagged Mig1 and analyzed by quantitative western blotting. The phosphorylation status of each protein was measured 0, 5, 10, 30 and 60 minutes after shifting cells to media with 0.05% glucose.
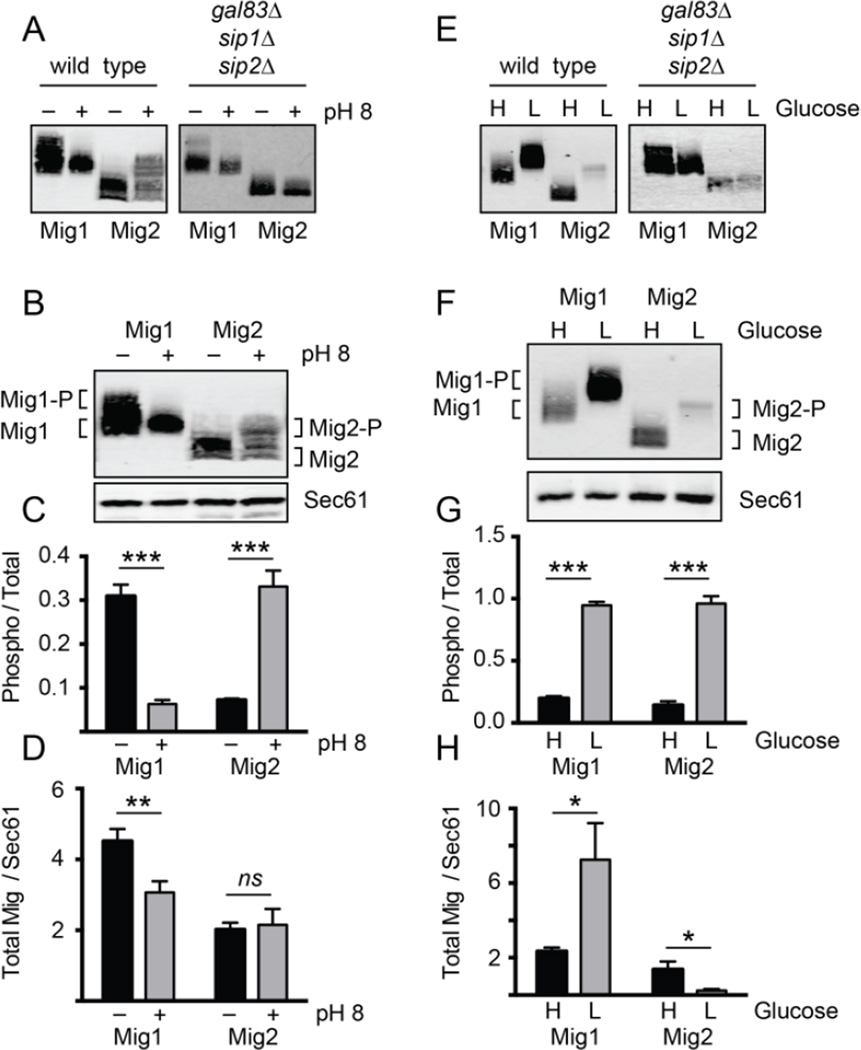
We next examined the effect of both glucose and alkaline stress on the phosphorylation state and abundance of Mig1 and Mig2. The phosphorylation of both Mig1 and Mig2 is Snf1-dependent since it is not observed in cells lacking Snf1 kinase activity due to the deletion of all three β subunits (Fig. 6A and E). These two Snf1 substrates responded very differently to these two stresses. Alkaline stress resulted in the increased phosphorylation of Mig2 but decreased phosphorylation of Mig1 (Fig 6B and C). Alkaline stress also caused a reduced abundance of Mig1 but no change in the abundance of Mig2 (Fig. 6D). In contrast, glucose stress caused an increased phosphorylation of both proteins (Fig. 6F and G) and the opposite effect on the abundance of these proteins. Glucose stress led to increased Mig1 abundance and decreased Mig2 abundance (Fig. 6H). These experiments were conducted using three independent cultures of each sample with the mean value ±SE plotted and statistical significance shown. These data show that two different signaling inputs show distinct Snf1-dependent responses for these proteins. Both stresses lead to increased Snf1-dependent changes in either abundance or phosphorylation. Thus, Snf1-dependent signaling can result in distinct outcomes depending on the substrate and the stimulus.
Fig 6.
