Summary
In many aromatic plants including spearmint (Mentha spicata), the sites of secondary metabolite production are tiny specialized structures called peltate glandular trichomes (PGT). Having high commercial values, these secondary metabolites are exploited largely as flavours, fragrances and pharmaceuticals. But, knowledge about transcription factors (TFs) that regulate secondary metabolism in PGT remains elusive. Understanding the role of TFs in secondary metabolism pathway will aid in metabolic engineering for increased yield of secondary metabolites and also the development of new production techniques for valuable metabolites. Here, we isolated and functionally characterized a novel MsYABBY5 gene that is preferentially expressed in PGT of spearmint. We generated transgenic plants in which MsYABBY5 was either overexpressed or silenced using RNA interference (RNAi). Analysis of the transgenic lines showed that the reduced expression of MsYABBY5 led to increased levels of terpenes and that overexpression decreased terpene levels. Additionally, ectopic expression of MsYABBY5 in Ocimum basilicum and Nicotiana sylvestris decreased secondary metabolite production in them, suggesting that the encoded transcription factor is probably a repressor of secondary metabolism.
Keywords: spearmint, secondary metabolism, terpene, sweet basil, transcription factor, YABBY
Introduction
The genus Mentha, a member of Lamiaceae family, includes species that are widely used as medicinal and aromatic herbs. The essential oils produced by these plants find wide usage in food, flavour, cosmetic and pharmaceutical industries (Champagne and Boutry, 2013; Sinha et al., 2013). Plants produce these volatile essential oils as secondary metabolites which have important roles in plant defence, plant‐to‐plant communication and pollination (Gershenzon et al., 2000). In mint, these essential oils are produced in specialized nonphotosynthetic glandular trichomes known as peltate glandular trichomes (PGT) which are found on the aerial surface of the plants. The PGT are dedicated to the production and storage of large amounts of volatile secretions (Champagne and Boutry, 2013; Croteau et al., 2000; Lange and Turner, 2013). In the case of spearmint (Mentha spicata), the essential oil is dominated mainly by two monoterpenes, limonene and carvone. Monoterpenes are the C10 type of terpenoids and are generally colourless, lipophilic and volatile. They are responsible for the characteristic aromas and flavours of essential oils, floral scents and resin of aromatic plants (Loza‐Tavera, 1999). Given their economic importance, strategies to metabolically engineer monoterpenes to increase yield is of considerable interest.
Varietal improvement in cultivated spearmint or peppermint varieties has been challenging because these varieties are sterile hybrids making classical breeding approach unfeasible. Hence, metabolic engineering provides an alternative method to improve essential oil yield and composition. Plants synthesize terpenes either by the mevalonate (MVA) pathway in the cytosol or by the 2‐C‐methyl‐d‐erythritol 4‐phosphate (MEP) pathway in plastids. Both pathways provide the precursors for terpene biosynthesis (Vranová et al., 2013) and have been well investigated. The MEP pathway in plastids is mainly responsible for producing monoterpenes and diterpenes, whereas the MVA pathway generates sesquiterpenes and triterpenes (Dubey et al., 2003). Studies have shown that under certain conditions an exchange of precursor metabolites can occur between the cytosolic MVA and plastid MEP pathways. Analysis of terpenoid production in many different plants has shown that under specific ecological conditions synthesis of monoterpenes, diterpenes, sesquiterpenes and polyterpenes can occur from precursors produced by both pathways. Metabolic intermediates like isopentenyl diphosphate, geranyl diphosphate or geranylgeranyl diphosphate can be transported across plastidial membranes (Hemmerlin et al., 2012; Vranová et al., 2013).
Apart from the precursor pathways, the downstream monoterpene biosynthetic pathways in both spearmint and peppermint are also well characterized (Lange et al., 2011). One of the strategies to increase yield in peppermint was to manipulate genes that code for enzymes involved in the monoterpene pathway, for example genes for limonene synthase and limonene hydroxylase (Diemer et al., 2001; Mahmoud et al., 2004), but overexpression of these genes did not enhance oil yields significantly. In their pioneering work, Mahmoud and Croteau (2001) and Lange et al. (2011) evaluated the efficacy of overexpressing genes encoding enzymes involved in precursor pathways on oil yields in peppermint (Mentha piperita). Most encouraging results were obtained in plants where two genes were manipulated simultaneously, the gene encoding 1‐deoxy‐d‐xylulose‐5‐phosphate reductoisomerase was overexpressed and the gene encoding menthofuran synthase was down‐regulated. Oil yields in these transgenic plants increased up to 61% over wild‐type controls while reducing the undesirable side‐products (+)‐menthofuran and its intermediate (+)‐pulegone (Lange et al., 2011). Recently, increase in monoterpene formation was achieved by introducing a noncanonical substrate neryl diphosphate (NPP), but the additional NPP had to be eliminated to avoid adverse impact on plant growth (Gutensohn et al., 2014).
It is increasingly evident that transcription factors (TFs) which are regulators of structural genes can activate or repress multiple genes in a metabolic pathway (Grotewold, 2008; Iwase et al., 2009). Manipulation of such TFs can be more effective for engineering pathways rather than changing the expression of genes for individual enzymes involved, because plant metabolic pathways are complex comprising of multiple genes encoding various enzymes (Broun and Somerville, 2001). The effectiveness of using TFs to modulate metabolic pathways has been validated in a few studies (Butelli et al., 2008; Luo et al., 2008; Schwinn et al., 2006). Although the enzymatic pathway leading to the synthesis of spearmint monoterpenes is well defined (Croteau et al., 1991; Lange et al., 2011; Muñoz‐Bertomeu et al., 2008), the developmental regulation of this secondary metabolite pathway still remains elusive. Few TFs have been reported from other plants that are involved in regulating terpene biosynthesis. They are Artemisia annua, AaWRKY1, AaERF1, AaERF2, AaORA1 and AabZIP1 (Lu et al., 2013; Ma et al., 2009; Yu et al., 2012; Zhang et al., 2015), cotton GaWRKY1 (Xu et al., 2004), TaWRKY1 from Taxus chinensis (Wang et al., 2001), rubber EREBP1 and HbWRKY1 (Chen et al., 2012; Zhou et al., 2012) and OsTGAP1 in rice (Miyamoto et al., 2014).
To investigate the genes involved in PGT formation and secondary metabolism in spearmint, we had performed comparative RNA‐Seq analysis of different tissues of spearmint, namely PGT, leaf devoid of PGT (leaf‐PGT) and leaf in a previous study (Jin et al., 2014). This led to the identification of many TF transcripts that were significantly more abundant in PGT when compared to leaf‐PGT and a YABBY transcript was among the top candidates. We cloned the full‐length cDNA of this transcript and sequence analysis showed that it is similar to YABBY5 subfamily of proteins. YABBY genes constitute a group of plant‐specific TFs that are known to play important roles in various aspects of vegetative and floral development in plants (Bonaccorso et al., 2012; Bowman, 2000). In this study, we report the engineering of spearmint plants for higher yields by suppressing this glandular trichome‐enriched TF MsYABBY5. The resulting MsYABBY5 RNAi lines showed an increase in monoterpene production which ranged from 20% to 77%. This is the first report of a transcription factor regulating monoterpene production in mint plants and assigns a new role for YABBY genes in plant secondary metabolism. Ectopic expression of MsYABBY5 in sweet basil (Ocimum basilicum), an aromatic herb similar to mint, and in Nicotiana sylvestris resulted in decreased secondary metabolite production in them. Essential oil of sweet basil has compounds derived from both terpene and phenylpropanoid pathways, whereas N. sylvestris produces mainly diterpenes. As MsYABBY5 could affect metabolites derived from different metabolic pathways, it suggests that it regulates an upstream step in plant secondary metabolism. We further found that MsYABBY5 probably regulates terpene synthesis through a regulatory network that involves MsWRKY75.
