Abstract
Background & Aims
ABT‐530 is a next‐generation hepatitis C virus (HCV) NS5A inhibitor with potent pangenotypic antiviral activity in vitro. Paritaprevir is an NS3/4A protease inhibitor codosed with ritonavir that displays in vitro activity against HCV genotypes 1–4 and 6.
Methods
Efficacy, pharmacokinetics and safety of ABT‐530 with paritaprevir/ritonavir and ribavirin were evaluated in this phase 2, open‐label, multicentre study in treatment‐naïve non‐cirrhotic patients with genotype 3 infection. Ten patients, all genotype 3a, received 120 mg ABT‐530 and 150/100 mg paritaprevir/ritonavir once daily with ribavirin for 12 weeks.
Results
Nine (90%) patients achieved a sustained virological response at post‐treatment weeks 12 and 24. One patient experienced virological failure at treatment week 6. Sequence analyses for HCV variants in samples from this patient identified A166S in NS3 at baseline and after breakthrough, as well as A30K at baseline and linked S24F+M28K+A30K variants in NS5A after breakthrough. Neither genotype 3 NS3 A166S nor NS5A A30K variant confers any resistance to paritaprevir or ABT‐530 respectively. However, genotype 3 NS5A S24F+M28K+A30K‐linked variant confers a >5000‐fold increase in ABT‐530 EC 50 relative to that of the wild‐type replicon. This patient's ABT‐530 exposure was comparable to the cohort, while paritaprevir and ritonavir exposures were the lowest of all patients. No serious or severe adverse events and adverse events leading to early discontinuation were reported.
Conclusions
Results from this study show that ABT‐530 holds promise as part of a direct‐acting antiviral treatment regimen for HCV genotype 3 infection.
Keywords: direct‐acting antiviral, next generation, pharmacokinetics, resistant variant, sustained virological response
Abbreviations
- BID
twice daily
- Ctrough
trough plasma concentration
- DAA
direct‐acting antiviral
- HCV
hepatitis C virus
- PTV/r
paritaprevir/ritonavir
- QD
once daily
- RBV
ribavirin
- SVR12
sustained virological response at post‐treatment week 12
- SVR24
Sustained virological response at post‐treatment week 24
Keypoints.
Hepatitis C virus genotype 3 infection accounts for 30% of all cases and is associated with an increased risk for liver steatosis, hepatocellular carcinoma and fibrosis progression.
ABT‐530 is a next‐generation NS5A inhibitor that demonstrated pangenotypic antiviral activity in vitro and maintained potent activity against common NS5A single‐position variants that confer resistance to other NS5A inhibitors
ABT‐530 dosed with paritaprevir/ritonavir and ribavirin for 12 weeks provided sustained virological response in 90% of treatment‐naïve, non‐cirrhotic patients with genotype 3 infection.
Treatment was generally well tolerated with no serious or severe adverse events reported.
Hepatitis C virus (HCV) infection presents a high healthcare burden, with 184 million people affected worldwide 1. HCV exhibits vast genetic diversity, with genotype 3 accounting for approximately 30% of all infections 2. Genotype 3 infection is particularly common in Europe, including Greece, Poland and the Netherlands, where it can be found in up to 30% of cases 3, as well as in South Asia, where 72% of HCV cases are genotype 3 in India 2. In the USA, genotype 3 makes up 8–13% of infections 4. Overall, there is a higher prevalence of genotype 3a infection worldwide compared with other subgenotypes, which is associated with injection drug use 5. Genotype 3 infection results in the highest rates of liver steatosis among HCV genotypes 6, 7 and increases the risk for hepatocellular carcinoma and hepatic fibrosis progression 7, 8. Antiviral treatment resulting in HCV clearance is associated with decreases in morbidity and mortality and improvement in liver histology 9.
With genotype 3 infection considered the more difficult‐to‐cure in the era of interferon‐free therapies 10, there is high demand for newer direct‐acting antivirals (DAAs) that can overcome the limitations of the current treatment regimens. Several first‐generation DAA regimens are approved for the treatment of genotype 3 infection, including sofosbuvir, an HCV NS5B inhibitor, plus ribavirin for 24 weeks 11. For patients with genotype 3 infection, daclatasvir, an NS5A inhibitor, plus sofosbuvir/ribavirin for 24 weeks is approved in the EU for patients with cirrhosis or who are treatment‐experienced, and a ribavirin‐free daclatasvir/sofosbuvir combination for 12 weeks is approved in the USA 12, 13. The regimen of ledipasvir, another NS5A inhibitor, and sofosbuvir/ribavirin for 24 weeks is also approved in the EU for genotype 3‐infected patients with cirrhosis and/or who are treatment‐experienced 14, 15. Though these treatment options have higher overall rates of sustained virological response at post‐treatment week 12 (SVR12) than the pegylated interferon/ribavirin (73–89% vs 68%) 16, 17, 18, 19, their SVR rates do not match the high rates reported for treatments approved for HCV genotype 1 infection. Therefore, efficacy of treatment for patients with genotype 3 infection still has room for improvement.