Differential effects of glucose and alkaline stress response signaling to the Mig1 and Mig2 proteins. The phosphorylation (gel mobility) and abundance of the Mig1 and Mig2 proteins was examined by quantitative western blotting of extracts prepared before or after alkaline stress (A–D) or low glucose stress (E–H). (A and E) β subunit requirement for Mig1 and Mig2 phosphorylation during the alkaline and glucose stress responses. (B and F) Representative western blot of Mig1 and Mig2 proteins before and after alkaline stress and glucose stresses. (C and G) Quantification of phosphorylation status of Mig1 and Mig2 from triplicate extracts. Mean value ±SE is plotted for phosphorylated/total protein. (D and H) Abundance of Mig1 and Mig2 proteins from triplicate extracts before and after alkaline or glucose stress. Mean values ±SE are plotted for total Mig1/ control protein Sec61.
3.5 All three isoforms of Snf1 can mediate phosphorylation of Mig1
We have previously shown that Snf1 isoforms with any one of the β subunits are able to promote phosphorylation of Mig1 [12]. This result is puzzling in light of the observations that unphosphorylated Mig1 is localized to the nucleus [37] and only the Gal83 isoform of Snf1 is able to efficiently enter the nucleus upon glucose limitation [17,31]. However, our earlier study relied on the use of gene deletions [12]. The complete absence of a protein might allow paralogous proteins to interact with partners that they would not normally interact with and to localize to sites they would not normally occupy. Here we decided to re-examine this question using cells that express all three β subunits using mutant versions that associate with Snf1 and Snf4 but lack the ability to support Snf1 activation. The non-functional alleles used here were Gal83-H384A, Sip2-H380A and Sip1-Q798. The phosphorylation state of Mig1 protein was examined using cells that were grown in either high glucose or 30 minutes after shifting to low glucose (Fig. 7A). In cells expressing wild type version of all three β subunits, the shift of Mig1 to the slower migrating, phosphorylated form is readily apparent upon glucose limitation. Cells with complete deletions of all three β subunits lack Snf1 kinase function and were unable to phosphorylate the Mig1 protein. Cells expressing one or two non-functional β subunits were all fully able to promote Mig1 phosphorylation. Cells lacking a functional Gal83 show no defect in Mig1 phosphorylation. Thus any one of the Snf1 isoforms is capable phosphorylating Mig1 even when other isoforms were present though non-functional. This result suggests that either all Snf1 isoforms can equivalently target nuclear substrates or that the nucleo-cytoplasmic shuttling of Mig1 [38] allows it to be accessed by all Snf1 isoforms regardless of their subcellular distribution.
3.6 The Gal83 isoform is necessary and sufficient for phosphorylation of Mig2
We next tested the Snf1 isoform specificity of the alkaline stress response by examining the phosphorylation of Mig2 in cells expressing mutant versions of the β subunits that lack the ability to support Snf1 activation (Fig 7B). Cells expressing all three β subunits show increased phosphorylation of Mig2 ten minutes after exposure to alkaline stress. The shift in Mig2 mobility is not observed in cells lacking all three β subunits. In cells expressing two functional β subunits and one mutant variant, we found that cells lacking a functional Gal83 isoform were unable to promote Mig2 phosphorylation while loss of Sip1 or Sip2 function did not affect Mig2 phosphorylation. In cells lacking two functional β subunits, phosphorylation of Mig2 was only observed in cells expressing Gal83 as the functional β subunit. Therefore, the Gal83 isoform of Snf1 kinase is both necessary and sufficient for the phosphorylation of Mig2 in response to alkaline stress. Consistent with this observation, we monitored β subunit localization in response to alkaline stress. We found that only Gal83-GFP concentrated in the nucleus in response to alkaline stress (Fig 7E, red arrows), whereas in unstressed cells it localized to the cytosol and appeared to be largely excluded from the nucleus (white arrows). Both Sip1 and Sip2 were largely excluded from the nucleus under all conditions, including alkaline stress (Figs 7C–D, white arrows). Thus for Mig2, the ability of the Gal83 isoform of Snf1 to phosphorylate Mig2 correlates with its ability to concentrate in the nucleus.
