Abstract
In flowering plants, pollen tube growth is essential for delivery of male gametes into the female gametophyte or embryo sac for double fertilization. Although many genes have been identified as being involved in the process, the molecular mechanisms of pollen tube growth remains poorly understood. In this study, we identified that the Arabidopsis Transmembrane Protein 18 (AtTMEM18) gene played important roles in pollen tube growth. The AtTMEM18 shares a high similarity with the Transmembrane 18 proteins (TMEM18s) that are conserved in most eukaryotes and may play important roles in obesity in humans. Mutation in the AtTMEM18 by a Ds insertion caused abnormal callose deposition in the pollen grains and had a significant impact on pollen germination and pollen tube growth. AtTMEM18 is expressed in pollen grains, pollen tubes, root tips and other vegetative tissues. The pollen‐rescued assays showed that the mutation in AtTMEM18 also caused defects in roots, stems, leaves and transmitting tracts. AtTMEM18‐GFP was located around the nuclei. Genetic assays demonstrated that the localization of AtTMEM18 around the nuclei in the generative cells of pollen grains was essential for the male fertility. Furthermore, expression of the rice TMEM18‐homologous protein (OsTMEM18) driven by LAT52 promoter could recover the fertility of the Arabidopsis attmem18 mutant. These results suggested that the TMEM18 is important for plant growth in Arabidopsis.
Keywords: Arabidopsis, male gametophyte, plant, pollen, TMEM18, transmembrane protein
Edited by: Mengxiang Sun, Wuhan University, China
INTRODUCTION
In higher plants, pollen tube elongation in the pistil is a crucial step for sexual reproduction (Xia et al. 2010). The process begins with deposition of pollen grains on the stigmatic tissue. The compatible interaction between pollen and stigmatic cells triggers the hydration and germination of the pollen grains (Jiang et al. 2005). The resulting pollen tube invades the stigmatic tissue, penetrates the style, navigates through the transmitting tract, and then is guided toward the ovule to deliver the sperm cells into the embryo sac for fertilization (Hülskamp et al. 1995; Wilhelmi and Preuss 1996; Ray et al. 1997; Lennon and Lord 2000; Palanivelu and Preuss 2000; Johnson and Preuss 2002; Lord and Russell 2002; Preuss 2002; Kim et al. 2003; Palanivelu et al. 2003).
Pollen tube elongation is a polar growth event, namely the growth exclusively occurs at the tip (Bedinger 1992; Krichevsky et al. 2007; Chebli et al. 2012). Many cell biological processes are involved in pollen tube growth, such as calcium gradients (Iwano et al. 2009), vesicular trafficking (Šamaj et al. 2006), actin cytoskeleton dynamics (Wilsen et al. 2006), ion fluxes (Michard et al. 2009), energy metabolism (Hepler et al. 2013) and cell wall synthesis (Chebli et al. 2012; Guan et al. 2013).
Cell wall plays a crucial role in maintaining cell integrity under high turgor pressure (Chebli et al. 2012; Guan et al. 2013). Cell wall synthesis mainly occurs in the tip region of growing pollen tubes. The newly formed cell wall at the pollen tube tip is mainly composed of pectins (Lancelle and Hepler 1992; Edlund et al. 2004; Geitmann and Steer 2006; Dardelle et al. 2010; Chebli et al. 2012), which are synthesized in the Golgi apparatus and secreted by exocytosis, and are highly methyl‐esterified. In the subapical and distal region of the pollen tubes, the pectins are de‐esterified by pectin methylesterase (PME) to confer the rigidity (Jiang et al. 2005; Harholt et al. 2010). Behind the tip of the pollen tube, a secondary layer is formed beneath the outer pectic layer (Ferguson et al. 1998; Li et al. 1999a). The most abundant component in this secondary wall layer is callose. Cellulose is also present in this layer, but at a low level (Dong et al. 2005; Nishikawa et al. 2005; Geitmann and Steer 2006). Callose deposition was first detected immediately after germination of pollen grains in liquid medium, at the rim of the germination aperture (Li et al. 1999b). In pollen tube, callose can be steadily detected in the distal region, which is confined to the inner wall layer and to transverse walls named plugs (Ferguson et al. 1998; Li et al. 1999b). The plugs were consistent with its role in resisting tension stresses (Chebli et al. 2012). However, how the genes regulate the callose deposition in pollen tube remains poorly understood.
The tip growth events of cells include pollen tube and root hair growth in higher plants, nerve growth and axon guidance in animals, and hyphal growth in fungi (Li et al. 1999b). Many genes have been identified as being involved in tip growth, including the Transmembrane protein 18 (TMEM18) in animals. The TMEM18 is predicted to consist of α helices and has three transmembrane domains (Speakman 2013). Phylogenetic analysis has revealed that TMEM18 has an ancient origin, being found in plants and most other eukaryotic organisms (Almén et al. 2010; Speakman 2013). TMEM18 is located in the nuclear envelope in neural stem cells (Jurvansuu et al. 2008) and has been proposed to be involved in cell migration as demonstrated by overexpression of the protein increasing the migration of neural stem cells towards glioma in the rat brain (Jurvansuu et al. 2008). The TMEM18 is expressed ubiquitously with differences in expression levels in the different tissues (Speakman 2013). In mouse brain, its highest expression occurred in the hypothalamus, thalamus and substantia nigra. The hypothalamus is the region that is responsible for the control of energy homeostasis. Recently TMEM18 was identified to be involved in obesity and body mass index (BMI) (Almén et al. 2010). Nevertheless, so far, little is known about the function of TMEM18. Especially, the roles and functions of the TMEME18 protein in plant species remain unknown.
In this study, the Arabidopsis thaliana TMEM18 (AtTMEM18) was identified by a screening for male gametophyte‐defective mutant from the gene‐ and enhancer‐trap Ds insertional mutant library (Sundaresan et al. 1995; Yang et al. 1999). AtTMEM18 is encoded by At1g34350 and shares a higher similarity with the TMEM18 proteins from other species. The insertion of the Ds in At1g34350 caused an unusual deposition of callose in the germinating pollen grains and affected the growth of pollen tubes and vegetative organs, indicating that AtTMEM18 plays important roles in the pollen tube and cell morphogenesis.
RESULTS
Isolation and genetic analysis of the attmem18 mutant
The attmem18 mutant was isolated in a genetic screen for male gametophyte‐defective mutants from a collection of gene‐trap and enhancer‐trap Ds insertion lines in Arabidopsis ecotype Landsberg erecta (Ler) (Sundaresan et al. 1995). The Ds insertion created a genetic tag of kanamycin resistance in the mutant plant. At first, a genetic analysis of the heterozygous attmem18‐1 (attmem18/+) plants was performed (Table 1). The progeny from self‐pollinated attmem18/+ plants exhibited a segregation ratio of approximately 1 kanamycin‐resistant (KanR): 1 kanamycin‐sensitive (KanS) (1282:1278), much lower than the typical Mendelian segregation ratio of 3 to 1. Furthermore, no homozygous KanR plants (attmem18/‐) were identified in the progeny. This result indicates that the attmem18 mutant is defective in gametophytic function.
Table 1.
Genetic analysis of the attmem18/+ mutant
| Crosses (Female × Male) | Number of KanR seedlings | Number of KanS seedlings | KanR/ KanS |
|---|---|---|---|
| attmem18/+ × attmem18/+ | 1,282 | 1,278 | 1.003 |
| Wild type × attmem18/+ | 0 | 2,237 | 0 |
| attmem18/+ × wild type | 710 | 690 | 1.029 |
KanR, kanamycin‐resistant; KanS, knanamycin‐sensitive.
To determine how the mutation affected male or female gametophytic functions, the attmem18/+ plants were used as males or females to cross with wild type plants. As shown in Table 1, when the attmem18/+ plants were used as female parents, 50.71% (710/1400) of the resulting F1 progeny were KanR. In contrast, when attmem18/+ pollen grains were used to pollinate wild type plants, no KanR plants in the F1 progeny were obtained (0:2237). Thus, the attmem18 mutation disrupted male gametophytic function completely, but did not affect female gametophytic function.