Results
MsYABBY5 shows high expression in spearmint PGT
Mint leaves have PGT on both surfaces (Figure 1A). From the RNA‐Seq data of leaves, we identified four YABBY‐like transcripts that showed high expression levels. Of these, only MsYABBY5 was preferentially expressed in PGT, whereas the others were more enriched in leaf tissues. The differential expression pattern of these transcripts as observed by RNA Seq was further validated by quantitative RT‐PCR (qRT‐PCR) (Figure 1B). Full‐length open reading frames (ORFs) of all these four YABBYs including MsYABBY5 were amplified from leaf cDNA using RACE. All the four cloned ORFs contained a conserved C2C2 zinc finger domain located at N‐terminus and a helix‐loop‐helix motif (YABBY domain) at the C terminus which is similar to the HMG box motif. These two domains are highly conserved among all YABBY proteins (Figure 2A). As we were interested in TFs involved in regulating secondary metabolism in mint, we focussed on MsYABBY5. In situ hybridization also confirmed the PGT‐specific expression of MsYABBY5, as no signal was observed in the leaf tissue (Figure 1C). The ORF of MsYABBY5 encoded a polypeptide of 190 amino acids. BLAST analysis showed that MsYABBY5 has highest sequence similarity to Antirrhinum PROLONGATA YABBY‐like transcription factor. We generated a phylogenetic tree based on amino acids sequences of YABBY proteins from different plants. The results revealed that MsYABBY5 and MsYABBY6 belonged to the YABBY5 subfamily, whereas the other two, MsYABBY2 and MsYABBY4, are members of the YABBY2 subfamily (Figure 2B).
Figure 1.

Validation of MsYABBY genes expression pattern in spearmint. (A) Spearmint leaf showing peltate glandular trichome (PGT) on upper leaf surface as visualized under scanning electron microscope. (B) qRT‐PCR analysis of MsYABBY genes in different tissues. PGT, peltate glandular trichome; leaf‐PGT, leaves where PGT were brushed away. The housekeeping gene elongation factor 1 (ef1) was used as control. (C) In situ hybridization: antisense (a) and sense (b) probe detection of MsYABBY5.
Figure 2.

Amino acid sequence alignment (A) and phylogenetic tree analysis (B) of MsYABBYs.
Subcellular localization of MsYABBY5 protein
To examine the subcellular localization patterns of YABBY proteins, cDNAs of all the four MsYABBYs were fused in‐frame to cDNA encoding the yellow‐fluorescent protein (YFP) and the fusion genes were transiently expressed in tobacco by agroinfiltration. All the MsYABBYs except MsYABBY5 showed nuclear localization. Interestingly, MsYABBY5 showed both nuclear and cytoplasmic localization (Figure 3A). Online software prediction programs indicated that MsYABBY5 contained a potential transmembrane domain (http://dgpred.cbr.su.se/index.php?p=fullscan) at the amino terminal and participated in the secretory pathway (http://www.cbs.dtu.dk/services/TargetP-1.1/output.php). To investigate this, Golgi markers were used for colocalization experiment which showed that MsYABBY5 localized to Golgi (Figure 3B). To further assess this localization pattern, tobacco leaves were treated with Brefeldin A (BFA). BFA treatment in tobacco results in the complete disappearance of Golgi apparatus and disrupts the secretory system (Robinson and Ritzenthaler, 2006). After treatment with 50 μg/mL BFA for 3 h, MsYABBY5 was found to exhibit nuclear localization only, while both nuclear and cytosolic distribution was still observed in the control plants (treated with 1 : 1000 dilution of DMSO in ddH2O) (Figure 3C).
Figure 3.

Subcellular localization of MsYABBYs in Nicotiana benthamiana. (A) MsYABBY5 showed both nuclear and cytoplasmic expression, while other MsYABBYs were found in nucleus only. (a) MsYABBY5 was localized to both nucleus and cytoplasm. (b) MsYABBY6. (c) MsYABBY2. (d) MsYABBY4. (B) MsYABBY5 protein colocalization with Golgi marker. (C) BFA treatment leads to nuclear localization of MsYABBY5 protein in N. benthamiana. (a) Mock group treated with DMSO. (b) Test group treated with 50 μg/mL BFA for 3 h.
To directly observe the distribution pattern of MsYABBY5 protein, we performed immunogold labelling of native MsYABBY5 in PGT of spearmint. A 14‐amino acid peptide that is presumably located on the surface of the predicted three‐dimensional structure of MsYABBY5 was used as an antigen to raise specific antibody. Western blot analysis showed that the antibody exclusively bound to MsYABBY5 but not to MsYABBY2, MsYABBY4 or MsYABBY6 (Figure S1A,B). MsYABBY5 proteins conjugated with gold particles were observed to accumulate inside the nucleus and also present in the cytoplasm. As the cell organelles were not clearly distinguishable in the TEM sections, the localization to cell organelles could not be verified by immunostaining (Figure S1C).
Analysis of the promoter region of MsYABBY5
A 1116‐bp genomic DNA fragment upstream of the putative start codon of MsYABBY5 was cloned by genome walking. Bioinformatics analysis of this region revealed the presence of many cis‐acting regulatory elements apart from the common TATA and CAAT box (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Figure 4A). Four light‐responsive motifs, including two Box4, one TCT and one Sp1 motifs, were found in the promoter sequence. Regulation of terpene biosynthesis by light is known in many plants (Cordoba et al., 2009). With respect to hormones, two cis‐acting elements, CGTCA‐motif and TGACG‐motif, involved in the MeJA‐responsiveness were found and one for gibberellin cis elements, TATC box, was found within the sequence (Zhou et al., 2012; Zhu et al., 2014). Further, tissue‐specific expression pattern of the cloned promoter was analysed. The 1116‐bp promoter fragment was fused with a β‐glucuronidase (GUS) reporter gene and transformed into Nicotiana benthamiana plants. The transgenic plants showed trichome‐specific expression pattern in leaves and stems of tobacco plants (Figure 4B,C). No staining was observed in flowers or roots. Hence, this promoter is potentially a glandular trichome‐specific promoter. Additionally, this promoter was used to drive MsYABBY5 cDNA fused to a cyan‐fluorescent protein reporter gene in basil and tobacco. The fluorescence was observed exclusively in PGT of basil plants and head cells of the glandular trichomes of tobacco, but subcellular localization was difficult to decipher (Figure 5).
Figure 4.
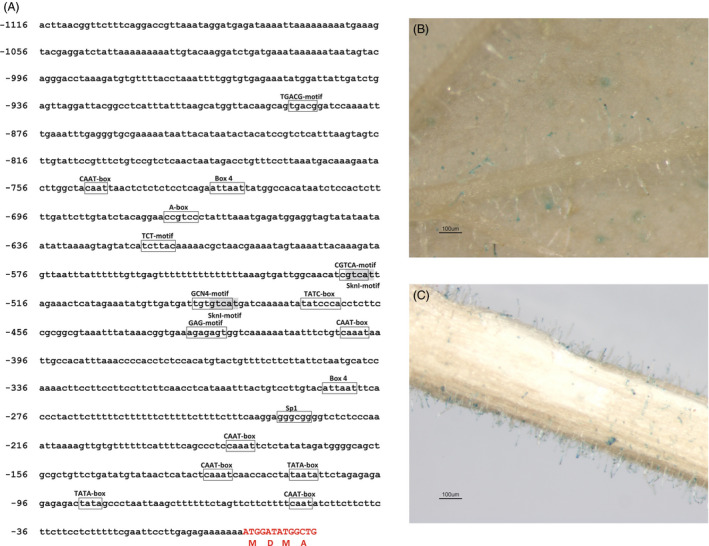
Ms YABBY5 promoter analysis and expression pattern. (A) cis‐acting regulatory elements in the 5′UTR (−1116 bp) region of MsYABBY5. (B and C) Trichome‐specific GUS expression pattern observed in Nicotiana benthamiana leaves and stems of plants transformed with pM sYABBY5::GUS.
Figure 5.

Localization of MsYABBY5 under its native promoter in sweet basil (A) and tobacco (B). pM sYABBY5:: MsYABBY5‐CFP showed exclusive expression in PGT of plants.
Silencing of MsYABBY5 Increases monoterpene production in spearmint
To examine the function of MsYABBY5 in spearmint PGT, an RNAi construct targeting a specific region of MsYABBY5 was generated and transformed into wild‐type spearmint using Agrobacterium tumefaciens‐mediated T‐DNA transfer. Many transgenic lines were generated, of which four independent transgenic lines analysed by Southern blotting for transgene integration were selected for further characterization (Figure S2). All these RNAi plants showed a reduction in MsYABBY5 transcripts (Figure 6A). No significant changes were observed in the expression of other leaf‐specific YABBY genes (MsYABBY2, MsYABBY4 and MsYABBY6), suggesting that the RNAi construct was specific to MsYABBY5 and did not target other YABBY transcripts (Figure S3). The RNAi transgenic plants appeared phenotypically similar to WT plants. Scanning electron microscopy analysis revealed no phenotypical changes in either the number or the structure of PGT.