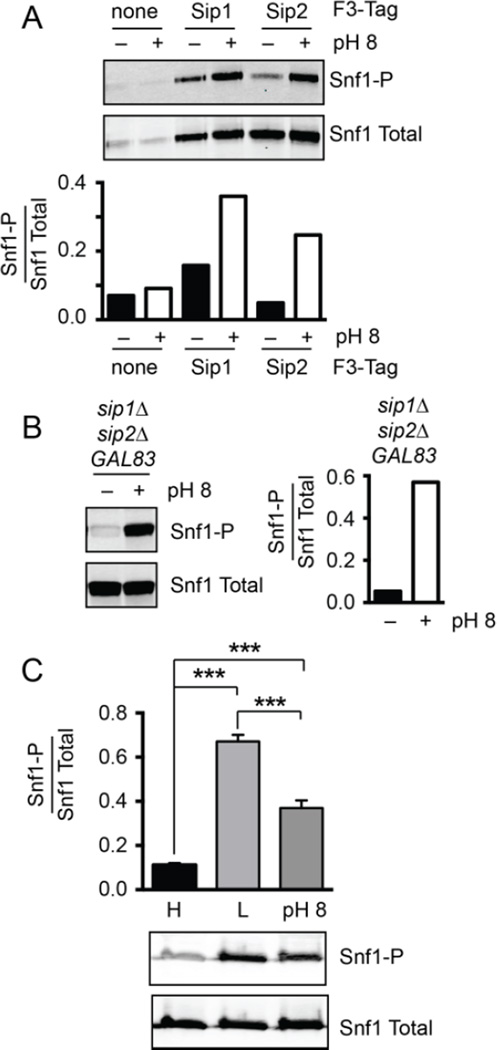
3.7 Alkaline stress activates all three isoforms of Snf1
We next investigated the mechanism by which the alkaline stress signaling to the Mig2 protein was restricted to Gal83 isoform of Snf1 kinase. We considered the possibility that alkaline stress might lead to the selective activation of only the Gal83 isoform of Snf1. To test this idea, we measured the phosphorylation of the Snf1 activation loop in the different isoforms of the kinase following alkaline stress. Our strategy was to prepare extracts from cells expressing all three Snf1 isoforms, two isoforms with untagged β subunits and one β subunit with a triple Flag-tag at its N-terminus. The activation state of the single tagged isoform of Snf1 kinase could then be measured following immunoprecipitation of the Flag-tagged β subunit followed by western blotting with antibodies that detect total and activated Snf1. When either Sip1 or Sip2 were the Flag-tagged β subunit, the Snf1 complex isolated by immunoprecipitation showed increased phosphorylation of Snf1 in response to alkaline pH stress (Fig. 8A). In the absence of a Flag tag, very little total Snf1 or phospho-Snf1 were detected demonstrating that the western signal observed in the pull-downs with Flag-tagged beads were in fact isoform specific. Thus, alkaline stress leads to activation of the Sip1 and Sip2 isoforms of Snf1 yet does not produce detectable phosphorylation of Mig2. For technical reasons, we were unable to consistently immunoprecipitate Flag-tagged Gal83 complexes. However, the Gal83 isoform of Snf1 must be activated in response to alkaline stress since it is required for Mig2 phosphorylation. When we examined Snf1 phosphorylation in cells expressing only the Gal83 isoform, we found increased Snf1 phosphorylation following alkaline stress (Fig. 8B). These data demonstrate that alkaline stress does not lead to selective activation of only the Gal83 isoform of Snf1 but rather leads to the activation of all three isoforms of Snf1 kinase. Therefore, selective activation of the Snf1 isoforms does not provide substrate specificity.
Fig. 8.
Isoform selectivity and amplitude of Snf1 activation in response to alkaline stress. (A) Activation of the Sip1 and Sip2 isoforms of Snf1 kinase in response to alkaline stress was measured by immunoprecipitation of Flag-tagged Sip1 or Flag-tagged Sip2 followed by western blotting for total Snf1 and phosphorylated Snf1. (B) Activation of the Gal83 isoform of Snf1 kinase in response to alkaline stress was measured in GAL83 sip1Δ sip2Δ cells. Total and phosphorylated Snf1 was assessed by western blotting. (C) Amplitude of Snf1 kinase activation in response to glucose and alkaline stress. Extracts were prepared from triplicate cultures of cells grown in 2% glucose (High, H), 30 minutes after shifting to 0.05% glucose (L) or 10 minutes after shift to pH 8. Mean values of the ratio of phosphorylated Snf1 over total Snf1 are plotted ±SE.
