Abstract
Reverse genetics and next‐generation sequencing unlocked a new era in biology. It is now possible to identify an animal(s) with the unique biology most relevant to a particular question and rapidly generate tools to functionally dissect that biology. This review highlights the rise of one such novel model system, the starlet sea anemone Nematostella vectensis. Nematostella is a cnidarian (corals, jellyfish, hydras, sea anemones, etc.) animal that was originally targeted by EvoDevo researchers looking to identify a cnidarian animal to which the development of bilaterians (insects, worms, echinoderms, vertebrates, mollusks, etc.) could be compared. Studies in Nematostella have accomplished this goal and informed our understanding of the evolution of key bilaterian features. However, Nematostella is now going beyond its intended utility with potential as a model to better understand other areas such as regenerative biology, EcoDevo, or stress response. This review intends to highlight key EvoDevo insights from Nematostella that guide our understanding about the evolution of axial patterning mechanisms, mesoderm, and nervous systems in bilaterians, as well as to discuss briefly the potential of Nematostella as a model to better understand the relationship between development and regeneration. Lastly, the sum of research to date in Nematostella has generated a variety of tools that aided the rise of Nematostella to a viable model system. We provide a catalogue of current resources and techniques available to facilitate investigators interested in incorporating Nematostella into their research. WIREs Dev Biol 2016, 5:408–428. doi: 10.1002/wdev.222
For further resources related to this article, please visit the WIREs website.
INTRODUCTION
Many EvoDevo researchers wished to understand how evolutionary radiation gave rise to the impressive array of biodiversity present in a group of animals called the bilaterians. To answer this question, biologists sought to infer the bilaterian molecular and morphological ‘starting point’ by identifying ancestral traits likely present in the ancestor of all bilaterians (the urbilaterian) through comparisons to non‐bilaterian species. Cnidarian animals (hydras, jellyfish, corals, siphonophores, and sea anemones) are widely accepted to be the sister taxon to bilaterians1, 2, 3 (Figure 1(b)). The sister relationship makes cnidarians the most closely related non‐bilaterians and the obvious group to target for developing a model animal. The Cnidaria have two main clades, the Medusazoa (hydras, jellyfish, and siphonophores) and the Anthozoa (e.g., corals, sea anemones) (Figure 1(b)). Hydra had long been used as an animal model to better understand regeneration and principles of patterning; however, they produce small numbers of embryos in culture, and those embryos are poorly accessible during most of their development.4, 5 In 1992, Hand and Uhlinger reported a protocol to maintain the complete life cycle of Nematostella in culture, which allowed for robust investigation of cnidarian development.4 As Nematostella, a number of other cnidarian species such as Clytia and Hydractinia have been developed as models.5, 6 However, Nematostella is the most developed and commonly used anthozoan cnidarian model. Together investigations in multiple cnidarian systems will complement and strengthen our understanding of cnidarian biology, and thus provide insight into the ancestral tool kit that gave rise to the complex bilaterian body plans. Here we focus on Nematostella and provide brief summaries of the current state of research on three well‐studied aspects of Nematostella development in regards to bilaterians (axial patterning, emergence of mesoderm, and nervous system evolution). We also provide references to the tools available for researchers interested in introducing Nematostella into their research program, and briefly comment on additional areas, such as regeneration, where Nematostella offers a novel model system to better understand animal biology.
Figure 1.
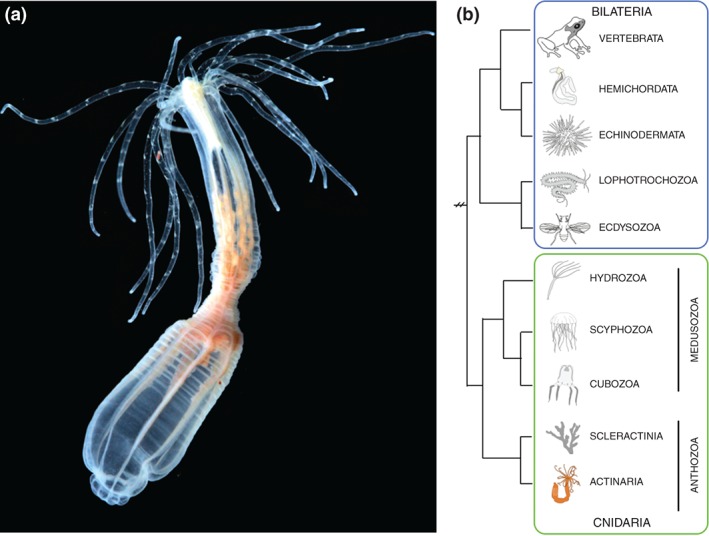
Nematostella vectensis is an anthozoan cnidarian sea anemone. (a) Image of adult Nematostella polyp. Image taken by Eric Röttinger. (b) Phylogeny showing the sister relationship of cnidarians to bilaterians and Nematostella's position within the cnidarians.
OVERVIEW OF NEMATOSTELLA VECTENSIS BIOLOGY
Morphology and Life Cycle of Nematostella
Nematostella vectensis lives in estuarine environments like salt marshes or brackish water pools, and is thus normally exposed to significant fluctuations in temperature and salinity. The physiological tolerance required for life in an unsteady environment may contribute to the relative ease with which Nematostella can be cultured. The life cycle of Nematostella takes approximately 12 weeks in culture.4 They are dioecious, and fertilization occurs in the external environment after spawning. Zygotes initially undergo a series of reductive cleavages to form a blastula. Gastrulation occurs via invagination and initiates at the animal pole (Figure 2(a)). The gastrula stage is followed by the planula stage, which lasts from approximately 24 h post fertilization (hpf) to 96 hpf (temperature dependent, unpublished observations). Planulae are free swimming and during this stage the developing anemone undergoes a slight elongation and formation of pharynx and mesenteries occurs.7 The planula undergoes a subtle type of metamorphosis, which is first apparent by the arrest of swimming movement as well as the formation of tentacle buds, and results in formation of the four tentacle juvenile polyp. At this point growth and maturation of the polyp occurs in a nutrient dependent manner until they reach sexual maturity. Development from egg to juvenile polyp occurs over roughly 6 days.4, 7, 8, 9, 10
Figure 2.
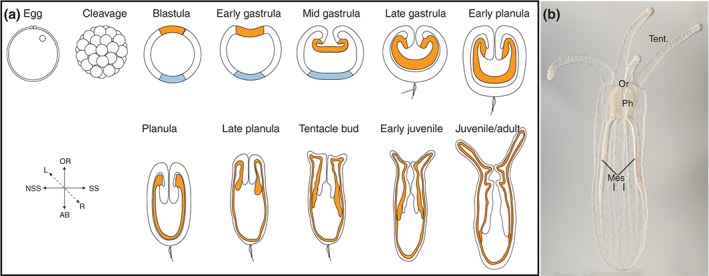
Development and morphology of Nematostella. (a) Schematic of Nematostella developmental stages. Modified from Ormestad et al. Ref 90. Orange represents presumptive endoderm in blastula and the endoderm in all subsequent stages. The outer ectoderm is white, and the aboral ectodermal domain is in light blue. (b) DIC image taken by Aldine Amiel of juvenile polyp. The mouth (oral opening) is indicated by (Or), the tentacles by (tent), the pharynx by (Ph), and the mesenteries by (mes). Oral is up in all images.