The attmem18 pollen grains exhibited an abnormal callose deposition
To investigate how attmem18 affected male gametophytic function, observation of attmem18 male gametophyte development was performed. Scanning electron microscopy (SEM) showed that most of the pollen grains from the attmem18/+ plants were as normal as wild type pollen grains in morphology (Figure 1A, B). 4′6‐diamidino‐2‐phenylindole (DAPI) staining showed that the pollen grains from the attmem18/+ plants had two sperm nuclei and a vegetative nucleus like wild type pollen grains (Figure 1C, D). These results indicated that the attmem18 mutation did not affected pollen formation.
Figure 1.
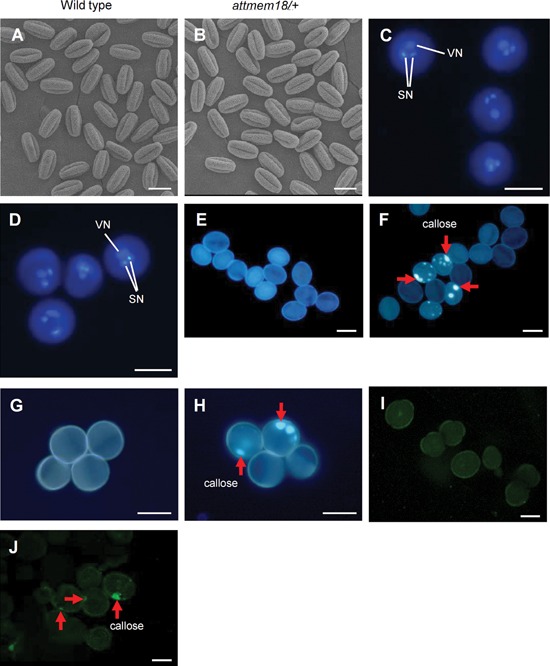
Phenotypic characterization of attmem18 pollen grains (A, B) SEM images of pollen grains from wild type (A) and attmem18/+ plants (B). (C, D) DAPI‐stained pollen grains from wild type (C) and attmem18/+ plants (D). (E–H) Aniline blue‐stained pollen grains from wild type (E), attmem18/+ (F), qrt1/qrt1 (G) and attmem18/+; qrt1/qrt1 plants (H). (I, J) Section and immunofluorescence assays of pollen grains from wild type (I) and attmem18 /+ plants (J). The red arrows indicate the dotted callose deposition in the pollen grains. SN, sperm cell nuclei; VN, vegetative cell nuclei. Bars = 20 µm.
Aniline blue, which is a specific dye for callose, was used to study the callose deposition pattern in the attmem18 pollen by comparison with wild type pollen. No obvious fluorescence signal was observed in wild type pollen grains (Figure 1E) as expected. In contrast, 23.2% of pollen grains from attmem18/+ plants exhibited the dotted fluorescence signals (Table S1, Figure 1F), indicating that a number of attmem18/+ pollen grains had an abnormal callose deposition. Because no homozygous attmem18 mutant could be generated for the assays, attmem18 mutant was introgressed into a background of quartet1‐2 (qrt1‐2) mutant to perform quartet assay (Preuss et al. 1994). The qrt1‐2 mutation blocks physical separation of the tetrad pollen grains, but does not affect the germination and growth of the pollen tubes (Preuss et al. 1994). Therefore, a tetrad from the attmem18/+; qrt1/qrt1 mutant plants had two qrt1‐2 (presenting wild type) pollen gains and two qrt1‐2; attmem18 (presenting attmem18 mutant) pollen grains. When the tetrads pollen grains from qrt1‐2 and attmem18/+; qrt1/qrt1 plants were stained with aniline blue, no obvious fluorescence could be seen in qrt1‐2 pollen grains from the qrt1/qrt1 mutant plant, while 24.1% of pollen grains from the attmem18/+; qrt1‐2/qrt1‐2 plants showed dotted callose deposition patterns (Table S1, Figure 1G, H), indicating that the pollen grains carrying the attmem18 in the tetrads exhibited an unusual callose deposition.
To verify the location of callose deposition in the attmem18 pollen grains, callose‐specific antibody was used to locate the callose in the section of attmem18 pollen grains from the attmem18/+ plants. As shown in Figure 1J, the punctate callose signals appeared nearby the inner wall in the attmem18 pollen grain sections, compared to that no obvious fluorescence signals were observed in the sections of the wild type pollen grains (Figure 1I). These results showed that the attmem18 pollen grains had an abnormal callose deposition during germination process.
The attmem18 pollen is defective in pollen germination and pollen tube growth
To study how attmem18 mutation affected the male gametophytic function, we further examined the germination of attmem18 pollen grains. The mature pollen grains from attmem18/+ plant were cultured in vitro. 39.8% of the pollen grains from attmem18/+ plants germinated in vitro, compared to that 76.2% of the pollen grains from wild type plants could germinate under the same culture condition (Figure 2A–C), indicating that the attmem18 significantly reduced pollen germination in vitro.
Figure 2.

The attmem18 affected in vitro and in vivo germination of pollen grains (A, B) The in vitro germination of pollen grains from wild‐type (A) and attmem18/+ plants (B). (C) Germination rates of pollen grains from wild type (WT) and attmem18/+ plants. (D, E) SEM images showing in vivo germination of the pollen grains from wild type (C) and attmem18/+ plants (D). (F–I) The in vivo germination of pollen grains from qrt1/qrt1 plants (F), wild type (G), attmem18/+ (H) and attmem18/+; qrt1/qrt1 plants (I). Pg, pollen grain; Pt, pollen tubes; Stg, stigmatic cells. White arrows indicate the ungerminated pollen grains. The red arrows indicate abnormal callose deposition in the pollen grains and pollen tubes. Bars = 20 µm. * P < 0.05.
Then we examined the pollen germination efficiency of the attmem18 in vivo. After the pollen grains from attmem18/+ and wild type plants were pollinated to the emasculated wild type flowers for 12 h, they were observed by SEM, respectively. All the wild type pollen tubes could normally grow into the stigma (Figure 2D). In contrast, some of the pollen tubes generated by the pollen grains from attmem18/+ plants were short and swollen (Figure 2E). Among the pollen grains from attmem18/+ plants, 57.0% could grow morphologically normal pollen tubes, 18.4% did not germinate, and 24.6% produced shorter and swollen pollen tubes. In comparison, 95.9% of wild type pollen tubes could grow normally through the stigma (Table S2). The quartet assays also achieved a similar result. In most tetrads from the attmem18/+; qrt1/qrt1 plants, only 1 or 2 pollen grains could germinate pollen tubes. In general, 36.2% pollen grains produced normal pollen tubes, 35.0% pollen grains did not germinate, and 28.7% of pollen grains generated swollen pollen tubes (Table S2). In the control, most of the qrt1‐2 tetrads could have three or four pollen grains produce pollen tubes (Figure 2F). The germination rate was 68.1% (Table S2). Aniline blue staining showed that the callose deposition pattern in attmem18 pollen tube inner walls was obviously different from that in wild type pollen tubes. In the wild type, the callose exhibited as strong and round fluorescent signals, and uniformly distributed on the inner wall of pollen tubes. Callose plugs were formed in the elongating wild type pollen tubes (Figure 2G). In the attmem18 mutant pollen grains from attmem18/+ and attmem18/+; qrt1/qrt1 plants, callose exhibited as a punctate distribution pattern (Figure 2H, I) and distributed irregularly in the swollen pollen tube inner walls. The punctate distribution pattern was not found in the morphologically normal pollen tubes (Figure 2G, H). This result indicated that the abnormal callose deposition was likely related to the defect of the attmem18 pollen tubes.