3.8 Amplitude of Snf1 activation following alkaline and glucose stresses
Another signaling variable that could dictate substrate selectivity is the amplitude of kinase activation. In this scenario, one stress could activate a small fraction of Snf1 kinase enzymes thus limiting its potential signaling outputs while another stress might activate a large fraction of Snf1 kinase enzymes thus producing a distinct output. We compared the amplitude of Snf1 activation following alkaline stress and glucose stress using quantitative western blotting of extracts prepared in triplicate (Fig. 8C). Glucose stress leads to a 6-fold increase in the phosphorylation of Snf1 defined as the mean value of the western signal for phosphorylated Snf1 divided by the western signal for total Snf1 (Snf1-P/Snf1 Total). In contrast, alkaline stress leads to only a 3-fold increase in Snf1 phosphorylation. The differences in Snf1 activation following these two stresses were statistically significant (p<0.001). Thus, the amplitude of Snf1 activation varies depending on the stimulus and may play a role in determining downstream signaling outcomes.
3.9 All three Snf1 isoforms can phosphorylate Mig2 in vitro
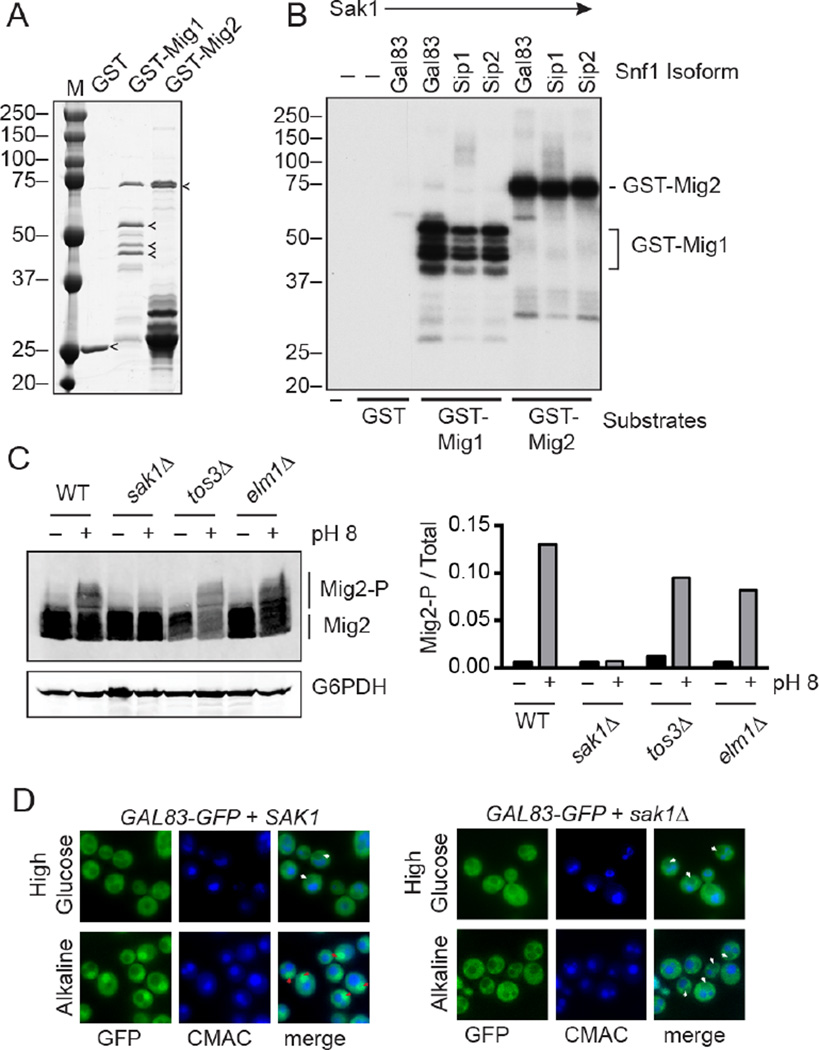
One possible mechanism for isoform specificity could be direct protein-protein interaction between substrates and the different β subunits of the Snf1 isoforms. To test whether all three isoforms can recognize and phosphorylate the Mig2 protein, we purified GST fusions of Mig1 and Mig2 expressed in bacteria (Fig. 9A). These proteins were then used as substrates in an in vitro kinase assay using purified preparations of the three Snf1 isoforms (Fig 9B). Each isoform was activated in vitro by Sak1 kinase TAP-purified from yeast [39]. We found that all three Snf1 isoforms were catalytically active and able to phosphorylate both Mig1 and Mig2 proteins in vitro. These results indicate that direct protein-protein interactions between the β subunits and the substrate proteins are not likely to dictate Snf1 isoform specificity.
Fig. 9.
