Abstract
Lineages of hypervirulent Aeromonas hydrophila (vAh) are the cause of persistent outbreaks of motile Aeromonas septicemia in warm-water fishes worldwide. Over the last decade, this virulent lineage of A. hydrophila has resulted in annual losses of millions of tons of farmed carp and catfish in the People's Republic of China and the United States (US). Multiple lines of evidence indicate US catfish and Asian carp isolates of A. hydrophila affiliated with sequence type 251 (ST251) share a recent common ancestor. To address the genomic context for the putative intercontinental transfer and subsequent geographic spread of this pathogen, we conducted a core genome phylogenetic analysis on 61 Aeromonas spp. genomes, of which 40 were affiliated with A. hydrophila, with 26 identified as epidemic strains. Phylogenetic analyses indicate all ST251 strains form a coherent lineage affiliated with A. hydrophila. Within this lineage, conserved genetic loci unique to A. hydrophila were identified, with some genes present in consistently higher copy numbers than in non-epidemic A. hydrophila isolates. In addition, results from analyses of representative ST251 isolates support the conclusion that multiple lineages are present within US vAh isolated from Mississippi, whereas vAh isolated from Alabama appear clonal. This is the first report of genomic heterogeneity within US vAh isolates, with some Mississippi isolates showing closer affiliation with the Asian grass carp isolate ZC1 than other vAh isolated in the US. To evaluate the biological significance of the identified heterogeneity, comparative disease challenges were conducted with representatives of different vAh genotypes. These studies revealed that isolate ZC1 yielded significantly lower mortality in channel catfish, relative to Alabama and Mississippi vAh isolates. Like other Asian vAh isolates, the ZC1 lineage contains all core genes for a complete type VI secretion system (T6SS). In contrast, more virulent US isolates retain only remnants of the T6SS (clpB, hcp, vgrG, and vasH) which may have functional implications. Collectively, these results characterize a hypervirulent A. hydrophila pathotype that affects farmed fish on multiple continents.
Keywords: Aeromonas hydrophila, pathogenesis, comparative genomics, emerging disease, bacteria, catfish, carp
Introduction
Aquaculture industries across the world have been decimated by epidemics of a hypervirulent pathotype of A. hydrophila (vAh) (Nielsen et al., 2001; Hemstreet, 2010). A. hydrophila is ubiquitous within warm-water environments and has a diverse host range (i.e., amphibians, birds, fishes, reptiles, and mammals) with equally diverse diseases that include motile Aeromonas septicemia (MAS) in fish. MAS produces significant internal and external hemorrhage and exophthalmia, often followed by mortality within several hours of manifestation of disease (Xu et al., 1993; Camus et al., 1998; Nielsen et al., 2001; da Silva et al., 2012; Beaz-Hidalgo et al., 2013).
The first report of the vAh pathotype was A. hydrophila J-1 in 1989, which was isolated from epizootics of MAS in China's Jiangsu Province (Chen and Lu, 1991). This strain was categorized as sequence type 251 (ST251) and recognized as being capable of causing high mortality in grass carp (Ctenopharyngodon idella). Outbreaks of MAS in farmed carp have persisted in China, resulting in losses estimated at 2,200 tons of dead fish per year (Chen and Lu, 1991; Nielsen et al., 2001; Zhang et al., 2002; Pang et al., 2015). Epidemics of ST251-associated MAS occurred within the same province in 2010, with vAh isolate A. hydrophila NJ-35 identified as the etiologic agent (Pang et al., 2012). Another vAh isolate from China (A. hydrophila ZC1) was isolated from grass carp exhibiting signs of hemorrhagic septicemia from an aquaculture farm in China's Guangdong Province (Deng et al., 2009). In general, MAS is a regular occurrence each summer, resulting in significant economic losses in the Chinese aquaculture industry, with estimates exceeding five billion yuan per year (Prof. Hui Chen, personal communication).
In 2004, the first reported case of ST251-related MAS in the US arose when A. hydrophila S04-690 was isolated from diseased channel catfish (Ictalurus punctatus) from a catfish farm in Washington County, Mississippi (MS) (Hossain et al., 2014). Beginning in 2009, vAh strains were consistently recovered from recurring outbreaks of MAS in aquaculture ponds in western Alabama (AL) with a reported 2,000 tons of dead fish in the first year (Hemstreet, 2010). To date, this number has grown to exceed an estimated 10,500 tons, with vAh isolates representing the largest percentage (35%) of disease cases at the Alabama Fish Farming Center (Hemstreet, 2015). Although representative data on production losses attributed solely to vAh are difficult to attain, vAh clearly represents a significant threat to warm-water aquaculture industries.
Previous comparative genomic analyses of vAh strains isolated from catfish in the US and carp in China indicated these strains share a recent common ancestor (Hossain et al., 2014). Within this monophyletic clade, vAh strains isolated from carp and catfish have unique phenotypes and genotypes (L-fucose metabolism, an inducible prophage and the ability to use myo-inositol as a sole carbon source) that distinguish them from more typical strains of A. hydrophila not associated with epizootics (non-vAh) (Hossain et al., 2013, 2014; Pang et al., 2015). These studies also demonstrated that MS vAh strain S04-690 is more similar to the carp isolate ZC1 than to other vAh strains from AL (Hossain et al., 2014). Since this study was published, MAS outbreaks attributed to vAh have spread to the Delta region of west Mississippi, with anecdotal reports from the industry suggesting annual losses in MS now exceed 150 tons. The purpose of this study was to characterize the vAh pathotype by examining vAh strains collected from farmed catfish in AL and MS in recent years and compare genome sequences of all available ST251 strains together with other Aeromonas spp. genomes available in GenBank by phylogenomic analysis, determine the presence of putative virulence factors in vAh and non-vAh lineages and assess the relative capacity of selected strains to cause MAS in channel catfish.
Methods
Bacterial strains–disease isolates and catfish challenge
A. hydrophila isolates for disease challenge were recovered from diseased channel catfish from commercial aquaculture operations in western Alabama and the Delta region of west Mississippi. Briefly, catfish demonstrating symptoms typical of MAS were collected in a moribund state and submitted for diagnostic evaluation and necropsy at Auburn University in Auburn, AL or Mississippi State University's Aquatic Research and Diagnostic Laboratory at the Thad Cochran National Warmwater Aquaculture Center (NWAC) in Stoneville, MS. Liver and kidney tissues were sampled for aerobic bacterial cultures. Samples of tissue were homogenized in sterile phosphate buffered saline and portions of the homogenate streaked onto tryptic soy agar (TSA; Beckton Dickinson, Franklin Lakes, NJ) or brain heart infusion (BHI; Beckton Dickinson) for bacterial isolation. Pure cultures were identified as vAh strains by the vAh-specific qPCR method previously described and/or utilization of myo-inositol as a sole carbon source (Griffin et al., 2013; Hanson et al., 2014). From this sampling design, A. hydrophila isolates ML09-119, ML10-51K, S04-690, S14-296, and S14-452 were cryogenically preserved (mixed with 50% glycerol, stored at −80°C) and subsequently used in catfish immersion challenges.
Infection and histopathology of channel catfish using an immersion challenge model
Channel catfish were obtained as fry from the USDA-ARS Warmwater Aquaculture Research Unit housed at NWAC and reared to experimental size in 340 liter troughs supplied with 26 ± 2°C dechlorinated municipal water under pathogen-free conditions. All animal experiments were approved by, and conducted in compliance with, regulations of the Institutional Animal Care and Use Committee of the Aquatic Animal Health Research Unit (USDA-ARS) in Auburn, Alabama. Water temperature was maintained at 26 ± 2°C with a centralized heater. Prior to trials, heart, liver, head kidney, trunk kidney, spleen, brain, and skeletal muscle tissues were collected from 10 randomly sampled fish and sampled by culturing on BHI to verify fish were not presently infected with vAh.
Two hundred catfish fingerlings, with a mean weight of 111 ± 47 g and length of 19 ± 3 cm, were acclimated for 12 days in 56-L glass aquaria (10 fish per tank, 3 tanks per isolate, and 2 mock infected control tanks) containing about 50-L water prior to challenge. The immersion challenge was conducted using the recently described fin clip method (Zhang et al., 2016). At the time of infection, water volume was reduced to 15-L per tank. To sedate animals for handling, fish were netted from individual aquaria and placed into a container filled with 20-L of dechlorinated water containing 150 mg/L of buffered Tricaine-S (tricaine methanesulfonate; Western Chemical, Inc., Ferndale, WA). Once fish were anesthetized, the adipose fin was clipped at its base and fish were returned to respective aquaria for recovery from anesthesia.
For the bacterial challenge, 100 mL of tryptic soy broth (TSB) containing approximately 3.0 × 109 CFU/mL of the respective A. hydrophila strains (ML-09-119, ML10-51K, S04-690, S14-296, S14-452, and ZC1) was added to each of three aquaria, resulting in an approximate challenge dose of ~2.0 × 107 CFU/mL. Two tanks served as mock infected controls, receiving only 100 mL sterile TSB. After 1-h exposure, water flow to aquaria (0.5 L/min) was resumed. Fish mortality was monitored daily for 7 days. At least 50% of dead fish were sampled for confirmation for the presence of vAh in liver and kidney tissues using M9 minimal medium containing 0.3% (w/v) myo-inositol (M9I) agar (Hanson et al., 2014). Moribund fish were removed from aquaria daily and surviving fish were euthanized by at least 15 min exposure to 300 mg/L buffered Tricaine-S solution, then necropsied. Heart, liver, head kidney, trunk kidney, spleen, brain, and skeletal muscle tissues were harvested and fixed in 10% buffered formalin for histopathology. Tissues were also collected and used for bacterial identification and quantitation. Formalin fixed tissues were processed and embedded in paraffin. The tissues were cut in four micron sections, stained with hematoxylin and eosin and evaluated for microscopic lesions by light microscopy.
Bacterial strains–comparative genomics
In total, 61 complete and draft Aeromonas spp. genomes were included in this study (Table 1) which included A. hydrophila representative genomes as well as Aeromonas spp. genomes that are now recognized as having had an erroneous affiliation with A. hydrophila (Figueras et al., 2014; Beaz-Hidalgo et al., 2015a). These genomes were retrieved from the US National Center for Biotechnology Information (NCBI) GenBank database and included two A. caviae isolates, one A. dhakensis isolate, one A. enteropelogenes isolate, one A. media isolate, one A. molluscorum isolate, one A. taiwanensis isolate, one Aeromonas sp. isolate, 26 A. hydrophila fish vAh disease isolates, and 28 non-vAh A. hydrophila isolates.