Figure 6.

Transcript level of MsYABBY5 and monoterpene production in MsYABBY5 RNAi and overexpression plants. (A) MsYABBY5 transcripts level in RNAi plants. (B) Limonene production in RNAi plants. (C) Carvone production in RNAi plants. (D) MsYABBY5 transcripts level in overexpression plants. (E) Limonene production in overexpression plants. (F) Carvone production in overexpression plants. Gene expression is presented as relative to ef1. Leaves from the second node (2–3 cm) were harvested and used for analysis. Results of terpene production are presented as mean ± SD. *P < 0.05; **P < 0.01.
Gas chromatography–mass spectrometry (GC–MS) analysis was performed on these transgenic plants to evaluate the quality and quantity of the volatiles produced. Young WT spearmint leaves contain an abundance of both limonene and carvone monoterpenes. Limonene is the first committed step towards carvone production. Limonene is converted to carvone by a two‐step reaction. In our greenhouse conditions, we observed that in WT spearmint, the productions of limonene and carvone were about 1.47 ± 0.11 and 2.10 ± 0.25 μg/mg fresh leaf, respectively. Upon GC–MS analysis, all the four transgenic lines showed a significant increase (20%–77%) in total monoterpene production (limonene and carvone) (Figure 6B,C). The RNAi lines were tested for the expression levels of enzymes involved in carvone production (limonene synthase, limonene 6‐hydroxylase and carveol dehydrogenase) by qRT‐PCR; however, no major changes were observed. Transcripts for all enzymes of the MEP precursor pathways including geranyl diphosphate synthase small and big subunits were also investigated, but no significant changes in their expression levels were found. These results suggest that increase in monoterpene production is probably not due to the transcriptional activation of biosynthetic genes. MsYABBY5 might be acting upstream to regulate flux into the terpene pathway.
Overexpression of MsYABBY5 results in decrease in monoterpene production
To gain further insight into the role of MsYABBY5 in secondary metabolism, we overexpressed this gene in spearmint under the control of a CaMV 35S promoter. Four independent lines confirmed by southern hybridization were selected for further characterization. The results of qRT‐PCR showed high MsYABBY5 expression levels in all the transgenic plants (Figure 6D). GC–MS analysis of young leaves showed a reduction in total monoterpene production, which ranged from 23% to 52% (Figure 6E,F). The observed phenotype in RNAi and overexpression transgenic lines suggests that MsYABBY5 might be a repressor of secondary metabolism in spearmint.
Possible downstream target of MsYABBY5 to regulate secondary metabolism
To better understand MsYABBY5's biological functions and signalling pathways, it is essential to know its downstream target genes. There is not much known about the molecular mechanism of regulation by YABBYs and their direct target genes. As genes encoding enzymes in the biosynthetic pathways leading to monoterpene production were not significantly changed, we decided to investigate the expression of genes for transporters. Transporters play a key role in plant cellular metabolism (Fischer, 2011; Flügge et al., 2011). From our previous RNA‐Seq data, we had identified several transcripts in PGT which were involved in transport of carbon and ATP (Jin et al., 2014). One such transcript MsNTT similar to the plastidic ATP/ADP transporter‐like proteins showed differential expression in overexpression and RNAi plants. All the RNAi lines showed increased MsNTT transcript levels compared with WT plants or 35S::GFP control plants and reduced expression was observed in the overexpression plants (Figure 7A,B). Plastidic ATP/ADP transporter facilitates the movement of ATP across the plastid inner membrane and can determine the rate of metabolic activity in plastids (Flügge et al., 2011; Neuhaus et al., 1997). MsNTT was seen localized to the plastid membrane (Figure 7C). Enhanced energy import into the plastids can be one of the reasons the knock‐down plants showed a higher metabolic activity.
Figure 7.

MsNTT expression and localization. Transcript levels of MsNTT in MsYABBY5 RNAi (A) and overexpression plants (B) Leaves from the second node (2–3 cm) were harvested and used for qPCR analysis. Gene expression was normalized against the house keeping gene ef1. *P < 0.05; **P < 0.01. (C) MsNTT was localized to the chloroplast membrane in Nicotiana benthamiana.
In a recent study, ChIP‐Seq and RNA‐Seq methods were used to identify YABBY‐regulated genes during various stages of soya bean seedling development. The major candidate genes regulated by YABBY were found to be fatty acid desaturase, APETALA2 (AP2) and WRKY transcription factor (Shamimuzzaman and Vodkin, 2013). Recent research shows the emergence of WRKY TFs as key regulators of terpene production (Patra et al., 2013). From our RNA‐Seq data, we identified a WRKY transcript MsWRKY75, which was enriched in PGTs (Figure 8A). The level of this transcript showed reduction in MsYABBY5 RNAi lines, but no significant increase was observed in overexpression lines (Figure 8B). To check whether MsYABBY5 can bind to the promoter regions of MsNTT and MsWRKY75, we performed EMSA using the purified recombinant His tagged MsYABBY5 protein (Figure 8C). About ~1 kb promoter regions of both the genes were cloned and they were divided into four overlapping fragments and screened by EMSA. No binding was observed with MsNTT promoter, but a protein–DNA complex with reduced mobility was observed when recombinant MsYABB5 was incubated with MsWRKY75 probe (Figure 8D). Interestingly, only the fragment of −909 to −555 bp region of MsWRKY75 promoter was found to bind with MsYABBY5. DNA binding specificity was further confirmed by competition experiments using 10–100‐fold excess unlabelled probe which led to the disappearance of DNA/protein complex. To further determine whether MsYABBY5 protein can regulate the MsWRKY75 promoter in plants, transient expression assays in N. benthamiana were performed. Leaves were coinfilterated with reporter MsWRKY75 promoter::GUS and effector 35S::YFP or 35S::MsYABBY5. Promoter activity of MsWRKY75::GUS was significantly enhanced in 35S::MsYABBY5 expressing leaves when compared to 35S::YFP (Figure 8E). This suggests that MsYABBY5 activates MsWRKY75 which probably represses terpene production in spearmint.
Figure 8.

Transactivation of MsWRKY75 by MsYABBY5. (A) Expression pattern of MsWRKY75 in different tissues of wild‐type spearmint. (B) Transcript levels of MsWRKY75 in MsYABBY5 RNAi plants. (C) Purification of recombinant His tagged MsYABBY5 expressed in Escherichia coli. (D) EMSA assay of MsYABBY5 binding ability to MsWRKY75 promoter. Upper panel shows the fragment −909 to −555 of MsWRKY75 promoter region, (E) transactivation of MsWRKY75 by MsYABBY5 in Nicotiana Benthamiana. Results are presented as mean ± SD. **, P < 0.01.
Ectopic expression of MsYABBY5 affects secondary metabolism in tobacco and sweet basil
To understand the functions of MsYABBY5 in other plants, the gene was ectopically expressed in tobacco and sweet basil. High transcript levels of MsYABBY5 were detected in transgenic tobacco and sweet basil plants (Figure S4). Nicotiana sylvestris glandular trichomes mainly produce diterpenes which are generally derived from the same MEP pathway as the monoterpenes. Ectopic expression of MsYABBY5 was found to reduce cembranoids (CBT‐diol) production in N. sylvestris by 29.5%–47.1% (Figure 9A). Sweet basil essential oil produced in PGT consists of both terpenes and phenylpropanoids. To explore whether MsYABBY5 has an effect on secondary metabolites originating from different metabolic pathways in PGT, we ectopically expressed MsYABBY5 in sweet basil. Three independent sweet basil transgenic lines confirmed by southern hybridization were selected for further characterization. The results of qRT‐PCR showed high expression levels of MsYABBY5 in all the transgenic plants (data not shown). GC–MS analysis on T‐2 plants showed that the total production of both monoterpene (eucalyptol, β‐ocimene and linalool) and sesquiterpene (α‐bergamotene, γ‐muurolene and copaene) decreased (Figure 9B,C). Besides terpene production, phenylpropanoid production was also affected. Eugenol, which is the dominant compound, with a production of 1.93 ± 0.58 μg/mg fresh leaf, showed a significant reduction (P < 0.05) in transgenic plants (Figure 9D). This suggests that transcriptional regulators can govern fluxes in multiple metabolic pathways. Additionally, the sweet basil transgenic lines also showed curled leaf and delayed flowering (about 2–3 weeks delay) when compared to WT plants sown at the same time (Figure S5).