The morphology of Nematostella is superficially simple. They are diploblastic animals, meaning that they consist of two germ layers, a bifunctional internal endoderm also referred to as the entoderm, endomesoderm, or gastrodermis as well as an outer ectoderm (Figure 2(a)). They possess a sac‐like gut with a single oral opening that corresponds to the site of gastrulation7, 8 (Figure 2(b)). The adult mouth is surrounded by as many as 16 tentacles11 (Figure 1(a)). They have a noticeable pharynx and eight radially repeated mesentery structures that run along the long oral–aboral axis12 (Figure 2(b)). The mesenteries are comprised of gonads, cnidocytes (stinging cells), and myoepithelial cells that allow the animal to quickly contract along the long axis.12, 13, 14 The two earliest forming mesenteries are termed the primary (previously termed directive) mesenteries; they are situated ca 160° from one another.15 There are two main axes to the anthozoan body plan, the oral–aboral axis, which is the primary axis and runs from the oral opening to the opposite aboral side, and the directive axis. The directive axis is oriented orthogonal to the oral–aboral axis. Although, cnidarians have been described as radially symmetric around the long oral–aboral axis of the animal, is has long been known that this is not the case for anthozoans. The arrangement of retractor muscles on the mesenteries and the presence of a ciliated groove (the siphonoglyph) on one side of the pharynx establish a clear, although subtle bilateral symmetry (summarized in Ref 16). The exact relationship between oral–aboral and directive axes and the anterior–posterior and dorsal‐ventral axes are still unclear, but numerous investigations have shed a great deal of light on this issue, and they are discussed below.
The ectoderm and endoderm have differentiated epithelial cells and specialized cells (neurons, gland cells, myoepithelial cells, etc.) intermixed with each other. This is in contrast to most bilaterians, which organize specialized cell types into organs with a dedicated function. However, it should be noted that distinct regions of the animal are enriched for particular types of differentiated cells. For example, tentacles, which serve to capture and move prey to the mouth, are highly enriched for offensive cnidocyte stinging cells, which envenomate prey.17 In addition, regional patterning is observed for neural markers.18, 19, 20 The ectoderm serves as a protective barrier between the animal and environment. Differentiated cell types found in the ectoderm include sensory neurons, cnidocytes stinging cells, gland cells, and interneurons with as of yet unknown function. The endoderm contains absorptive cells that serve in digestion and nutrient uptake. The non‐mesentery endoderm contains neurons, myoepithelial cells (musculature), and gland cells.12, 14, 18 Between the endoderm and ectoderm lies a mostly acellular mesoglea. The mesoglea contains extracellular matrix as well as some neurites extending from both the endodermal and ectodermal neurons.14, 21 The mesoglea also contains amoebocyte cells that appear to be migratory, but as of yet their function is unknown.14
Nematostella Genome and Experimental Tools
In 2007, the genome of Nematostella was released. Based on early comparisons between protostome and deuterostome genomes, it was believed that the cnidarian genome would be relatively simple (e.g., lacking many genes or gene families present in bilaterians). Surprisingly, the Nematostella genome is incredibly complex. Nearly all of the gene families present in bilaterians were found in Nematostella. Of note, is a nearly complete catalog of the Wnt ligands present in deuterostome bilaterians.22, 23 In addition, there was considerable syntenic conservation between stretches of cnidarian and vertebrate genomes and high conservation of intron‐exon boundaries with vertebrate genomes.23 All told at the genomic level, vertebrates and Nematostella are more similar to each other than either group is to ecdysozoan model animals (Drosophila and nematodes).24 Thus, the first insight about evolution of bilaterian complexity was that complex molecular architecture predated the emergence of bilaterians and that there is no simple correlation between organismal and genomic complexity. The quest for such a correlation is still attracting attention and has driven the adaptation of new experimental tools for Nematostella: microRNAs and other smallRNAs have been isolated systematically.25, 26 ChIP‐seq has been used to identify epigenetic marks and predict regulatory elements,27 and improved transcriptome resources can provide insight into the prevalence of alternative splicing and long non‐coding RNAs.28, 29, 30
Because of the sequencing of the Nematostella genome, tool development has occurred on a relatively rapid scale providing new investigators an array of mechanisms for incorporating Nematostella into their research. Table 1 highlights all the tools and resources available for Nematostella and provides references and hyperlinks to resource pages for protocols and genomic tools. First, the sequenced annotated genome offers the ability to rapidly identify gene homologs.23 Although a second generation genome is still in the works, transcriptome studies provide refined gene models.28, 29 Tools for visualizing gene expression and protein localization using transgenics, mRNA in situ hybridization, injection of mRNA fusion constructs, and antibody staining have been developed.12, 31, 38, 42 The ability to knockdown gene function using morpholinos or knockout gene function using CRISPR/Cas9 or TALEN‐Fok approaches is now available.35, 36, 37, 38 Overexpression via mRNA injection allows for misexpression in early embryos.19, 30, 38, 39 Recently, a heat‐shock responsive promoter was identified paving the way for more complex inducible constructs and the ability to test gain of function phenotypes at later stages of development.35 Technology improving conditional gene regulation would strengthen Nematostella’s utility as a model system. Protocols for ChIP‐seq and RNA‐seq have also been developed.27, 28, 29 To date Nematostella has proven amenable for the adaption of methods deployed in other animal systems to disrupt gene function and analyze phenotypes in the sea anemone, and with advances like CRISPR/Cas9 technology it seems reasonable that tool development in Nematostella will continue to be relatively straight‐forward.
Table 1.
List of Techniques and Tools Available for Culturing and Experimental Manipulation of Nematostella
OVERVIEW OF AXIAL PATTERNING
Formation and Patterning of the Oral–Aboral Axis
How the oral–aboral primary body axis of cnidarians relates to the anterior–posterior axis of bilaterians is a longstanding question. Cnidarian polyps lack a brain‐like concentration of the nervous system at one pole of the axis (see below) and a through gut in which the mouth would be located anterior to the anus, which are key morphological features that correlate with the orientation of the anterior–posterior axis in most bilaterians. The lack of these features is usually considered to represent an evolutionarily early body plan, and accordingly studies on extant cnidarian polyps can provide insights into the ground state from which traits like cephalization and a through gut evolved. Despite the dual function as mouth and anus, the single body opening of cnidarian polyps is by convention called oral opening and the opposite end of the primary body axis is called the aboral pole or physa (Figure 2). Labeling experiments in Nematostella demonstrate that the blastopore and thus the body opening derive from the animal pole of the egg, i.e., the site where the polar bodies are extruded and where the first cleavage initiates.7, 8 This had previously been observed in hydrozoans,43 which suggests that the relation between oocyte polarity and the oral–aboral axis is conserved in different cnidarians clades. Gastrulation from the animal hemisphere is found in cnidarians and ctenophores, a second non‐bilataterian clade, but it is strikingly different from bilaterians, which gastrulate from the vegetal hemisphere. Despite this difference a general similarity exists in the molecular systems that control the development of axial polarity and patterning in cnidarian and bilaterian lineages.