Nuclear localization of Gal83 isoform is required for alkaline stress response. (A) Coomassie stained gel of kinase substrates purified from bacteria. (B) In vitro kinase assay of the different Snf1 isoforms incubated with [γ-32P]ATP and GST, GST-Mig1 or GST-Mig2 as the substrates. Sak1 kinase was present in all reactions as indicated. (C) Western blot of HA-tagged Mig2 before and 10 minutes after alkaline stress in wild type cells (WT) and cells lacking one of the three Snf1-activating kinases. The mobility of Mig2 and phosphorylated Mig2 (Mig2-P) is indicated. Ratio of phospho-Mig2 over total Mig2 is plotted on the right. (D) Localization of Gal83-GFP before and after alkaline stress in wild type cells and cells lacking Sak1. Position of the vacuoles was determined by staining with CMAC. White arrows indicate the fluorescent signal is excluded from the nucleus and red arrows indicate the fluorescent is enriched in the nucleus.
3.10 Nuclear localization of Gal83 isoform is necessary for alkaline stress response
We next sought to determine if nuclear localization of Gal83 was required for its selective activation of Mig2 in response to alkaline stress. Earlier studies of Gal83 isoform localization in response to glucose limitation demonstrated that activation by the Sak1 kinase was required for Gal83 nuclear localization. When the Gal83 isoform was activated by the other Snf1-activating kinases, Tos3 and Elm1, the Gal83 isoform was unable to translocate to the nucleus. We sought to test whether phosphorylation of Mig2 also showed Snf1-activating kinase specificity. Cells expressing HA-tagged Mig2 but lacking one of the Snf1-activating kinases were exposed to alkaline stress. Wild type cells showed an increase in the phosphorylation of Mig2 protein following alkaline stress (Fig. 9C). Cells lacking Tos3 and Elm1 also showed phosphorylation of Mig2. In contrast, cells lacking the Sak1 kinase were unable to phosphorylate Mig2. Localization of Gal83-GFP following alkaline stress in cells with and without the Sak1 kinase showed that the Sak1 was required for nuclear localization (Fig. 9D). Therefore, the nuclear localization of the Gal83 isoform of Snf1 is necessary for its ability to phosphorylate Mig2, and this shift in localization may be responsible for the isoform selectivity of Mig2 phosphorylation.
4. Discussion
A first step toward understanding the distinct roles played by the different isoforms of the Snf1 kinase is to determine the relative abundance of the isoforms. Earlier studies using immunoblotting [17] and biochemical purification [40] have supported the idea that the Sip1 isoform of the Snf1 kinase complex is by far the least abundant isoform. The low expression level of the Sip1 isoform might explain why cells expressing only this isoform manifest a severe growth defect on non-fermentable carbons sources [12]. Alternatively, localization studies of the different Snf1 isoforms have reported that the Sip1 isoform is localized to the outer surface of the vacuole [17], and it is possible that the localization of the Sip1 isoform limits its signaling potential. A similar membrane association of mammalian AMPK has been reported and suggests conserved functions of the vacuolar (yeast) and lysosomal (mammal) localized AMPK isoforms [41]. Recent studies in our lab with membrane-associated proteins [42] have taught us that the method of protein extraction can greatly influence protein recovery. Therefore we tested whether protein extraction methods have affected estimations of the abundance of the Sip1 isoform of Snf1. Indeed, we found that using a standard glass bead extraction protocol underestimates the abundance of the Sip1 protein relative to a TCA extraction protocol Membrane association may have also affected our low recovery of the Sip1 isoform during biochemical purification [40]. Using immunofluorescence as a measure of protein abundance eliminates the variable effects of protein extraction methods. In high glucose, we found that the Gal83 isoform was the most abundant while the Sip2 isoform was the least abundant (Fig. 2D). A recent study using qPCR to measure protein abundance also found that Sip2 was the least abundant of the β subunits in cells growing in media with high glucose [29]. Sip1 and Sip2 are expressed at similar levels yet they display large differences in their ability to confer growth on non-fermentable carbon sources. Based on these data, we suggest that the inability of the Sip1 isoform to support growth on non-fermentable carbon sources more likely reflects limited signaling potential due to localization as opposed to limitations derived from its low abundance.