Phenotypes of attmem18 could be complemented by wild type AtTMEM18
Thermal asymmetric interlaced PCR (TAIL‐PCR) (Liu et al. 1995; Liu and Whittier 1995) was applied to obtain the flanking sequences adjacent to the Ds element in attmem18. Sequence analysis showed that the Ds element was inserted into the first exon of gene At1g34350, 261 bp downstream from start codon ATG (Figure 3A). The Ds insertion site was further confirmed by conventional PCR using the gene‐specific (AtTMEM18‐Ds‐3′) and Ds‐5′ border (Ds5‐3) primers (Table S4).
Figure 3.
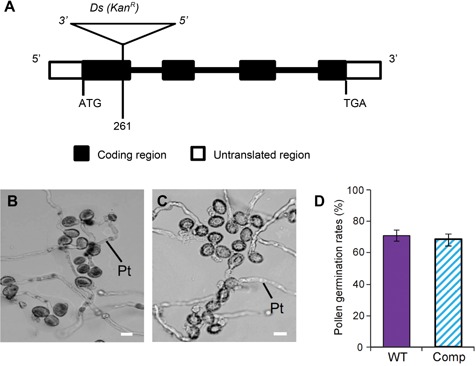
Molecular characterization of attmem18 mutant (A) The organization of the AtTMEM18 gene, showing the Ds element insertion site. (B, C) The in vitro germination of pollen grains from wild type (B) and complemented plants (C). (D) Pollen germination rates of the wild type (WT) and the complemented (Comp) plants. Bars = 20 µm.
To verify the phenotype of attmem18/+ was caused by the Ds insertion in AtTMEM18 (At1g34350), the genomic sequence of AtTMEM18, which included the 1.0‐kb promoter, 1.5‐kb coding region and 3′‐terminal region of AtTMEM18, was cloned into Ti‐derived vector pCAMBIA1300 (CAMBIA, Australia) and introduced into the heterozygous attmem18 (attmem18/+) plants via Agrobacterium tumefaciens‐mediated infiltration transformation (Bechtold and Pelletier 1998). The seeds from the T1 transgenic attmem18/+ plants were plated on kanamycin‐containing agar plates to analyze the Ds segregation ratio. The progeny from self‐pollinated 34 out of 40 transformant lines segregated in an increased ratio of KanR: KanS, compared to those from self‐crossed attmem18/+ controls (Table S3). The aniline blue staining assays showed that the transgenic attmem18 pollen grains exhibited a normal callose distribution pattern like wild type pollen grains (Figure S1). Furthermore, the in vitro pollen germination rates of the transformed attmem18 lines were increased (Figure 3B–D) to the values similar to that of wild type pollen. These results demonstrated that the defects in pollen tube growth could be complemented by the introduction of the wild type AtTMEM18 genomic DNA fragment.
AtTMEM18 is expressed in the tissues active in cell growth and differentiation
The expression pattern of AtTMEM18 was first analyzed by RT‐PCR and Northern blotting hybridization using the RNAs extracted from different wild type plant tissues including seedlings, roots, stems, leaves, flowers, and siliques. The expression of AtTMEM18 were detected mainly in roots and inflorescences, weaker in seedlings, leaves and siliques (Figure 4A, B), suggesting that AtTMEM18 was a constitutive expression gene.
Figure 4.

Expression patterns of AtTMEM18 (A, B) RT‐PCR assay and Northern‐blotting, comparing the AtTMEM18 mRNA levels in seedlings, roots, stems, leaves, flowers and siliques. (C–I) GUS activity in seedling (C), root (D), leaf (E), inflorescence (F), anther (G), pollen tube (H) and silique (I) from transgenic wild type plants carrying the pAtTMEM18:GUS construct. Bars = 1 mm in (C) and (E), 0.5 mm in (F) and (I), 0.1 mm in (D) and (G), 10 µm in (H).
The expression pattern of AtTMEM18 was further examined by promoter activity assay. The AtTMEM18 promoter was fused to GUS reporter gene and introduced into wild type plants. The GUS activity was detected mainly in root tips, the cotyledon of seedlings, mature anthers and pollen tubes (Figure 4C–I), consistent with the results from RT‐PCR and Northern blotting hybridization. These results indicated that AtTMEM18 is constitutively expressed in many tissues, with higher levels in the tissues active in cell growth and differentiation.
The attmem18 also affected the development of other vegetative tissues
As mentioned above, attmem18 disrupts the male gametophytic function. No attmem18 homozygous plant was obtained for further investigation of AtTMEM18 function. To further examine the impact of attmem18 on vegetative development, we performed the pollen rescue experiments using promoter of the pollen‐specific gene LAT52 (Muschietti et al. 1994) fused with AtTMEM18 coding region. The resulting pLAT52:AtTMEM18 construct was introduced into attmem18/+ mutant plants. The seeds from self‐pollinated T1 transgenic plants were planted on agar plates containing kanamycin. The progeny from the self‐pollinated pollen‐rescued plants (attmem18/+; pLAT52:AtTMEM18/+) segregated in a rate of approximately 2 KanR: 1 KanS, indicating that the construct (pLAT52:AtTMEM18) could restore attmem18 gametophytic function (Table S3). The pollen‐rescued lines, which were heterozygous for the Ds insertion and homozygous for the T‐DNA insertion (attmem18/+ pLAT52:AtTMEM18/pLAT52:AtTMEM18) were generated and selected for further assays. In the progeny from self‐pollinated pollen‐rescued plants, approximately 25% (247/1,014) of the seedlings were stunted, compared to wild type seedlings. The hypocotyls, roots and cotyledons of the pollen‐rescued seedlings were shorter (Figure 5B–D), compared to those of wild type. When these stunted seedlings developed into mature plants, the height of plants and length of siliques were shorter than those of wild type plants (Figure 5E). The clarified root tips and root paraffin cross sections of the pollen‐rescued attmem18 seedlings showed that the structures of the root cells were drastically disordered (Figure 5G, K) compared to those of wild type plants (Figure 5F, J). SEM and paraffin cross section observation showed that the epidermal cells of the hypocotyl in pollen‐rescued attmem18 seedlings were distorted (Figure 5I, M). The siliques of pollen‐rescued attmem18 plants were shorter, in which seed set often occurred only in the upper halves of the siliques (Figure 5N, O). When the pollen grains from pollen‐rescued attmem18 plants were pollinated to wild type pistils, the resulting pollen tubes could grow to the bottom of the pollinated wild type pistils (Figure 5P). In contrast, when the wild type pollen grains were pollinated to the pistils of the pollen‐rescued attmem18 plants, the wild type pollen tubes could grow into style, but could not elongate to the bottom of the pistil (Figure 5Q). These results suggested that the pLAT52:AtTMEM18 construct only specifically restored the gametophytic function, but did not restore silique development. Therefore, the pollen‐rescued mutant materials could be used to further study the impact of attmem18 on vegetative development.
Figure 5.