Hox Genes and Axial Patterning in Cnidarians
Initially, Hox genes were used to understand the relation of the oral–aboral axis and the anterior–posterior axis, but no consensus emerged from these studies. Cnidarians possess anterior hox and parahox genes, but the presence of central and posterior hox genes is controversial. Hox genes that do not belong to the anterior group have either been classified as posterior, as central or posterior, or as without affinity to anterior, central, or posterior groups.44, 45, 46, 47, 48 In Nematostella, one of the five anterior hox genes is expressed in the pharynx (which is derived from the oral domain) at the planula stage,44 whereas three other anterior hox genes are expressed in a unilateral domain in the endoderm.44, 45 Of the two potential posterior hox genes, one is expressed at the aboral pole and the other one in a unilateral endodermal domain of the same oral–aboral extension as the three anterior hox genes.44 Interestingly, the domains of the endodermally expressed genes have clearly different extensions along the secondary body axis (see also below).45 These expression patterns suggest that hox genes are involved in axial patterning in Nematostella; however, they also show that they are not a reliable tool to clarify the relation of the cnidarian and bilaterian primary body axes. This is further illustrated by the observation that orthologs of the aborally expressed putative posterior hox gene are expressed at the opposite pole in the hydrozoans Clytia hemispherica and Podocoryne carnea.47, 49 Also, it should be noted that further functional characterization of cnidarian hox genes coupled with improved sampling of cnidarian genomes for putative hox homologs will likely prove insightful in regards to understanding how hox genes became so prominent in patterning the bilaterian anterior–posterior axis.
Wnt Signaling and the Formation of Embryonic Polarity
In most bilaterians, intracellular components of the Wnt/β‐catenin signaling pathway are key players for the establishment of axial polarity and consequently, the role of this pathway has been studied extensively in Nematostella. In the unfertilized egg, NvDsh protein localizes predominantly to the cell cortex adjacent to the pronucleus and after fertilization it associates with the zygotic nucleus (Figure 3(a)). NvDsh remains enriched at the first cleavage furrow and on one side (presumably the animal domain) of the blastula.8 Consistent with an involvement of the β‐catenin branch of Wnt signaling (canonical Wnt), an antibody against Xenopus β‐catenin and injection of mRNA encoding a Nvβ‐catenin‐GFP fusion construct revealed preferential stabilization and nuclearization on one side of the embryo from the 32‐cell stage on and later in the invaginating part of the gastrula.39 Interference with the function of Nvdsh, Nvtcf, and Nvβ‐catenin led to conflicting results about their requirement for the development of axial polarity. Depending on the experimental approach, either complete absence of morphological polarity and failure to gastrulate; or only a defect in the specification of endoderm and pharynx was observed.8, 39, 40, 50 Interestingly, a component of the non‐canonical Wnt/PCP signaling pathway, NvStrabismus (NvStbm), is also enriched at the animal pole from zygote to gastrula stage (Figure 3(a)). Knockdown of Nvstbm by morpholino injection prevented gastrulation, but not the localized expression of endodermal markers,50 indicating that this branch of Wnt signaling is specifically required for the morphogenetic movements at gastrulation.
Figure 3.
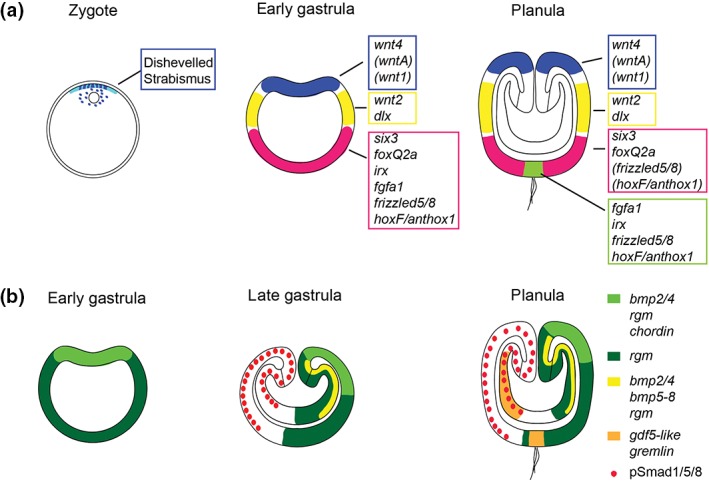
Gene expression during axial patterning. Localization of transcripts (in italic) and proteins involved in the patterning of the oral–aboral (a) and directive (b) axis, respectively. The developmental stages are indicated above each cartoon. Note that not all details of the expression patterns are captured. In (a), the expression domain of wnt4 is broader than that of wntA and wnt1. frizzled5/8 is at planula stage strongly expressed at the aboral pole (green) and weakly in a broader aboral domain (pink). hoxF/anthox1 is at planula stage expressed in the small aboral pole domain (green) and in individual cells within the broader aboral domain (pink). It is not clear whether the expression domains depicted in blue, yellow, and pink are directly abutting each other. In the zygote, Disheveled protein is localized at the cortex and around the nucleus. Disheveled and Strabismus protein remain preferentially localized to the animal/blastoporal region at least until gastrula stage. In (b), BMP5‐8 is at gastrula stage co‐expressed in the ectoderm with bmp2/4, chordin, and rgm (i.e., the domain in light green). pSmad1/5/8 staining forms a gradient with highest levels on the side opposite to the bmp expressing side and low levels on the bmp expressing side. See main text for references.
Wnt signaling appears to be important in the development of early asymmetry, but despite the animal pole localization of NvDsh and Nvβcat it is still not clear how the polarity of the oocyte is initially determined. Future work focused on identification of the mechanisms that stabilize NvDsh at the animal pole will likely yield critical cues as to the most upstream cues that regulate oral–aboral patterning in Nematostella.
Wnt Signaling and the Patterning of the Oral–Aboral Axis
While the role of Wnt/β‐catenin signaling in the initial formation of the oral–aboral axis remains unclear, several studies support a role for this pathway in the subsequent patterning along this axis. Starting from mid‐blastula stage on, the expression of individual Wnt genes demarcates distinct, staggered domains within the oral half of the embryo22, 40, 51 (Figure 3(a)). Both in the ecto‐and endoderm, these expression domains show little overlap and could in principle determine separate territories along this part of the oral–aboral axis. Over activation of Wnt/β‐catenin signaling by GSK3 inhibitors has shown that different levels of β‐catenin can indeed elicit shifts in the expression of regional markers, indicating that high levels of Wnt/β‐catenin signaling promote oral pole identity.20, 40, 52 Consistently, knockdown of Nvtcf leads to an expansion of the aboral marker gene Nvfgfa1 and to the loss of the oral ectoderm markers Nvwnt2 and Nvchordin.40 Surprisingly, there are no reports on the function of individual Wnt ligands in Nematostella, but it will be important to analyze these molecules in an endogenous context to understand potential differences in their signaling mechanisms. These studies together with data from other cnidarian species53, 54, 55 clearly suggest that Wnt signaling has ancient functions in the determination of the site of gastrulation and the patterning of the oral–aboral axis.
The Development of the Aboral Domain—Clues to the Evolution of Brain Formation?
