Abstract
Background
Pentraxin 3 (PTX3) is expressed in the heart under inflammatory conditions and plays an important role in atherogenesis. Patients with increased PTX3 levels may suffer from higher rates of cardiac events. Regulation of specific genes by promoter methylation is important in atherogenesis. The factors influencing PTX3 levels and the association between epigenetics and PTX3 levels have not been investigated.
Methods
Blood samples were collected from 64 patients admitted to the Department of Cardiology, 35 who had coronary artery disease (CAD), and 29 who were CAD-free. Plasma levels of PTX3 were measured by ELISA. PTX3 promoter methylation was evaluated via methyl-specific PCR. The severity of coronary artery lesion was evaluated by angiography.
Results
The level of PTX3 promoter methylation in the CAD group was 62.69% ± 20.57%, significantly lower than that of the CAD-free group, which was 72.45% ± 11.84% (P = 0.03). Lower PTX3 promoter methylation levels in the CAD group were associated with higher plasma PTX3 concentrations (r = −0.29, P = 0.02). Furthermore, lower PTX3 promoter methylation levels were associated with higher neutrophil to lymphocyte ratio (NLR) in men (r = −0.58, P = 0.002).
Conclusions
The present study provides new evidence that methylation of the PTX3 promoter is associated with PTX3 plasma levels and NLR in coronary artery disease. This study also shows that modification of epigenetics by chronic inflammation might be a significant molecular mechanism in the atherosclerotic processes that influence plasma PTX3 concentrations.
Keywords: Coronary artery disease, Lymphocyte, Methylation, Neutrophil, Pentraxin 3
1. Introduction
Pentraxin 3 (PTX3) is a member of the pentraxin superfamily, a family of acute phase proteins that includes two classical short pentraxins, C reactive protein (CRP) and serum amyloid-P (SAP). In contrast to hepatically produced CRP and SAP, PTX3 is produced at sites of inflammation by several cell types, primarily dendritic cells, macrophages, neutrophils, fibroblasts, activated endothelial cells, and renal cells.[1],[2] PTX3 is a type of soluble pattern recognition receptor that protects against extracellular microbes and against autoimmunity.[3]
Mounting evidence indicates that PTX3 is involved in atherosclerotic diseases. PTX3 is clearly expressed in the setting of advanced atherosclerotic lesions.[4] Plasma PTX3 levels correlate positively with lipid volume percentages and correlate negatively with fibrous volume percentages.[5] PTX3 may be a potent and independent predictor of subsequent cardiac events in patients hospitalized for coronary artery disease (CAD).[6],[7]
Several genes have been implicated in CAD susceptibility.[8] Studies have reported the effects of PTX3 genetic variants in human diseases, although these were limited to infections and female fertility.[9]–[11] One recent study reported that PTX3 plasma levels were influenced by three PTX3 polymorphisms. However, a genetic predisposition to high PTX3 levels does not influence the risk for AMI.[12]
Gene expression is influenced by epigenetic mechanisms as well as transcription and translation.[13] Hypermethylation of gene promoter regions can result in transcriptional silencing of the gene. The epigenetic control of PTX3 gene expression in relation to atherosclerotic disease has not been investigated. Chronic inflammation has been proposed to induce changes in the epigenetic program.[14] Therefore, the present study was designed to determine the relationship between PTX3 promoter methylation and plasma PTX3 levels, as well as the chronic inflammatory biomarker neutrophil to lymphocyte ratio (NLR) in CAD.
2. Methods
2.1. Ethics statement
Written informed consent to participate in the study, including blood sampling, was provided by each subject before the beginning of the study. The investigation conformed to the principles outlined in the Declaration of Helsinki and was approved by the Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology (IORG No.: IORG0003571).