Figure 9.

Ectopic expression of MsYABBY5 decreased terpene production in Nicotiana sylvestris and sweet basil. (A) CBT‐diol production in wild‐type and transgenic N. sylvestris overexpressed with MsYABBY5. Volatiles production of total monoterpenes (B), sesquiterpenes (C) and eugenol (D) in wild‐type and transgenic basil overexpressed with MsYABBY5. Leaves from the second node (2–4 cm) were harvested and used for GC–MS analysis. Results of terpenes and phenylpropanoid production are presented as mean ± SD. *P < 0.05; **P < 0.01.
Discussion
Glandular trichomes are found on the aerial surface of approximately 30% of vascular plants. As they can synthesize and store a large amount of secondary metabolites, they are aptly termed as ‘tiny chemical factories’ of plants. But very few studies have focussed on TFs that regulate glandular trichome‐specific metabolic pathways (Wang, 2014), which will greatly facilitate metabolic engineering efforts to increase yield or develop plant platforms to produce high value compounds. Studies in understanding the transcriptional control of secondary metabolite production show the expression of both activators and repressors is necessary to fine‐tune the flux, timing and the level of structural gene expression in a pathway (Albert et al., 2014; Cavallini et al., 2015; Patra et al., 2013). In this study, we isolated a YABBY gene that is preferentially expressed in spearmint PGT and appears to be a negative regulator of secondary metabolism. This is a novel function for the plant‐specific YABBY family of TFs. In Arabidopsis, YABBY gene family promotes several aspects of leaf, shoot and flower development (Eshed et al., 1999; Goldshmidt et al., 2008; Golz et al., 2004; Stahle et al., 2009), but how they mediate these effects at the molecular level largely remains unknown. Single mutants of yabby5 in Arabidopsis showed no morphological defects, but significantly enhanced the phenotype of yab1yab3 double mutant (Sarojam et al., 2010). In rice, the functions of YABBY genes are divergent from their Arabidopsis homologs. Rice YAB1 is required for gibberellin‐mediated repression of GA3ox2 gene which is involved in the synthesis of gibberellin (Dai et al., 2007).
Studies have revealed that YABBYs are bifunctional TFs acting as either repressors or activators (Bonaccorso et al., 2012; Stahle et al., 2009), but their direct downstream target genes are not well known. The increase in monoterpene production observed in MsYABBY5 RNAi lines was not due to an increase in transcripts level of the structural genes involved in the pathway. Additionally, no significant changes were observed in transcript level of genes encoding enzymes in the precursor MEP pathway. The difference between metabolite and transcript levels can be attributed to either post‐transcriptional modification, protein stability or enhanced flux into the metabolic pathway (Xie et al., 2008). In peppermint, it was shown that most of the biosynthetic enzymes leading to monoterpene production including limonene synthase were regulated at the level of gene expression (McConkey et al., 2000). Many primary metabolic pathways like glycolysis, the TCA cycle, pentose phosphate pathway and shikimate pathway provides carbon, ATP and precursors compounds to diverse secondary metabolic pathways. Transcription factors can affect the synthesis of a particular secondary metabolite by regulating metabolic enzymes in these primary pathways (Aharoni and Galili, 2011). Both mint and basil PGT are nonphotosynthetic organs and need to import ATP and carbon to sustain their high metabolic activities, which too can be possibly controlled by TFs. The fact that MsYABBY5 expression was able to affect metabolite production in tobacco and sweet basil plants suggests that this gene might be probably functioning upstream regulating flux into metabolic pathways. The MEP pathway and shikimate pathway leading to mono/diterpene and phenylpropanoid precursor production, respectively, are both localized in plastids making direct interactions between these pathways possible. Production of anthocyanin pigment (PAP1), a MYB transcription factor from Arabidopsis, is an activator of the phenylpropanoid pathway (Li et al., 2010). Recently it was shown that ectopic expression of PAP1 in rose plants, led to an increase in floral volatile compounds originating from both phenylpropanoid and terpenoid pathways. Transcriptional activation of only few biosynthetic genes was observed, whereas the rest of the increase was attributed to enhanced flux in both pathways (Ben Zvi et al., 2012). Interactions between phenylpropanoid and terpenoid pathways have also been shown in tomato mutants (Enfissi et al., 2010), as well as in Ipomoea flowers (Majetic et al., 2010), but the mechanism still remains to be elucidated.
In a recent study, ChIP‐Seq and RNA‐Seq methods were used to identify YABBY‐regulated genes during various stages of soya bean seedling development. About 96 potential genes were found to be either up‐regulated or down‐regulated by YABBY. One of the major candidate genes regulated by YABBY was found to be WRKY transcription factor (Shamimuzzaman and Vodkin, 2013). WRKY TFs play important roles in regulation of plant stress response and secondary metabolism both as activators and repressors (Schluttenhofer and Yuan, 2015). Research has shown that WRKYs can activate structural genes involved in monoterpene (Spyropoulou et al., 2014), sesquiterpene (Ma et al., 2009) and diterpene (Qiu et al., 2008), as well as phenylpropanoid production (Wang et al., 2007). On the other hand, they can act as negative regulators too. In rice, OsWRKY76 repressed terpene and phenylpropanoid synthesis but increased cold stress tolerance (Yokotani et al., 2013). In rubber, HbWRKY1 negatively regulates a gene involved in natural rubber synthesis (Zhou et al., 2012).
In our study, we found that MsYABBY5 negatively regulates the process of terpene biosynthesis and activates MsWRKY75 in spearmint. MsWRKY75 may be a negative transcriptional regulator of genes involved in terpene synthesis. MsYABBY5 expression overlaps with MsWRKY75 in PGTs. Our RNA seq was performed on PGTs isolated from young leaves at a stage where terpene production is very active. The high expression of both of these TFs at this stage does indicate an important role for them. In MsYABBY5 overexpression lines, no significant increase in MsWRKY75 was observed; this can be due to a threshold of minimal expression required for MsWRKY75 activation. MsWRKY75 probably is one of the many downstream targets regulated by MsYABBY5 to control secondary metabolism. Transcriptome analysis of MsYABBY5 RNAi lines can provide us with more candidate genes that are potentially regulated by MsYABBY5. Our data reveal a regulatory network involving YABBY5 and WRKY that controls terpene production in spearmint. As information regarding terpene pathway regulation is minimal, where YABBY5 and WRKY75 fall in the regulatory hierarchy to govern terpene biosynthesis and how they function with other TFs in the network remains to be explored. Considering the phenotypes observed in transgenic plants, it seems plausible that MsYABBY5 controls an upstream event in metabolite production.
We found increased levels of transcript for the plastidic ATP/ADP transporter in transgenic lines making more secondary metabolites and decreased levels in transgenic lines producing less metabolite. In potato, overexpression and suppression of the plastidic ATP/ADP transporter led to an increase and decrease, respectively, in the amount of tuber starch produced (Tjaden et al., 1998). Further overexpression of both NTT and glucose‐6‐phosphate/phosphate translocator which supplies carbon skeletons to the plastids significantly increased total starch content in potato (Zhang et al., 2008). This suggested that import of both energy and carbons into amyloplasts is a rate‐limiting step for starch formation. Most of the plastid proteins are encoded in the nucleus and transported to plastids. MsYABBY5 does not seem to directly regulate the expression of MsNTT to control secondary metabolite production, but the effect can be indirect due to changes in secondary metabolism brought by MsYABBY5.