Brain‐like nervous system centralization occurs in most bilaterian clades close to the anterior end of the body,56 but a readily comparable neural structure is not present in cnidarian polyps. Whether a molecular program that resembles anterior patterning is present in cnidarians, and thus whether they have a body region from which cephalization might have evolved, has been an important question in studies on the molecular control of cnidarian development.57
Identification of unambiguous markers to compare the anterior end of the bilaterian body axis to the oral or aboral pole of cnidarians is complicated; however, the transcription factors six3, foxQ2, and the Wnt regulator SFRP1 are among the best candidates currently available. In Nematostella, these genes are expressed at the aboral pole20, 41 (Figure 3(a)), and Nvsix3/6 has been shown to be a key regulator of the development of the aboral territory. Knockdown of Nvsix3/6 leads to a progressive respecification of the aboral area to a more oral identity, as indicated by the expanded expression of Nvwnt2 toward the aboral pole and the failure of the aboral ectodermal cells to acquire their typical columnar shape. While the available functional and expression data suggest that the aboral pole of Nematostella develops under the control of anterior patterning genes, there are also noticeable differences at a more detailed level, for example in the exact relation of the expression domains, which likely reflect differences in the regulatory interactions between the conserved aboral/anterior genes.58, 59, 60, 61, 62
Taken together, the development of the oral and aboral domains in Nematostella is regulated by conserved patterning genes and signaling pathways with clear similarities of oral to bilaterian posterior, and aboral to bilaterian anterior development. Despite this overall conservation many questions remain: How is Disheveled protein localized to the animal pole and how does this compare to bilaterians? What is the function of the different Wnt genes along the axis and what are their signaling mechanisms? Are other bilaterian anterior genes integrated in the aboral patterning system in Nematostella? Addressing these questions will allow a better understanding of the evolution of basic features of animal body plans and they will provide new insights into the logic and the diversity of patterning systems.
BMP Signaling and the Secondary Body Axis
Traditionally, cnidarians have been described as radially symmetric animals, although bilaterally arranged internal structures have been observed in anthozoans already more than a century ago.16, 63, 64 After the discovery of BMP signaling as a unifying mechanism for the patterning of the bilaterian dorso‐ventral axis and the re‐positioning of the anthozoans as the sister group to the other cnidarian clades, it became conceivable that bilaterality was already present in the last common ancestor of cnidarians and bilaterians. Studies in Nematostella and in the coral Acropora millepora indeed showed that BMP pathway genes are expressed asymmetrically along the secondary axis, the so‐called directive axis.37, 44, 65, 66, 67 Functional studies in Nematostella revealed that the establishment and patterning of the directive axis depend on BMP signaling, but that this pathway is employed in an unconventional manner. Nvbmp2/4, Nvbmp5/8, and the extracellular BMP regulator Nvchordin (Nvchd) are expressed radially around the blastopore at early gastrula stage, but their expression then quickly becomes restricted to one side of the mid‐gastrula embryo. Surprisingly, all three genes are expressed on the same side, in partially overlapping domains in the ectoderm. The BMPs are in addition expressed in an elongated endodermal domain on the same side65, 68 and another BMP‐related molecule, Nvgdf5‐like, and a BMP inhibitor, Nvgremlin, start being expressed in similar endodermal domains on the opposite side,44, 68 leading to a complex arrangement of BMPs and BMP inhibitors along the directive axis. Knockdown of the Nvbmps or of Nvchd abolishes all asymmetric gene expression, but in contrast to the situation in many bilaterians, it revealed a negative feedback of the NvBMPs on their own expression, i.e., knockdown led to strong radial expression around the blastopore.69 This suggested that the domain of Nvbmp transcription is actually characterized by low BMP signaling activity, which was later confirmed by immunohistochemical detection of the activated form of the intracellular BMP signal transducer Smad1/5/8.15, 70 Interestingly, hox genes are expressed along the directive axis in a pattern reminiscent of bilaterian anterior–posterior patterning and their expression is controlled by BMP signaling, the major bilaterian dorso‐ventral patterning system.15, 45, 70 As for the patterning of the oral–aboral axis, these findings identify an interesting mix of conserved and divergent features. While a role of BMP/Chordin signaling in secondary axis patterning is common to Nematostella and bilaterians, the roles of individual players and their positions in this signaling network vary considerably. These studies have revealed a similar molecular basis for the patterning of the secondary body axis; however, because the structures that are arranged in a bilateral manner in anthozoans have no counterpart in bilaterians, it remains unclear whether the directive axis of anthozoans is homologous to the dorso‐ventral axis of bilaterians.
Axial patterning in Nematostella is directed by conserved developmental regulators with overall similar functions compared to bilaterian model systems, but there are plenty of differences in the exact deployment of these regulators. Expression profiling of animals with manipulated axial patterning and detection of direct targets of key transcription factors by ChIP‐seq will likely lead to a detailed picture of the transcriptional control of axial patterning in Nematostella, and this will continue to provide insights into the principles of axial patterning systems and their evolution.
OVERVIEW OF NEMATOSTELLA GERM LAYER SPECIFICATION
Triploblastic animals are characterized by the presence of three distinct germ layers called the endo‐, meso‐, and the ectoderm, which form during embryogenesis. During bilaterian gastrulation, the ectoderm first separates from the endomesoderm (or mesendoderm), which subsequently segregates into endoderm and mesoderm.71, 72, 73 These tissues are arranged such that ectoderm surrounds the exterior of the animal and interfaces with the environment, endoderm is the innermost germ layer, and the mesoderm lies between the endoderm and ectoderm. The position of each germ layer is critical because each germ layer generates a specific set of specialized tissues during development. For example, the epidermis derives from ectoderm, the gut arises from endoderm, and blood and musculature arise from mesoderm.
Most non‐bilaterian animals, with exception to ctenophores and medusa stages of a sub‐group of hydrozoan cnidarians are diploblastic animals that possess only two germ layers, the inner endoderm and outer ectoderm. As a result, the evolutionary origin of bilaterian mesoderm and its derivatives is intensely debated and investigated (reviewed in Ref 74, 75, 76, 77, 78, 79, 80). As the closest diploblastic relative to the bilaterians, cnidarians can offer key insights as to the origin of the bilaterian mesoderm.
Nematostella (and cnidarians in general) generate an epidermis derived from the ectoderm and a gastrodermis derived from the endoderm. There are no mesenchymal muscle cells or other mesodermally derived tissues in cnidarians.75, 81 However, the gastrodermis is bifunctional in that it possesses absorptive cells associated with nutrient uptake and various myoepthelial cells with contractile functions.82 Thus, the cnidarian endoderm has both properties and cell types that segregate into distinct endodermal and mesodermal tissues in bilaterians. Consistent, with having traditional functions associated with both the bilaterian endoderm and mesoderm, the Nematostella endoderm expresses a large number of genes that are historically associated with either bilaterian endodermal or mesodermal formation in triploblastic organisms such as otx and gata, respectively.74, 83 These observations suggest that the precursor cells of the cnidarian gastrodermis form an endomesodermal germ layer that fails to segregate into formal endoderm and mesoderm. One prediction that arises from this is that the ancestral gene regulatory network (GRN) that describes the development of cnidarian endoderm/gastrodermis gave rise to the specific regulatory network that we associate with specification of endomesoderm in bilaterian animals. Testing a hypothesis as complex as the Nematostella endodermal GRN potentially being homologous to the bilaterian endomesodermal GRN would have been more difficult in the pre‐‘omics’ era. However, by exploiting tools developed for studying Nematostella development and the ability to rapidly establish novel tools for ‘omics’ level studies in non‐traditional model systems, researchers are building an endodermal GRN in Nematostella to determine if and how the cnidarian endodermal GRN gave rise to the endomesodermal GRNs of bilaterians. Addressing this question will allow researchers to better infer the origins and possible mechanisms that gave rise to the bilaterian mesodermal layer.