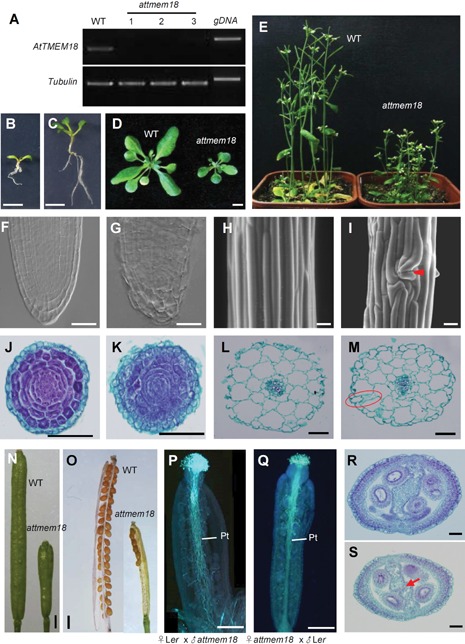
Phenotypic characterization of the pollen‐rescued attmem18 mutants (A) Expression levels of AtTMEM18 were drastically reduced in the pollen‐rescued attmem18 homozygous seedlings as revealed by RT‐PCR. (B, C) A 10‐d‐old pollen‐rescued attmem18 homozygous seedling (B), compared with the 10‐d‐old wild type seedling (C) under the same growth conditions. (D) A mature pollen‐rescued attmem18 homozygous plant compared with wild type plant (WT) under the same growth conditions. (E) The flowering pollen‐rescued attmem18 homozygous plants compared with wild type plants (WT) under the same growth conditions. (F) A clarified wild type root tip, showing the typical cell organization pattern. (G) A clarified pollen‐rescued attmem18 homozygous root tip, showing the defect in the cell morphology and organization. (H, I) The SEM images of hypocotyl epidermal cells in wild type seedlings (H) and pollen‐rescued attmem18 homozygous seedling (I). The red arrow indicates the distorted epidermal cell organization. (J, K) The cross sectioning images of wild type root (J) and pollen‐rescued attmem18 homozygous root (K). (L, M) Comparison of the paraffin cross‐sections between wild type hypocotyl (L) and pollen‐rescued attmem18 homozygous hypocotyl (M). The red oval indicates irregularly hypocotyl cells. (N) Comparison of the siliques at the 7th day after pollination (DAP) between wild type and pollen‐rescued attmem18 homozygous plants. The silique of homozygous attmem18 mutant was much shorter than that of wild type. (O) Mature siliques from wild type plants and attmem18 plants. (P) The growth of attmem18 pollen tubes in the wild type (Ler) at 12 DAP. (Q) The growth of wild type pollen tubes in the attmem18 silique at 12 DAP. (R, S) Comparison of paraffin cross section in wild type pistil (R) and attmem18 pistil (S). The red arrow indicates the unfused style diaphragm in attmem18 pistil. Bars = 500 µm in (B) to (D), (N) and (O), 50 µm in (F) to (M), (P) to (S).
To examine the impact of the attmem18 on silique development, the paraffin section of transmitting tissue of pollen‐rescued attmem18 plants were observed. The results showed that the style diaphragm of pollen‐rescued attmem18 siliques was not fused, compared to the wild type siliques (Figure 5R, S). These results indicated that the cell morphogenesis in pollen‐rescued attmem18 mutant siliques were affected. PCR assays using DNA from the defective pollen‐rescued attmem18 seedlings indicated that they were homozygous for the attmem18 mutation. RT‐PCR assays using the RNAs extracted from the pollen‐rescued attmem18 homozygous seedlings showed that expression of AtTMEM18 was dramatically reduced in the pollen‐rescued attmem18 seedlings (Figure 5A). Transgenic wild type plants with pLAT52:AtTMEM18 coding region construct were examined as a negative control, no defective plants were found in these transgenic plants in wild type background. Therefore, the attmem18 mutation also affected vegetative development in Arabidopsis.
AtTMEM18 protein is located around nucleus
AtTMEM18 protein has 163 amino acid residues in length and contains three predicted transmembrane domains (http://www.cbs.dtu.dk/services). It shares similarities with the proteins from many species in eucaryon except yeast and nematode, but not in bacteria. In particular, AtTMEM18 shares 72% similarity with LOC100527513 (Glycine max) and POPTRDRAFT_287702 (Populus trichoca), 59% similarity with LOC100283849 (Zea mays), 53% similarity with Os05g0432500 (Oryza sativa) and 34% similarity with Transmembrane 18 (Mus musculus) and TMEM18 (Homo sapiens) (Figure 6A). The phylogenetic tree analysis showed that TMEM18 proteins could be clustered into animal and plant groups, respectively (Figure 6B). AtTMEM18 belongs to the plant group.
Figure 6.
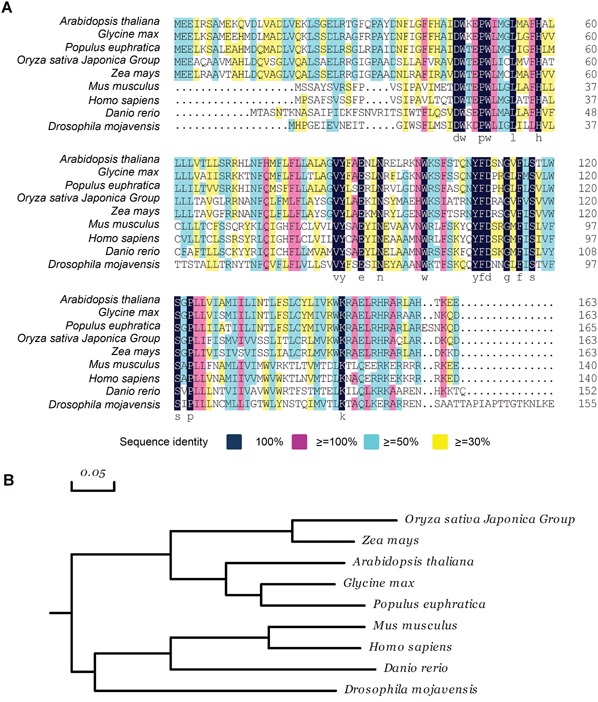
AtTMEM18 shares sequence similarity with many TMEM18 from other species (A) Sequence alignment of AtTMEM18‐homologous proteins. (B) Phylogenetic analysis of AtTMEM1‐homologous proteins. The scale bar represents genetic distance. Species and accession number: Arabidopsis thaliana, AT1G34350; Glycine max, NP_001237116; Populus euphratica, XP_011038158; Oryza sativa Japonica Group, Os05g0432500; Zea mays, AFW61473; Mus musculus, NP_742046; Homo sapiens, NP_690047; Danio rerio, NP_001005601; Drosophila mojavensis, EDW08271.
To investigate the subcellular localization of AtTMEM18, 1.0‐kb promoter of AtTMEM18 was fused to the AtTMEM18 genomic coding region and the coding sequence of green fluorescent protein (GFP). The resulting construct pAtTMEM18:gAtTMEM18‐GFP was introduced into wild type and heterozygous attmem18/+ plants, respectively. Genetic analysis showed that the progeny from self‐pollination of the resulting transgenic heterozygous (attmem18/+) segregated in a KanR: KanS ratio of up to 2:1 (Table S3), indicating that the AtTMEM18‐GFP fusion protein could function normally. The GFP signals were observed in roots, pollen grains at the unicellular to tricellular stages and pollen tubes. In unicellular pollen grains, the GFP signals were detected around the unique nuclei (Figure 7A). In the bicellular pollen grains, the GFP signals appeared around the vegetative and generative nuclei (Figure 7B). In the tricellular pollen grains, the GFP signals were located around the vegetative and two sperm nuclei (Figure 7C). In the germinating pollen grains, the GFP signals were detected around the vegetative and sperm nuclei in the growing pollen tubes (Figure 7D). In the root hair, the GFP signals were expressed around the nuclei (Figure 7E).
Figure 7.
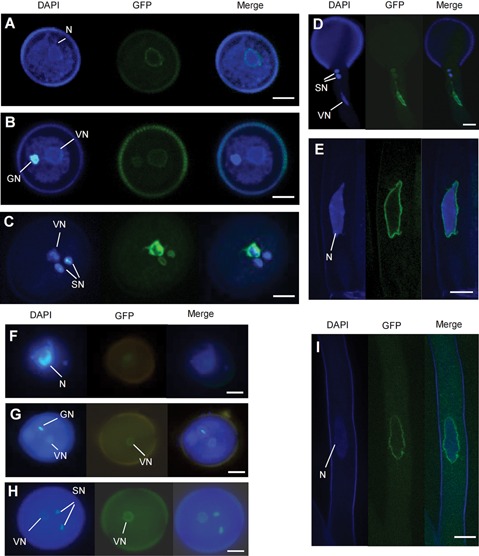
Subcellular localization of AtTMEM18‐GFP (A–E) The GFP signals were located around the nuclei in unicellular pollen grain (A), bicellulare pollen grain (B), tricellulare pollen grain (C), pollen tube (D) and root hair (E) from the pAtTMEM18:gAtTMEM18‐GFP transgenic plants. (F–I) The GFP signals were located around the nuclei in the unicellular pollen grain (F), bicellulare pollen grain (G), tricellulare pollen grain (H) and root hair (I) from pAtTMEM18:cAtTMEM18‐GFP transgenic plants. GN, generative nuclei; N, nuclei; VN, vegetative nuclei; Bars = 5 µm.