Immunostaining and fluorescence assays showed MsYABBY5 localized to both nucleus and cytoplasm. Cell fractionation and protein gel blot assays can be further performed to test the association of MsYABBY5 with intracellular membranes. There are several reports of TFs especially those involved in stress response to rapidly translocate from the cytoplasm to the nucleus in response to external signals. Some examples from the mammalian systems are STAT (signal transducer and activator of transcription), NF‐κB (nuclear factor of immunoglobulin kappa B cells), NFAT (nuclear factor of activated T cells) and steroid receptors proteins (Beals et al., 1997; McBride et al., 2002). In the case of plants, ER membrane‐associated bZIP and NAC089 TFs which are responsible for mediating ER‐related plant immunity and abiotic stress responses, respectively, were detected in the cytoplasm (Che et al., 2010; Moreno et al., 2012; Yang et al., 2014). Abscisic acid (ABA) is a vital plant hormone that plays important roles in stress response. In Arabidopsis, a WRKY protein, AtWRKY40 functions as negative regulator of ABA signalling by directly repressing expression of many ABA‐responsive genes. WRKY40 was shown to localize to both nucleus and cytosol in wild‐type plant cells that has ABA at physiological concentrations and ABA was essential for its cytosolic distribution (Shang et al., 2010). Secondary metabolites produced by plants are also related to abiotic and biotic stress responses of plants. Further studies are required to assess the change in expression levels and localization patterns of MsYABBY5 and WRKY75 in response to stress conditions and its significance.
Experimental procedures
Plant material and transformation
Commercial spearmint variety (M. spicata) and sweet basil (O. basilicum) were tested for their secondary metabolites by GC–MS and grown in greenhouse under natural light conditions. Spearmint plants were propagated using stem cuttings, whereas basil plants were propagated from seeds. Agrobacterium‐mediated transformation of spearmint was performed according to the previously published protocol (Niu et al., 1998, 2000). Agrobacterium‐mediated transformation of sweet basil was performed by the following procedure. O. basilicum seeds were sterilized by washing in 40% Clorox for 3 min followed by several rinses with sterile water. The sterile seeds were imbibed overnight and kept at 4 °C. The following day the seeds were dissected under a dissection microscope to harvest the mature embryos. The dissected embryos were precultured in dark for 1 day in cocultivation medium (CC). Agrobacterium EHA105 strain was used for transformation. Enhanced green‐fluorescent protein gene along with kanamycin was used as a selection marker. The precultured embryos were immersed in agrobacterium culture and sonicated for 15 s, four times. After sonication, the embryos were immersed in fresh Agrobacterium solution and vacuum‐infiltrated for 3 min. After infection for 30 min, the embryos were placed in CC medium [MS salts + myo‐inositol 100 mg/L sucrose (30 g/L) + BA (0.4 mg/L) + IBA (0.4 mg/L) + cefotaxime (150 mg/l)] for 3 days. After 3 days, the embryos were washed multiple times with sterile‐distilled water containing cefotaxime (150 mg/L). The washed embryos were kept in CC media for 3–4 weeks in dark for shoot induction. After 3–4 weeks, GFP‐positive shoots were selected and transferred to light. The well‐grown shoots were transferred to elongation media [MS salts + sucrose (30 g/L) + BA (3 mg/L) + IAA (0.5 mg/L) + cefotaxime (150 mg/L)] and kept for 2–3 weeks. The shoots were hardened on basal media and allowed for root formation. Plantlets with well‐developed roots were transferred to soil and grown under glasshouse conditions before further analysis. Tobacco transformation was done as previously described by Gallois and Marinho (1995).
RNA extraction and quantitative RT‐PCR
Total RNA was extracted from different tissues (PGT, leaf‐PGT, leaf and root) of spearmint using an RNeasy® Plus Mini kit from Qiagen. Reverse transcription reaction and quantitative RT‐PCR (qRT‐PCR) were carried out as described in Jin et al. (2014). Expression levels of target genes were represented as mean ± SD. Approximately 1 μg RNA was employed to synthesize first strand cDNA.
In situ hybridization
In situ hybridization assay was performed according to the method described by Javelle et al. (2011) with some minor modifications. Briefly, samples were fixed in 4% paraformaldehyde fixative and subjected to vacuum for 30 min on ice. After that, the vials were kept overnight at 4 °C. Next day, samples were dehydrated with ethanol series and embedded in Paraplast (McCormick Scientific, St Louis, MO) until use. The blocks were sectioned at 10 μm and mounted on Probe‐on Plus slides (Fisher Scientific, singapore). For probe synthesis, the MsYABBY5 cDNA was inserted into a pGEM®‐T vector (Promega, Wisconsin). Sense and antisense probes were synthesized by T7 and SP6 RNA polymerase (Roche, Basel, Switzerland), respectively.
Cloning and vector construction
Promoter cloning of MsYABBY5
Genomic DNA was isolated from young leaves of spearmint using CTAB method. The flanking sequence of MsYABBY5 gene was amplified using a GenomeWalker™ Universal kit. The −1116 bp flanking region of the gene was ligated with pGEM®‐T vector. The resulting product was transformed into Escherichia coli XL1‐Blue and sequenced. The promoter was amplified with Phusion® High‐Fidelity DNA Polymerase (NEB) and subcloned into a gateway donor vector pENTR™/D‐TOPO® (Invitrogen, California). Then, the recombinant plasmid was introduced into destination vectors pBGWFS7 by LR recombination. The destination plasmid was further transformed into A. tumefaciens EHA105 by heat shock and used to generate transgenic tobacco lines. Sequences of all primers used in this study are listed in Table S1.
Full‐length cloning of all four YABBYs
Full length of all four YABBYs cDNAs was obtained by performing 3′ and 5′ RACE using the SMARTer™ RACE cDNA amplification kit from Clontech. For sequencing full‐length ORFs, the purified fragments were ligated with pGEM®‐T vector. The resulting product was transformed into E. coli XL1‐Blue.
Overexpression and RNAi vector construction
To overexpress or silence MsYABBY5, sequences were amplified with Phusion® High‐Fidelity DNA Polymerase (NEB). The purified fragments were inserted into a gateway donor vector pENTR™/D‐TOPO® (Invitrogen). Then, the recombinant plasmids were introduced into destination vectors pK7WG2D for overexpression in spearmint and sweet basil by LR recombination. For MsYABBY5 RNAi, four primers with restriction enzymes located at flanking region were used to amplify the fragment showing low similarity to other YABBY genes. The purified PCR product was cloned into the donor vector and subsequently introduced into pK7WG2D by LR recombination. The MsYABBY5 gene was driven by 35S promoter in both overexpression and RNAi plants. All destination plasmids harbouring the target genes were transformed into A. tumefaciens EHA105 by heat shock and used for spearmint and basil transformation.
Subcellular localization
YABBY and MsNTT ORFs were amplified and inserted into the pENTR™/D‐TOPO®. The donor vectors harbouring ORFs were introduced into pBADC/YFP vector by LR recombination. For testing expression pattern of MsYABBY5 promoter, the 5′UTR sequence was amplified and inserted into pENTR™/D‐TOPO®. Subsequently, the plasmid was transformed into pBGWFS7 by LR recombination. All destination plasmids harbouring target genes were transformed into A. tumefaciens EHA105 by heat shock. The recombinant A. tumefaciens EHA strains were used for plant transformation. Subcellular localization pattern of YABBY proteins was performed as described in Jin et al. (2014). Briefly, the recombinant A. tumefaciens EHA strains were grown in LB medium overnight at 28 °C. After centrifugation at 4000 × g , 4 °C for 15 min, cell pellets were collected and resuspended in MMA solution (10 mm MES, 10 mm MgCl2, 100 μm acetosyringone) to OD600 = 1. The solution was then injected into N. benthamiana leaves. After that, plants were kept at 28 °C for 2 days. Leaf samples were collected and viewed with an upright confocal microscope (Zeiss, Jena, Germany).
Southern blotting
A total of 15 μg genomic DNA was digested overnight with EcoRI at 37 °C. Next day, digestion product was electrophoresed on a 1.2% (w/v) agarose gel at 50 V for 4 h. After that, the gel was transferred to a nylon membrane and hybridized by the CaMV 35S promoter probe using a DIG DNA labelling and detection kit (Roche). DNA probe against 35S promoter was generated using PCR DIG probe synthesis kit from Roche (Hart and Basu, 2009).