Table 1.
Bacterial genomes used in comparative genomic analyses.
| Strain | Phenotype | Isolation source | GenBank species assignation | Species based on phylogeny and ANI | Accession | Reference |
|---|---|---|---|---|---|---|
| Ae398 | Non-vAh | Human | A. caviae | A. caviae | SAMEA2272404 | Beatson et al., 2011 |
| YL12 | Non-vAh | Compost | A. caviae | A. caviae | SAMN02870964 | Lim et al., 2014 |
| AAK1 | Non-vAh | Clinical | A. dhakensis | A. dhakensis | SAMD00036618 | Martínez-Murcía et al., 2008 |
| 1999lcr | Non-vAh | Clinical | A. trota | A. molluscorum | SAMN02732394 | Dallagassa et al., Unpublished |
| 116 | Non-vAh | Clinical | A. hydrophila | A. dhakensis | NZ_ANPN00000000.1 | Chan et al., 2011 |
| 14 | Non-vAh | Clinical | A. hydrophila | A. dhakensis | NZ_AOBM00000000.1 | Chan et al., 2011 |
| 173 | Non-vAh | Clinical | A. hydrophila | A. dhakensis | NZ_AOBN00000000.1 | Chan et al., 2011 |
| 187 | Non-vAh | Clinical | A. hydrophila | A. dhakensis | NZ_AOBO00000000.1 | Chan et al., 2011 |
| 226 | Non-vAh | Clinical | A. hydrophila | A. hydrophila | NZ_JEML00000000.1 | Chan et al., 2011 |
| 259 | Non-vAh | Clinical | A. hydrophila | A. dhakensis | NZ_AOBP00000000.1 | Chan et al., 2011 |
| 277 | Non-vAh | Clinical | A. hydrophila | A. dhakensis | NZ_AOBQ00000000.1 | Chan et al., 2011 |
| 4AK4 | Non-vAh | Industrial | A. hydrophila | A. sp. nov. | NZ_CP006579.1 | Gao et al., 2013 |
| AD9 | Non-vAh | Soil | A. hydrophila | A. hydrophila | NZ_JFJO00000000.1 | Lenneman and Barney, 2014 |
| Ae34 | Non-vAh | Koi carp | A. hydrophila | A. hydrophila | NZ_BAXY00000000.1 | Jagoda et al., 2014 |
| AH10 | Non-vAh | Grass carp | A. hydrophila | A. hydrophila | NZ_CP011100.1 | Xu et al., 2013 |
| AL06-01 | Non-vAh | Bluegill | A. hydrophila | A. hydrophila | SAMN01085623 | Hossain et al., 2013 |
| AL06-06 | Non-vAh | Goldfish | A. hydrophila | A. hydrophila | NZ_CP010947.1 | Tekedar et al., 2015 |
| AL10-121 | Non-vAh | Channel catfish | A. hydrophila | A. hydrophila | NZ_LRRW00000000.1 | Hossain, 2012 |
| AL97-91 | Non-vAh | Channel catfish | A. hydrophila | A. hydrophila | SAMN04967787 | Hossain, 2012 |
| ATCC7966T | Non-vAh | Milk tin | A. hydrophila | A. hydrophila | NC_008570.1 | Seshadri et al., 2006 |
| BWH65 | Non-vAh | Clinical | A. hydrophila | A. caviae | NZ_LESK00000000.1 | Earl et al., 2015 |
| E1 | Non-vAh | Clinical | A. hydrophila | A. hydrophila | SAMN01886638 | Grim et al., 2013 |
| E2 | Non-vAh | Clinical | A. hydrophila | A. hydrophila | SAMN01886639 | Grim et al., 2013 |
| GA97-22 | Non-vAh | Rainbow trout | A. hydrophila | Aeromonas spp. | SAMN01085627 | Hossain, 2012 |
| HZM | Non-vAh | Soil | A. hydrophila | A. caviae | SAMN02596469 | Chua et al., 2015 |
| KOR1 | Non-vAh | Mangrove | A. hydrophila | A. dhakensis | NZ_LJOE00000000.1 | Yin et al., 2015 |
| MN98-04 | Non-vAh | Tilapia | A. hydrophila | A. hydrophila | SAMN04967900 | Hossain, 2012 |
| RB-AH | Non-vAh | Soil | A. hydrophila | A. hydrophila | NZ_JPEH00000000.1 | Rheault et al., 2015 |
| S14-230 | Non-vAh | Tilapia | A. hydrophila | A. hydrophila | SAMN05292364 | This study |
| SSU | Non-vAh | Clinical | A. hydrophila | A. dhakensis | NZ_AGWR00000000.1 | Ribeiro et al., 2012 |
| TN97-08 | Non-vAh | Bluegill | A. hydrophila | A. hydrophila | NZ_LNUR00000000.1 | Hossain, 2012 |
| YL17 | Non-vAh | Compost | A. hydrophila | A. dhakensis | NZ_CP007518.2 | Lim et al., 2016 |
| WS | Non-vAh | Water sample | A. media | A. media | SAMN02472129 | Chai et al., 2012 |
| 848T | Non-vAh | Wedge-shells | A. molluscorum | A. molluscorum | SAMN02471397 | Spataro et al., 2013 |
| LMG24683T | Non-vAh | Unknown | A. taiwanensis | A. taiwanensis | SAMEA2752407 | Colston et al., 2014 |
| MDS8 | Non-vAh | Dairy sludge | Aeromonas sp. | A. dhakensis | SAMN02472124 | Raychaudhuri et al., 2013 |
| Ahy_Idx71 | vAh | Channel catfish | A. hydrophila | A. hydrophila | SAMN05292361 | This study |
| AL09-71 | vAh | Channel catfish | A. hydrophila | A. hydrophila | NZ_CP007566.1 | Pridgeon et al., 2014 |
| AL09-79 | vAh | Channel catfish | A. hydrophila | A. hydrophila | NZ_LRRV00000000.1 | Hossain, 2012 |
| ALG15-098 | vAh | Channel catfish | A. hydrophila | A. hydrophila | SAMN05223361 | This study |
| IPRS15-28 | vAh | Channel catfish | A. hydrophila | A. hydrophila | SAMN05223362 | This study |
| J-1 | vAh | Crucian carp | A. hydrophila | A. hydrophila | NZ_CP006883.1 | Pang et al., 2015 |
| JBN2301 | vAh | Crucian carp | A. hydrophila | A. hydrophila | NZ_CP013178.1 | Yang et al., 2016 |
| ML09-119 | vAh | Channel catfish | A. hydrophila | A. hydrophila | NC_021290.1 | Liles et al., 2011 |
| ML09-121 | vAh | Channel catfish | A. hydrophila | A. hydrophila | NZ_LRRX00000000.1 | Hossain, 2012 |
| ML09-122 | vAh | Channel catfish | A. hydrophila | A. hydrophila | NZ_LRRY00000000. | Hossain, 2012 |
| ML10-51K | vAh | Channel catfish | A. hydrophila | A. hydrophila | SAMN05223363 | This study |
| NJ-35 | vAh | Crucian carp | A. hydrophila | A. hydrophila | NZ_CP006870.1 | Pang et al., 2015 |
| PB10-118 | vAh | Channel catfish | A. hydrophila | A. hydrophila | SAMN01085622 | Hossain, 2012 |
| pc104A | vAh | Soil | A. hydrophila | A. hydrophila | NZ_CP007576.1 | Pridgeon et al., 2014 |
| S04-690 | vAh | Channel catfish | A. hydrophila | A. hydrophila | SAMN02404466 | Hossain et al., 2014 |
| S13-612 | vAh | Channel catfish | A. hydrophila | A. hydrophila | SAMN05292362 | This study |
| S13-700 | vAh | Channel catfish | A. hydrophila | A. hydrophila | SAMN05292363 | This study |
| S14-296 | vAh | Channel catfish | A. hydrophila | A. hydrophila | SAMN05292365 | This study |
| S14-452 | vAh | Channel catfish | A. hydrophila | A. hydrophila | SAMN05256776 | This study |
| S14-458 | vAh | Channel catfish | A. hydrophila | A. hydrophila | SAMN05223364 | This study |
| S14-606 | vAh | Channel catfish | A. hydrophila | A. hydrophila | SAMN05292366 | This study |
| S15-130 | vAh | Channel catfish | A. hydrophila | A. hydrophila | SAMN05223365 | This study |
| S15-242 | vAh | Channel catfish | A. hydrophila | A. hydrophila | SAMN05223366 | This study |
| S15-400 | vAh | Channel catfish | A. hydrophila | A. hydrophila | SAMN05223367 | This study |
| S15-591 | vAh | Channel catfish | A. hydrophila | A. hydrophila | SAMN05223368 | This study |
| ZC1 | vAh | Grass carp | A. hydrophila | A. hydrophila | SAMN02404465 | Hossain et al., 2014 |
Strains are indicated as virulent A. hydrophila (vAh) or other Aeromonas spp. based on their phylogenetic affiliation (Figure 1).
Genome sequencing
Strains representative of different vAh lineages were selected for Illumina sequencing based on results of vAh genotype-specific PCR (see below) from a total of 38 suspected vAh isolates recovered from diagnostic cases in MS between 2013 and 2015. Strains were selected so that isolates represented MAS outbreaks from multiple geographically discrete operations. Genome sequencing with 250 bp read-length using paired-end sequencing was performed on the Illumina MiSeq platform using the Nextera XT kit (Illumina, San Diego, CA) to prepare bar-coded fragment libraries according to the manufacturer's protocol. Sequence reads were trimmed and quality sequence reads were assembled de novo using the CLC Genomics Workbench (Qiagen, Redwood City, CA) using default settings. vAh strain draft genomes (n = 14) were generated for strains Ahy_Idx7_1, ALG15-098, IPRS-15-28, ML10-51K, S13-612, S13-700, S14-230, S14-296, S14-458, S14-606, S15-130, S15-242, S15-400, and S15-591 (Supplemental Table 1). In addition to a standard Illumina MiSeq run for vAh strain S14-452, a NxSeq 20 kb mate pair library was constructed and sequenced using an Illumina MiSeq at the Lucigen Corporation (Middleton, WI). The de novo assembly from the standard Illumina MiSeq sequences resulted in 13 contigs (80.15 average coverage) whereas the combination of these sequences together with the mate pair-derived sequences using de novo assembly with SPAdes (v3.5.0) resulted in a complete genome sequence.