To examine whether the protein localization was related to the introns of the gene, another construct was generated by replacing the genomic coding sequence (gAtTMEM18) with the AtTMEM18 cDNA. The resulting pAtTMEM18:cAtTMEM18‐GFP construct was also introduced into heterozygous attmem18 (attmem18/+) plants. The progeny from the self‐pollinated transgenic attmem18/+ plants segregated in a ratio of 1 KanR:1 KanS, like the progeny from the self‐pollinated nontransgenic attmem18/+ plants, indicating that pAtTMEM18:cAtTMEM18‐GFP could not restore the fertility of attmem18 mutant. The GFP signal was detected in the pAtTMEM18:cAtTMEM18‐GFP transgenic pollen grains and roots (Figure 7F–I) roots and pollen grains. In the transgenic root hairs, the GFP signals could be detected around the nucleus like those in the pAtTMEM18:gAtTMEM18‐GFP transgenic plants (Figure7I). During the periods of pollen development, however, the GFP signals were appeared only around the vegetative cell nuclei (Figure 7F–H), differently from those in the pAtTMEM18:gAtTMEM18‐GFP transgenic pollen grains in which GFP signals were located not only around the vegetative nuclei but also around the generative nuclei at the bicellular stage or around the sperm nuclei at the mature pollen stage. These results indicated that the introns of the AtTMEM18 was related to its expression in generative and sperm nuclei in Arabidopsis.
To further examine which intron was involved in its expression regulation in AtTMEM18, deletion assays were performed. AtTMEM18 gene has three introns (Figure S2A). As shown in Figure S2A, six constructs were generated for the assays. The resulting six constructs were introduced into attmem18/+ plants, respectively. The genetic analyses showed that the progeny from all the six construct transgenic attmem18/+ plants segregated in a ratio of approximately 2 KanR: 1 KanS (Table S3), indicating that all six constructs could complement the fertility of attmem18 mutant. The GFP signals were located around the vegetative and sperm nuclei of the transgenic tricellular pollen grains (Figure S2B). Thus, any one intron of AtTMEM18 could be enough to enable the correct expression of AtTMEM18 in the pollen grains.
Expression of the rice TMEM18 (Os05g0432500) in attmem18 could complement the attmem18 mutant phenotype
BlastP search showed that rice (Oryza sativa) also has a TMEM18‐homologous protein, namely Os05g0432500. To examine whether Os05g0432500 had the same function as AtTMEM18 in Arabidopsis, the construct pLAT52:Os05g0432500 was generated and introduced into attmem18/+ mutant plants. The seedlings from the self‐pollinated T1 transgenic plants segregated in a ratio of 2 KanR: 1 KanS (Table S3), compared to the 1 KanR: 1 KanS segregation of the seedlings from the self‐pollinated non‐transgenic attmem18/+ mutant plants. The result showed that the rice Os05g0432500 protein could complement the functional loss of AtTMEM18 in Arabidopsis.
DISCUSSION
AtTMEM18 is important for pollen tube growth
We report here the functional characterization of AtTMEM18 that is involved in pollen tube growth. AtTMEM18 encodes a transmembrane protein, which shares higher similarity with the TMEM18 proteins found in many animal and plant species. In the rat brain, the TMEM18 is located in the nuclear envelope of the neural stem cells (Jurvansuu et al. 2008). The GFP fusion protein assays demonstrated that AtTMEM18 is located around nuclei, implying that it might be also located in the nuclear envelope. However, more direct evidence still is required to address this question.
AtTMEM18 is expressed in the developing pollen grains at the stages from unicellular to mature pollen grains, indicating that it is related to male gametophytic function. The Ds insertion in AtTMEM18 caused defect in pollen germination and pollen tube growth, but did not affect the division of generative cells and morphology of mature pollen grains. Thus, AtTMEM18 protein is important for pollen germination and pollen tube growth, but may not be essential for pollen formation.
The attmem18 pollen tubes became swollen. It has been reported that defects in pollen tube wall components can cause the pollen tube wall to be unstable, resulting in swollen pollen tubes (Wang et al. 2013). Our histological assays showed that the attmem18 pollen grains exhibited a dotted callose distribution pattern before germination, different from those in wild type pollen grains. In wild type, callose appears prolifically at two stages, namely tetrad stage and pollen tube growth stage after germination. At the tetrad stage, the callose walls were formed to separate and protect the microspores. During emergence of the pollen tubes, the callose deposition could be detected at the rim of the germination aperture (Li et al. 1999b). Then, in pollen tube, callose can be steadily detected in the distal region, which is confined to the inner wall layer and to transverse walls named a plug (Ferguson et al. 1998; Li et al. 1999b). The callose plug is consistent with its role in resisting tension stresses (Chebli et al. 2012). According to these results, the mature pollen grains did not synthesize callose before germination. The irregular callose deposition in the inner wall of attmem18 pollen gains suggested AtTMEM18 may play roles in the time‐control of callose synthesis. Therefore, we speculate that the premature callose synthesis in ungerminated pollen grain may be the major cause for the abnormal germination. Furthermore, our results also showed that the callose distribution in the swollen attmem18 pollen tubes were significantly different from those of wild type pollen tube. When attmem18 pollen grains were cultured in vitro, no callose plugs were formed in the swollen mutant pollen tubes after germination although the dotted callose deposition still appeared in the inner wall of the pollen grains. Moreover, the pollen tube walls lacked or had less callose deposition. Therefore, we speculated that AtTMEM18 may be involved in pollen tube growth through controlling deposition or synthesis of callose in pollen grains and pollen tubes. There could be two possibilities for the mechanism that AtTMEM18 controls deposition or synthesis of the callose. One is that AtTMEM18 may be involved in the regulation of expression of the genes related to callose biosynthesis. The data supporting this hypothesis is that the C‐terminal of the TMEM18 proteins in animal could form homo‐polymer, bind to DNA of the chromosome and repress transcription of certain genes in the nucleus (Jurvansuu and Goldman 2011). Another possibility is that AtTMEM18 may participate in the transportation of the proteins related to callose biosynthesis or deposition. Nevertheless, more studies are still required to address how AtTMEM18 controls the deposition of callose to the walls of pollen grains and pollen tubes.
AtTMEM18 may influence the morphogenesis of the vegetative cells in Arabidopsis
The pollen‐rescue assays showed that the attmem18 also severely affected the development of the vegetative tissues, including hypocotyls, cotyledon, roots, leaves, siliques, resulting in shorter plants and reduction in seed set. The sectioning of the tissues showed that the diaphragm was not fused in the mutant siliques. The shapes of the cells in the distorted tissues including siliques and roots were altered. These results indicate that TMEM18 is important for normal plant cell growth, especially for cell morphogenesis.