Immunogold labelling
A 14‐AA peptide of MsYABBY5 showing low similarity to other MsYABBYs was used as antigen for antibody synthesis (GenScript, Piscataway, NJ). Specificity test of the antibody was performed by Western blotting). Leaf samples from spearmint were fixed for 3 h in 4% paraformaldehyde/0.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) and rinsed in 0.1 M phosphate buffer (pH 7.2) for three times followed by dehydration in ethanol. After that, the samples were infiltrated with and embedded in LR White. Ultrathin sections around 90 nm were prepared with Leica Ultracut UCT microtome equipped with diamond knives and collected on uncoated, 300‐mesh nickel grids. The procedure of labelling and washing was performed according to the protocol described by Skepper and Powell (2008) with some minor modifications. Briefly, the sections were incubated for 4 h on drops of antimint YAB5 antibody (produced in rabbit) with 1 : 100 dilutions in PBSG buffer [1% (w/v) gelatin in PBS buffer]. After that, sections were rinsed on drops of TBST (50 mm Tris, 150 mm NaCl, 0.05% Tween 20) for ten times, 2 min for each time. Then, sections were incubated for 1 h on drops of goat anti‐rabbit antibody conjugated with 10 nm gold particles with 1 : 100 dilutions in PBSG buffer [1% (w/v) gelatin in PBS buffer]. The sections were further rinsed in TBST buffer for 10 times, 2 min each time, and in ddH2O for 30 s. Subsequently, samples were counterstained by applying the grid on drops of uranyl acetate and lead citrate. Finally, sections were extensively rinsed in ddH2O and viewed at 120 kV with a transmission electron microscope (JEOL JEM‐1230, Japan).
Electrophoretic mobility shift assay (EMSA)
MsYABBY5 was expressed in E. coli BL21 (DE3) and induced by 1 mm isopropyl‐β‐thiogalactopyranoside (IPTG) for 6 h. Then, the recombinant protein was purified using 6× His tagged Ni‐NTA agarose (Qiagen, Hilden, Germany) and used for EMSA. The 5′ ends of probes used for EMSA were labelled with biotin (Table S1). The assay was performed using a LightShift Chemiluminescent EMSA kit (Thermo, Waltham, MA) according to the manufacturer's instructions.
Transactivation activity assay
The ~1 kb promoter region of MsWRKY75 was amplified and inserted into pENTR™/D‐TOPO®. The resulting plasmid was transformed into pBGWFS7 by LR recombination and further introduced into A. tumefaciens EHA. Leaves of N. benthamiana were agroinfiltrated with effector and reporter at a ratio of 1 : 1. Two days after infiltration, leaves were harvested to isolate crude protein. GUS quantitative assay was performed as described by Li et al. (2014). Each assay was performed in triplicate.
Gas chromatography–mass spectrometry analysis
In case of mint, each transgenic plant was propagated clonally before GC–MS studies were conducted on them. For basil and tobacco plants, the analysis was performed on T‐1 plants. Terpene and phenylpropanoid production in leaves of spearmint, sweet basil and tobacco were determined using a GC–MS method as described in Jin et al. (2014). Camphor was used as an internal standard.
Statistical analysis
Data are indicated as ‘mean ± SD’ of three biological replicates each performed in triplicates. Statistical significance between transgenic plants and WT was analysed using a two‐tailed Student's t‐test and indicated by asterisks. *P < 0.05; **P < 0.01.
Competing interests
The authors declare that they have no competing interests.
Supporting information
Figure S1 MsYABBY5 protein was observed in both nucleus (N) and cytoplasm (C) in peltate glandular trichome (PGT) of spearmint.
Figure S2 Southern blotting analysis of transgenic plants.
Figure S3 Transcript levels of other MsYABBYs in RNAi plants.
Figure S4 Transcript level of MsYABBY5 in MsYABBY5 overexpression sweet basil (A) and Nicotiana sylvestris (B).
Figure S5 Ectopic expression of MsYABBY5 caused leaf curling and flowering time delay in sweet basil.
Table S1 Sequences of the primers used in this study.
Acknowledgements
This research was funded by a grant from Singapore National Research Foundation (Competitive Research Programme Award No: NRF‐CRP8‐2011‐02). We thank Dr Jin Jingjing for transcriptome data analysis, Mr. Xuezhi Ouyang for TEM sample preparation and Dr Jun‐Lin Yin for support in GC–MS analysis.
References
- Aharoni, A. and Galili, G. (2011) Metabolic engineering of the plant primary–secondary metabolism interface. Curr. Opin. Biotechnol. 22, 239–244. [DOI] [PubMed] [Google Scholar]
- Albert, N.W. , Davies, K.M. , Lewis, D.H. , Zhang, H. , Montefiori, M. , Brendolise, C. , Boase, M.R. et al. (2014) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell, 26, 962–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beals, C.R. , Sheridan, C.M. , Turck, C.W. , Gardner, P. and Crabtree, G.R. (1997) Nuclear export of NF‐ATc enhanced by glycogen synthase kinase‐3. Science, 275, 1930–1933. [DOI] [PubMed] [Google Scholar]
- Ben Zvi, M.M. , Shklarman, E. , Masci, T. , Kalev, H. , Debener, T. , Shafir, S. , Ovadis, M. et al. (2012) PAP1 transcription factor enhances production of phenylpropanoid and terpenoid scent compounds in rose flowers. New Phytol. 195, 335–345. [DOI] [PubMed] [Google Scholar]
- Bonaccorso, O. , Lee, J.E. , Puah, L. , Scutt, C.P. and Golz, J.F. (2012) FILAMENTOUS FLOWER controls lateral organ development by acting as both an activator and a repressor. BMC Plant Biol. 12, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L. (2000) The YABBY gene family and abaxial cell fate. Curr. Opin. Plant Biol. 3, 17–22. [DOI] [PubMed] [Google Scholar]
- Broun, P. and Somerville, C. (2001) Progress in plant metabolic engineering. Proc. Natl Acad. Sci. USA, 98, 8925–8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli, E. , Titta, L. , Giorgio, M. , Mock, H.P. , Matros, A. , Peterek, S. , Schijlen, E.G. et al. (2008) Enrichment of tomato fruit with health‐promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26, 1301–1308. [DOI] [PubMed] [Google Scholar]
- Cavallini, E. , Matus, J.T. , Finezzo, L. , Zenoni, S. , Loyola, R. , Guzzo, F. , Schlechter, R. et al. (2015) The phenylpropanoid pathway is controlled at different branches by a set of R2R3‐MYB C2 repressors in grapevine. Plant Physiol. 167, 1448–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne, A. and Boutry, M. (2013) Proteomic snapshot of spearmint (Mentha spicata L.) leaf trichomes: a genuine terpenoid factory. Proteomics, 13, 3327–3332. [DOI] [PubMed] [Google Scholar]
- Che, P. , Bussell, J.D. , Zhou, W. , Estavillo, G.M. , Pogson, B.J. and Smith, S.M. (2010) Signaling from the endoplasmic reticulum activates brassinosteroid signaling and promotes acclimation to stress in Arabidopsis . Sci. Signal. 3, ra69. [DOI] [PubMed] [Google Scholar]
- Chen, Y.Y. , Wang, L.F. , Dai, L.J. , Yang, S.G. and Tian, W.M. (2012) Characterization of HbEREBP1, a wound‐responsive transcription factor gene in laticifers of Hevea brasiliensis Muell. Arg. Mol. Biol. Rep. 39, 3713–3719. [DOI] [PubMed] [Google Scholar]
- Cordoba, E. , Salmi, M. and León, P. (2009) Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J. Exp. Bot. 60, 2933–2943. [DOI] [PubMed] [Google Scholar]
- Croteau, R. , Karp, F. , Wagschal, K.C. , Satterwhite, D.M. , Hyatt, D.C. and Skotland, C.B. (1991) Biochemical characterization of a spearmint mutant that resembles peppermint in monoterpene content. Plant Physiol. 96, 744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau, R. , Kutchan, T.M. and Lewis, N.G. (2000) Natural products (secondary metabolites). In Biochemistry and Molecular Biology of Plants. ( Buchanan, B. , Gruissem, W. and Jones, R. , eds), pp. 1250‐1268. Rockville, MD: American Society of Plant Physiologists. [Google Scholar]