Pathotype-specific PCR for vAh genotypes
Evaluation of the previously described vAh-specific qPCR primer set (listed as 2986L and 2986R in Table 2) (Griffin et al., 2013) was performed in silico using Geneious v. R9 with a maximum mismatch setting of two bases and a band prediction interval of between 100 and 1000 bases, which predicted that numerous vAh isolates (Ahy_Idx7_1, J-1, AL09-79, AL10-121, JBN2301, ML09-121, ML09-122, NJ-35, PB10-118, S13-612, and S15-591) would not produce an amplicon. To address this potential pitfall, we evaluated the primer set using touchdown PCR on all available and relevant vAh and non-vAh isolates (data not shown). Touchdown PCR was performed on an Eppendorf Mastercycler Gradient S with 50 ng of template gDNA extracted from each isolate (E.Z.N.A.® Bacterial DNA Kit; Omega Biotek, Georgia, USA), 13 μl of Econotaq Plus Green 2x MasterMix (Lucigen, Madison, Wisconsin, USA), and 20 picomoles of each primer in a 25-μl reaction. Cycling parameters comprised of an initial denaturation of 94°C for 3 min; 10 cycles of 94°C for 30 s, 68°C for 30 s (−1oC per cycle), and 72°C for 1 min; followed by 25 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min; and a final extension at 72°C for 5 min. Amplicons were resolved on a 1% agarose gel and stained with ethidium bromide.
Table 2.
Oligonucleotide primers specific to members of the vAh pathotype (vAh-SerF and vAh-SerR), previously described qPCR vAh primers (2986F and 2986R), and primers used to screen for unique isolates used in this study (ML09-119F, ML09-119R, S14-452F, S14-452R, ZC1F, and ZC1R).
| Primer name | Direction | Sequence | Amplicon size (bp) |
|---|---|---|---|
| vAh-SerF | Forward | 5′-AG′CATCACCAGCGTTGGCCC-3′ | 502 |
| vAh-SerR | Reverse | 5′-GCCGGGCTGAACTTCCGCAT-3′ | |
| 2986F | Forward | 5′-CTATTACTGCCCCCTCGTTC-3′ | 167 |
| 2986R | Reverse | 5′-ATTGAGCGGTATGCTGTCG-3′ | |
| ML09-119F | Forward | 5′-GTTCCGTTCCATCTGTTCGTGA-3′ | 246 |
| ML09-119R | Reverse | 5′-CAACCATCTTGGTCGCAATC-3′ | |
| S14-452F | Forward | 5′-CAGAACGTGCTGCAGAGATTGA-3′ | 350 |
| S14-452R | Reverse | 5′-TCCGAGAATTCGATGACGAAGG-3′ | |
| ZC1F | Forward | 5′-GCAATTCTGCGGTCACTTCTCG-3′ | 400 |
| ZC1R | Reverse | 5′-AGCGTACCGTCTCGTCGATATG-3′ |
In order to have a PCR assay that differentiates vAh from non-vAh strains while encompassing both US and Asian vAh isolates, new vAh genotype-specific primers (listed as vAh-SerF and vAh-SerR in Table 2) were designed using comparative analysis of annotations produced by the Rapid Annotations using Subsystems Technology (RAST) v2.0 server. These primers target a serine protease gene sequence and are predicted to produce a 502 bp amplicon (data not shown). Touchdown PCR conditions and validation methods were identical to those described previously.
In addition to primers that help to identify vAh strains, A. hydrophila lineage-specific primers were developed that were specific to vAh lineages represented by strains ML09-119 (NC_021290), S14-452 (SAMN05256776), and ZC1 (SAMN02404465) (Table 2). The ML09-119 lineage-specific primer set targets the tnsA endonuclease gene and produces a 246 bp amplicon, the S14-452 lineage-specific primer set targets the COG3339 genetic locus and produces a 350 bp amplicon, and the ZC1 lineage-specific primer set targets a hypothetical protein and produces a 400 bp amplicon.
To determine the vAh genotype affiliation of disease isolates in AL and MS, genomic DNA was isolated from each isolate (Gentra Puregene DNA isolation kit; Qiagen, Hilden, Germany) and used as a template in a 25-μl PCR that comprised of 13 μl of Econotaq Plus Green 2x MasterMix (Lucigen, Madison, Wisconsin, USA), 20 picomoles of each oligonucleotide primer and 50 ng of template gDNA. Samples were run on a C1000 Touch thermal cycler (BioRad, Hercules, California, USA) with an initial denaturation of 94°C for 3 min followed by 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 5 min and amplicon verification was performed as previously described.
Core genome analyses
A core genome was created using both coding and non-coding sequences of 61 genomes labeled as A. hydrophila within GenBank, some of which were revealed to be misclassified. Specifically, any contigs less than 10 Kbp in size were first filtered from draft genomes to increase computational efficiency and decrease false positives by limiting the mapping of smaller fragments to non-homologous and/or multiple regions. In general, smaller fragments contribute less to the production of core genomes as a function of being flanked by areas of heavily repetitive sequences. In the assembly stage, these regions stopped growing and remained short. While core genomes produced by this method are often smaller, the removal of smaller contigs did not compromise the ability to form a phylogenetically-informative core genome.
Filtered sequence data were then submitted as FASTA files to the multiple whole genome alignment tool Mugsy v1.2.3 (Angiuoli and Salzberg, 2011) under default parameters. The resulting alignment was subsequently processed with GBLOCK v0.91b (Castresana, 2000) in order to identify regions of high conservation across all isolates. Parameters for retention by GBLOCK are dictated by the input alignment and were: A minimum of 31 and 51 sequences for conserved and flanked positions, respectively, a maximum of 8 contiguous, but non-conserved positions, a minimal block length of 10, and one-half of the sequences were allowed to possess gapped positions within a block. From the final alignment, a maximum likelihood (ML) phylogeny for the 61 Aeromonas spp. isolates, including 54 isolates labeled in GenBank as A. hydrophila, was inferred using RAxML v8.2.8 (Stamatakis, 2014) under the General Time Reversible model of evolution with estimated proportions of invariable sites and rate variation among sites (i.e., GTR+I+G) and 1000 bootstrap replicates to determine branch supports. Trees were visualized using Archaeopteryx v.beta 0.9901.
Following generation of a consensus sequence, the National Microbial Pathogen Data Resource (NMPDR) Rapid Annotations using Subsystems Technology (RAST) v2.0 server was used in conjunction with the SEED v2.0 algorithm to annotate the core genome and generate metabolic models (Aziz et al., 2008; Overbeek et al., 2014). These predictive models were evaluated using both protein-protein Basic Local Alignment Search Tool (e.g., BLASTp and BLASTx) algorithms through GenBank as well as the Joint Genome Institute's Genomes OnLine Database (GOLD) v5.0.
Calculating average nucleotide identity
To assess overall genetic similarity, the average nucleotide identity (ANI) comparison of 61 Aeromonas spp. genomes was evaluated using JSpecies (v1.2.1) and cross-validated with the Konstantinidis lab ANI calculator (Richter and Rosselló-Móra, 2009; Rodriguez and Konstantinidis, 2014). According to criteria used for the genus Aeromonas, ANI-values >96% indicated strains belong to the same species (Colston et al., 2014; Beaz-Hidalgo et al., 2015a).
vAh differential gene identification
To evaluate differences in gene content among the 61 genomes, data from the PAthogen Resource Integration Center (PATRIC) protein family sorter tool, RAST/SEED gene annotations, the Pathogen Host Interaction database (http://www.phi-base.org), and the Virulence Factors of Pathogenic Bacteria databases (http://www.mgc.ac.cn/VFs) were combined with copy number data for previously identified virulence genes (Rasmussen-Ivey et al., 2016). Results were evaluated by comparing predicted virulence-associated genes with closely and disparately related isolates to identify genes that were differentially present. Notably, genes shared between vAh and non-vAh isolates or those unique to an individual strain were removed from downstream analyses. These data were subsequently coupled with screening of virulence factors and vAh-associated genes using searches of the NCBI GenBank database with MegaBLAST and BLASTn algorithms (Wheeler et al., 2003). Thresholds for absence were specific to the respective gene and were restricted to mutations altering the predicted functional domains of proteins in which the protein sequence in question returned a functionally divergent protein. Once validated, these gene clusters and virulence factors were transformed for statistical analyses using Orange Data Mining software v.3.3.5 and/or R Studio v.0.99.896. After pre-processing in R Studio using packages MuMIn v.1.15.6, randomForest v.4.6-12, and k-means v.0.1.1, heat maps of resultant data were generated using Orange Data Mining software. To analyze subclade differences, vAh and non-vAh strains with known virulence properties (n = 25) were evaluated using k-means at 20 clusters (100% between sum of squares/total sum of squares). In addition to these analyses, the T346 Secretion System Hunter (version is not published) was used to identify T6SS-associated gene clusters using Glimmer v3.02, and HMMER3 v3.1b2 (Martínez-García et al., 2015).
Results
Identifying new vAh isolates
The S04-690 genome was previously found to be the genetic intermediate (raw genetic distance) between US catfish vAh isolates (represented by strain ML09-119) and the Asian carp vAh isolate ZC1 (Hossain et al., 2014). A total of 38 vAh isolates recovered from MAS outbreaks in Mississippi from 2013 to 2015 and confirmed as vAh by phenotypic (myo-inositol usage) and/or genomic tests (qPCR) (Griffin et al., 2013; Hanson et al., 2014) were subjected to vAh lineage-specific PCR. Testing using lineage-specific primer sets based on representative isolates ML09-119, S14-452, and ZC1 showed that of the 38 MS isolates recovered from MAS outbreaks between 2013 and 2015, 20 isolates from a single farm were positive for the ML09-119 lineage-specific amplicon, while 18 isolates across five geographically discrete farms were positive for the S14-452 lineage-specific amplicon. No isolates produced amplicons with the ZC1 lineage-specific primer set (data not shown). Isolates that produced an amplicon with either the ML09-119 or S14-452 lineage-specific primer sets did not produce amplicons when assayed with other lineage-specific primer sets (data not shown).