2.2. Patients
We studied 64 unrelated subjects of both genders who were recruited from the Department of Cardiology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology (China). Patients were required to undergo diagnostic coronary angiography. Individuals were classified into two groups (CAD or CAD-free) based on their medical records and evidence of diagnostic electrocardiographic and enzyme changes, coronary angiography results or clinical symptoms. CAD inclusion criteria included coronary arteries with stenosis greater than 50% of the luminal diameter (angiographic). Only five of the CAD patients had been diagnosed with acute coronary syndrome. Patients with infectious diseases or other inflammatory diseases were excluded from this study.[5],[15]
2.3. Collection and processing of the blood samples
Venous blood was collected from CAD and control patients in a vacuum tube with pre-cooled EDTA and aprotinin. DNA was prepared from a population of blood cell types, incubated with proteinase K, and treated with RNase A, followed by the TIANamp Genomic DNA Kit [DP304, Tiangen Biotech (Beijing) CO.,LTD]. Plasma was separated within 2 h of blood collection and kept at −80 °C until it was used further for analysis.
2.4. Epigenetic analysis: methylation of the PTX3 gene promoter
DNA was amplified using a methylation-specific primer set, PTX3-MF: 5′-CGTTTGCGGTTAGGAGTATTC-3′ and PTX3-MR: 5′-CAAAACGTCGTCCGTAACTTA-3′, or a non-methylation-specific primer set, PTX3-UF: 5′-TGTGT TTGTGGTTAGGAGTATTTG-3′ and PTX3-UR: 5′-CAA AACATCATCCATAACTTA-3′,[16] in a total volume of 20 µL, using 0.5 units of hot-start Taq-polymerase (Takara, Japan) per reaction. The size of the non-methylated amplicon was 105 bp, and the methylated amplicon was 103 bp. The region where these primers bind or amplify is as follows: CGTTTGCGGTTAGGAGTATTCGGAATGGGAT AAGTTTTTTATTATGTTGGAGAATTCGTAGATGAG AGAGCGTATGTTGTTGTAAGTTACGGACGACGTTT TG. Cycle conditions were as follows: initial denaturation and hot start for 10 min at 95°C, then 35 cycles consisting of 30 s at 94°C, 30 s at 59°C (for methylated reactions) or 50°C (for unmethylated reactions), and 45 s at 72°C, and a final extension of 10 min at 72°C.[16] Each methyl- or non-methyl band was visualized using an Alpha Imager 2200 system (Alpha Innotech Co.). The densities of each band were calculated with the Alpha Imager 2200 system. The indices of methylation were expressed as percent densities of methyl band/(methyl band + non-methyl band).[17]
2.5. PTX3 assays
This assay utilized the quantitative sandwich enzyme immunoassay technique. Plasma PTX3 levels were measured via the ELISA assay from R&D Systems, Inc. (Minneapolis, America) in a subset of subjects with available samples of whole-blood. A streptavidin-coated plate was incubated with biotinylated monoclonal antibodies specific for human PTX3. The minimum detectable dose (MDD) of human PTX3 ranged from 0.007−0.116 ng/mL. The mean MDD was 0.025 ng/mL.
2.6. Statistical analysis
All analyses were performed with R (http://www.R-project.org) and EmpowerStats software (http://www.empowerstats.com, X&Y solutions, Inc., Boston, MA, USA). We firstly compared the data distribution of each covariate between the CAD and the CAD-free groups, using Kruskal-Wallis rank sum tests for continuous variables and chi-square tests for categorical data, as shown in Table 1. One-way ANOVA was used to compare the difference of PTX3 plasma concentrations and PTX3 methylation levels between CAD-free and CAD patients. Pearson's correlation was used to assess the association between PTX3 methylation levels and PTX3 plasma concentrations, as well as NLR. Data were expressed as the mean ± SD. Statistical significance was defined as P < 0.05.