The Endodermal GRN of Nematostella
Bilaterian presumptive endomesoderm cells are demarcated by nuclear β‐catenin, which is in cooperation with the transcription factor TCF, the effector of canonical Wnt signaling.84, 85, 86, 87, 88 Because previous reports implicated canonical Wnt (cWnt) signaling and nuclear β‐catenin in Nematostella endoderm formation,8, 39 researchers sought to identify components of the Nematostella endodermal specification pathway by pharmacologically expanding nuclear β‐catenin and thus the presumptive endoderm. Animals with increased nuclear β‐catenin were subjected to microarray analysis to identify likely targets of β‐catenin.40 This study identified over 100 transcription factors and signaling molecules potentially downstream of nuclear β‐catenin.40 Many key genes associated with bilaterian endoderm and mesoderm formation that were previously shown to be expressed in presumptive endoderm and/or bodywall endoderm in Nematostella such as snailA,B, otx, foxA,B, twist, wnts, tcf, and bmp were identified in this array.22, 36, 68, 74, 83, 89, 90 Identified targets were further analyzed by an mRNA in situ screen to characterize their wild type spatiotemporal expression patterns. The genes were expressed in one of three distinct expression domains within the animal hemisphere prior to the onset of gastrulation (Figure 4(a)–(c)). The majority of the analyzed genes were expressed in the ‘central domain’ that corresponds to cells around the animal pole of the blastula (Figure 4). Surrounding this central domain were two rings of expression, an inner ‘central ring’ that borders or partially overlaps with edges of the central ring, and the ‘external ring’ surrounding the central ring.40 The gathered spatial expression patterns are deposited in an online database91 where they are mined to predict elements of the gene regulatory network that govern endoderm formation and gastrulation movements.92, 93 Current fate mapping experiments clearly demonstrate that the animal pole derivatives are internalized during gastrulation.8 However, it is not yet clear if cells in the central and external ring domains are also internalized. Regardless, the majority of genes identified in the microarray represent genes expressed in presumptive endoderm. At first pass it is clear that (1) cells that internalize during gastrulation (cnidarian endoderm and bilaterian endomesoderm) share characteristic nuclear β‐catenin, which is often linked to cWnt signaling and (2) the cnidarian endoderm and bilaterian mesoderm both express a highly overlapping suite of genes during development.
Figure 4.
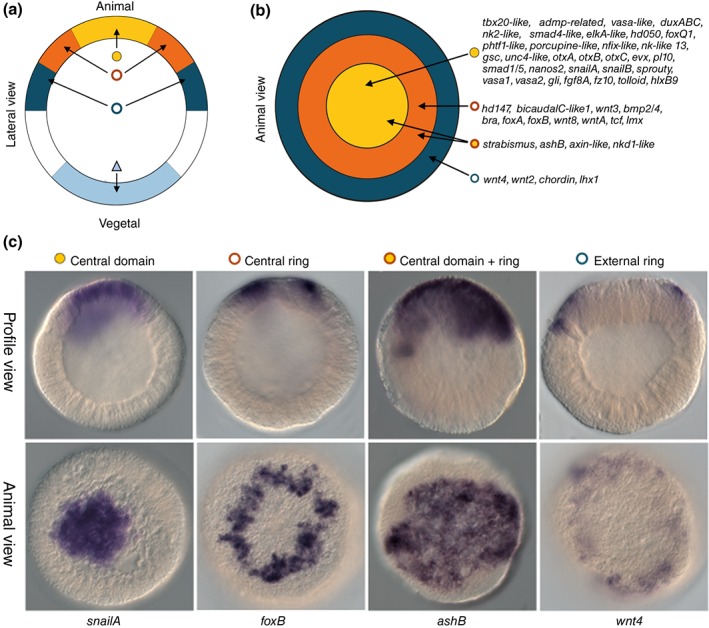
(GRN) Co‐expression domains within the animal hemisphere of the Nematostella blastula. (a, b) Diagram illustrating four co‐expression domains within the animal hemisphere defined by differential spatial expression of the genes indicated in (b). (c) Examples of genes expression in either the central ring (NvsnailA), the central domain (NvfoxB), the central domain + ring (NvashB), or the external ring (Nvwnt4). In situ hybridization expression profiles are shown in either lateral view (a, top row of c) or animal view (b, bottom row of c) to distinguish expression domains. (Reprinted with permission from Ref 40. Copyright 2012 PLoS Genetics)
By coupling the spatiotemporal gene expression analysis with known and additional functional experiments with the 100 genes identified above researchers were able to generate a preliminary GRN describing endodermal specification at the blastula stage40 for Nematostella. Ectopic activation of cWnt signaling induced the formation of an excess of endoderm and exogastrulation, the knockdown of NvTcf, the potential effector of this pathway, showed that NvTcf is not required for gastrulation movements but for pharynx formation.40 The molecular analysis of the obtained phenotypes at the blastula stage, revealed that cWnt signaling is required for proper gene expression in all three domains of the animal hemisphere and to restrict expression of aboral gene expression.40 These results suggest that the canonical Wnt pathway in Nematostella plays a crucial role in launching a GRN in a broad domain within the animal hemisphere (endodermal + presumptive ectodermal tissues). This is similar to what is observed in both echinoderms and Xenopus.94 At later developmental stages, other pathways may be required to segregate ectodermal from endodermal fates within the animal hemisphere. Interestingly, the Fgf/Erk and Bmp/Smad pathways are expressed in the central domain and central ring respectively and could play a role in this process.40
While there are tremendous similarities between the Nematostella endoderm and bilaterian mesendoderm in terms of expression patterns and upstream regulators, key differences exist that need to be considered. Most of our understanding about bilaterian endomesodermal GRNs comes from work in echinoderms, where blimp1, otx, bra, foxA, and gataE form a so‐called ‘kernel’ that is critical for the endomesoderm GRN.84, 95, 96, 97 The ‘kernel’ genes are not only expressed within the endomesodermal domain of the developing sea urchin embryo, but also crucial for specification of the endo and mesodermal germ layer precursors.84, 95, 96, 97 Thus, we would expect these genes to be expressed in the central domain in Nematostella. The current assumption is that the central domain will mainly give rise to endoderm derivatives /gastrodermis, while all other domains will give rise to ectodermal derivatives, such as the pharynx.36 Two genes, Nvotx and Nvsnail, are expressed in the central domain. However, Nvwnt8, NvfoxA, and Nvbrachyury are expressed in the central ring at the blastula stage. In Nematostella, a blimp‐like gene expressed in the animal hemisphere prior to gastrulation has yet to be identified, and Nvgata is expressed in individual cells throughout the ectoderm74 suggesting that neither of these critical ‘kernel’ genes are involved in cnidarian endodermal specification. Based on expression, the only ‘kernel’ gene definitively expressed in the correct region to be specifying the endoderm in Nematostella is Nvotx, but the central ring expression of NvfoxA and Nvbraychury at the blastula stage does not exclude them as possible endodermal GRN regulators during earlier developmental steps. Are these genes also part of the ancestral ‘endomesodermal’ GRN or do they belong to a GRN that specifies ectodermal domains in the future mouth/pharynx? Functional studies in conjunction with modern cell or tissue tracking techniques (photoconversion, tissue‐specific inducible GFP expression) are required to determine the fate of the above described expression domains and to link them to specific GRNs.