Paritaprevir is an HCV NS3/4A protease inhibitor that is codosed with ritonavir to increase peak, trough and overall drug exposures 20. The regimen of coformulated paritaprevir/ritonavir and ombitasvir, an NS5A inhibitor, dosed with NS5B non‐nucleoside polymerase inhibitor dasabuvir (with/without ribavirin) provides high cure rates and is approved for the treatment of HCV genotypes 1 and 4 (without dasabuvir and ribavirin) 21. However, results from a pilot study showed that ombitasvir/paritaprevir/ritonavir with ribavirin (dasabuvir has no antiviral activity against genotype 3) provided suboptimal efficacy in genotype 3‐infected patients 22. These results are consistent with higher EC50 values of paritaprevir and ombitasvir against genotype 3 compared with those against genotype 1 23, 24.
ABT‐530 is a next‐generation NS5A inhibitor that demonstrated pangenotypic activity in vitro and maintained potent antiviral activity against common HCV NS5A single‐position variants that confer resistance to first‐generation NS5A inhibitors, including daclatasvir, ledipasvir and ombitasvir 25. For genotype 3, in vitro results showed that ABT‐530 had an EC50 value that was 10 times lower than that of ombitasvir and selected fewer drug‐resistant colonies 25. ABT‐530 maintains activity against Y93H, the most common resistance‐associated variant detected at virological failure in patients with genotype 3 infection who failed a regimen containing an NS5A inhibitor 13, 26, which confers ~500‐fold resistance to elbasvir 26, >2000‐fold resistance to daclatasvir 13 and >6000‐fold resistance to ombitasvir 25.
Based on these improved in vitro antiviral characteristics of ABT‐530 for genotype 3, the present study explored the efficacy, pharmacokinetics and safety of the combination of ABT‐530, paritaprevir/ritonavir and ribavirin in treatment‐naïve patients with HCV genotype 3 infection.
Patients and methods
Study population
Treatment‐naïve, non‐cirrhotic patients with chronic HCV genotype 3 infection were screened for enrolment in this study. HCV genotype was determined by Versant® HCV Genotype Inno‐LiPA Assay during screening and later confirmed by phylogenetic analysis based on NS5A sequence of the samples 27. Eligible genotype 3‐infected patients were 18–70 years of age, with a body mass index between 18 and 38 kg/m2, who had plasma HCV RNA >10 000 IU/ml at screening. Chronic HCV infection was determined by presence of anti‐HCV antibodies or HCV RNA at least 6 months before screening and positive results for anti‐HCV antibodies and HCV RNA at screening, or by presence of anti‐HCV antibodies and HCV RNA at screening with a liver biopsy consistent with chronic infection. Absence of cirrhosis was documented based on one of the following: liver biopsy (e.g. METAVIR score ≤3, Ishak score ≤4); screening FibroTest score ≤0.72 and aspartate aminotransferase to platelet ratio index ≤2 or screening FibroScan score <12.5 kPa. Key exclusion criteria were a positive test at screening for hepatitis B or human immunodeficiency virus, coinfection with any other HCV genotype, an alanine aminotransferase or aspartate aminotransferase level of more than 5 times the upper limit of normal, calculated creatinine clearance <60 ml/min (according to the Cockcroft–Gault formula), albumin level below the lower limit of normal, prothrombin‐time international normalized ratio >1.5, haemoglobin level below the lower limit of normal, platelet count <120 000 cells/mm3, neutrophil count <1500 cells/mm3 and direct bilirubin level above the upper limit of normal.
Study design and conduct
This phase 2 open‐label, multicentre study (NCT02068222) was designed to explore the efficacy, pharmacokinetics and safety of ABT‐530 coadministered with paritaprevir/ritonavir and ribavirin for 12 weeks in treatment‐naïve, non‐cirrhotic adults with genotype 3 infection. Patients received 120 mg ABT‐530 administered once daily (QD), 150/100 mg paritaprevir/ritonavir QD and a total daily dose of 1000 mg or 1200 mg ribavirin if the patient's body weight was <75 kg or ≥75 kg, respectively, divided twice daily (BID). Based on a previous study that evaluated 3‐day monotherapy with a range of ABT‐530 doses from 15 mg to 400 mg in genotype 1‐infected patients, 120 mg ABT‐530 was selected for this study as it provided optimal viral load reductions 28. A 150/100 mg paritaprevir/ritonavir dose is recommended in a regimen including ombitasvir and dasabuvir 21, which achieved SVR rates of 92–100% across a range of genotype 1‐infected patients 29. In healthy volunteers, exposure of 120 mg ABT–530 is boosted around two‐fold by 150/100 mg paritaprevir/ritonavir, while exposures for paritaprevir/ritonavir are unchanged by ABT‐530 (data on file). In this present study, patients received ABT‐530, paritaprevir/ritonavir and ribavirin for 12 weeks and were followed throughout a 24‐week post‐treatment period (Fig. 1).
Figure 1.