Table 1. Clinical and biochemical characteristics of the study subjects.
| Parameters | CAD-free | CAD | P-value |
| n = 29 | n = 35 | ||
| Age, yrs | 57.5 ± 10.8 | 63.1 ± 10.0 | 0.034 |
| FBG, mmol/L | 5.2 ± 0.8 | 5.1 ± 0.5 | 0.627 |
| Cholesterol, mmol/L | 4.0 ± 1.1 | 4.4 ± 0.9 | 0.085 |
| LDL-C, mmol/L | 2.0 ± 0.6 | 2.3 ± 0.6 | 0.058 |
| TG, mmol/L | 1.7 ± 1.7 | 1.5 ± 0.7 | 0.592 |
| HDL-C, mmol/L | 1.2 ± 0.4 | 1.3 ± 0.4 | 0.406 |
| Gender | 0.094 | ||
| Female | 16 (55.2%) | 12 (34.3%) | |
| Male | 13 (44.8%) | 23 (65.7%) | |
| Smoking | 0.783 | ||
| No | 20 (69.0%) | 23 (65.7%) | |
| Yes | 9 (31.0%) | 12 (34.3%) | |
| Hypertension | 0.665 | ||
| No | 14 (48.3%) | 15 (42.9%) | |
| Yes | 15 (51.7%) | 20 (57.1%) | |
| History of statin use | 0.362 | ||
| No | 19 (65.5%) | 19 (54.3%) | |
| Yes | 10 (34.5%) | 16 (45.7%) |
Results are shown as the mean ± SD or n (%). CAD: coronary artery disease; CHOL: cholesterol; FBG: fasting blood glucose; HDL: high density lipoprotein; LDL: low density lipoprotein; TG: triglycerides.
3. Results
Primary clinical and biochemical characteristics are presented in Table 1. Except for age, there were no differences in the clinical and biochemical characteristics between the CAD and CAD-free groups, such as gender, smoking, hypertension, fasting blood glucose, cholesterol, low density lipoproteins, high density lipoproteins, triglycerides or the history of statin use.
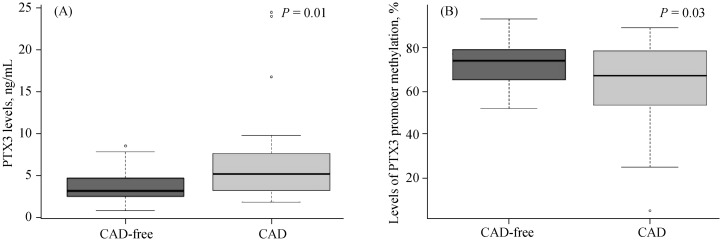
As shown in Figure 1, the PTX3 plasma concentration in the CAD group was 6.57 ± 5.45 ng/mL, higher than the CAD-free group, which was 3.65 ± 2.14 ng/mL (P = 0.01, Figure 1A). Moreover, the mean PTX3 promoter methylation in the CAD group was 62.69% ± 20.57%, significantly lower than the level in the CAD-free group, which was 72.45% ± 11.84% (P = 0.03, Figure 1B).
Figure 1. PTX3 levels and levels of PTX3 promoter methylation according to CAD occurrence.
(A): PTX3 levels in the CAD-free group were 3.65 ± 2.14 ng/mL (n = 25), lower than the CAD group (n = 39); (B): the levels of PTX3 promoter methylation in the CAD-free group were 72.45% ± 11.84% (n = 25), higher than the CAD group (62.69% ± 20.57%, n = 39). CAD: coronary artery disease; PTX3: Pentraxin 3.