Taken together, it is reasonable to conclude that the bilaterian endomesodermal GRN is derived from an ancestral GRN that also gave rise to the cnidarian endodermal GRN. Nuclear β‐catenin and/or canonical Wnt activity is a critical upstream regulator in both GRNs, and they have a highly overlapping suite of genes that define or are functionally required for formation of the endoderm or endomesodermal tissue. It is not yet clear if differences between the bilaterian endomesodermal GRN and cnidarian preliminary endodermal GRN are going to be sufficient to explain why endomesoderm separates into two distinct tissues and the cnidarian endoderm does not. To address this question in more detail, gene‐specific functional studies of theses pathways as well as of other transcription factors such as Nvbrachury and NvfoxA are required to better understand the architecture and the molecular mechanism underlying this cnidarian endodermal GRN and its developmental outputs. For instance do subsets of the later forming bilaterian mesodermal GRN regulate myoepithelial cells. While we begin to get a basic understanding of the wiring of this GRN, future work needs to take into account the dynamics of the regulatory state over time as well as the cis‐regulation that underlies this process. This will enable the community to compare these findings with described GRNs from echinoderms and Xenopus and determine the mechanistic (i.e., presence of positive/negative feedback loops, lock‐down mechanisms of given regulatory states, etc.) similarities or differences that led to the evolution of distinct endodermal and mesodermal germ layers in bilaterians.
OVERVIEW OF NEMATOSTELLA NERVOUS SYSTEM
Most agree that the nervous systems of bilaterians (the central and peripheral nervous systems) are derived from a nerve net‐like ancestral nervous system.98, 99 Thus, better understanding development of nerve nets in Nematostella and other cnidarians will inform us about the structure and molecular make up of the ancestral nervous system that gave rise to the cnidarian nerve net and the bilaterian nervous systems. Here we will discuss the architecture and development of the Nematostella nervous system (Figure 5).
Figure 5.
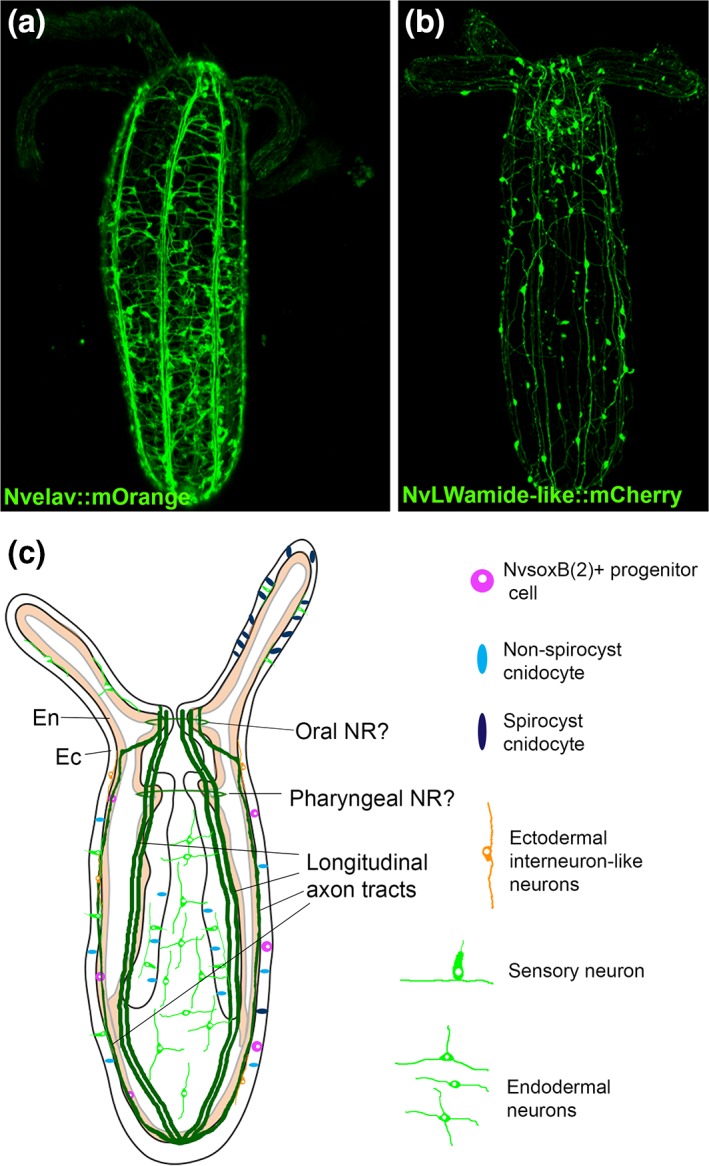
Morphology of Nematostella nervous system in juvenile polyps. (a, b) 3D reconstruction of confocal Z‐series acquired for two Nematostella transgenic lines that label the nervous system (a) NvElav::mOrange and (b) NvLWamide::mCherry. (c) Schematic of juvenile polyp nervous system highlights key structures and provides examples of neuronal morphologies observed in transgenic animals.
Neurons in Nematostella are born in both ectoderm and endoderm,19, 21 and neural soma are intermixed with other cell types. Although, cnidarian nervous systems are described as being a nerve net, there are clear examples of neural condensations and neuropil‐like structures in multiple cnidarian species. For example, longitudinal tracts of neurites run along the mesenteries in Nematostella 18, 21 (Figure 5(a)–(c)). In addition, oral and pharyngeal nerve rings have been reported18 (Figure 5(c)). It should be noted that while cross reactive antibodies used to identify the pharyngeal and oral nerve rings showed a slightly higher concentration of neurons in a location consistent with that of a putative oral and pharyngeal nerve ring18 recent data with transgenic lines, such as Nvelav::morange, find relatively uniform neural concentrations along the length of the oral–aboral axis, and thus it is not yet definitive that oral and pharyngeal nerve rings are present. Nematostella possesses sensory and interneurons as well as myoepithelial cells18, 21 suggesting that dedicated inter, sensory, and muscle innervating neurons are present in the Nematostella nerve net. In addition, Nematostella possesses neurosecretory‐like gland cells,100 and a cnidarian‐specific neural cell type the cnidocyte.17 Cnidocytes are considered neural cells because they display mechanosensory and calcium dependent ultrafast exocytosis neural‐like properties.101 Cnidocytes are unique to cnidarians and offer an opportunity to investigate the evolution novel cell types (Box 1). Identification and characterization of the neuronal classes in Nematostella will provide better understanding as to which neurons are interneurons, sensory, motoneurons, and what distinctive properties each class has (neurotransmitter type, receptors, molecular markers, etc.).