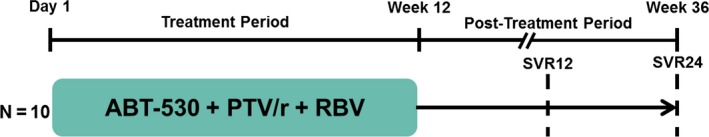
Study design. This study consisted of a 12‐week treatment period followed by a 24‐week post‐treatment period. PTV/r, paritaprevir/ritonavir; RBV, ribavirin; SVR12, sustained virological response at post‐treatment week 12 (primary endpoint); SVR24, sustained virological response at post‐treatment week 24.
All patients provided written informed consent. The study was designed by the study investigators and sponsor according to Good Clinical Practice guidelines, the Declaration of Helsinki and applicable regulations, with Quorum institutional review board approval for all study sites. The sponsor conducted all data analyses, and investigators had full access to data for review. The first draft of the manuscript was written by a sponsor‐employed medical writer, with input from authors. The authors confirm that the results presented are accurate and that the study was conducted and reported according to the protocol.
Efficacy assessments
The primary efficacy endpoint was the percentage of patients who achieved SVR12 (HCV RNA <25 IU/ml at post‐treatment week 12). Secondary assessments included the percentage of patients who achieved SVR at post‐treatment week 24 (SVR24), with on‐treatment breakthrough, or who relapsed following treatment.
Plasma samples for HCV RNA measurements were collected at screening; days 1, 2 and 3; treatment weeks 1, 2, 3, 4, 5, 6, 8, 10 and 12 and post‐treatment weeks 2, 4, 8, 12 and 24. Plasma HCV RNA levels were measured using the COBAS TaqMan® real‐time reverse transcriptase‐PCR assay with High Pure System v2.0 (Roche Molecular Diagnostics, Pleasanton, CA, USA), with a lower limit of detection of 15 IU/ml and lower limit of quantification of 25 IU/ml 30. Patients were required to stop treatment if they had a confirmed increase from nadir in HCV RNA (defined as two consecutive HCV RNA measurements >1 log10 IU/ml above nadir) at any time point during treatment, failed to achieve unquantifiable levels of HCV RNA by week 6 or had a confirmed quantifiable HCV RNA level at any point after documented unquantifiable levels.
Resistance testing
For DNA sequencing, only samples with HCV RNA ≥1000 IU/ml were amplified in order to reduce the chance of sampling bias. A baseline plasma sample was collected from each patient on day 1 before dosing of study drugs. The relevant targets, full‐length genotype 3 NS3/4A (1‐685 amino acids) and NS5A (1‐452 amino acids), were amplified from each baseline sample with an HCV RNA level of ≥1000 IU/ml and analysed by population sequencing to identify variants in each target. For the single patient who experienced virological failure, the sample closest in time after virological failure with an HCV RNA level of ≥1000 IU/ml was also analysed by population sequencing. The presence of amino acid variants at NS3 positions 56, 155, 156, 166 and 168 31, 32, as well as NS5A positions 24, 28, 29, 30, 31, 32, 58, 92 and 93, was included in the sequence analyses 24, 25, 33, 34. Phenotypic resistance of genotype 3a NS3 variants to paritaprevir was determined with a chimeric replicon that had the variant engineered in the genotype 3a NS3 gene in the backbone of a genotype 1b‐Con 1 replicon 23. Similarly, phenotypic resistance of genotype 3a NS5A variants to ABT‐530 was determined with a chimeric replicon that had the variants engineered in the genotype 3a NS5A gene in the backbone of a genotype 1b‐Con 1 replicon 24.
Pharmacokinetic assessments
On day 1, blood samples were collected immediately prior to the morning dose of study drug and at 2, 4 and 6 h post dose. A single sample was collected at each subsequent study visit, regardless of the time of dosing. Plasma concentrations were measured by validated methods including liquid chromatography‐tandem mass spectrometry, and pharmacokinetic parameters were evaluated by non‐compartmental analysis.
Safety and tolerability assessments
Safety and tolerability assessments were conducted at screening and throughout the study. Evaluations included adverse event monitoring, vital signs, 12‐lead electrocardiography, physical examinations and clinical laboratory tests (e.g. haematology, serum chemistry and urinalysis). Adverse events were graded by severity based on NCI CTCAE (v4.0) criteria 35 and were assessed for causality as related to study drugs. Data on serious adverse events were collected up till 30 days after the last dose.
Statistical analysis
All statistical summaries and analyses were performed with sas ® software, version 9.3 (SAS Institute, Inc., Cary, NC, USA). Efficacy, safety and demographic analyses included all patients who received at least one dose of study drugs. Descriptive statistics are summarized for all evaluations.
Results
Patient demographics and baseline characteristics
Of the 14 patients screened, 10 genotype 3‐infected patients enrolled, received at least one dose of study drug and completed the study. Demographics and baseline characteristics are summarized in Table 1. All patients were white, with 2 identifying as Hispanic/Latino ethnicity, and 8 patients were males (80%). All patients had HCV genotype 3a infection and nine had IL28B non‐CC genotype (90%). Based on data collected from an electronic Medication Event Monitoring System (MEMS), all patients were considered compliant to treatment, defined as the percentage of capsules/tablets taken relative to total capsules/tablets equalling between 80% and 120%.
Table 1.