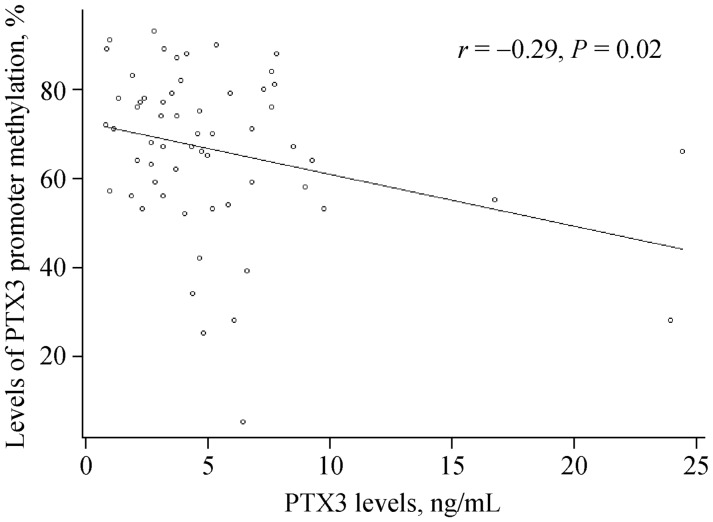
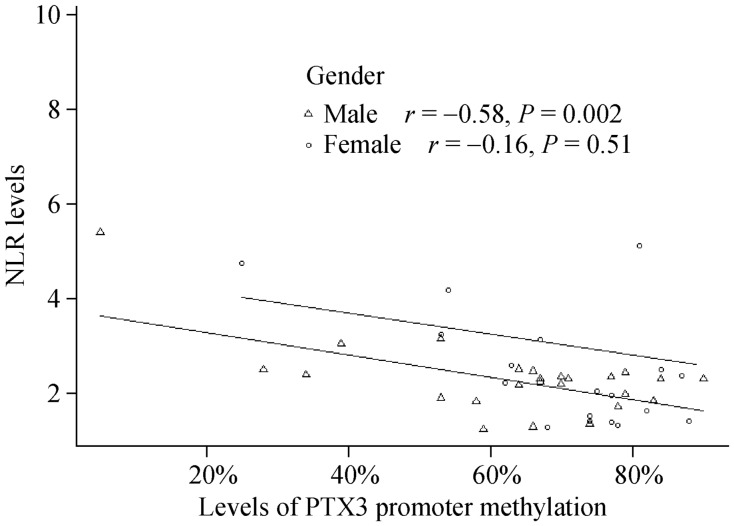
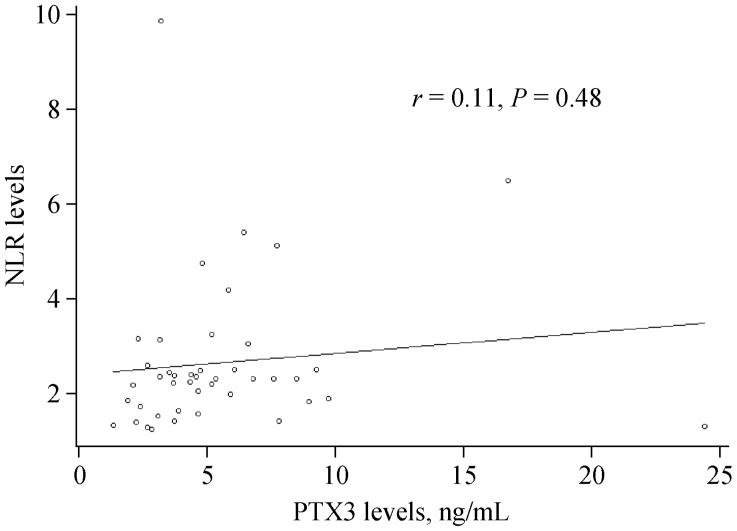
We used Pearson's correlation analyses to determine the relationship between PTX3 promoter methylation and plasma PTX3 levels as well as between plasma PTX3 levels and NLR. Our results demonstrate that the level of PTX3 promoter methylation negatively correlated with plasma PTX3 levels (r = −0.29, P = 0.02, Figure 2). We also found that PTX3 promoter methylation levels negatively correlated with NLR in men (r = −0.58, P = 0.001, Figure 3). However, NLR was not correlated with PTX3 levels regardless of gender (r = 0.11, P = 0.48, Figure 4).
Figure 2. Association between PTX3 levels and levels of PTX3 promoter methylation.
Levels of PTX3 promoter methylation are negatively correlated with plasma PTX3 levels (r = −0.29, P = 0.02). PTX3: Pentraxin 3.
Figure 3. Association between the NLR and levels of PTX3 promoter methylation.