The activation of the Snf1 kinase requires two steps: phosphorylation of the activation loop and a second step that requires the regulatory β and γ subunits [22]. Structural studies of the mammalian AMPK have proposed a model in which the active conformation of the kinase domain is stabilized by its interaction with the C-terminal AG domain of the β subunit present in the heterotrimer core [3,6]. The phosphorylated activation loop of the kinase domain forms a large fraction of the interaction surface between the kinase domain and heterotrimer core. The activation loop makes direct contact with the NHVxNHL motif in the AG domain that is conserved in both yeast and mammalian β subunits (Fig. 3A). Mutations in these conserved histidine residues reduce kinase activity in the yeast and mammalian enzymes [2,3]. Here we show that substitution of the second histidine (H384 in Gal83 and H380 in Sip2) with alanine results in a dramatic reduction in Snf1 kinase function without affecting β subunit expression or association with the Snf1 complex (data not shown and [2]). In contrast, this same histidine to alanine substitution in the Sip1 protein (H772) had only a minor effect on Snf1 kinase activity. Substitutions of additional nearby histidine residues in Sip1 had no effect on the activity of the Sip1 isoform. Therefore, the residue contacts between the Snf1 kinase domain and the Sip1 AG domain that lead to activation of the kinase must be distinct from those occurring in the Gal83 and Sip2 complexes. This result was somewhat surprising since the C-terminal AG domain is the domain that is most highly conserved between β subunits from diverse species. Structural models of the active form Snf1-Snf4-Sip1 isoform will be needed to understand how the phosphorylated activation loop is stabilized and what β subunit residues contribute to this conformation.
Unraveling the distinct functional roles played by the different β subunits of Snf1/AMPK is a subject of great interest. In mammalian cells, only AMPK isoforms containing the β-1 subunit are efficiently activated by the small molecule activator known as A-769662 [10]. In yeast, the different isoforms display distinct subcellular localizations [17] and distinct functionality [12]. In an earlier paper, we had proposed two mechanisms to explain the distinct functions of the Snf1 isoforms [12]. In a localization model, the Snf1 isoform localization restricts by proximity which substrates it can phosphorylate. In a substrate binding model, the direct interaction between the substrate and the β subunit determines specificity. Of course, these models need not be mutually exclusive. Our earlier studies with the Mig1 transcription factor did not allow us to distinguish between these models. Using gene deletions, we have found that any one of the three Snf1 isoforms can phosphorylate the transcription factor Mig1 in vitro [40] and in vivo [12]. Here, we re-examined this question using inactivating mutations in the β subunits rather than gene deletions. Using these new reagents, we found once again that any one of the Snf1 isoforms was able to phosphorylate Mig1 in response to glucose limitation. However, when we measured Mig2 phosphorylation in response to alkaline stress, we found that the nuclear isoform of Snf1 containing Gal83 as the β subunit was both necessary and sufficient for the phosphorylation of Mig2. Further, we showed that activation of Snf1 by Sak1 kinase was necessary for nuclear translocation of the Gal83 isoform in response to alkaline stress and that Sak1 was necessary for signaling to Mig2. These data strongly support the localization model to explain the isoform specificity of signaling to Mig2. The lack of isoform specificity for Mig1 might be explained by the fact that Mig1 is known to cycle back and forth between the nuclear and cytoplasmic compartments [38]. The different isoform requirements for signaling to Mig1 and Mig2 are more likely reflecting differences in the localization dynamics of Mig1 and Mig2 rather than differences in the properties of the Snf1 isoforms.
Finally, we showed that the amplitude of Snf1 activation varies in response to different stimuli. Quantitative immunoblotting revealed that glucose stress results in much higher levels of activated Snf1 than does alkaline stress. We and others have previously reported that glucose stress results in greater activation of Snf1 when compared to sodium ion stress [22,43]. Work from the Hohmann lab has also found that activation of Snf1 in high glucose does not result in the increased phosphorylation of Mig1 [29,43]. Thus the greater amplitude of Snf1 activation does not always directly correlate with the phosphorylation of Mig1.
5. Conclusions
Snf1 kinase responds to multiple environmental stresses each generating a distinct signaling output. Glucose stress leads to the largest amplitude of Snf1 activation and any one of the three Snf1 isoforms can promote Mig1 phosphorylation. Alkaline stress leads to a smaller amplitude of Snf1 activation and displays a high degree of isoform specificity. Nuclear translocation of the Gal83 isoform of Snf1 is necessary for signaling to the Mig2 protein, suggesting that Snf1 isoform localization plays a critical role in substrate selection.
Highlights.
Membrane association limits extraction and recovery of Snf1 kinase isoforms.
The Gal83 isoform of Snf1 is necessary and sufficient for the phosphorylation of Mig2 in response to alkaline stress.
All three isoforms of Snf1 can phosphorylate Mig2 in vitro.
Nuclear translocation of the Gal83 isoform of Snf1 is necessary for Mig2 phosphorylation.