Patient demographics and baseline characteristics
| ABT‐530 + PTV/r + RBVN = 10 | |
|---|---|
| Male, n (%) | 8 (80) |
| White, n (%) | 10 (100) |
| Hispanic or Latino, n (%) | 2 (20) |
| Age, mean (min, max), years | 53 (34, 69) |
| BMI, mean (min, max), kg/m2 | 27 (22, 34) |
| HCV genotype 3a, n (%) | 10 (100) |
| IL28B non‐CC genotype, n (%) | 9 (90) |
| HCV RNA, mean (min, max), log10 IU/ml | 5.8 (1.4, 7.3) |
| Baseline fibrosis stage, n (%) | |
| F0–F1 | 7 (70) |
| F2 | 2 (20) |
| F3 or greater | 1 (10) |
BMI, body mass index; HCV, hepatitis C virus; max, maximum; min, minimum; PTV/r, paritaprevir/ritonavir; RBV, ribavirin.
Virological response and resistance analyses
The mean baseline HCV RNA level for all patients was 5.8 log10 IU/ml with all patients reaching undetectable HCV RNA levels by treatment week 4 (Fig. 2). Of the 10 patients treated, 9 (90%) achieved both SVR12 and SVR24. One patient experienced on‐treatment breakthrough at week 6. Population sequencing was performed on 9 baseline samples. Titre of the baseline sample from a tenth patient who achieved SVR was too low for viral sequence analysis. None of the sequenced baseline samples from patients who achieved SVR harboured any NS3 or NS5A variants at amino acid positions for key resistance‐associated variants. In samples from the patient who experienced breakthrough, an A30K variant in NS5A was identified at baseline, and variants S24F, M28K and A30K in NS5A were detected after breakthrough. Results of population sequencing, with a limit of detection of 15–20%, suggested that these 3 NS5A variants were probably linked because there were no mixtures detected at the corresponding nucleotide positions in the sequencing chromatograms. The genotype 3 NS5A A30K variant alone does not confer any resistance to ABT‐530, but genotype 3‐linked NS5A variants S24F+M28K+A30K confer a >5000‐fold increase in the ABT‐530 EC50 value relative to the wild‐type genotype 3 HCV replicon. In NS3, the genotype 3 A166S variant, which does not confer any resistance to paritaprevir, was identified at baseline and after breakthrough in this patient.
Figure 2.
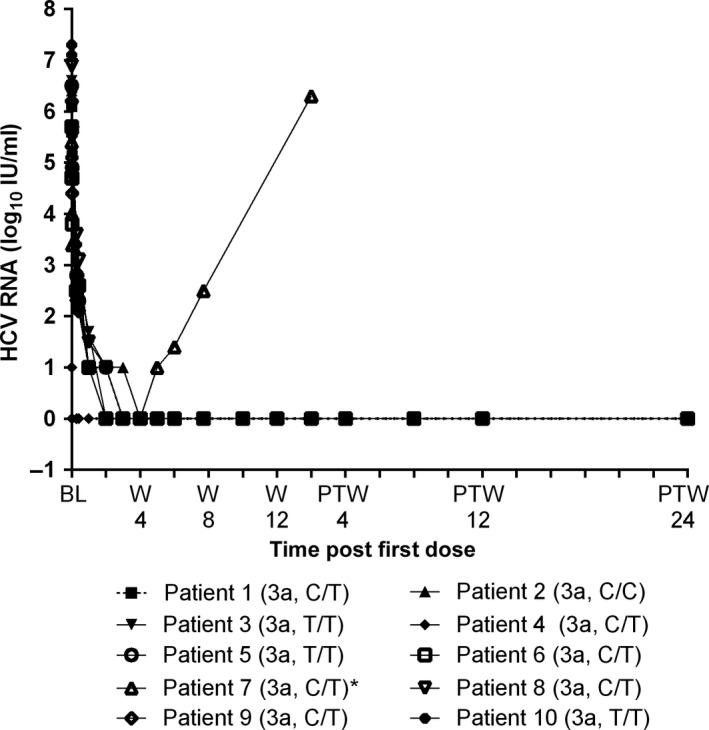
Mean hepatitis C virus (HCV) RNA levels for patients who received ABT‐530 in combination with paritaprevir/ritonavir and ribavirin for 12 weeks. Presented are the mean HCV RNA levels (log10 IU/ml) evaluated during the M14‐213 study for each of the 10 patients who received ABT‐530 in combination with paritaprevir/ritonavir and ribavirin. The genotype 3 subtype and IL28B genotype are described for each patient in parentheses. Dotted line at y‐axis 0 indicates HCV RNA below the lower limit of detection (15 IU/ml). * indicates the one patient with on‐treatment breakthrough at treatment week 6; this patient did not have assessments following post‐treatment week 2. BL, baseline; W, treatment week; PTW, post‐treatment week.
Pharmacokinetics
Paritaprevir/ritonavir boosted ABT‐530 120 mg exposures by around two‐fold compared with exposure of ABT‐530 120 mg during 3‐day monotherapy as studied in patients with genotype 1 infection 36. Paritaprevir, ritonavir and ribavirin exposures in patients with genotype 3 infection appear to be comparable to historical data observed in patients with genotype 1 infection who received the regimen of ombitasvir/paritaprevir/ritonavir and dasabuvir with ribavirin (data on file). The patient who experienced breakthrough had similar ABT‐530 trough levels compared with patients who achieved SVR, though this patient had the lowest trough levels for paritaprevir and ritonavir and second lowest ribavirin trough levels among all patients (Fig. 3).