Levels of PTX3 promoter methylation are negatively correlated with NLR in male patients (r = −0.58, P = 0.002). NLR: neutrophil to lymphocyte ratio; PTX3: Pentraxin 3.
Figure 4. Association between the NLR and PTX3 levels.
NLR was not correlated with PTX3 levels (r = 0.11, P = 0.48). NLR: neutrophil to lymphocyte ratio; PTX3: Pentraxin 3.
4. Discussion
This study has revealed that the patients suffering from CAD had lower levels of PTX3 promoter methylation than CAD-free patients. Lower PTX3 promoter methylation levels in CAD patients were associated with higher plasma PTX3 concentrations. This is the first study, to our knowledge, to address the association of the PTX3 promoter methylation with CAD and its relationship with plasma PTX3 levels.
Epigenetic modifications play a fundamental role in the inhibition of gene expression. They involve specific histone tail posttranslational modifications, promoter DNA methylation and non-coding RNAs, particularly microRNAs.[18] However, promoter methylation is the most widely studied and the best characterized epigenetic marker. Changes in promoter methylation may contribute to disease phenotypes. Specific genes regulated by promoter methylation are important in atherogenesis, and important examples of promoter methylation-regulated genes include genes encoding interferons, platelet-derived growth factors, MMP-2, MMP-7, MMP-9, tissue inhibitor of metalloproteinases, intracellular adhesion molecules, oestrogen receptor α, extracellular superoxide dismutase and p53.[19] Hypermethylation of gene promoter regions can result in silencing of the gene.[20]
A significant association between PTX3 plasma concentrations and the risk of all-cause mortality at three years has been shown in AMI patients. However, genetic predisposition to high PTX3 levels does not influence the risk of acute AMI.[12] There was an inter-relationship of genetic and epigenetic modifications at the DNA promoter site. Further studies are necessary to explore the relationship between PTX3 genetic polymorphisms (SNPs) and promoter methylation. Genetic predisposition to high PTX3 levels could be reduced by hypermethylation. It has been reported that PTX3 SNPs are related to high plasma levels of a functionally less active protein. The SNP (rs3816527) could cause an amino acid change (Asp48Ala) in a strategic position of the PTX3 primary structure. This could potentially interfere with the N-terminally mediated binding of PTX3 to its ligands. Moreover, amino acid 48 is located between two cysteine residues at positions 47 and 49, which are involved in the formation of the inter-chain disulphide bonds required for the tetrameric arrangement of four PTX3 protomers.[21] This provides a possible explanation for the genetic predisposition to high PTX3 levels not influencing the risk of AMI.
We also show here that lower PTX3 promoter methylation levels are associated with higher NLR in men. NLR, an inflammatory biomarker, may be of predictive and prognostic value for cardiovascular events.[22] A higher NLR may reflect a low-grade inflammation status. Chronic inflammation could induce a change of the epigenetic program.[14] PTX3 is produced at sites of inflammation by several cells, such as neutrophils and monocytes.[23],[24] This provides a novel hypothesis whereby proinflammatory factors not only increase leukocytes but also modify methylation of the PTX3 promoter. Further studies are needed to explore the mechanism underlying the involvement of inflammation in the change of PTX3 promoter methylation.
We found that PTX3 promoter methylation levels were associated with NLR in males but not females. The X-chromosome inactivation that is characterized by a hypomethylated inactive X-chromosome, as confirmed by previous reports may underlie this finding.[25] However, from the data presented in this study, we cannot determine whether this observed gender difference in methylation levels could originate from the inactivated X-chromosome. A detailed future study of the methylation of X-linked or autosomal specific PTX3 regions are necessary to clarify this phenomenon.
The NLR did not correlate with PTX3 levels (r = 0.11, P = 0.48). PTX3 levels may be affected by PTX3 genetic polymorphisms. A previous study has shown that three PTX3 SNPs (rs2305619, rs3816527, and rs1840680) were significantly associated with different PTX3 plasma levels.[12] Therefore, PTX3 expression is not only associated with environment factors but also with genetic factors.