Acknowledgments
This work was supported by NIH R01 research grant GM46443 to MS and NSF MCB Career grant 1553143 to AO. We thank Jeff Brodsky for the generous gift of Sec61 antibodies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement
The authors declare that there are no conflicts of interests.
References
- 1.Hardie DG. AMPK: positive and negative regulation, and its role in whole-body energy homeostasis. Curr. Opin. Cell Biol. 2015;33:1–7. doi: 10.1016/j.ceb.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Mayer FV, Heath R, Underwood E, et al. ADP regulates SNF1, the Saccharomyces cerevisiae homolog of AMP-activated protein kinase. Cell metabolism. 2011;14:707–714. doi: 10.1016/j.cmet.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiao B, Sanders MJ, Underwood E, et al. Structure of mammalian AMPK and its regulation by ADP. Nature. 2011;472:230–233. doi: 10.1038/nature09932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polekhina G, Gupta A, van Denderen BJ, Feil SC, Kemp BE, Stapleton D, Parker MW. Structural basis for glycogen recognition by AMP-activated protein kinase. Structure. 2005;13:1453–1462. doi: 10.1016/j.str.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Amodeo GA, Rudolph MJ, Tong L. Crystal structure of the heterotrimer core of Saccharomyces cerevisiae AMPK homologue SNF1. Nature. 2007;449:492–495. doi: 10.1038/nature06127. [DOI] [PubMed] [Google Scholar]
- 6.Li X, Wang L, Zhou XE, et al. Structural basis of AMPK regulation by adenine nucleotides and glycogen. Cell Res. 2015;25:50–66. doi: 10.1038/cr.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao B, Sanders MJ, Carmena D, et al. Structural basis of AMPK regulation by small molecule activators. Nature Com. 2013;4:3017. doi: 10.1038/ncomms4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawley SA, Fullerton MD, Ross FA, et al. The ancient drug salicylate directly activates AMP-activated protein kinase. Science. 2012;336:918–922. doi: 10.1126/science.1215327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calabrese MF, Rajamohan F, Harris MS, et al. Structural basis for AMPK activation: natural and synthetic ligands regulate kinase activity from opposite poles by different molecular mechanisms. Structure. 2014;22:1161–1172. doi: 10.1016/j.str.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Scott JW, van Denderen BJ, Jorgensen SB, et al. Thienopyridone drugs are selective activators of AMP-activated protein kinase beta1-containing complexes. Chem. Biol. 2008;15:1220–1230. doi: 10.1016/j.chembiol.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Ward P, Equinet L, Packer J, Doerig C. Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC Genomics. 2004;5:79. doi: 10.1186/1471-2164-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt MC, McCartney RR. beta-subunits of Snf1 kinase are required for kinase function and substrate definition. Embo J. 2000;19:4936–4943. doi: 10.1093/emboj/19.18.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 14.Mangat S, Chandrashekarappa D, McCartney RR, Elbing K, Schmidt MC. Differential roles of the glycogen-binding domains of beta subunits in regulation of the Snf1 kinase complex. Eukaryot. Cell. 2010;9:173–183. doi: 10.1128/EC.00267-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iseli TJ, Walter M, van Denderen BJ, Katsis F, Witters LA, Kemp BE, Michell BJ, Stapleton D. AMP-activated protein kinase beta subunit tethers alpha and gamma subunits via its C-terminal sequence (186–270) J. Biol. Chem. 2005;280:13395–13400. doi: 10.1074/jbc.M412993200. [DOI] [PubMed] [Google Scholar]
- 16.Chandrashekarappa DG, McCartney RR, Schmidt MC. Ligand binding to the AMP-activated protein kinase active site mediates protection of the activation loop from dephosphorylation. The Journal of biological chemistry. 2013;288:89–98. doi: 10.1074/jbc.M112.422659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vincent O, Townley R, Kuchin S, Carlson M. Subcellular localization of the Snf1 kinase is regulated by specific beta subunits and a novel glucose signaling mechanism. Genes Dev. 2001;15:1104–1114. doi: 10.1101/gad.879301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vyas VK, Kuchin S, Berkey CD, Carlson M. Snf1 kinases with different beta-subunit isoforms play distinct roles in regulating haploid invasive growth. Mol. Cell. Biol. 2003;23:1341–1348. doi: 10.1128/MCB.23.4.1341-1348.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winzeler EA, Shoemaker DD, Astromoff A, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 20.Rose MD, Winston F, Hieter P, editors. Methods in Yeast Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor; 1990. [Google Scholar]