Figure 3.
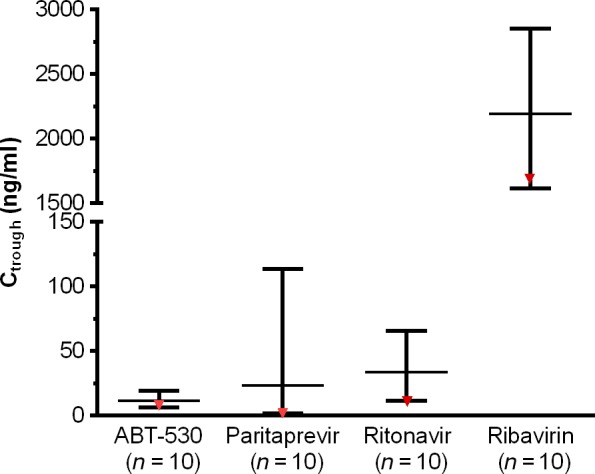
Distribution of trough plasma concentrations measured for patients who received ABT‐530 in combination with paritaprevir/ritonavir and ribavirin for 12 weeks. Presented is the distribution of trough plasma concentrations for the 10 patients who received ABT‐530 in combination with paritaprevir/ritonavir and ribavirin for 12 weeks. The middle line indicates the mean value, and top and bottom lines show the maximum and minimum values, respectively, for each compound. Data for the patient who experienced virological failure at treatment week 6 are indicated in red. Ctrough, trough plasma concentrations, is estimated based on concentrations measured at 22–26 h post dosing for ABT‐530, paritaprevir and ritonavir and at 10–14 h post dosing for ribavirin.
Safety
Nine (90%) patients experienced adverse events (Table 2). All adverse events were classified as grade 1 (mild) or 2 (moderate) in severity, with no events reported as grade 3 (severe) or greater 35. Adverse events occurring with a frequency greater than 10% among patients included fatigue, dyspnoea, dizziness, myalgia, nausea and rash (Table 2). No serious adverse events, events leading to early discontinuation, or deaths were reported. One patient experienced fatigue and a decrease in haemoglobin levels that were deemed possibly related to ribavirin, and the ribavirin dose was modified for this patient. One patient had a total bilirubin level greater than 2 times the upper limit of normal, which was considered clinically significant by the investigator. No laboratory abnormalities were categorized as grade 4 (life threatening or disabling). Overall, there were no clinically meaningful changes in vital sign assessments.
Table 2.
Summary of adverse events
| ABT‐530 + PTV/r + RBVN = 10 | |
|---|---|
| Any AE, n (%) | 9 (90) |
| AEs leading to study discontinuation, n (%) | 0 (0) |
| Any serious AE, n (%) | 0 (0) |
| AEs reported at grade 3 (severe) or greater,a n (%) | 0 (0) |
| Common AEs,b n (%) | |
| Fatigue | 4 (40) |
| Dyspnoea | 3 (30) |
| Dizziness | 2 (20) |
| Myalgia | 2 (20) |
| Nausea | 2 (20) |
| Rash | 2 (20) |
| AEs with a reasonable possibility of being related to ABT‐530 treatment,b n (%) | |
| Fatigue | 3 (30) |
| Dyspnoea | 3 (30) |
| Dizziness | 2 (20) |
| Nausea | 2 (20) |
| Rash | 2 (20) |
Based on NCI CTAE (v4.0) criteria 35.
Reported in >10% of patients.
AE, adverse event; PTV/r, paritaprevir/ritonavir; RBV, ribavirin.
Discussion
The combination of ABT‐530, paritaprevir/ritonavir and ribavirin displayed robust antiviral potency in patients with genotype 3 infection, with nine (90%) patients achieving SVR12 and one patient experiencing breakthrough at week 6. The pharmacokinetic profile for ABT‐530 was comparable in the patient who experienced breakthrough compared with the mean of the cohort, though paritaprevir and ritonavir concentrations were lower relative to the mean. This regimen was well tolerated and adverse events were predominately mild in severity. No severe or serious adverse events were reported. Laboratory data were favourable with no new safety concerns identified. No clinically meaningful changes in vital sign assessments were reported.
No baseline variants in NS3 or NS5A were detected at the designated resistance‐associated positions in sequenced samples from patients who achieved SVR. In the patient who experienced breakthrough, A166S was detected in NS3 at baseline and after breakthrough, whereas A30K and linked S24F+M28K+A30K variants were detected in NS5A at baseline and after breakthrough respectively. ABT‐530 maintains activity against NS5A single‐position variants across HCV genotypes, including genotype 3 25; however, the genotype 3 triple variant S24F+M28K+A30K resulted in a high level of resistance to ABT‐530. As the genotype 3 A166S variant in NS3 detected at baseline and breakthrough does not confer any resistance to paritaprevir, other factors may have contributed to virological failure. Although paritaprevir exposure in this patient should have been adequate to suppress the genotype 3 wild‐type virus, low paritaprevir concentrations and the low compliance rates for ribavirin reported for this patient (data not shown) may have decreased the probability to achieve SVR.