Our sample size, despite being relatively small for association studies, did not influence the present conclusion. We tested the different factors that may influence the results between CAD and CAD-free groups. Except for age, there were no differences in the clinical and biochemical characteristics, such as gender, smoking, hypertension, fasting blood glucose, cholesterol, low density lipoproteins, high density lipoproteins, triglycerides or history of statin use. The age difference between CAD and CAD-free groups was not associated with PTX3 promoter methylation in men. We conclude that PTX3 promoter methylation is associated with PTX3 plasma levels and NLR and that this association is independent of age and other clinical and biochemical factors.
5. Conclusions
This study confirms that lower PTX3 promoter methylation levels are associated with higher plasma PTX3 levels and higher NLR. These findings could help our understanding of a novel epigenetic molecular mechanism involved in the pathophysiological processes leading to CAD, in addition to genetic predisposition.
Acknowledgments
We acknowledge Dr. Lei Ye who made substantial contributions to statistical analysis.
References
- 1.Kravitz MS, Pitashny M, Shoenfeld Y. Protective molecules--c-reactive protein (CRP), serum amyloid p (SAP), pentraxin3 (PTX3), mannose-binding lectin (MBL), and apolipoprotein a1 (Apo A1), and their autoantibodies: Prevalence and clinical significance in autoimmunity. J Clin Immunol. 2005;25:582–591. doi: 10.1007/s10875-005-7828-2. [DOI] [PubMed] [Google Scholar]
- 2.Razvina O, Jiang S, Matsubara K, et al. Differential expression of pentraxin 3 in neutrophils. Exp Mol Pathol. 2015;98:33–40. doi: 10.1016/j.yexmp.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Baruah P, Propato A, Dumitriu IE, et al. The pattern recognition receptor ptx3 is recruited at the synapse between dying and dendritic cells, and edits the cross-presentation of self, viral, and tumor antigens. Blood. 2006;107:151–158. doi: 10.1182/blood-2005-03-1112. [DOI] [PubMed] [Google Scholar]
- 4.Savchenko A, Imamura M, Ohashi R, et al. Expression of pentraxin 3 (ptx3) in human atherosclerotic lesions. J Pathol. 2008;215:48–55. doi: 10.1002/path.2314. [DOI] [PubMed] [Google Scholar]
- 5.Iwata A, Miura S, Tanaka T, et al. Plasma pentraxin-3 levels are associated with coronary plaque vulnerability and are decreased by statin. Coron Artery Dis. 2012;23:315–321. doi: 10.1097/MCA.0b013e328352ffec. [DOI] [PubMed] [Google Scholar]
- 6.Latini R, Maggioni AP, Peri G, et al. Prognostic significance of the long pentraxin ptx3 in acute myocardial infarction. Circulation. 2004;110:2349–2354. doi: 10.1161/01.CIR.0000145167.30987.2E. [DOI] [PubMed] [Google Scholar]
- 7.Matsui S, Ishii J, Kitagawa F, et al. Pentraxin 3 in unstable angina and non-ST-segment elevation myocardial infarction. Atherosclerosis. 2010;210:220–225. doi: 10.1016/j.atherosclerosis.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 8.Lusis AJ, Fogelman AM, Fonarow GC. Genetic basis of atherosclerosis: Part I: New genes and pathways. Circulation. 2004;110:1868–1873. doi: 10.1161/01.CIR.0000143041.58692.CC. [DOI] [PubMed] [Google Scholar]
- 9.May L, Kuningas M, van Bodegom D, et al. Genetic variation in pentraxin (ptx) 3 gene associates with ptx3 production and fertility in women. Biol Reprod. 2010;82:299–304. doi: 10.1095/biolreprod.109.079111. [DOI] [PubMed] [Google Scholar]