- Dai, M. , Zhao, Y. , Ma, Q. , Hu, Y. , Hedden, P. , Zhang, Q. and Zhou, D.X. (2007) The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol. 144, 121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diemer, F. , Caissard, J.‐C. , Moja, S. , Chalchat, J.‐C. and Jullien, F . (2001) Altered monoterpene composition in transgenic mint following the introduction of 4S‐limonene synthase. Plant Physiol. Biochem. 39, 603–614. [Google Scholar]
- Dubey, V.S. , Bhalla, R. and Luthra, R. (2003) An overview of the non‐mevalonate pathway for terpenoid biosynthesis in plants. J. Biosci. 28, 637–646. [DOI] [PubMed] [Google Scholar]
- Enfissi, E.M. , Barneche, F. , Ahmed, I. , Lichtlé, C. , Gerrish, C. , McQuinn, R.P. , Giovannoni, J.J. et al. (2010) Integrative transcript and metabolite analysis of nutritionally enhanced DE‐ETIOLATED1 downregulated tomato fruit. Plant Cell, 22, 1190–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed, Y. , Baum, S.F. and Bowman, J.L. (1999) Distinct mechanisms promote polarity establishment in carpels of Arabidopsis . Cell, 99, 199–209. [DOI] [PubMed] [Google Scholar]
- Fischer, K. (2011) The import and export business in plastids: transport processes across the inner envelope membrane. Plant Physiol. 155, 1511–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flügge, U.I. , Häusler, R.E. , Ludewig, F. and Gierth, M. (2011) The role of transporters in supplying energy to plant plastids. J. Exp. Bot. 62, 2381–2392. [DOI] [PubMed] [Google Scholar]
- Gallois, P. and Marinho, P. (1995) Leaf disk transformation using Agrobacterium tumefaciens‐expression of heterologous genes in tobacco. In Plant gene transfer and expression protocols, Methods in Molecular Biology, Vol. 49 ( Jones, H. , ed.), pp. 39–48. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- Gershenzon, J. , McConkey, M.E. and Croteau, R.B. (2000) Regulation of monoterpene accumulation in leaves of peppermint. Plant Physiol. 122, 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldshmidt, A. , Alvarez, J.P. , Bowman, J.L. and Eshed, Y. (2008) Signals derived from YABBY gene activities in organ primordia regulate growth and partitioning of Arabidopsis shoot apical meristems. Plant Cell, 20, 1217–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golz, J.F. , Roccaro, M. , Kuzoff, R. and Hudson, A. (2004) GRAMINIFOLIA promotes growth and polarity of Antirrhinum leaves. Development, 131, 3661–3670. [DOI] [PubMed] [Google Scholar]
- Grotewold, E. (2008) Transcription factors for predictive plant metabolic engineering: are we there yet. Curr. Opin. Biotechnol. 19, 138–144. [DOI] [PubMed] [Google Scholar]
- Gutensohn, M. , Nguyen, T.T. , McMahon, R.D. , Kaplan, I. , Pichersky, E. and Dudareva, N. (2014) Metabolic engineering of monoterpene biosynthesis in tomato fruits via introduction of the non‐canonical substrate neryl diphosphate. Metab. Eng. 24, 107–116. [DOI] [PubMed] [Google Scholar]
- Hart, S.M. and Basu, C. (2009) Optimization of a digoxigenin‐based immunoassay system for gene detection in Arabidopsis thaliana . J. Biomol. Tech. 20, 96–100. [PMC free article] [PubMed] [Google Scholar]
- Hemmerlin, A. , Harwood, J.L. and Bach, T.J. (2012) A raison d'être for two distinct pathways in the early steps of plant isoprenoid biosynthesis? Prog. Lipid Res. 51, 95–148. [DOI] [PubMed] [Google Scholar]
- Iwase, A. , Matsui, K. and Ohme‐Takag, M. (2009) Manipulation of plant metabolic pathways by transcription factors. Plant Biotechnol. 26, 29–38. [Google Scholar]
- Javelle, M. , Marco, C.F. and Timmermans, M. (2011) In situ hybridization for the precise localization of transcripts in plants. J. Vis. Exp. 57, 3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, J. , Panicker, D. , Wang, Q. , Kim, M.J. , Liu, J. , Yin, J.L. , Wong, L. et al. (2014) Next generation sequencing unravels the biosynthetic ability of Spearmint (Mentha spicata) peltate glandular trichomes through comparative transcriptomics. BMC Plant Biol. 14, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, B.M. and Turner, G.W. (2013) Terpenoid biosynthesis in trichomes – current status and future opportunities. Plant Biotechnol. J. 11, 2–22. [DOI] [PubMed] [Google Scholar]
- Lange, B.M. , Mahmoud, S.S. , Wildung, M.R. , Turner, G.W. , Davis, E.M. , Lange, I. , Baker, R.C. et al. (2011) Improving peppermint essential oil yield and composition by metabolic engineering. Proc. Natl Acad. Sci. USA, 108, 16944–16949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Gao, M.J. , Pan, H.Y. , Cui, D.J. and Gruber, M.Y. (2010) Purple canola: Arabidopsis PAP1 increases antioxidants and phenolics in Brassica napus leaves. J. Agric. Food Chem. 58, 1639–1645. [DOI] [PubMed] [Google Scholar]
- Li, R. , Weldegergis, B.T. , Li, J. , Jung, C. , Qu, J. , Sun, Y. , Qian, H. , et al. (2014) Virulence factors of Geminivirus interact with MYC2 to subvert plant resistance and promote vector performance. Plant Cell, 26, 4991–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loza‐Tavera, H. (1999) Monoterpenes in essential oils – biosynthesis and properties. Adv. Exp. Med. Biol. 464, 49–62. [DOI] [PubMed] [Google Scholar]
- Lu, X. , Jiang, W.M. , Zhang, L. , Zhang, F. , Zhang, F.Y. , Shen, Q. , Wang, G.F. et al. (2013) AaERF1 positively regulates the resistance to Botrytis cinerea in Artemisia annua . PLoS ONE, 8, e57657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, J. , Butelli, E. , Hill, L. , Parr, A. , Niggeweg, R. , Bailey, P. , Weisshaar, B. et al. (2008) AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: expression in fruit results in very high levels of both types of polyphenol. Plant J. 56, 316–326. [DOI] [PubMed] [Google Scholar]
- Ma, D. , Pu, G. , Lei, C. , Ma, L. , Wang, H. , Guo, Y. , Chen, J. et al. (2009) Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha‐4,11‐diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 50, 2146–2161. [DOI] [PubMed] [Google Scholar]
- Mahmoud, S.S. and Croteau, R.B. (2001) Metabolic engineering of essential oil yield and composition in mint by altering expression of deoxyxylulose phosphate reductoisomerase and menthofuran synthase. Proc. Natl Acad. Sci. USA, 98, 8915–8920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud, S.S. , Williams, M. and Croteau, R. (2004) Cosuppression of limonene‐3‐hydroxylase in peppermint promotes accumulation of limonene in the essential oil. Phytochemistry, 65, 547–554. [DOI] [PubMed] [Google Scholar]
- Majetic, C.J. , Rausher, M.D. and Raguso, R.A. (2010) The pigment‐scent connection: do mutations in regulatory vs. structural anthocyanin genes differentially alter floral scent production in Ipomoea purpurea? S. Afr. J. Bot. 76, 632–642. [Google Scholar]
- McBride, K.M. , Banninger, G. , McDonald, C. and Reich, N.C. (2002) Regulated nuclear import of the STAT1 transcription factor by direct binding of importin‐α. EMBO J. 21, 1754–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConkey, M.E. , Gershenzon, J. and Croteau, R.B. (2000) Developmental regulation of monoterpene biosynthesis in the glandular trichomes of peppermint. Plant Physiol. 122, 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, K. , Matsumoto, T. , Okada, A. , Komiyama, K. , Chujo, T. , Yoshikawa, H. , Nojiri, H. et al. (2014) Identification of target genes of the bZIP transcription factor OsTGAP1, whose overexpression causes elicitor‐induced hyperaccumulation of diterpenoid phytoalexins in rice cells. PLoS ONE, 9, e105823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, A.A. , Mukhtar, M.S. , Blanco, F. , Boatwright, J.L. , Moreno, I. , Jordan, M.R. , Chen, Y. et al. (2012) IRE1/bZIP60‐mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS ONE, 7, e31944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz‐Bertomeu, J. , Ros, R. , Arrillaga, I. and Segura, J. (2008) Expression of spearmint limonene synthase in transgenic spike lavender results in an altered monoterpene composition in developing leaves. Metab. Eng. 10, 166–177. [DOI] [PubMed] [Google Scholar]