BOX 1. CNIDOCYTES AND THE EVOLUTION OF NOVEL CELL TYPES.
Cnidocytes are stinging cells found only within the cnidarians. They represent one of the few examples of an unequivocal evolutionary novelty, and thus investigating their composition and development can inform us about the mechanisms by which novel cell types arise. Cnidocytes possess a unique organelle called a cnidocyst that is comprised of a harpoon wound up inside of a capsule. The capsule is generated by a number of cnidarian‐specific minicollagen genes. To date functional dissection of the molecular programs that generate cnidocytes is limited. Disruption of Notch signaling inhibits proper differentiation of cnidocytes in Hydra 126 and reduces the number of mature cnidocytes in Nematostella.111 In Nematostella the neural transcription factors Nvsoxb(2) and a unique splice isoform of the mesodermal transcription factor Nvmef2 are required for proper specification of cnidocytes.30, 103 From these limited studies, we can infer that the evolution of this novel cell type likely required the emergence of a novel gene family (the minicollagens) and co‐option of pre‐existing developmental transcription factors. Interestingly, there are three classes of cnidocytes and some studies suggest species‐specific cnidocyte molecular components. This cell type is ripe for further studies focused on understanding the make up and development of cnidocytes which will provide insight into the how novel proteins emerge, and how pre‐existing developmental programs interact with new genes to generate novel cell types.
Mechanisms of Neurogenesis
Neural Induction
Understanding neural induction in Nematostella is challenging, because no distinct neural structure like a central nervous system (CNS) exists, and we do not know what the earliest definitively neural gene expression is yet. As a result, how to assay induction phenotypes is unclear. Current data argue that Nematostella and bilaterian neural induction occurs via distinct mechanisms. Inhibition of bmp2/4 (also called Dpp) in the neural ectoderm via Nog and Chd is the most conserved neural induction mechanism in bilaterians. Thus bmp2/4 homologs suppress central nervous system formation in bilaterians. Nvbmp2/4 patterns the directive axis and together with Nvbmp5/8 patterns endoderm. However, loss or gain of Nvbmp2/4 doesn't disrupt embryonic neural expression in the ectoderm.69, 102 On the other hand, loss or gain of Nvbmp2/4 activity results in loss of at least a subset of endodermal neurons in planulae.69, 102 In Nematostella, it appears NvBmp2/4 is required for endodermal neural development, but can act as an inhibitor at high doses. One caveat to these studies is that loss of later neural expression is more likely due to broader defects in patterning resulting from loss of Nvbmp2/4 at earlier stages. Truly understanding neural induction will require identification of the earliest markers that distinguish committed neural cell types followed by a systematic assessment of potential upstream inputs that regulate their expression.
Embryonic Neurogenesis
Many of the core neurogenic programs acting downstream of induction in bilaterians function during Nematostella neurogenesis. Neurons arise in the ectoderm at early gastrula stages.18, 19, 21 NvashA and NvsoxB(2), homologs of the bilaterian neural transcription factors, promote development of embryonic ectodermal neurogenesis.19, 103 NvashA is a homolog of achaete‐scute family bHLH proneural transcription factors. bHLH proneural genes encompass both atonal and achaete‐scute family transcription factors. Bilaterian proneural genes are necessary and sufficient to induce neural development (summarized in the study of Bertrand, 2002).104 Nematostella has four achaete‐scute genes and at least seven atonal‐like genes encoded in its genome.105 NvsoxB(2) is a HMG‐box sox family transcription factor related to the bilaterian soxB1 and soxB2 families. sox genes are expressed in developing neurons in bilaterians.106, 107, 108 In particular, sox gene expression in neural stem and progenitor cells is well known in neurogenesis of multiple bilaterian species as well as in other cnidarians.109 There are multiple sox gene families that function in neurogenesis as well as development of other tissues. Nematostella has 14 sox genes, two of which have reported roles in neural development.102, 103, 110 Morpholino knockdown of either NvashA or NvsoxB(2) results in a reduction of NvanthoRFamide and Nvelav1expression, and NvsoxB(2) morphant animals also show reduced numbers of cnidocytes.19, 103 Global misexpression of NvashA induces ectopic expression of neural subtype markers, but interestingly only within their respective domains.19 Additional work has shown that Notch activity can act to suppress neural development by suppressing NvashA, and that Notch activity also suppresses expression of NvsoxB(2).111, 112 These data broadly suggest that neurogenic transcription factors in Nematostella function similarly to their bilaterian homologs. Teasing out detailed regulatory networks that promote general neuronal properties and neural subtype development will improve our understanding of the level of conservation between molecular mechanisms governing neurogenesis in cnidarians and bilaterians.
Analysis of NvsoxB(2) reporter transgenes determined that as many as 30% NvsoxB(2) expressing cells incorporate the thymidine analog EdU in pulse labeling experiments and form clones suggesting that a subset of the NvsoxB(2) + cells are proliferative neural progenitor cells.103 However, it is not entirely clear how widespread deployment of neural progenitor cells is, or how many divisions progenitors are capable of undergoing. Further advances in techniques to label and track individual clones in vivo will provide greater detail in future studies. Still missing from our understanding is the origin of the neural progenitor cells. Are neural progenitor cells derived from a dedicated neural stem cell, a multipotent stem cell, or from epithelial cells (like Drosophila sensory organ precursors)? Regardless, the presence of progenitor cells in Nematostella suggests that not only are the molecular mechanisms of neurogenesis conserved between bilaterians and Nematostella, but also the cellular dynamics are also conserved between these groups.
Larval Neurogenesis
Beginning at late gastrula and early larval stages neural marker gene expression is detected in the endoderm. Throughout larval stages neurogenesis occurs in endoderm and ectoderm. Here too sox and bHLH proneural genes have demonstrated roles in neural development. Morpholino mediated knockdown of NvsoxB(2) also reduced the number of endodermal neurons labeled by the Nvelav1 transgenic reporter.21, 103 A more recent study identified an oral population of ectodermal NvanthoRFamide labeled neurons and a small population of asymmetrically positioned NvGLWamide oral endodermal neurons.102 These neurons are patterned by NvsoxB(2a) activating expression of NvashB, which in turn activates expression of an atonal‐like bHLH proneural gene. The NvGLWamide neurons specifically require additional positive inputs from Nvarp6, which is asymmetrically expressed in the domain that gives rise to the NvGLWamide positive cells. Notch activity was also shown to suppress endodermal expression of NvashA suggesting that Notch also acts upstream of neurogenic transcription factors during larval endodermal neurogenesis.112 NvsoxB(2) + precursors were identified in the larval endoderm and ectoderm, suggesting that larval neurogenesis occurs via specification of neural progenitor cells.103 In total the current data argue that molecular mechanisms responsible for neurogenesis are the same in both endoderm and ectoderm. Thus understanding neural development in either tissue will inform about the ancestral neurogenic program that gave rise to the bilaterian nervous systems.