- 21.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCartney RR, Schmidt MC. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 2001;276:36460–36466. doi: 10.1074/jbc.M104418200. [DOI] [PubMed] [Google Scholar]
- 23.Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 24.Fisher CL, Pei GK. Modification of a PCR-based site-directed mutagenesis method. BioTechniques. 1997;23:570–574. doi: 10.2144/97234bm01. [DOI] [PubMed] [Google Scholar]
- 25.Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Nijbroek G, Sullivan ML, McCracken AA, Watkins SC, Michaelis S, Brodsky JL. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol Biol Cell. 2001;12:1303–1314. doi: 10.1091/mbc.12.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 28.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Salcedo R, Lubitz T, Beltran G, et al. Glucose de-repression by yeast AMP-activated protein kinase SNF1 is controlled via at least two independent steps. FEBS J. 2014;281:1901–1917. doi: 10.1111/febs.12753. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, McCartney RR, Chandrashekarappa DG, Mangat S, Schmidt MC. Reg1 protein regulates phosphorylation of all three Snf1 isoforms but preferentially associates with the Gal83 isoform. Eukaryot. Cell. 2011;10:1628–1636. doi: 10.1128/EC.05176-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hedbacker K, Hong SP, Carlson M. Pak1 protein kinase regulates activation and nuclear localization of Snf1-Gal83 protein kinase. Mol. Cell. Biol. 2004;24:8255–8263. doi: 10.1128/MCB.24.18.8255-8263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin SS, Manchester JK, Gordon JI. Sip2, an N-myristoylated beta subunit of Snf1 kinase, regulates aging in Saccharomyces cerevisiae by affecting cellular histone kinase activity, recombination at rDNA loci, and silencing. The Journal of biological chemistry. 2003;278:13390–13397. doi: 10.1074/jbc.M212818200. [DOI] [PubMed] [Google Scholar]
- 33.Xiao B, Sanders MJ, Carmena D, et al. Structural basis of AMPK regulation by small molecule activators. Nat Commun. 2013;4:3017. doi: 10.1038/ncomms4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 35.Treitel MA, Kuchin S, Carlson M. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Molecular & Cellular Biology. 1998;18:6273–6280. doi: 10.1128/mcb.18.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serra-Cardona A, Petrezselyova S, Canadell D, Ramos J, Arino J. Coregulated expression of the Na+/phosphate Pho89 transporter and Ena1 Na+-ATPase allows their functional coupling under high-pH stress. Mol. Cell. Biol. 2014;34:4420–4435. doi: 10.1128/MCB.01089-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeVit MJ, Johnston M. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr. Biol. 1999;9:1231–1241. doi: 10.1016/s0960-9822(99)80503-x. [DOI] [PubMed] [Google Scholar]
- 38.Bendrioua L, Smedh M, Almquist J, Cvijovic M, Jirstrand M, Goksor M, Adiels CB, Hohmann S. Yeast AMP-activated protein kinase monitors glucose concentration changes and absolute glucose levels. The Journal of biological chemistry. 2014;289:12863–12875. doi: 10.1074/jbc.M114.547976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elbing K, McCartney RR, Schmidt MC. Purification and characterization of the three Snf1-activating kinases of Saccharomyces cerevisiae. Biochem. J. 2006;393:797–805. doi: 10.1042/BJ20051213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nath N, McCartney RR, Schmidt MC. Purification and characterization of Snf1 kinase complexes containing a defined Beta subunit composition. The Journal of biological chemistry. 2002;277:50403–50408. doi: 10.1074/jbc.M207058200. [DOI] [PubMed] [Google Scholar]
- 41.Zhang CS, Jiang B, Li M, et al. The lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell metabolism. 2014;20:526–540. doi: 10.1016/j.cmet.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 42.O’Donnell AF, McCartney RR, Chandrashekarappa DG, Zhang BB, Thorner J, Schmidt MC. 2-Deoxyglucose impairs Saccharomyces cerevisiae growth by stimulating Snf1-regulated and alpha-arrestin-mediated trafficking of hexose transporters 1 and 3. Mol. Cell. Biol. 2015;35:939–955. doi: 10.1128/MCB.01183-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye T, Elbing K, Hohmann S. The pathway by which the yeast protein kinase Snf1p controls acquisition of sodium tolerance is different from that mediating glucose regulation. Microbiology. 2008;154:2814–2826. doi: 10.1099/mic.0.2008/020149-0. [DOI] [PubMed] [Google Scholar]