The introns of AtTMEM18 is essential for its expression in generative cells of pollen grains
In the GFP fusion protein assays for AtTMEM18 protein localization, the pAtTMEM18:cAtTMEM18‐GFP could not complement the male sterile phenotype of attmem18 mutant, but the pAtTMEM18:gAtTMEM18‐GFP could. In pAtTMEM18:cAtTMEM18‐GFP, the AtTMEM18 cDNA was used, which lacked the three introns of AtTMEM18, whereas in pAtTMEM18:gAtTMEM18‐GFP, the genomic coding sequences was fused to GFP, which contained three introns. Therefore, the results indicated that the introns are essential for the expression of AtTMEM18 function on male fertility. In the pAtTMEM18:gAtTMEM18‐GFP transgenic attmem18/+ plants, the AtTMEM18‐GFP was expressed around the nuclei of both vegetative cells and two sperm cells, while in pAtTMEM18:cAtTMEM18‐GFP transgenic attmem18/+ plants, the AtTMEM18‐GFP was expressed only around the nucleus of the vegetative cell, but not in those of the sperm cells. Therefore, the introns of AtTMEM18 are essential for its expression in sperm cells. Further deletion assays for the introns demonstrated that either of the three introns is enough to ensure the expression of AtTMEM18 in sperm cells, which could restore the male gametophytic function of attmem18 mutant. Taken together, expression of AtTMEM18 in sperm cells is essential for its function on male gametophytic function, and the introns of AtTMEM18 are required to ensure its expression in sperm cells.
It is still worthy to point out that when the pollen specific promoter LAT52 was used to drive the expression of AtTMEM18 in pollen grain and pollen tubes, the progeny from the self‐pollinated pLAT52:cAtTMEM18‐GFP transgenic attmem18/+ mutant plants segregated in ratio of approximately 2 KanR: 1 KanS (data not shown), indicating that pLAT52:cAtTMEM18‐GFP also could restore the male gametophytic function of attmem18. The green fluorescence of AtTMEM18‐GFP was distributed in the whole pollen grains, included the vegetative cell and sperm cells (Figure S3). This result demonstrated that use of LAT52 promoter also could complement the roles of AtTMEM18 introns.
The TMEM18 genes in plants might be different from those in animals
TMEM18 protein is remarkably well conserved across this 1,500 million year evolutionary history (Almén et al. 2010). Surprisingly it has no clear homologs in either yeast or Caenorhabditis elegans (Almén et al. 2010). AtTMEM18 shares 72%, 53% and 34% similarity with the homologous proteins of Populus trichoca, Oryza sativa and Homo sapien, respectively. The homologous analysis showed the AtTMEM18 was clustered with the plant group. These results indicated that the functions of TMEM18 proteins might be different between plants and animals. The differences in the structure between TMEM18 and AtTMEM18 were mainly reflected in two aspects. First, the C‐terminal of TMEM18 protein has a typical nucleic acid‐binding motif (ERRKEKKRRRKED) and has been demonstrated to bind to DNA (Jurvansuu and Goldman 2011). The second significant difference is that there are four cysteine residues in the first and second transmembrane domain of TMEM18. With these four cysteine residues, TMEM18 proteins could form homo‐polymer and be located in nuclear envelope, and enable the C‐terminal of the protein to bind DNA on the chromosome and repress transcription of certain genes in the nucleus (Jurvansuu and Goldman 2011). In contrast, there were no any typical nucleic acid binding motif in the C‐terminal region of AtTMEM18 and other homologous proteins in plants. Furthermore, DNA binding assay showed that AtTMEM18 could not bind to DNA. Moreover, BIFC assay showed that AtTMEM18 also could not form homodimers. In addition, expression of the mouse TMEM18 protein in attmem18 could not recover the male fertility of the mutant, but expression of the rice homologous gene (Os05G0432500) driven by the pollen‐specific LAT52 promoter could restore the male gametophytic fertility of attmem18. Taken together, the AtTMEM18 proteins may be functionally different from those in animals.
MATERIALS AND METHODS
Plant materials and mutant isolation
The Arabidopsis thaliana plants used in this study were all in Landsberg erecta background. The seeds were plated on MS‐salt agar plates with or without 50 mg/L kanamycin (Sigma‐Aldrich) or 20 mg/L hygromycin (Roche, Basel, Switzerland) at 4 °C for 2 d. Then the seeds were cultured at 22 °C under a light cycle of 16 h light/8 h dark for 10 d. The resulting 10‐d‐old seedlings were grown into soil in the same growth condition. The generation of Ds insertion lines and screen of mutants were performed as described by Sundaresan et al. (1995).
Characterization of attmem18 mutant phenotype
Morphological observations of pollen grains by SEM were performed as described by Jiang et al. (2005). Pollen viability was examined using Alexander staining (Alexander 1969). DAPI staining of pollen grains was performed as described by Yang et al. (2009).
To examine in vitro pollen germination, the pollen grains were plated on the agar‐solid pollen germination medium containing 1 mM KCl, 10 mM CaCl2, 0.8 mM MgSO4, 0.01% H3BO3 and 18% sucrose. Then the pollen plates were incubated at 22–24 °C for 6 h. At least 300 pollen grains were examined to calculate the average germination rates for each sample.
To examine in vivo pollen tube growth, the flowers were emasculated one day before anthesis. The being cultured at 22 °C for 6 h, the pollinated pistils were mounted onto the sample platform and quickly examined for the pollen tube growth patterns on the stigma by SEM (HITACHI, S‐3400N). On the other hand, the pollinated pistils were collected and fixed in fixing solution (ethanol:acetic acid = 3:1) for 6 h. Then the fixed pistils were softened in 8 M NaOH overnight. After washed with water for several times, the pistils were stained with aniline blue buffer (0.1% aniline blue in 0.1 M K2HPO4‐KOH buffer with pH = 11.0) for 2 h in the dark. The pollen tube growth patterns inside the pistils were observed using fluorescence microscopy (LEICA, DM2500, Germany).
Pollen grain immunocytochemistry assay was performed as described by Geitmann et al. (1995).
Localization of Ds insertion site in attmem18 mutant
The flanking sequence adjacent to the Ds element was obtained by TAIL‐PCR (Liu et al. 1995; Liu and Whittier 1995). Extraction of the attmem18 genomic DNA and TAIL‐PCR were performed using Ds/AD primer sets, as described previously (Yang et al. 1999). The Ds insertion was confirmed by conventional PCR using Ds‐specific primers Ds5‐3 (Table S4) paired with the gene‐specific primer AtTMEM18‐Ds‐3′ (Table S4). The Ds insertion site was determined by sequencing of the resulting flanking DNA fragments.
Complementation experiments
The 5′ upstream sequence and genomic DNA of AtTMEM18 was amplified using rTaq (TaKaRa, Japan) with primers AtTMEM18‐g‐5′ and AtTMEM18‐g‐3′ (Table S4) and verified by sequencing. The resulting DNA fragment was cloned into the Ti‐derived binary vector pCAMBIA1300 (CAMBIA, Australia) and introduced into the attmem18 heterozygous mutant plants by Agrobacterium tumefaciens (strain GV3101)‐mediated infiltration method (Bechtold and Pelletier 1998). The transformant plants were selected on MS medium supplemented with 50 mg/L kanamycin and 20 mg/L hygromycin. The selected transgenic plants were transferred into soil to produce T2 seeds. The T2 seeds were germinated on kanamycin‐containing MS agar plates to evaluate the complementation efficiency by analyzing the segregation ratio of kanamycin resistance.
The pollen rescue experiment was performed by pollen‐specific expression of AtTMEM18 coding region in attmem18/+ plants. The coding region of AtTMEM18 was amplified using rTaq with primers AtTMEM18‐c‐5′ and AtTMEM18‐c‐3′ (Table S4). The LAT52 promoter fragment was obtained from the plasmid pUCLNGFP2 (a gift from Dr. Zhenbiao Yang, Department of Botany and Plant Science, University of California at Riverside, CA). The resulting DNA fragment was cloned into Ti‐derived binary vector pCAMBIA1300. The resulting constructs were transformed into attmem18/+ plants. The selection and segregation analysis of transformant plants was performed as described above.
The total RNA of rice (Oryza sativa) seedlings was extracted with cetyltrimethylammonium bromide (CTAB) solution (Yu and Goh 2000). Reverse transcription was performed using alfalfa mosaic virus reverse transcriptase (AMV, TaKaRa, http://www.takara-bio.com/). The coding region of Os05g0432500 was amplified using rTaq with primers Os‐c‐5′ and Os‐c‐3′ (Table S4). The pLAT52:Os05g0432500 construct was transformed into attmem18/+ plants. The selection and genetic analysis of transformant plants was performed as described above.