This study was exploratory in nature with a small sample size. All patients had genotype 3a infection and none had cirrhosis; thus, these data may not be readily extrapolated to other genotype 3 subgenotypes or subpopulations of patients with HCV.
Similar to the 90% SVR rate reported here with an ABT‐530‐containing regimen, studies with other NS5A inhibitors in development reported improved efficacy against genotype 3 infection. NS5A inhibitor velpatasvir with sofosbuvir, with or without ribavirin, for 12 weeks provided SVR12 rates of 88–100%; two (8%) patients experienced post‐treatment relapse 37. An ongoing study assessing the combination of NS5A inhibitor elbasvir dosed with grazoprevir (NS3 inhibitor) and sofosbuvir for genotype 3 infection reported SVR4 rates of 100% in 10 patients without cirrhosis and 90% in 10 cirrhotic patients 38.
Both daclatasvir and ledipasvir are two NS5A inhibitors approved to treat chronic genotype 3 infection as part of combination DAA regimens 13, 14. In patients with genotype 3 infection, daclatasvir/sofosbuvir, with or without ribavirin, for 24 weeks provided an SVR12 rate of 89% 12, 17, 18. Ledipasvir with sofosbuvir/ribavirin for 24 weeks provided SVR12 rates of 73% and 89% in genotype 3‐infected patients, with and without cirrhosis, respectively 19.
Compared with ombitasvir, the first‐generation NS5A inhibitor, in vitro results suggest that ABT‐530 has improved antiviral characteristics for genotype 3 infection 25. This study explored the efficacy of 120 mg ABT‐530 dosed with 150/100 mg paritaprevir/ritonavir and ribavirin in patients with HCV genotype 3, while the NAVIGATOR study (NCT 01458535) with similar study design evaluated 25 mg ombitasvir with 200/100 mg paritaprevir/ritonavir and ribavirin also in patients with genotype 3 infection 22. Despite the higher 200 mg paritaprevir dose used with ombitasvir, comparison of SVR12 data between these two studies suggests that a regimen containing ABT‐530 is more potent than the ombitasvir‐containing regimen as it reduced the occurrence of virological failure. Since ABT‐530, paritaprevir/ritonavir and ribavirin demonstrated greater efficacy against genotype 3 compared with an ombitasvir‐containing combination, it provides a rationale for developing ABT‐530 in combination with other potent DAAs as a viable option for genotype 3‐infected patients.
Although the combination evaluated here demonstrated high efficacy, ABT‐530 with paritaprevir/ritonavir and ribavirin will not be evaluated further. Instead, ABT‐530 coadministered with ABT‐493, a next‐generation NS3/4A protease inhibitor identified by AbbVie and Enanta, is currently being investigated to treat all six major HCV genotypes. ABT‐493 has potent pangenotypic antiviral activity, including genotype 3 31, 33. When compared with paritaprevir, ABT‐493 has a higher barrier to resistance and improved pharmacokinetic profile, not requiring pharmacokinetic enhancement with ritonavir 36. Compared with ABT‐530 with paritaprevir/ritonavir and ribavirin, ABT‐530 with ABT‐493 optimizes treatment by providing an interferon‐, ribavirin‐ and ritonavir‐free option that has pangenotypic HCV coverage.
With HCV genotype 3 infection having emerged as difficult‐to‐cure 10, development of new treatments that address this unmet medical need is important and timely. Based on the results from this study, ABT‐530 holds promise as an efficacious NS5A inhibitor for use with other potent DAAs to treat chronic HCV genotype 3 infection.
Acknowledgements
AbbVie sponsored the study (NCT02068222); contributed to its design and participated in the collection, analysis and interpretation of the data and in the writing, review and approval of the article. Medical writing support was provided by Sharanya Ford, PhD, of AbbVie.
Financial support: This study was funded by AbbVie Inc.
Conflict of interest: Fred Poordad: Grant/Research Support – AbbVie, Achillion Pharmaceuticals, Anadys Pharmaceuticals, Biolex Therapeutics, Boehringer Ingelheim, Bristol‐Myers Squibb, Genentech, Gilead Sciences, GlaxoSmithKline, GlobeImmune, Idenix Pharmaceuticals, Idera Pharmaceuticals, Intercept Pharmaceuticals, Janssen, Medarex, Medtronic, Merck, Novartis, Santaris Pharmaceuticals, Scynexis Pharmaceuticals, Vertex Pharmaceuticals, ZymoGenetics; speaker – Gilead, Kadmon, Merck, Onyx/Bayer, Genentech, GSK, Salix and Vertex; consultant/advisor – AbbVie, Achillion Pharmaceuticals, Anadys Pharmaceuticals, Biolex Therapeutics, Boehringer Ingelheim, Bristol‐Myers Squibb, Gilead Sciences, GlaxoSmithKline, GlobeImmune, Idenix, Merck, Novartis, Tibotec/Janssen, Theravance and Vertex. Charles S. Landis: Grant/Research Support – AbbVie, Bristol‐Myers Squibb, Gilead Sciences. Daniel F. Jackson III: No relevant conflicts of interest to disclose. Armen Asatryan, Teresa I. Ng, Bo Fu, Chih‐Wei Lin, Betty Yao, Jens Kort: Employees of AbbVie and may hold AbbVie stock or options.