- Neuhaus, H.E. , Thom, E. , Möhlmann, T. , Steup, M. and Kampfenkel, K. (1997) Characterization of a novel eukaryotic ATP/ADP translocator located in the plastid envelope of Arabidopsis thaliana L. Plant J. 11, 73–82. [DOI] [PubMed] [Google Scholar]
- Niu, X. , Lin, K. , Hasegawa, P.M. , Bressan, R.A. and Weller, S.C. (1998) Transgenic peppermint (Mentha× piperita L.) plants obtained by cocultivation with Agrobacterium tumefaciens . Plant Cell Rep. 17, 165–171. [DOI] [PubMed] [Google Scholar]
- Niu, X. , Li, X. , Veronese, P. , Bressan, R.A. , Weller, S.C. and Hasegawa, P.M. (2000) Factors affecting Agrobacterium tumefaciens‐mediated transformation of peppermint. Plant Cell Rep. 19, 304–310. [DOI] [PubMed] [Google Scholar]
- Patra, B. , Schluttenhofer, C. , Wu, Y. , Pattanaik, S. and Yuan, L. (2013) Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim. Biophys. Acta, 1829, 1236–1247. [DOI] [PubMed] [Google Scholar]
- Qiu, D. , Xiao, J. , Xie, W. , Liu, H. , Li, X. , Xiong, L. and Wang, S. (2008) Rice gene network inferred from expression profiling of plants overexpressing OsWRKY13, a positive regulator of disease resistance. Mol. Plant, 1, 538–551. [DOI] [PubMed] [Google Scholar]
- Robinson, D.G. and Ritzenthaler, C. (2006) Imaging the early secretory pathway in BY‐2 cells. In Tobacco BY‐2 Cells: From Cellular Dynamics to Omics, Vol. 58 ( Nagata, T. , Matsuoka, K. and Inzé, D. , eds), pp. 135–151. Biotechnology in Agriculture and Forestry. Heidelberg: Springer‐Verlag. [Google Scholar]
- Sarojam, R. , Sappl, P.G. , Goldshmidt, A. , Efroni, I. , Floyd, S.K. , Eshed, Y. and Bowman, J.L. (2010) Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell, 22, 2113–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluttenhofer, C. and Yuan, L. (2015) Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 167, 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwinn, K. , Venail, J. , Shang, Y. , Mackay, S. , Alm, V. , Butelli, E. , Oyama, R. et al. (2006) A small family of MYB‐regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum . Plant Cell, 18, 831–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamimuzzaman, M. and Vodkin, L. (2013) Genome‐wide identification of binding sites for NAC and YABBY transcription factors and co‐regulated genes during soybean seedling development by ChIP‐Seq and RNA‐Seq. BMC Genom. 14, 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, Y. , Yan, L. , Liu, Z.Q. , Cao, Z. , Mei, C. , Xin, Q. , Wu, F.Q. et al. (2010) The Mg‐chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA‐responsive genes of inhibition. Plant Cell, 22, 1909–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, R. , Bhattacharyya, D. , Majumdar, A.B. , Datta, R. , Hazra, S. and Chattopadhyay, S. (2013) Leaf proteome profiling of transgenic mint infected with Alternaria alternata . J. Proteomics, 93, 117–132. [DOI] [PubMed] [Google Scholar]
- Skepper, J.N. and Powell, J.M. (2008) Immunogold staining of epoxy resin sections for transmission electron microscopy (TEM). Cold Spring Harbor Protocols, 6, pdb‐prot5015. [DOI] [PubMed] [Google Scholar]
- Spyropoulou, E.A. , Haring, M.A. and Schuurink, R.C. (2014) RNA sequencing on Solanum lycopersicum trichomes identifies transcription factors that activate terpene synthase promoters. BMC Genom. 15, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahle, M.I. , Kuehlich, J. , Staron, L. , von Arnim, A.G. and Golz, J.F. (2009) YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis . Plant Cell, 21, 3105–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden, J. , Möhlmann, T. and Kampfenkel, K. (1998) Altered plastidic ATP/ADP‐transporter activity influences potato (Solanum tuberosum L.) tuber morphology, yield and composition of tuber starch. Plant J. 16, 531–540. [Google Scholar]
- Vranová, E. , Coman, D. and Gruissem, W. (2013) Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 64, 665–700. [DOI] [PubMed] [Google Scholar]
- Wang, G. (2014) Recent progress in secondary metabolism of glandular trichome. Plant Biotechnol. 31, 353–361. [Google Scholar]
- Wang, C. , Wu, J. and Mei, X. (2001) Enhancement of taxol production and excretion in Taxus chinensis cell culture by fungal elicitation and medium renewal. Appl. Microbiol. Biot. 55, 404–410. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Hao, J. , Chen, X. , Hao, Z. , Wang, X. , Lou, Y. , Peng, Y. et al. (2007) Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol. Biol. 65, 799–815. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Guo, D. , Li, H.L. and Peng, S.Q. (2013) Characterization of HbWRKY1, a WRKY transcription factor from Hevea brasiliensis that negatively regulates HbSRPP . Plant Physiol. Biochem. 71, 283–289. [DOI] [PubMed] [Google Scholar]
- Xie, Z. , Kapteyn, J. and Gang, D.R. (2008) A systems biology investigation of the MEP/terpenoid and shikimate/phenylpropanoid pathways points to multiple levels of metabolic control in sweet basil glandular trichomes. Plant J. 54, 349–361. [DOI] [PubMed] [Google Scholar]
- Xu, Y.H. , Wang, J.W. , Wang, S. , Wang, J.Y. and Chen, X.Y. (2004) Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)‐δ‐cadinene synthase‐A. Plant Physiol. 135, 507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z.T. , Wang, M.J. , Sun, L. , Lu, S.J. , Bi, D.L. , Sun, L. , Song, Z.T. et al. (2014) The membrane‐associated transcription factor NAC089 controls ER‐stress‐induced programmed cell death in plants. PLoS Genet. 10, e1004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani, N. , Sato, Y. , Tanabe, S. , Chujo, T. , Shimizu, T. , Okada, K. , Yamane, H. et al. (2013) WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 64, 5085–5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Z.X. , Li, J.X. , Yang, C.Q. , Hu, W.L. , Wang, L.J. and Chen, X.Y. (2012) The jasmonate‐responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant, 5, 353–365. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Häusler, R.E. , Greiten, C. , Hajirezaei, M.R. , Haferkamp, I. , Neuhaus, H.E. , Flügge, U.I. et al. (2008) Overriding the co‐limiting import of carbon and energy into tuber amyloplasts increases the starch content and yield of transgenic potato plants. Plant Biotechnol. J. 6, 453–464. [DOI] [PubMed] [Google Scholar]
- Zhang, Q. , Zhu, J. , Ni, Y. , Cai, Y. and Zhang, Z. (2012) Expression profiling of HbWRKY1, an ethephon‐induced WRKY gene in latex from Hevea brasiliensis in responding to wounding and drought. Trees, 26, 587–595. [Google Scholar]
- Zhang, F.Y. , Fu, X.Q. , Lv, Z.Y. , Lu, X. , Shen, Q. , Zhang, L. , Zhu, M.M. et al. (2015) A basic leucine zipper transcription factor, Aabzip1, connects abscisic acid signaling with artemisinin biosynthesis in Artemisia annua . Mol. Plant, 8, 163–175. [DOI] [PubMed] [Google Scholar]
- Zhou, M. , Wu, L. , Liang, J. , Shen, C. and Lin, J. (2012) Expression analysis and functional characterization of a novel cold‐responsive gene CbCOR15a from Capsella bursa‐pastoris . Mol. Biol. Rep. 39, 5169–5179. [DOI] [PubMed] [Google Scholar]
- Zhu, W. , Zhang, L. , Lv, H. , Zhang, H. , Zhang, D. , Wang, X. and Chen, J. (2014) The dehydrin wzy2 promoter from wheat defines its contribution to stress tolerance. Funct. Integr. Genomics, 14, 111–125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 MsYABBY5 protein was observed in both nucleus (N) and cytoplasm (C) in peltate glandular trichome (PGT) of spearmint.
Figure S2 Southern blotting analysis of transgenic plants.
Figure S3 Transcript levels of other MsYABBYs in RNAi plants.
Figure S4 Transcript level of MsYABBY5 in MsYABBY5 overexpression sweet basil (A) and Nicotiana sylvestris (B).
Figure S5 Ectopic expression of MsYABBY5 caused leaf curling and flowering time delay in sweet basil.
Table S1 Sequences of the primers used in this study.