Pathotype-specific PCR for vAh genotypes
Results from touchdown PCR assays demonstrate that the original 2968 primer set (Griffin et al., 2013) reliably distinguishes US vAh from non-vAh isolates (data not shown). However, the Asian carp isolates J-1 and NJ-35 were unable to be experimentally evaluated and in silico analysis predicts that Asian isolates J-1 and NJ-35 would not produce a PCR amplicon using the 2968 primer set. To address this possible limitation, a new vAh genotype-specific primer set (listed as vAh-SerF and vAh-SerR in Table 2) was designed to be inclusive of all Asian and US isolates, based on in silico analyses of vAh and non-vAh isolates. The vAh-SerF/R primer set was empirically confirmed to produce a 502 bp amplicon in all available US vAh isolates while returning a negative result for non-vAh isolates (data not shown).
Core genome analysis
Alignment of the complete and filtered draft genomes of the 61 Aeromonas spp. genomes via Mugsy produced a matrix of 19,817,762 positions. Following processing with GBLOCK, the core genome of these 61 strains contained 32,401 blocks and a consensus of 3,776,490 bp. This included 120,049 variable (>2 different nucleotides) and 79,507 phylogenetically informative sites (nucleotides that contributed to sorting phylogenetic groups), with percent G+C composition of 62.6%. Notably, these conserved regions collectively have a higher percent G+C content than the 61-isolate average of 61.1% (with a range of 60% for the genome of the type strain of A. molluscorum 848T to 63.2% for the one of A. taiwanensis LMG24683T; p-value = 0.003). Across the core analysis, within complete sites, the average transition to transversion ratio was 1.437 for all sequence pairs, with a minimum of 0 transitions and 1 transversion (A. hydrophila S04-690 and S13-700) and a maximum of 13 transitions and 2 transversions (A. hydrophila 173 and 14).
Core genome phylogeny
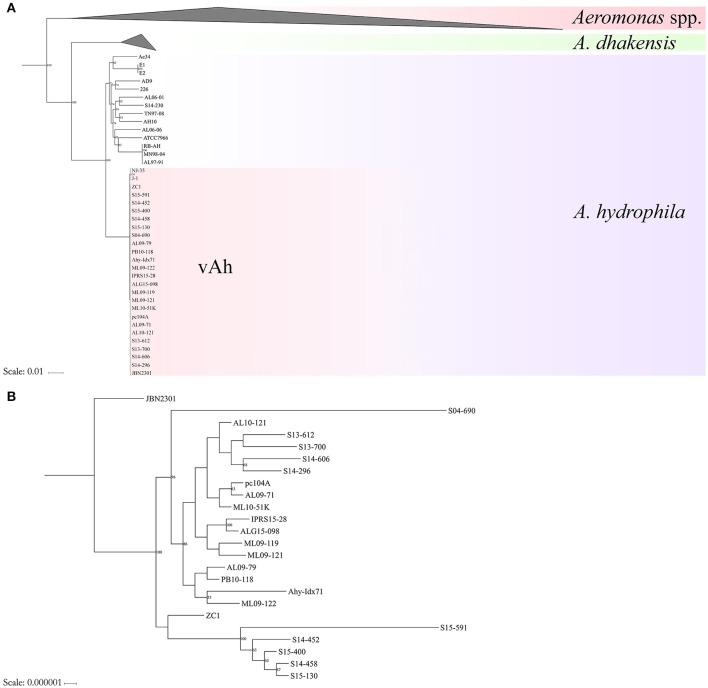
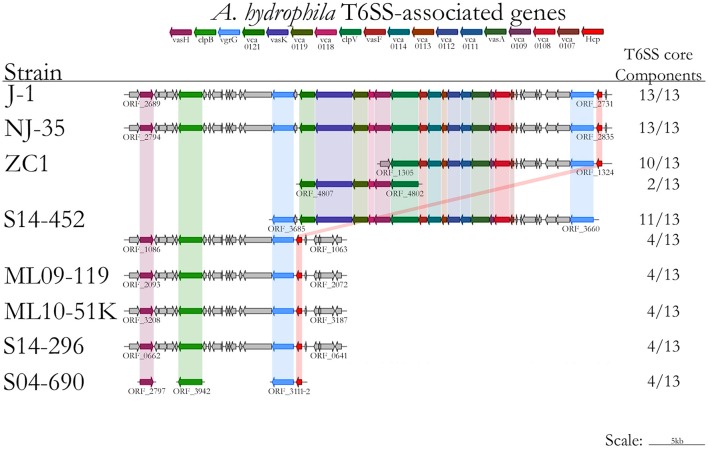
Conserved sequences from the core genome analysis were used to infer phylogenetic relationships among the 61 Aeromonas spp. isolates (Figure 1A). The core genome phylogeny indicates with 100% bootstrap support that vAh strains form a monophyletic group that is fundamentally distinct from other A. hydrophila (Figure 1A). In addition to supporting the monophyly of these ST251 isolates, this phylogenetic analysis also revealed support for distinct sub-clades among vAh isolates, with 2660 single nucleotide polymorphisms identified between the vAh sub-clades. For example, A. hydrophila ZC1, isolated from a grass carp in China (Figure 1B), is closely affiliated with vAh strains isolated from catfish in MS (S14-452, S14-458, S15-130, S15-400, and S15-591). A key genotype that differentiates the vAh sub-clades is the presence and genetic organization of T6SS components (Figure 2). Each of the ZC1-affiliated strains, as well as the other strains isolated from carp in China (i.e., J-1 and NJ-35), were found to contain at least 80% of the core proteins necessary for a complete T6SS, with ZC1 and S14-452 having two separate T6SS-associated gene clusters, whereas J-1 and NJ-35 each have a single complete T6SS cluster (Figure 2). In contrast, vAh isolates from catfish in AL as well as other MS isolates (i.e., S13-612, S13-700, S14-606, and S14-296) formed a distinct subclade that lack the majority of the core T6SS genes (Figures 1B, 2). Notably, while AL and MS vAh strains lacked the majority of T6SS components, they consistently retain other genes that are involved in T6SS in other bacteria such as a valine-glycine repeat protein G (VgrG), a T4 bacteriophage tail-like hole forming protein (Leiman et al., 2009); hemolysin coregulated protein (Hcp), a repetitive tubular protein that is similar to the phage major tail protein GpV (Pell et al., 2009); the chaperone protein ClpB, a chaperone and ATPase that interacts with Hcp to translocate effectors (Shrivastava and Mande, 2008); and VasH, a putative transcriptional regulator (Kitaoka et al., 2011) (Figure 2).
Figure 1.
Maximum likelihood (ML) phylogeny of (Panel A) Aeromonas spp. and (Panel B) vAh isolates based on the core genome of 3.78 Mb conserved among these bacterial strains.
Figure 2.
Type VI secretion system gene prediction using the T346 Secretion System Hunter, with results including strains included in the immersion catfish challenge (ML09-119, MNL10-51K, S04-690, S14-296, S14-452, and ZC1) and representatives from Chinese strains (J-1, NJ-35, and ZC1).
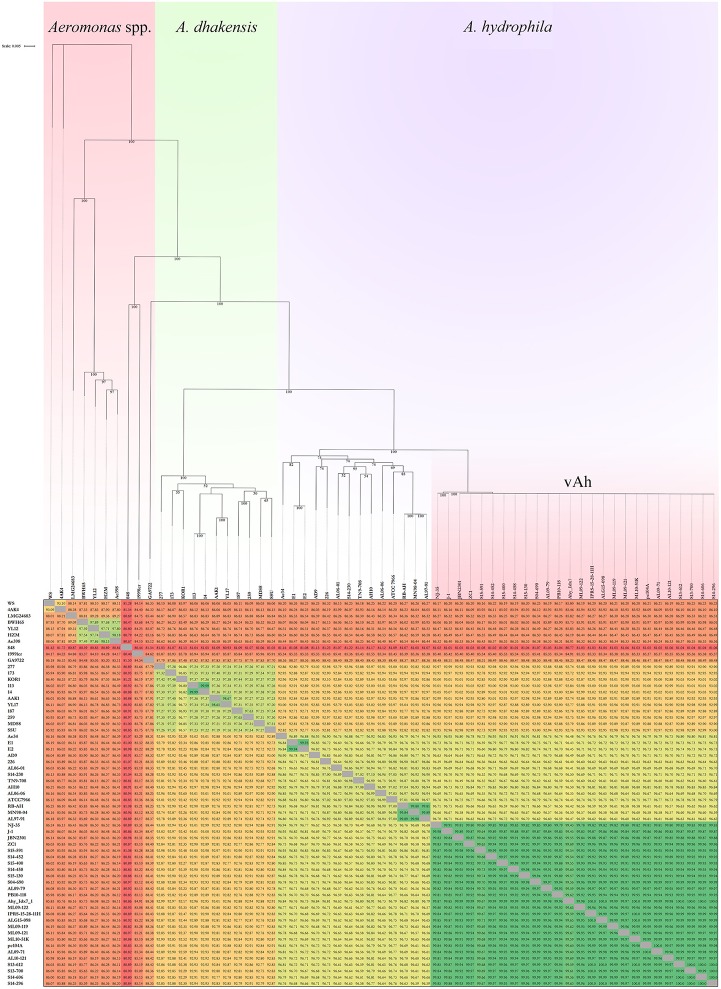
Average nucleotide identity
The average nucleotide identity values of the 61 Aeromonas spp. genomes was determined (Figure 3). High ANI-values (>99%) were found for all vAh-vAh pairwise comparisons, supporting the core genome phylogeny (Figures 1B, 3). In contrast, all vAh comparisons with non-vAh isolates possessed ANI-values less than 97%. In accordance with previous results, 14 strains appear to have a discrepancy between the species classification listed in GenBank and the species affiliation indicated by ANI-values (Table 1).
Figure 3.
Average nucleotide identities (ANI) among Aeromonas spp. strains and an associated maximum likelihood phylogram based on a core genome phylogeny (Figure 1). Please note that the branch length of strains 848 and 1999lcr were reduced to improve readability and pairwise ANI-values are color-coded according to percent identity.