Sponsor role: AbbVie sponsored the study; contributed to its design; participated in the collection, analysis and interpretation of the data and in the writing, review and approval of the article. This manuscript contains information about the investigational product ABT‐530, as well as the investigational use of paritaprevir (formerly ABT‐450), which was identified by AbbVie and Enanta Pharmaceuticals.
Trial registration number: ClinicalTrials.gov: NCT02068222.
Liver Int. 2016; 36: 1125–1132. DOI: 10.1111/liv.13067
Handling editor: Luca Valenti
References
- 1. Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age‐specific antibody to HCV seroprevalence. Hepatology 2013; 57: 1333–42. [DOI] [PubMed] [Google Scholar]
- 2. Messina JP, Humphreys I, Flaxman A, et al Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 2015; 61: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ampuero J, Romero‐Gomez M, Reddy KR. Review article: HCV genotype 3 – the new treatment challenge. Aliment Pharmacol Ther 2014; 39: 686–98. [DOI] [PubMed] [Google Scholar]
- 4. Kohli A, Shaffer A, Sherman A, Kottilil S. Treatment of hepatitis C: a systematic review. JAMA 2014; 312: 631–40. [DOI] [PubMed] [Google Scholar]
- 5. Pybus OG, Cochrane A, Holmes EC, Simmonds P. The hepatitis C virus epidemic among injecting drug users. Infect, Genet Evol 2005; 5: 131–9. [DOI] [PubMed] [Google Scholar]
- 6. Jhaveri R, McHutchison J, Patel K, Qiang G, Diehl AM. Specific polymorphisms in hepatitis C virus genotype 3 core protein associated with intracellular lipid accumulation. J Infect Dis 2008; 197: 283–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goossens N, Negro F. Is genotype 3 of the hepatitis C virus the new villain? Hepatology 2014; 59: 2403–12. [DOI] [PubMed] [Google Scholar]
- 8. Nkontchou G, Ziol M, Aout M, et al HCV genotype 3 is associated with a higher hepatocellular carcinoma incidence in patients with ongoing viral C cirrhosis. J Viral Hepatitis 2011; 18: e516–22. [DOI] [PubMed] [Google Scholar]
- 9. Wartelle‐Bladou C, Le Folgoc G, Bourliere M, Lecomte L. Hepatitis C therapy in non‐genotype 1 patients: the near future. J Viral Hepatitis 2012; 19: 525–36. [DOI] [PubMed] [Google Scholar]
- 10. Tapper EB, Afdhal NH. Is 3 the new 1: perspectives on virology, natural history and treatment for hepatitis C genotype 3. J Viral Hepatitis 2013; 20: 669–77. [DOI] [PubMed] [Google Scholar]
- 11. SOVALDI (sofosbuvir) . Prescribing Information. Gilead Sciences; Foster City, CA, 2014. [Google Scholar]
- 12. DAKLINZA (daclatasvir) . Summary of Product Characteristics. Bristol‐Myers Squibb; Uxbridge, UK, 2014. [Google Scholar]
- 13. DAKLINZA (daclatasvir) . Prescribing Information. Bristol‐Myers Squibb, New York City, NY, 2015. [Google Scholar]
- 14. HARVONI (ledipasvir and sofosbuvir) . Summary of Product Characteristics. Gilead Sciences, Cambridge, UK, 2015. [Google Scholar]
- 15. Foster GR, McLauchlan J, Irving W, et al Treatment of decompensated HCV cirrhosis in patients with diverse genotypes: 12 weeks of sofosbuvir and NS5A inhibitors with/without ribavirin is effective in genotypes 1 and 3. J Hepatol 2015; 62: S187–212. [Google Scholar]
- 16. Andriulli A, Mangia A, Iacobellis A, et al Meta‐analysis: the outcome of anti‐viral therapy in HCV genotype 2 and genotype 3 infected patients with chronic hepatitis. Aliment Pharmacol Ther 2008; 28: 397–404. [DOI] [PubMed] [Google Scholar]
- 17. Nelson DR, Cooper JN, Lalezari JP, et al All‐oral 12‐week treatment with daclatasvir plus sofosbuvir in patients with hepatitis C virus genotype 3 infection: ALLY‐3 phase III study. Hepatology 2015; 61: 1127–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sulkowski MS, Gardiner DF, Rodriguez‐Torres M, et al Daclatasvir plus sofosbuvir for previously treated or untreated chronic HCV infection. N Engl J Med 2014; 370: 211–21. [DOI] [PubMed] [Google Scholar]
- 19. Keating GM. Ledipasvir/Sofosbuvir: a review of its use in chronic hepatitis C. Drugs 2015; 75: 675–85. [DOI] [PubMed] [Google Scholar]
- 20. Menon RM, Klein CE, Lawal AA, et al Pharmacokinetics and Tolerability of the HCV Protease Inhibitor ABT‐450 Following Single Ascending Doses in Healthy Adult Volunteers With and Without Ritonavir. HepDART, Kohala Coast, HI, 2009. [Google Scholar]