- 10.Chiarini M, Sabelli C, Melotti P, et al. Ptx3 genetic variations affect the risk of pseudomonas aeruginosa airway colonization in cystic fibrosis patients. Genes Immun. 2010;11:665–670. doi: 10.1038/gene.2010.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olesen R, Wejse C, Velez DR, et al. Dc-sign (cd209), pentraxin 3 and vitamin D receptor gene variants associate with pulmonary tuberculosis risk in west africans. Genes Immun. 2007;8:456–467. doi: 10.1038/sj.gene.6364410. [DOI] [PubMed] [Google Scholar]
- 12.Barbati E, Specchia C, Villella M, et al. Influence of pentraxin 3 (PTX3) genetic variants on myocardial infarction risk and ptx3 plasma levels. PLoS One. 2012;7:e53030. doi: 10.1371/journal.pone.0053030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibney ER, Nolan CM. Epigenetics and gene expression. Heredity (Edinb) 2010;105:4–13. doi: 10.1038/hdy.2010.54. [DOI] [PubMed] [Google Scholar]
- 14.Abu-Remaileh M, Bender S, Raddatz G, et al. Chronic inflammation induces a novel epigenetic program that is conserved in intestinal adenomas and in colorectal cancer. Cancer Res. 2015;75:2120–2130. doi: 10.1158/0008-5472.CAN-14-3295. [DOI] [PubMed] [Google Scholar]
- 15.Muller B, Peri G, Doni A, et al. Circulating levels of the long pentraxin ptx3 correlate with severity of infection in critically ill patients. Crit Care Med. 2001;29:1404–1407. doi: 10.1097/00003246-200107000-00017. [DOI] [PubMed] [Google Scholar]
- 16.Wang JX, He YL, Zhu ST, et al. Aberrant methylation of the 3q25 tumor suppressor gene ptx3 in human esophageal squamous cell carcinoma. World J Gastroenterol. 2011;17:4225–4230. doi: 10.3748/wjg.v17.i37.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friso S, Lotto V, Choi SW, et al. Promoter methylation in coagulation f7 gene influences plasma fvii concentrations and relates to coronary artery disease. J Med Genet. 2012;49:192–199. doi: 10.1136/jmedgenet-2011-100195. [DOI] [PubMed] [Google Scholar]
- 18.Borghini A, Cervelli T, Galli A, Andreassi MG. DNA modifications in atherosclerosis: From the past to the future. Atherosclerosis. 2013;230:202–209. doi: 10.1016/j.atherosclerosis.2013.07.038. [DOI] [PubMed] [Google Scholar]
- 19.Hiltunen MO, Yla-Herttuala S. DNA methylation, smooth muscle cells, and atherogenesis. Arterioscler Thromb Vasc Biol. 2003;23:1750–1753. doi: 10.1161/01.ATV.0000092871.30563.41. [DOI] [PubMed] [Google Scholar]
- 20.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 21.Inforzato A, Rivieccio V, Morreale AP, et al. Structural characterization of ptx3 disulfide bond network and its multimeric status in cumulus matrix organization. J Biol Chem. 2008;283:10147–10161. doi: 10.1074/jbc.M708535200. [DOI] [PubMed] [Google Scholar]
- 22.Verdoia M, Barbieri L, Di Giovine G, et al. Neutrophil to lymphocyte ratio and the extent of coronary artery disease: Results from a large cohort study. Angiology. 2016;67:75–82. doi: 10.1177/0003319715577529. [DOI] [PubMed] [Google Scholar]
- 23.Jaillon S, Peri G, Delneste Y, et al. The humoral pattern recognition receptor ptx3 is stored in neutrophil granules and localizes in extracellular traps. J Exp Med. 2007;204:793–804. doi: 10.1084/jem.20061301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma YJ, Doni A, Romani L, et al. Ficolin-1-ptx3 complex formation promotes clearance of altered self-cells and modulates il-8 production. J Immunol. 2013;191:1324–1333. doi: 10.4049/jimmunol.1300382. [DOI] [PubMed] [Google Scholar]
- 25.Hellman A, Chess A. Gene body-specific methylation on the active x chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]