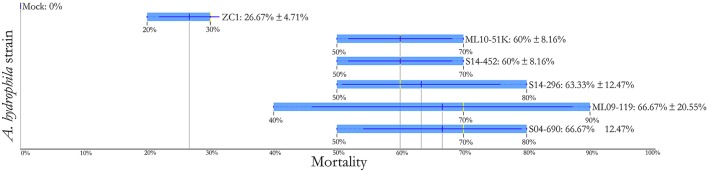
Virulence of vAh strains in channel catfish using an immersion challenge model
Challenge of channel catfish with vAh isolates from AL and MS resulted in ≥ 60% mortality (Figure 4). Within these US vAh isolates tested, there were no observed differences in virulence among ML09-119, ML10-51K, S04-690, S14-296, and S14-452, based on Duncan's multiple range test (p-value > 0.05). The carp isolate ZC1 was less virulent than AL and MS isolates with only 26.7% mortality observed (Figure 4). Most mortality (~96%) occurred within 48 h post challenge for all isolates, including ZC1. All dead fish (100%) sampled for confirmation were positive for the presence of vAh in liver tissue. Control fish yielded no mortality from the mock challenge.
Figure 4.
Comparative assessment of the relative virulence of vAh isolates in channel catfish using 1 h immersion exposure with fin clip (ANOVA = 7.628, p-value = 0.001).
Pathology of epidemic A. hydrophila infections
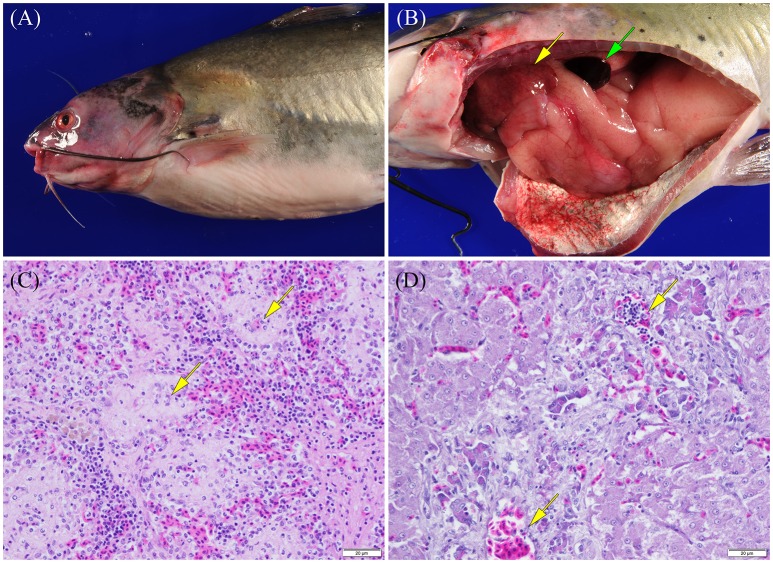
Cutaneous lesions observed in fish infected with vAh by immersion challenge included extensive hyperemia over the pale ventrum of the fish, in tissues surrounding and within the mouth and at fin bases (Figure 5A). There was also extensive hyperemia around the eyes and exophthalmos in some fish. Cutting into the muscle of the lateral body wall revealed multifocal to coalescing foci of congestion/hemorrhage. Gill lesions were variable; gills were pale in some fish and reddened in others. Internally, there was widespread hyperemia of abdominal organs as well as petechial and ecchymotic hemorrhages scattered over mesenteric tissues. The spleen was moderately to severely swollen and dark red (Figure 5B). Head and trunk kidneys were moderately edematous and red and friable when harvested. The intestinal tract was mildly to moderately dilated and red (Figure 5B). The liver was mildly to moderately swollen with slightly rounded edges (Figure 5B). Glisson's capsule was peppered with variable numbers of petechial and ecchymotic hemorrhages. The atrial chamber of the heart was dilated and filled with blood.
Figure 5.
Photographs of channel catfish infected by vAh showing (A) external surfaces that are exhibiting congestion/hemorrhage around the head/pectoral fin and within the eye and (B) the celomic cavity that has internal organs moderately congested and enlarged, a congested/hemorrhagic spleen (green arrow), and multifocal pale foci corresponding to areas of necrosis (yellow arrow) scattered over the liver (photographs courtesy of Dr. Wes Baumgartner, Mississippi State University) as well as photomicrographs of a channel catfish infected by vAh strain ML09-119 showing (C) a section of spleen with splenic ellipsoids (arrows) that are edematous and ellipsoidal arteries that are lined by degenerating as well as necrotic endothelial cells and (D) a section of liver with edema and necrosis of pancreatic acinar tissue surrounding branches of the hepatic portal vein (arrows).
Histopathologic lesions observed were strongly suggestive of a septic disease and included edema and necrosis in many internal organs. Splenic lesions included ellipsoidal necrosis and congestion/hemorrhage in the splenic red pulp (Figure 5C). In scattered necrotic ellipsoids variable numbers of short, rod-shaped bacteria were observed. In the liver there was often necrosis of the acinar pancreatic tissue surrounding hepatic vessels (Figure 5D). Scattered acinar cells were rounded and necrotic and extracellular zymogen granules could be observed in the necrotic exudate. Small numbers of inflammatory cells including macrophages, lymphocytes, and neutrophils were observable in scattered areas of pancreatic acinar necrosis. Multifocally within the hepatic parenchyma were variable sized foci of hepatocellular necrosis characterized by the presence of small aggregates of degenerating and necrotic hepatocytes. Hematopoietic cells in the renal interstitum and renal epithelial cells lining scattered renal tubules were undergoing degeneration and necrosis. In sections of head kidney, the tissue was edematous and congested/hemorrhagic. There was scattered degeneration and necrosis of red and white cell precursors. Intestinal lesions were minimal in fish infected in this fin clip model and were limited to mild congestion and hemorrhage of the vessels in the lamina propria and vessels of the muscularis and serosa. In some sections of heart there was mild necrosis of myofibers in the myocardium. Vessel of the brain were often moderately dilated and congested but the neuropil of the cerebrum, cerebellum, and brain stem was normal. The epithelium of the gills was normal but branchial capillaries were sometimes dilated and congested with erythrocytes.
Differentiating pathotypes
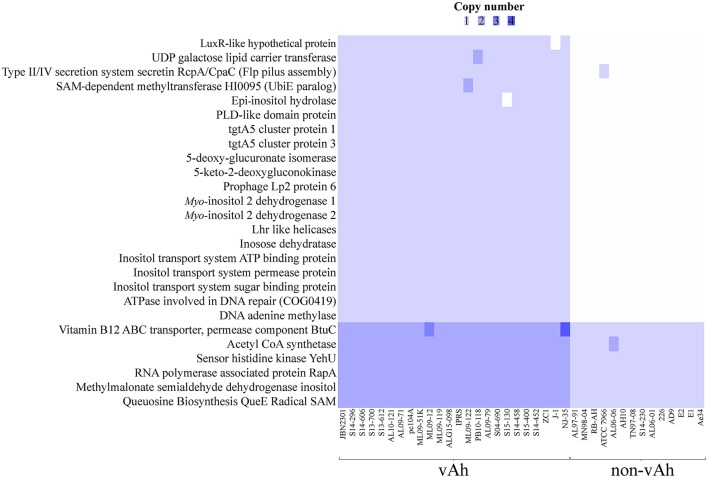
In order to robustly define the vAh pathotype-specific loci in this study, previous results on established A. hydrophila virulence factors as well as genes unique to vAh strains were used in combination with a clustering approach and a random forest decision tree to identify gene products that may contribute to functional differences in virulence. This approach confirmed previously described gene clusters for L-fucose and O-antigen biosynthesis as well as myo-inositol catabolism. Additionally, we identified predicted virulence factors conserved within all vAh strains (Table 3) which includes virulence factors well-known to be important in A. hydrophila pathogenesis (Rasmussen-Ivey et al., 2016). Among predicted virulence factors conserved among vAh strains, there were many genes uniquely associated with vAh strains that were not present in other sequenced A. hydrophila strains, including a L-serine dehydratase, a N-acetylmannosamine kinase, a N-acetylneuraminate lyase, and a gene product predicted to be required for queuosine biosynthesis (Table 4). In a more comprehensive approach, all RAST/SEED predicted genes from the 41 confirmed A. hydrophila genomes (26 vAh and 15 non-vAh) included in this study were evaluated on the basis of linkage with the vAh pathotype using exhaustive iterations of random forest modeling, which resulted in 26 genes uniquely associated with vAh by either presence/absence or by differential copy number when compared to non-vAh (Figure 6).
Table 3.
Predicted virulence factors that are conserved within vAh strains (not unique to), based on a comparison of significant BLASTn hits between vAh isolates against the VFDB (virulence factors with additional results are marked with an asterisk and are available in supplementary data).
| Putative virulence factor | Gene | Uniprot ID | GI | Reference bacterium |
|---|---|---|---|---|
| 3-oxoacyl-acyl carrier protein synthase II | fabB | A0A0H2V610 | 77416726 | Escherichia coli O6:H1 |
| Acriflavine resistance protein AcrB | acrB | P31224 | 25009252 | Escherichia coli K12 |
| Aerolysin/hemolysin/cytolytic enterotoxin | ahh | Q06303 | 89276735 | Aeromonas hydrophila AH-1 |
| Asparaginyl-tRNA synthetase | asnS | Q56112 | 16502162 | Salmonella enterica serovar Typhi CT18 |
| Cephalosporinase; class C beta lactamase | ampC | Q8KU09 | 21311545 | Aeromonas caviae CIP 74.32 |
| Enterochelin esterase | fes | A0A0H2V760 | 26106962 | Escherichia coli O6:H1 |
| Ethanolamine utilization protein EutN | eutN | B7LTU3 | 984388511 | Escherichia fergusonii ATCC 35469 |
| Flagellar motor switch protein FliN | fliN | A0A0H3QVI1 | 674744044 | Pseudomonas aeruginosa Stone 130 |
| General secretion pathway protein PulF | pulF | P15745 | 149305 | Klebsiella pneuoniae oxytoca UNF5023 |
| Protein translocase subunit SecA | secA | Q8YJG2 | 672757090 | Brucella melitensis biotype 1 |
| Rod shape-determining protein MreB | mreB | P0A9X4 | 557273544 | Escherichia coli K12 |
| Sodium/proline symporter proline permease | putP | P07117 | 131658 | Escherichia coli K12 |
| Transcriptional activator NtrC | ntrC | O86057 | 5731350 | Herbaspirillum seropedicae DCP286A |
| Twitching motility protein PilU | pilU | G3XCX3 | 15595593 | Pseudomonas aeruginosa PAO1 |
Table 4.