Assays for AtTMEM18 expression pattern
The seedlings, roots, stems, leaves, flowers and siliques were harvested from 2‐week‐old seedlings and 4‐week‐old flowering plants, respectively. The total RNAs were extracted with cetyltrimethylammonium bromide (CTAB) solution (Yu and Goh 2000). Reverse transcription was performed using alfalfa mosaic virus reverse transcriptase (AMV, TaKaRa, http://www.takara-bio.com/). TUBULIN cDNA was used as an internal control to normalize the amount of cDNA templates. The primers TUBULIN‐RT‐5′ and TUBULIN‐RT‐3′ (Table S4) were used to amplify the TUBULIN cDNA fragment. The primers AtTMEM18–RT‐5′ and AtTMEM18–RT‐3′ (Table S4) were used to assess the expression level of AtTMEM18 as previously described by Yang et al. (2009).
In Northern blotting hybridization, AtTMEM18 gene‐specific probe was amplified by rTaq using the primers AtTMEM18–RT‐5′ and AtTMEM18–RT‐3′ (Table S4). The RNA probes were labeled using the DIG RNA labeling kit (Roche, Indianapolis, IN, USA) as described by the supplier. RNA gel blotting hybridizations were performed following the instructions of the DIG system and DIG application manual provided by the supplier.
The AtTMEM18 promoter was amplified using rTaq with primers AtTMEM18‐p‐5′ and AtTMEM18‐p‐3′ (Table S4). After verified by sequencing, the fragment was subcloned upstream of the GUS reporter gene in Ti‐derived binary vector pCAMBIA1300 to generate the expression cassette pAtTMEM18:GUS. The resulting construct was introduced into the wild type plants by Agrobacterium tumefaciens (strain GV3101)‐mediated infiltration method (Bechtold and Pelletier 1998). The transformant plants were selected on MS medium supplemented with 20 mg/L hygromycin. The selected transgenic plants were transferred into soil to produce T2 seeds. GUS staining of T2 plants tissues were performed as described previously (Sundaresan et al. 1995; Yang et al. 1999).
Phenotypic characterization of pollen‐rescued plants
For paraffin sectioning, the roots, stems, leaves and siliques of the pollen‐rescued attmem18 plants and wild type plants were fixed in 50% formalin‐acetic acid‐alcohol (FAA) solution and processed according to Shi et al. (2005) and Yang et al. (1999). The observation of the clarified root tips were as described by Yang et al. (1999).
Assays for protein subcellular localization
The AtTMEM18 promoter fragment was amplified using rTaq (TaKaRa) with primers AtTMEM18‐P‐5′ and AtTMEM18‐P‐3′ (Table S4). The AtTMEM18 genomic DNA fragment without the stop codon was amplified by rTaq using primers AtTMEM18‐g‐5′‐2 and AtTMEM18‐g‐3′‐2 (Table S4). The AtTMEM18 coding region fragment without the stop codon was amplified using rTaq with primers AtTMEM18‐c‐5′ and AtTMEM18‐c‐3′ (Table S4). After verified by sequencing, the AtTMEM18 promoter and genomic DNA fragments were subcloned in front of the NOS terminator sequence (TNOS) in the modified pCAMBIA1300 vector which contained GFP coding region, resulting in a pAtTMEM18:gAtTMEM18‐GFP:TNOS construct. The pAtTMEM18:cAtTMEM18‐GFP:TNOS construct was generated in the same way. The pAtTMEM18:gAtTMEM18‐GFP:TNOS and pAtTMEM18:cAtTMEM18‐GFP:TNOS constructs were introduced into attmem18/+ and wild type plants, respectively. The transformant plants were selected using 50 mg/L kanamycin and 20 mg/L hygromycin. The subcellular localization of AtTMEM18‐GFP fusion protein in the roots, pollen grains and pollen tubes was visualized under a confocal microscope (LSM510, Carl Zeiss, Germany, http://www.zeiss.com).
AUTHOR CONTRIBUTIONS
Conceived and designed the experiments: D.Y., X.Y.D. and K.Z.Y. Performed the experiments: X.Y.D., K.Z.Y., Z.X.M., L.Q.C. and X.Q.Z. Analyzed the data: X.Y.D., K.Z.Y. and D.Y. Wrote the paper: D.Y., X.Y.D., K.Z.Y., and J.R.B.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Table S1. Comparison of pollen grains with dotted callose deposition in wild type, mutant and complemented mutant plants
Table S2. The in vivo Germination of pollen grains from attmem18/+ mutant and wild type plants
Table S3. Genetic analyses of progeny from different transgenic attmem18/+ plants
Table S4. The sequences of primers used in this study
Figure S1. The full length wild type AtTMEM18 sequence could complement the phenotype of attmem18 pollen
Figure S2. The structure and location of the fused proteins expressed by the six constructs
Figure S3. Location of pLAT52:cAtTMEM18‐GFP construct in transgenic pollen grains
ACKNOWLEDGEMENTS
We thank Dr. V. Sundaresan, Dr. Wei‐Cai Yang and Mrs. Li‐Fen Xie for their kind help with the mutant screens. This work was supported by the research grants from the Ministry of Sciences and Technology (973 project number: 2007CB108700), the Natural Science Foundation of China (NSFC, project number: 30530060), the Ministry of Education (111 project numbered B06003) and the BJAST Youth Backbone Training Plan (No. 2015‐24).