- 21. VIEKIRA PAK (ombitasvir, paritaprevir/ritonavir, dasabuvir) . Prescribing Information. AbbVie Inc., North Chicago, IL, 2015. [Google Scholar]
- 22. Lawitz E, Sullivan G, Rodriguez‐Torres M, et al Exploratory trial of ombitasvir and ABT‐450/r with or without ribavirin for HCV genotype 1, 2, and 3 infection. J Infect 2015; 70: 197–205. [DOI] [PubMed] [Google Scholar]
- 23. Pilot‐Matias T, Tripathi R, Cohen D, et al In vitro and in vivo antiviral activity and resistance profile of the hepatitis C virus NS3/4A protease inhibitor ABT‐450. Antimicrob Agents Chemother 2015; 59: 988–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krishnan P, Beyer J, Mistry N, et al In vitro and in vivo antiviral activity and resistance profile of ombitasvir, an Inhibitor of HCV NS5A. Antimicrob Agents Chemother 2014; 59: 979–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ng T, Krishnan P, Kati W, et al ABT‐530, an HCV NS5A inhibitor with potent pangenotypic activity and high genetic barrier to resistance. Abstract 639. 21st Annual Conference on Retroviruses and Opportunistic Infections; Boston, 2014. [Google Scholar]
- 26. Gane E, Nahass R, Luketic V, et al P0776: Efficacy of 12 or 18 weeks of grazoprevir plus elbasvir with ribavirin in treatment‐naive, noncirrhotic HCV genotype 3‐infected patients. J Hepatol 2015; 62: S621. [DOI] [PubMed] [Google Scholar]
- 27. Schnell G, Tripathi R, Beyer J, et al Hepatitis C virus genotype 4 resistance and subtype demographic characterization of patients treated with ombitasvir plus paritaprevir/ritonavir. Antimicrob Agents Chemother 2015; 59: 6807–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lawitz EJ, O'Riordan WD, Asatryan A, et al Potent antiviral activity of direct‐acting antivirals, ABT‐493 and ABT‐530, with 3‐day monotherapy for hepatitis C genotype 1 infection. Antimicrob Agents Chemother 2015; 60: 1546–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. A 4‐drug combination (Viekira Pak) for hepatitis C. Med Lett Drugs Ther 2015;57:15–7. [PubMed] [Google Scholar]
- 30. Colucci G, Ferguson J, Harkleroad C, et al Improved COBAS TaqMan hepatitis C virus test (Version 2.0) for use with the high pure system: enhanced genotype inclusivity and performance characteristics in a multisite study. J Clin Microbiol 2007; 45: 3595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ng T, Reisch T, Middleton T, et al ABT‐493, a potent HCV NS3/4A protease inhibitor with broad genotypic coverage. Abstract 636. 21st Annual Conference on Retroviruses and Opportunistic Infections; Boston, 2014. [Google Scholar]
- 32. Halfon P, Locarnini S. Hepatitis C virus resistance to protease inhibitors. J Hepatol 2011; 55: 192–206. [DOI] [PubMed] [Google Scholar]
- 33. Ng T, Pilot‐Matias T, Liangjun L, et al A next generation HCV DAA combination: potent, pangenotypic inhibitors ABT‐493 and ABT‐530 with high barriers to resistance. Hepatology 2014; 60: 1142A. [Google Scholar]
- 34. Nakamoto S, Kanda T, Wu S, Shirasawa H, Yokosuka O. Hepatitis C virus NS5A inhibitors and drug resistance mutations. World J Gastroenterol 2014; 20: 2902–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. National Institutes of Health , National Cancer Institute. Common Terminology Criteria for Adverse Events (CTCAE), Version 4.03. 2009. Available at: http://evs.nci.nih.gov/ftp1/CTCAE/CTCAE_4.03_2010-06-14_QuickReference_5x7.pdf. Accessed 18 December 2014.
- 36. Lawitz EJ, O'Riordan WD, Freilich BF, et al Potent antiviral activity of ABT‐493 and ABT‐530 with 3‐day monotherapy in patients with and without compensated cirrhosis with hepatitis C virus (HCV) genotype 1 infection. Hepatology 2014; 60: 1128A–61A. [Google Scholar]
- 37. Gane E, Hyland RH, An D, et al Once daily sofosbuvir with GS‐5816 for 8 weeks with or without ribavirin in patients with HCV genotype 3 without cirrhosis result in high rates of SVR12: the ELECTRON2 study. Abstract #79. Hepatology 2014; 60: 236A. [Google Scholar]
- 38. Poordad F, Lawitz EJ, Gutierrez JA, et al C‐SWIFT: Grazoprevir/elbasvir + sofosbuvir in cirrhotic and noncirrhoitic, treatment‐naive patients with hepatitis C virus genotype 1 infection, for durations of 4, 6, or 8 weeks and genotype 3 infection for durations of 8 or 12 weeks. J Hepatol 2015; 62: S187–212. [Google Scholar]