Virulence factors that are unique to vAh strains, based on a comparison of significant BLASTn hits between A. hydrophila isolates against the RAST/SEED database.
| Subsystem | Role | GI |
|---|---|---|
| Glycine/serine Utilization | L-serine dehydratase | 958619257 |
| Inositol catabolism | 5-deoxy-glucuronate isomerase | 958618826 |
| Inositol catabolism | 5-keto-2-deoxygluconokinase | 827371814 |
| Inositol catabolism | Epi-inositol hydrolase | 612156152 |
| Inositol catabolism | Inositol transport system ATP-binding protein | 958621246 |
| Inositol catabolism | Inositol transport system permease protein | 958620586 |
| Inositol catabolism | Inositol transport system sugar-binding protein | 657060685 |
| Inositol catabolism | Inosose dehydratase | 507222178 |
| Inositol catabolism | Myo-inositol 2-dehydrogenase 1 | 656991783 |
| Inositol catabolism | Myo-inositol 2-dehydrogenase 2 | 827371809 |
| Inositol catabolism | Transcriptional regulator of the myo-inositol catabolic operon | 958618669 |
| Sialic Acid Metabolism | N-acetylmannosamine kinase | 827373367 |
| Sialic Acid Metabolism | N-acetylneuraminate lyase | 1043232173 |
| Sialic Acid Metabolism | Predicted sialic acid transporter | 446588390 |
| Sialic Acid Metabolism | Sugar isomerase involved in processing of sialic acid | 958620857 |
| Pyridoxin Biosynthesis | Predicted transcriptional regulator of pyridoxine metabolism | 16078013 |
| Phage DNA synthesis | DNA adenine methyltransferase, phage-associated | 67483065 |
| Phage capsid proteins | Phage capsid scaffolding protein | 516389014 |
| Phage capsid proteins | Phage major capsid protein | 507220251 |
| Phage lysis modules | Phage lysin, 1,4-beta-N-acetylmuramidase | 511291760 |
| Phage packaging machinery | Phage portal protein | 958618794 |
| Phage packaging machinery | Phage terminase small subunit | 759443491 |
| Phage packaging machinery | Phage terminase, large subunit | 958620694 |
| Queuosine-Archaeosine Biosynthesis | Queuosine biosynthesis QueD, PTPS-I | 1043232409 |
| Queuosine-Archaeosine Biosynthesis | archaeosine tRNA-ribosyltransferase type 5 | 507221161 |
Figure 6.
Comparative whole genome predicted gene-based analysis of all confirmed vAh (n = 26) and non-vAh isolates (n = 15).
Discussion
The core genome analysis of epidemic A. hydrophila strains obtained from diseased fish from several US states and Chinese provinces (identified within the literature as ST251/vAh), supports the genetic and functional unification of these hypervirulent bacteria within a monophyletic clade. These data are in agreement with previous reports based on single or multiple genetic loci from smaller numbers of vAh strains (Hossain et al., 2014; Pang et al., 2015). Interestingly, this study found evidence for genomic heterogeneity among the sampled vAh strains that may reflect geographic origin and/or host switching. For example, the isolates obtained from MS in 2013–2015 are affiliated with two different ST251/vAh clades, specifically the Asian carp-affiliated clade and the US catfish-affiliated clade. In contrast, the vAh isolates from diseased catfish in AL from 2009 to 2015 reflect a single, clonal clade. These data suggest that vAh has greater diversity within MS aquaculture ponds compared to those of AL. As previously hypothesized, this pattern would fit with a dissemination model in which carp or other fish or fish products from Asian source(s) were first introduced to the Mississippi delta region, after which a particularly more virulent vAh lineage spread among farmed catfish resulting in the initial epidemic outbreaks within AL.
Genomic comparisons indicate all members of the vAh pathotype strains share unique genetic loci that may be a result of their close genetic affiliation and may also contribute to their pathogenesis. The previously identified vAh-specific gene clusters of L-fucose and myo-inositol catabolism were also confirmed in this study. The use of myo-inositol as a sole carbon source is a rarely observed phenotype among Aeromonas species and, to our knowledge, has only been reported in vAh strains and strains of A. finlandiensis (Beaz-Hidalgo et al., 2015b). This study identified additional genetic loci that are present in all sequenced vAh strains and may have a contribution to virulence, such as a L-serine dehydratase, a N-acetylmannosamine kinase, a N-acetylneuraminate lyase, a sialic acid transporter, a transcriptional regulator of pyridoxine metabolism, an archaeosine tRNA-ribosyltransferase, a gene product required for queuosine biosynthesis (QueD), an acriflavine resistance protein A, and an IS5 transposase and transactivator. While the contribution of each of these gene products to vAh virulence has yet to be determined, studies in other bacterial pathogens suggest potential mechanisms that could enhance vAh virulence. For example, in Campylobacter jejuni a L-serine dehydratase is essential for gut colonization (Velayudhan et al., 2004). With regard to sialic acid, Vibrio cholera has been shown to use this system to evade the innate immune response (Severi et al., 2007), and promote the binding of cholera toxin to the host intestinal epithelium (Rohmer et al., 2011). In addition, increased acriflavine resistance may provide a selective advantage to vAh considering that acriflavine is a commonly used antiseptic in aquaculture (Martin, 1968).
All annotated genes were subsequently evaluated for linkage with the vAh genotypes, a method that removes the bias inherent in assuming that only known virulence factors contribute to vAh pathogenesis. This analysis revealed that 26 genes are synonymous with the vAh pathotype either by presence/absence or by copy number. Supporting the robust nature of this approach, previously described genes that are vAh-associated were also identified, including genes associated with the myo-inositol catabolic pathway. Of note, within this pathway a methylmalonate-semialdehyde dehydrogenase that is required for myo-inositol catabolism was identified in both vAh and non-vAh isolates; however, this enzyme is required for both myo-inositol and valine metabolism and there is no indication that non-vAh strains have the genetic capacity for myo-inositol catabolism. In addition to these results, this method also identified known Aeromonas spp. virulence factors, such as a DNA adenine methylase that has been identified as a regulator of virulence and required for viability in A. dhakensis SSU (Erova et al., 2006). From this approach other putative vAh virulence factors were identified, such as a hypothetical protein with a LuxR-like domain. LuxR has been previously shown to be a regulator of virulence factors and quorum sensing within Aeromonas spp. (Kirke et al., 2004). Lastly, other genetic loci were identified as being associated with vAh strains, such as the type II/IV system secretin RcpA/CpaC that is putatively involved in flp pilus assembly (Clock et al., 2008), but to our knowledge the contribution of this secretin to A. hydrophila pathogenesis has yet to be experimentally determined.
The histopathologic examination of tissues from vAh-infected farmed fish show a wide range of severity related to internal lesions, with some fish exhibiting minimal lesions and others having widespread sepsis with necrosis of spleen, liver, renal tissue, intestine, and brain tissues with subsequent high rates of mortality occurring throughout affected farms (W. Hemstreet, personal communication). For fish challenged with vAh strains in aquaria via intraperitoneal injection, rapid onset of mortality without these disease sequelae was observed (Hossain et al., 2014). In this study, fish were challenged with an immersion model and exhibited significant clinical signs including cutaneous and ocular hemorrhaging, splenic and renal congestion, and hemorrhage with mild to moderate necrosis of internal organs. Despite the high genetic similarity of the strains (ANIs > 99%), strain ZC1 had reduced virulence (~27% mortality) when compared with ML09-119; ML10-51K; S04-690; S14-296; and S14-452 which caused ≥ 60% mortality in channel catfish. The reduced virulence of strain ZC1, relative to strain ML09-119, in carp and in catfish was previously observed when fish were challenged intraperitoneally (Hossain et al., 2014). Interestingly, comparatively few reports of mortalities due to MAS have come from the state of MS, which may reflect a number of geographic differences, the heterogeneity of vAh strains present within MS aquaculture ponds, differences in management and production practices, and/or environmental conditions.
To identify genetic elements that may explain the disparity in virulence between the Asian carp isolate ZC1 and the US catfish isolates, genomic comparisons were conducted between ZC1 and the vAh strains included in the immersion challenge. These analyses revealed that all other vAh isolates tested in the disease challenge, but not strain ZC1, encode two putative transcription regulators: (1) a mobile element protein in the helix-turn-helix (HTH) superfamily and (2) a phage antirepressor protein in the antA superfamily present in Escherichia coli ECOR-9 (Sandt et al., 2002). The presence of these transcription factors within the highly virulent vAh strains could result in increased virulence factor expression in these strains. Furthermore, vAh isolates from Asian carp or the ZC1-affiliated clade from MS (including S14-452) were found to have a potentially functional T6SS, but the clade comprised solely of AL and MS catfish isolates (e.g., ML09-119, ML10-51K, S04-690, and S14-296) were found to consistently lack 9 of 13 core T6SS genes related to secretion of virulence factors such as Hcp. The observation that the isolates in the AL/MS vAh subclade lack a complete and/or functional T6SS would lead one to hypothesize that virulence is attenuated in these vAh strains; however, our experimental results indicate that these AL/MS disease isolates are highly virulent compared to strain ZC1. Previous disease challenges with strain ZC1 using intraperitoneal injection support the conclusion that this Asian carp strain ZC1 has reduced virulence relative to the AL/MS vAh clade in both catfish and carp (Hossain et al., 2014), indicating that the attenuated virulence observed in strain ZC1 is not solely attributable to host-specific differences. The vAh strains in the AL/MS subclade encode only a subset of T6SS components that include VgrG, Hcp, ClpB, and VasH. The absence of the other T6SS core genes, including vca0107-0109, vca0111-0114, vca0118, vca0119, vca0121, vasK, clpV, vasF, and vasA, largely accounts for the ~16 Kbp net difference in the genome sizes between strains ZC1 and ML09-119. Based on studies of T6SS in Aeromonas dhakensis SSU and in Salmonella enterica serovar Typhi (Suarez et al., 2008; Wang et al., 2011), it is hypothesized that US isolates lacking several T6SS proteins would exhibit better evasion of the host's immune response. Future research should aim to elucidate the role of these T6SS components, and the effect of a reduced or rearranged T6SS, on vAh virulence.