Dou XY, Yang KZ, Ma ZX, Chen LQ, Zhang XQ, Bai JR, Ye D ( 2016) AtTMEM18 plays important roles in pollen tube and vegetative growth in Arabidopsis . J Integr Plant Biol 58: 679–692
Available online on Dec. 24, 2015 at www.wileyonlinelibrary.com/journal/jipb
REFERENCES
- Alexander M ( 1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Almén MS, Jacobsson JA, Shaik JH, Olszewski PK, Cedernaes J, Alsiö J, Sreedharan S, Levine AS, Fredriksson R, Marcus C ( 2010) The obesity gene, TMEM18, is of ancient origin, found in majority of neuronal cells in all major brain regions and associated with obesity in severely obese children. BMC Med Genet 11: 58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G ( 1998) In planta Agrobacterium‐mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Bedinger P ( 1992) The remarkable biology of pollen. Plant Cell 4: 879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chebli Y, Kaneda M, Zerzour R, Geitmann A ( 2012) The cell wall of the Arabidopsis pollen tube — spatial distribution, recycling, and network formation of polysaccharides. Plant Physiol 160: 1940–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardelle F, Lehner A, Ramdani Y, Bardor M, Lerouge P, Driouich A, Mollet J‐C ( 2010) Biochemical and immunocytological characterizations of Arabidopsis pollen tube cell wall. Plant Physiol 153: 1563–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Hong Z, Sivaramakrishnan M, Mahfouz M, Verma DPS ( 2005) Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis . Plant J 42: 315–328 [DOI] [PubMed] [Google Scholar]
- Edlund AF, Swanson R, Preuss D ( 2004) Pollen and stigma structure and function: The role of diversity in pollination. Plant Cell 16: S84–S97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson C, Teeri T, Siika‐Aho M, Read S, Bacic A ( 1998) Location of cellulose and callose in pollen tubes and grains of Nicotiana tabacum . Planta 206: 452–460 [Google Scholar]
- Geitmann A, Hudák J, Vennigerholz F, Walles B ( 1995) Immunogold localization of pectin and callose in pollen grains and pollen tubes of Brugmansia suaveolens — implications for the self‐incompatibility reaction. J Plant Physiol 147: 225–235 [Google Scholar]
- Geitmann A, Steer M ( 2006) The architecture and properties of the pollen tube cell wall. The Pollen Tube Springer. pp. 177–200
- Guan Y, Guo J, Li H, Yang Z ( 2013) Signaling in pollen tube growth: Crosstalk, feedback, and missing links. Mol Plant 6: 1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülskamp M, Kopczak SD, Horejsi TF, Kihl BK, Pruitt RE ( 1995) Identification of genes required for pollen – stigma recognition in Arabidopsis thaliana . Plant J 8: 703–714 [DOI] [PubMed] [Google Scholar]
- Harholt J, Suttangkakul A, Scheller HV ( 2010) Biosynthesis of pectin. Plant Physiol 153: 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler PK, Rounds CM, Winship LJ ( 2013) Control of cell wall extensibility during pollen tube growth. Mol Plant 6: 998–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwano M, Entani T, Shiba H, Kakita M, Nagai T, Mizuno H, Miyawaki A, Shoji T, Kubo K, Isogai A ( 2009) Fine‐tuning of the cytoplasmic Ca2+ concentration is essential for pollen tube growth. Plant Physiol 150: 1322–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Yang SL, Xie LF, San Puah C, Zhang XQ, Yang WC, Sundaresan V, Ye D ( 2005) VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 17: 584–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Preuss D ( 2002) Plotting a course: Multiple signals guide pollen tubes to their targets. Dev Cell 2: 273–281 [DOI] [PubMed] [Google Scholar]
- Jurvansuu J, Zhao Y, Leung DS, Boulaire J, Yu YH, Ahmed S, Wang S ( 2008) Transmembrane protein 18 enhances the tropism of neural stem cells for glioma cells. Cancer Res 68: 4614–4622 [DOI] [PubMed] [Google Scholar]
- Jurvansuu JM, Goldman A ( 2011) Obesity risk gene TMEM18 encodes a sequence‐specific DNA‐binding protein. PLoS ONE 6: e25317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Mollet J‐C., Dong J, Zhang K, Park S‐Y, Lord EM ( 2003) Chemocyanin, a small basic protein from the lily stigma, induces pollen tube chemotropism. Proc Natl Acad Sci 100: 16125–16130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky A, Kozlovsky SV, Tian GW, Chen MH, Zaltsman A, Citovsky V ( 2007) How pollen tubes grow. Dev Biol 303: 405–420 [DOI] [PubMed] [Google Scholar]
- Lancelle SA, Hepler P ( 1992) Ultrastructure of freeze‐substituted pollen tubes of Lilium longiflorum . Protoplasma 167: 215–230 [Google Scholar]
- Lennon K, Lord E ( 2000) In vivo pollen tube cell of Arabidopsis thaliana I. Tube cell cytoplasm and wall. Protoplasma 214: 45–56 [Google Scholar]
- Li H, Bacic A, Read SM ( 1999a) Role of a callose synthase zymogen in regulating wall deposition in pollen tubes of Nicotiana alata Link et Otto. Planta 208: 528–538 [Google Scholar]
- Li H, Lin Y, Heath RM, Zhu MX, Yang Z ( 1999b) Control of pollen tube tip growth by a Rop GTPase‐dependent pathway that leads to tip‐localized calcium influx. Plant Cell 11: 1731–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF ( 1995) Efficient isolation and mapping of Arabidopsis thaliana T‐DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Liu YG, Whittier RF ( 1995) Thermal asymmetric interlaced PCR: Automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25: 674–681 [DOI] [PubMed] [Google Scholar]
- Lord EM, Russell SD ( 2002) The mechanisms of pollination and fertilization in plants. Annu Rev Cell Dev Biol 18: 81–105 [DOI] [PubMed] [Google Scholar]
- Michard E, Alves F, Feijó JA ( 2009) The role of ion fluxes in polarized cell growth and morphogenesis: The pollen tube as an experimental paradigm. Int J Dev Biol 53: 1609 [DOI] [PubMed] [Google Scholar]
- Muschietti J, Dircks L, Vancanneyt G, McCormick S ( 1994) LAT52 protein is essential for tomato pollen development: Pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J 6: 321–338 [DOI] [PubMed] [Google Scholar]
- Nishikawa S‐i, Zinkl GM, Swanson RJ, Maruyama D, Preuss D ( 2005) Callose (β‐1, 3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biol 5: 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanivelu R, Brass L, Edlund AF, Preuss D ( 2003) Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 114: 47–59 [DOI] [PubMed] [Google Scholar]
- Palanivelu R, Preuss D ( 2000) Pollen tube targeting and axon guidance: Parallels in tip growth mechanisms. Trends Cell Biol 10: 517–524 [DOI] [PubMed] [Google Scholar]
- Preuss D ( 2002) Sexual signaling on a cellular level: Lessons from plant reproduction. Mol Biol Cell 13: 1803–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D, Rhee SY, Davis RW ( 1994) Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264: 1458–1460 [DOI] [PubMed] [Google Scholar]
- Ray S, Park S‐S, Ray A ( 1997) Pollen tube guidance by the female gametophyte. Development 124: 2489–2498 [DOI] [PubMed] [Google Scholar]
- Šamaj J, Müller J, Beck M, Böhm N, Menzel D ( 2006) Vesicular trafficking, cytoskeleton and signalling in root hairs and pollen tubes. Trends Plant Sci 11: 594–600 [DOI] [PubMed] [Google Scholar]
- Shi DQ, Liu J, Xiang YH, Ye D, Sundaresan V, Yang WC ( 2005) SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17: 2340–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman JR ( 2013) Functional analysis of seven genes linked to body mass index and adiposity by genome‐wide association studies: A review. Hum Hered 75: 57–79 [DOI] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, Dean C, Ma H, Martienssen R ( 1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Wang L, Wang W, Wang YQ, Liu YY, Wang JX, Zhang XQ, Ye D, Chen LQ ( 2013) Arabidopsis galacturonosyltransferase (GAUT) 13 and GAUT14 have redundant functions in pollen tube growth. Mol Plant 6: 1131–1148 [DOI] [PubMed] [Google Scholar]
- Wilhelmi LK, Preuss D ( 1996) Self‐sterility in Arabidopsis due to defective pollen tube guidance. Science 274: 1535–1537 [DOI] [PubMed] [Google Scholar]
- Wilsen KL, Lovy‐Wheeler A, Voigt B, Menzel D, Kunkel JG, Hepler PK ( 2006) Imaging the actin cytoskeleton in growing pollen tubes. Sex Plant Reprod 19: 51–62 [Google Scholar]
- Xia C, Wang YJ, Li WQ, Chen YR, Deng Y, Zhang XQ, Chen LQ, Ye D ( 2010) The Arabidopsis eukaryotic translation initiation factor 3, subunit F (AteIF3f), is required for pollen germination and embryogenesis. Plant J 63: 189–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang KZ, Xia C, Liu XL, Dou XY, Wang W, Chen LQ, Zhang XQ, Xie LF, He L, Ma X ( 2009) A mutation in Thermosensitive Male Sterile 1, encoding a heat shock protein with DnaJ and PDI domains, leads to thermosensitive gametophytic male sterility in Arabidopsis . Plant J 57: 870–882 [DOI] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V ( 1999) The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev 13: 2108–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Goh CJ ( 2000) Identification and characterization of three orchid MADS‐box genes of the AP1/AGL9 subfamily during floral transition. Plant Physiol 123: 1325–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Table S1. Comparison of pollen grains with dotted callose deposition in wild type, mutant and complemented mutant plants
Table S2. The in vivo Germination of pollen grains from attmem18/+ mutant and wild type plants
Table S3. Genetic analyses of progeny from different transgenic attmem18/+ plants
Table S4. The sequences of primers used in this study
Figure S1. The full length wild type AtTMEM18 sequence could complement the phenotype of attmem18 pollen
Figure S2. The structure and location of the fused proteins expressed by the six constructs
Figure S3. Location of pLAT52:cAtTMEM18‐GFP construct in transgenic pollen grains