As new vAh isolates emerge and our collective knowledge of vAh genomic diversity grows, future research should investigate the spread of these pathogens as well as improve the design of more effective biosecurity strategies for the aquaculture industry. Toward this effort, we report a primer set that differentiates vAh from non-vAh strains and is inclusive of the known diversity of vAh strains in the US and Asia. The combination of tools available to differentiate vAh strains from non-vAh strains, including qPCR and the myo-inositol growth assay, are vital tools that can be used to map the global distribution of vAh in carp, catfish, and other warm-water fish species (e.g., tilapia). Disease control strategies should take into account the variability observed in this study among vAh strains and evaluate the efficacy of vaccination or other control measures against a panel of strains that represent the known diversity of this highly virulent A. hydrophila pathotype.
In conclusion, hypervirulent A. hydrophila within ST251 have emerged as pathogens of farmed warmwater fishes that are classified within the vAh pathotype based on strong phylogenetic evidence that includes a core genome phylogeny and ANI-values >99%; metabolic activities that are unique within this species, such as myo-inositol and sialic acid metabolism; a suite of conserved Aeromonas spp. virulence factors; 26 conserved genetic loci putatively linked with virulence; and the ability to induce motile Aeromonas septicemia, which is characteristically followed by rapid mortality in multiple species of farmed fish. Collectively, these traits distinguish vAh from non-epidemic A. hydrophila and define the vAh pathotype.
Author contributions
CR generated and analyzed genome sequence data; CR and SO analyzed gene copy number for putative virulence factors; JT and WH isolated vAh disease isolates from AL and collected data on fish disease; CS, DZ, and DX conducted vAh disease trials; CR and MH developed vAh-specific primer sets; MG collected disease isolates from Manuscript and typed them using different primer sets and bioassays, YL collected disease isolates from Chinese carp and analyzed data, MF contributed to the phylogenetic analysis and inositol utilization of Aeromonas isolates, SS conducted the core genome phylogenetic analysis, JN conducted the histology analysis and contributed to the writing and editing of the manuscript; and ML contributed to the genome sequence analysis, organizing the different research activities and to the writing and editing of the manuscript.
Funding
This work was supported by a grant from the United States Department of Agriculture's Agriculture and Food Research Initiative (Projects #2013-67015-21313 and #MIS-371530) and the USDA-ARS (CRIS Project No. 6010-32000-026-00D and the Catfish Health Initiative). MF thanks the support by the projects: Aquavalens from the European Union Seventh Framework Program (FP7/2007-2013) under Grant agreement No.: 311846, and by projects from Spanish Ministry of Science and Innovation: AGL2011-30461-C02-02 and JPIW2013-095-CO3.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01615
References
- Angiuoli S. V., Salzberg S. L. (2011). Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 27, 334–342. 10.1093/bioinformatics/btq665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatson S. A., das Graças de Luna M., Bachmann N. L., Alikhan N. F., Hanks K. R., Sullivan M. J., et al. (2011). Genome sequence of the emerging pathogen Aeromonas caviae. J. Bacteriol. 193, 1286–1287. 10.1128/JB.01337-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaz-Hidalgo R., Hossain M. J., Liles M. R., Figueras M. J. (2015a). Strategies to avoid wrongly labelled genomes using as example the detected wrong taxonomic affiliation for Aeromonas genomes in the genbank database. PLoS ONE 10:e0115813. 10.1371/journal.pone.0115813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaz-Hidalgo R., Latif-Eugenín F., Hossain M. J., Berg K., Niemi R. M., Rapala J., et al. (2015b). Aeromonas aquatica sp nov., Aeromonas finlandiensis sp nov and Aeromonas lacus sp nov isolated from Finnish waters associated with cyanobacterial blooms. Syst. Appl. Microbiol. 38, 161–168. 10.1016/j.syapm.2015.02.005 [DOI] [PubMed] [Google Scholar]
- Beaz-Hidalgo R., Martínez-Murcia A., Figueras M. J. (2013). Reclassification of Aeromonas hydrophila subsp. dhakensis Huys et al. 2002 and Aeromonas aquariorum Martinez-Murcia et al. 2008 as Aeromonas dhakensis sp. nov. comb nov. and emendation of the species Aeromonas hydrophila. Syst. Appl. Microbiol. 36, 171–176. 10.1016/j.syapm.2012.12.007 [DOI] [PubMed] [Google Scholar]
- Camus A. C., Durborow R. M., Hemstreet W. G., Thune R. L., Hawke J. P. (1998). Aeromonas Bacterial Infections: Motile Aeromonad Septicemia (No. 478). Southern Regional Aquaculture Center (Southern Regional Aquaculture Center collectively covers Alabama, Arkansas, Florida, Georgia, Kentucky, Louisiana, Mississippi, North Carolina, Oklahoma, Puerto Rico, South Carolina, Tennessee, Texas, U.S. Virgin Islands, and Virginia).
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Chai B., Wang H., Chen X. (2012). Draft genome sequence of high-melanin-yielding Aeromonas media strain WS. J. Bacteriol. 194, 6693–6694. 10.1128/JB.01807-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K. G., Puthucheary S. D., Chan X. Y., Yin W. F., Wong C. S., Too W. S., et al. (2011). Quorum sensing in Aeromonas species isolated from patients in Malaysia. Curr. Microbiol. 62, 167–172. 10.1007/s00284-010-9689-z [DOI] [PubMed] [Google Scholar]
- Chen H. Q., Lu C. P. (1991). Study on the pathogen of epidemic septicemia occurred in cultured cyprinoid fishes in southern China. J. Nanjing Agric. Univ. 14, 87–91. [Google Scholar]
- Chua P., Har Z. M., Austin C. M., Yule C. M., Dykes G. A., Lee S. M. (2015). Genome sequencing and annotation of Aeromonas sp. Genom. Data 5, 38–39. 10.1016/j.gdata.2015.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clock S. A., Planet P. J., Perez B. A., Figurski D. H. (2008). Outer membrane components of the tad (tight adherence) secreton of Aggregatibacter actinomycetemcomitans. J. Bacteriol. 190, 980–990. 10.1128/Jb.01347-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colston S. M., Fullmer M. S., Beka L., Lamy B., Gogarten J. P., Graf J. (2014). Bioinformatic genome comparisons for taxonomic and phylogenetic assignments using Aeromonas as a test case. mBio 5:e02136. 10.1128/mBio.02136-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva B. C., Mourino J. L. P., Vieira F. N., Jatoba A., Seiffert W. Q., Martins M. L. (2012). Haemorrhagic septicaemia in the hybrid surubim (Pseudoplatystoma corruscans x Pseudoplatystoma fasciatum) caused by Aeromonas hydrophila. Aquac. Res. 43, 908–916. 10.1111/j.1365-2109.2011.02905.x [DOI] [Google Scholar]
- Deng G. C., Jiang X. Y., Ye X., Liu M. Z., Xu S. Y., Liu L. H., et al. (2009). Isolation, identification and characterization of Aeromonas hydrophila from hemorrhagic grass carp. China 36, 1170–1177. [Google Scholar]
- Earl A. M., Onderdonk A. B., Kirby J., Ferraro M. J., Huang S., Spencer M., et al. (2015). The Genome Sequence of Aeromonas Hydrophila Strain BWH65. Cambridge, MA: EMBL/GenBank/DDBJ databases. [Google Scholar]
- Erova T. E., Pillai L., Fadl A. A., Sha J., Wang S., Galindo C. L., et al. (2006). DNA adenine methyltransferase influences the virulence of Aeromonas hydrophila. Infect. Immun. 74, 410–424. 10.1128/Iai.74.1.410-424.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueras M. J., Beaz-Hidalgo R., Hossain M. J., Liles M. R. (2014). Taxonomic affiliation of new genomes should be verified using average nucleotide identity and multilocus phylogenetic analysis. Genome Announc. 2, e00927–e00914. 10.1128/genomeA.00927-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Jian J., Li W. J., Yang Y. C., Shen X. W., Sun Z. R., et al. (2013). Genomic study of polyhydroxyalkanoates producing Aeromonas hydrophila 4AK4. Appl. Microbiol. Biotechnol. 97, 9099–9109. 10.1007/s00253-013-5189-y [DOI] [PubMed] [Google Scholar]
- Griffin M. J., Goodwin A. E., Merry G. E., Liles M. R., Williams M. A., Ware C., et al. (2013). Rapid quantitative detection of Aeromonas hydrophila strains associated with disease outbreaks in catfish aquaculture. J. Vet. Diagn. Invest. 25, 473–481. 10.1177/1040638713494210 [DOI] [PubMed] [Google Scholar]
- Grim C. J., Kozlova E. V., Sha J., Fitts E. C., van Lier C. J., Kirtley M. L., et al. (2013). Characterization of Aeromonas hydrophila wound pathotypes by comparative genomic and functional analyses of virulence genes. mBio 4, e00064–e00013. 10.1128/mBio.00064-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson L., Liles M. R., Hossain M. J., Griffin M., Hemstreet W. (2014). Motile aeromonas septicemia, in Fish Health Section Blue Book 2014 Edition. Bethesda, MD: American Fisheries Society - Fish Health Section; Available online at: http://www.afs-fhs.org/perch/resources/citation-guidelines-2014.pdf [Google Scholar]
- Hemstreet B. (2010). An update on Aeromonas hydrophila from a fish health specialist for summer 2010. Catfish J. 24, 4. [Google Scholar]
- Hemstreet W. (2015). Aeromonas summary. Fish Farming News: Alabama Fish Farming Center.
- Hossain M. J. (2012). Molecular Interactions between Phage and the Catfish Pathogen Edwardsiella ictaluri and Comparative Genomics of Epidemic Strains of Aeromonas hydrophila. Doctoral Dissertation, Auburn University. [Google Scholar]
- Hossain M. J., Sun D., McGarey D. J., Wrenn S., Alexander L. M., Martino M. E., et al. (2014). An asian origin of virulent Aeromonas hydrophila responsible for disease epidemics in United States-Farmed Catfish. mBio 5, e00848–e00814. 10.1128/mBio.00848-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M. J., Waldbieser G. C., Sun D., Capps N. K., Hemstreet W. B., Carlisle K., et al. (2013). Implication of lateral genetic transfer in the emergence of Aeromonas hydrophila isolates of epidemic outbreaks in channel catfish. PLoS ONE 8:e80943. 10.1371/journal.pone.0080943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagoda S. S., Tan E., Arulkanthan A., Kinoshita S., Watabe S., Asakawa S. (2014). Draft genome sequence of aeromonas hydrophila strain Ae34, isolated from a septicemic and Moribund Koi Carp (Cyprinus carpio koi), a freshwater aquarium fish. Genome Announc. 2:e00572-14. 10.1128/genomeA.00572-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirke D. F., Swift S., Lynch M. J., Williams P. (2004). The Aeromonas hydrophila LuxR homologue AhyR regulates the N-acyl homoserine lactone synthase, AhyI positively and negatively in a growth phase-dependent manner. FEMS Microbiol. Lett. 241, 109–117. 10.1016/j.femsle.2004.10.011 [DOI] [PubMed] [Google Scholar]
- Kitaoka M., Miyata S. T., Brooks T. M., Unterweger D., Pukatzki S. (2011). VasH Is a transcriptional regulator of the type VI secretion system functional in endemic and pandemic Vibrio cholerae. J. Bacteriol. 193, 6471–6482. 10.1128/Jb.05414-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman P. G., Basler M., Ramagopal U. A., Bonanno J. B., Sauder J. M., Pukatzki S., et al. (2009). Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U.S.A. 106, 4154–4159. 10.1073/pnas.0813360106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenneman E. M., Barney B. M. (2014). Draft genome sequences of the alga-degrading bacteria Aeromonas hydrophila strain AD9 and Pseudomonas pseudoalcaligenes strain AD6. Genome Announc. 2:e00709-14. 10.1128/genomeA.00709-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liles M., Hemstreet W., Waldbieser G., Griffin M., Khoo L., Bebak J., et al. (2011). Comparative Genomics of Aeromonas hydrophila Isolates from an Epidemic in Channel Catfish. Auburn, AL: American Society for Microbiology Meeting (1489). [Google Scholar]
- Lim Y. L., Ee R., Yin W. F., Chan K. G. (2014). Quorum sensing activity of Aeromonas caviae strain YL12, a bacterium isolated from compost. Sensors 14, 7026–7040. 10.3390/s140407026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y. L., Roberts R. J., Ee R., Yin W. F., Chan K. G. (2016). Complete genome sequence and methylome analysis of Aeromonas hydrophila strain YL17, isolated from a compost pile. Genome Announc. 4:e00060-16. 10.1128/genomeA.00060-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. L. (1968). Comparison of effects of concentrations of malachite green and acriflavine on fungi associated with diseased fish. Prog. Fish Culturist 30, 153–158. [Google Scholar]
- Martínez-García P. M., Ramos C., Rodriguez-Palenzuela P. (2015). T346Hunter: a novel web-based tool for the prediction of type III, type iv and type VI secretion systems in bacterial genomes. PLoS ONE 10:e0119317. 10.1371/journal.pone.0119317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Murcía A. J., Saavedra M. J., Mota V. R., Maier T., Stackebrandt E., Cousin S. (2008). Aeromonas aquariorum sp. nov., isolated from aquaria of ornamental fish. Int. J. Syst. Evol. Microbiol. 58(Pt 5), 1169–1175. 10.1099/ijs.0.65352-0 [DOI] [PubMed] [Google Scholar]
- Nielsen M. E., Høi L., Schmidt A. S., Qian D., Shimada T., Shen J. Y., et al. (2001). Is Aeromonas hydrophila the dominant motile Aeromonas species that causes disease outbreaks in aquaculture production in the Zhejiang Province of China? Dis. Aquat. Org. 46, 23–29. 10.3354/Dao046023 [DOI] [PubMed] [Google Scholar]
- Overbeek R., Olson R., Pusch G. D., Olsen G. J., Davis J. J., Disz T., et al. (2014). The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42, D206–D214. 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang M., Jiang J., Xie X., Wu Y., Dong Y., Kwok A. H. Y., et al. (2015). Novel insights into the pathogenicity of epidemic Aeromonas hydrophila ST251 clones from comparative genomics. Sci. Rep. 5:9833. 10.1038/Srep09833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang M. D., Lin X. Q., Hu M., Li J., Lu C. P., Liu Y. J. (2012). Tetrahymena: an alternative model host for evaluating virulence of Aeromonas strains. PLoS ONE 7:e48922. 10.1371/journal.pone.0048922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pell L. G., Kanelis V., Donaldson L. W., Howell P. L., Davidson A. R. (2009). The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. U.S.A. 106, 4160–4165. 10.1073/pnas.0900044106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pridgeon J. W., Zhang D., Zhang L. (2014). Complete genome sequence of a moderately virulent Aeromonas hydrophila strain, pc104A, Isolated from soil of a catfish pond in West Alabama. Genome Announc. 2:e00554-14. 10.1128/genomeA.00554-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen-Ivey C. R., Figueras M. J., McGarey D., Liles M. R. (2016). Virulence Factors of Aeromonas hydrophila: in the wake of reclassification. Front. Microbiol. 7:1337. 10.3389/fmicb.2016.01337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S., Saha A., Ghoshal T., Thakur A. R. (2013). Draft genome sequence of ammonia-producing Aeromonas sp. MDS8 strain MCC2167 from sludge of a dairy effluent treatment plant. Genome Announc. 1:e00710-13. 10.1128/genomeA.00710-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheault J. G., Vincent A. T., Trudel M. V., Brochu F., Boyle B., Tanaka K. H., et al. (2015). Variants of a genomic island in Aeromonas salmonicida subsp salmonicida link isolates with their geographical origins. Vet. Microbiol. 175, 68–76. 10.1016/j.vetmic.2014.11.014 [DOI] [PubMed] [Google Scholar]
- Ribeiro F. J., Przybylski D., Yin S., Sharpe T., Gnerre S., Abouelleil A., et al. (2012). Finished bacterial genomes from shotgun sequence data. Genome Res. 22, 2270–2277. 10.1101/gr.141515.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M., Róssello-Móra R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez L. M., Konstantinidis K. (2014). Kostas Lab | ANI Calculator.
- Rohmer L., Hocquet D., Miller S. I. (2011). Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 19, 341–348. 10.1016/j.tim.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandt C. H., Hopper J. E., Hill C. W. (2002). Activation of prophage eib genes for immunoglobulin-binding proteins by genes from the IbrAB genetic island of Escherichia coli ECOR-9. J. Bacteriol. 184, 3640–3648. 10.1128/Jb.184.13.3640-3648.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri R., Joseph S. W., Chopra A. K., Sha J., Shaw J., Graf J., et al. (2006). Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J. Bacteriol. 188, 8272–8282. 10.1128/JB.00621-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severi E., Hood D. W., Thomas G. H. (2007). Sialic acid utilization by bacterial pathogens. Microbiology 153, 2817–2822. 10.1099/mic.0.2007/009480-0 [DOI] [PubMed] [Google Scholar]
- Shrivastava S., Mande S. S. (2008). Identification and functional characterization of gene components of type VI secretion system in bacterial genomes. PLoS ONE 3:e2955. 10.1371/journal.pone.0002955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spataro N., Farfán M., Albarral V., Sanglas A., Lorén J. G., Fusté M. C., et al. (2013). Draft genome sequence of Aeromonas molluscorum strain 848TT, isolated from Bivalve Molluscs. Genome Announc. 1:e00382-13. 10.1128/genomeA.00382-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez G., Sierra J. C., Sha J., Wang S., Erova T. E., Fadl A. A., et al. (2008). Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 44, 344–361. 10.1016/j.micpath.2007.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekedar H. C., Karsi A., Akgul A., Kalindamar S., Waldbieser G. C., Sonstegard T., et al. (2015). Complete genome sequence of fish pathogen Aeromonas hydrophila AL06-06. Genome Announc. 3:e00368-15. 10.1128/genomeA.00368-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayudhan J., Jones M. A., Barrow P. A., Kelly D. J. (2004). L-serine catabolism via an oxygen-labile L-serine dehydratase is essential for colonization of the avian gut by Campylobacter jejuni. Infect. Immun. 72, 260–268. 10.1128/Iai.72.1.260-268.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Luo Z., Du H., Xu S., Ni B., Zhang H., et al. (2011). Molecular characterization of a functional type VI secretion system in Salmonella enterica serovar Typhi. Curr. Microbiol. 63, 22–31. 10.1007/s00284-011-9935-z [DOI] [PubMed] [Google Scholar]
- Wheeler D. L., Church D. M., Federhen S., Lash A. E., Madden T. L., Pontius J. U., et al. (2003). Database resources of the national center for biotechnology. Nucleic Acids Res. 31, 28–33. 10.1093/nar/gkg033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B. H., Yin Z., Wu Y. S., Cai T. Z. (1993). Studies on the taxonomy of pathogenic bacteria of the bacterial hemorrhagic septicemia in cultured fishes in freshwater. Acta Hydrobiol. Sin. 17, 259–266. [Google Scholar]
- Xu L., Wang H., Yang X., Lu L. (2013). Integrated pharmacokinetics/pharmacodynamics parameters-based dosing guidelines of enrofloxacin in grass carp Ctenopharyngodon idella to minimize selection of drug resistance. BMC Vet. Res. 9:126. 10.1186/1746-6148-9-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Li N., Li M., Zhang D., An G. (2016). Complete genome sequence of fish pathogen Aeromonas hydrophila JBN2301. Genome Announc. 4:e01615-15. 10.1128/genomeA.01615-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin M., Ma Z., Cai Z., Lin G., Zhou J. (2015). Genome sequence analysis reveals evidence of quorum-sensing genes present in Aeromonas hydrophila strain KOR1, isolated from a Mangrove Plant (Kandelia obovata). Genome Announc. 3:e01461-15. 10.1128/genomeA.01461-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Xu D.-H., Shoemaker C. A. (2016). Experimental induction of motile Aeromonas septicemia in channel catfish by water-borne challenge with virulent Aeromonas hydrophila. Aquac. Rep. 3, 18–23. 10.1016/j.aqrep.2015.11.003 [DOI] [Google Scholar]
- Zhang Y. L., Arakawa E., Leung K. Y. (2002). Novel Aeromonas hydrophila PPD134/91 genes involved in O-antigen and capsule biosynthesis. Infect. Immun. 70, 2326–2335. 10.1128/IAI.70.5.2326-2335.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.