Mechanisms of Neural Patterning
Neural subtype patterning likely occurs via the combinatorial action of regional patterning and neural differentiation pathways.18, 19, 20, 21, 37, 41, 52 This is most apparent in specification of the apical tuft sensory organ.20, 37, 41, 113, 114 Additional evidence comes from the regional distribution of distinct neural subtypes. Cnidocyte subtypes and molecular markers for ectodermal neurons are differentially distributed along the oral–aboral axis suggesting their identity is tied to regional patterning cues.17, 18, 19, 20, 102 This hypothesis is supported by the observation that ubiquitous misexpression of NvashA increases the number of neurons born throughout the embryo, but individual neural subtype markers only expand within the limits of their normal domain boundaries.19 Lastly, patterning of the directive axis restricts Nvarp6 to one side of the endoderm where it specifies NvGLWamide + neurons.102 Taken together, the current model is that neurogenic programs are induced more or less globally, but that the combinatorial action of regional patterning and neurogenic transcription factors act to produce distinct fates. This is similar to bilaterian CNS development in that a broad neural program is initiated to neuralize a subset of the ectoderm, which is followed by subdivision by regional patterning information to generate distinct domains that produce specific fates. To date our understanding about how regional patterning genes act to generate specific fates is relatively poor in Nematostella and further work is needed before we can compare details of neural patterning between Nematostella and bilaterians to each other.
The conserved activity of neurogenic transcription factors between Nematostella and bilaterian neural programs is clear, suggesting that the cnidarian nerve net and bilaterian nervous systems are derived from a common ancestor. There is a tremendous amount of speculation about how central nervous systems evolved from something that resembled a cnidarian nerve net. Ideas range from the oral nerve ring in cnidarians being a precursor to bilaterian nerve cords, the idea that the brain originated from the ectodermal nervous system in the aboral region of cnidarian embryos (see above),115 and possibly the most speculative that bilaterian nervous systems may have a common origin with the endodermal nerve net.116 To resolve these hypotheses, current research must build on the foundational studies and identify individual neuronal cell types within cnidarians and determine if definitive homology can be drawn between cnidarian and bilaterian neuronal classes. Also, a better characterization of the molecular basis of neural induction in cnidarians must be completed and compared in detail to the bilaterian neural inductive programs. Regardless, of the answers investigating the cnidarian nerve net will improve our understanding of how ancestral nervous systems were patterned and functioned during animal evolution.
OVERVIEW OF REGENERATION
Nematostella has the potential to distinguish itself as a model to investigate the relationship between development and regeneration. Increasing evidence suggests that regeneration does not faithfully recapitulate development.117, 118, 119 Not to say completely novel molecular programs are deployed during regeneration, but rather subtle differences that likely reflect the unique challenge of regeneration are involved. Being able to directly investigate and compare development and regeneration in the same species is critical to better understand why and how these processes vary from one another. One reason comparisons between development and regeneration are not more widespread is that most traditional model systems are ideal for studying development or regeneration, but not both. Hydra, planarians, and axolotls are the most well known regenerative animal models, but they lack the ability to reliably and cost effectively obtain large numbers of embryos or they lack critical tools to manipulate and/or visualize gene function. Traditional model systems (mouse, Caenorhabditis elegans, Drosophila, chicks, Xenopus, etc.) display little or no regenerative capacity. Zebrafish does offer a powerful developmental system that also displays impressive regenerative capacity.120 However, most vertebrates are not as regenerative as zebrafish, which suggests that they may have independently derived their regenerative ability. Thus, developing model systems that complement the current regenerative model animals including zebrafish will allow conserved mechanisms of regeneration in animals to be identified. Moreover, by comparing how regeneration and differentiation vary from one another in multiple species, it may be possible to better understand why regeneration does not recapitulate development and use that knowledge to improve design and deployment of regenerative medical therapies. The power of Nematostella as a developmental system has been discussed above, but they are also highly regenerative capable of whole body axis regeneration in less than 1 week (Figure 6) suggesting that it is a good choice as a model system to compare development and regeneration.34, 121
Figure 6.
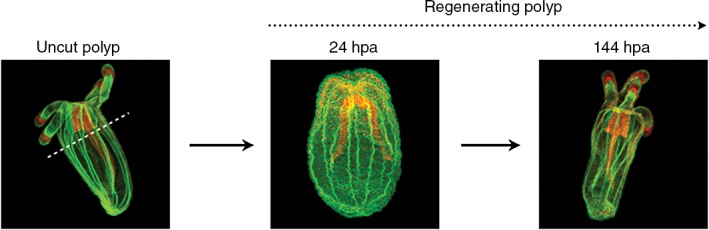
Oral regeneration of juvenile Nematostella takes place in 6 days. After sub‐pharyngeal bisection, missing oral part of the polyps regenerate within 6 days to reform a fully functional organism. Animals were fixed and stained with phalloidin (green) to show f‐actin filaments and propidium iodide (red) to visualize the nuclei. All images are lateral views with the oral part to the top.
Nematostella Regeneration
Nematostella regeneration is either experimentally or naturally induced by bisection of the animal into two halves or by physal pinching, respectively. The latter phenomenon is normally initiated by animals undergoing asexual reproduction via transverse fission of the most aboral region (the physa). Unlike Hydra, cell proliferation is required for Nematostella regeneration,121 and preliminary studies show that after bisection or physal pinching characteristic events occur during regenerative growth.34, 121 Although recent efforts have focused on characterization of morphological, cellular, and molecular events that occur during Nematostella wound‐healing and regeneration, these studies remain sparse and are heterogeneous in their experimental approaches (different ages of animals used, different sites of bisections, variable culture temperature, etc.).33, 121, 122, 123, 124 A universal protocol or normalized staging series would allow for direct and more accurate comparisons to be drawn between Nematostella regeneration, asexual reproduction, and regeneration in other animals. A first attempt to define a normalized staging series for adult Nematostella regeneration after supra‐physal amputation has been recently presented by Bossert and colleagues.34 While this study is an important resource, the chosen amputation site is unusual and adult tissue is more difficult to analyze due to the size and opacity of tissue compared to juveniles. In an effort to have a common foundation for comparing experimental results for scientists working on Nematostella regeneration, Amiel and colleagues have compared sub‐pharyngeal oral regeneration in juveniles and adults and shown that there are no age but temperature‐specific variations in the timing of this process. The reassessment of cell proliferation revealed that mitotic cells are detected as soon as 12 hpa at the amputation site in juveniles and that cellular proliferation is also required for oral regeneration in adults suggesting that the cellular mechanisms underlying this process may be shared between juveniles and adults.32 In addition, the authors have developed an in vivo assay to analyze the wound‐healing success and showed that after amputation the wound is completely healed 6 hpa. A detailed morphological analysis of regenerating juveniles revealed a characteristic four‐step process (Figure 7).32 After amputation (1) the mesenteries fuse to each other and enter in contact to the amputation site. When the tentacle buds become visible, (2) the mesenteries are pulled/pushed down to create a gap between the amputation site and the mesenteries where high proliferation is detected and where (3) the pharyngeal lip starts to form, followed by (4) the completion of pharynx formation. Interestingly, the beginning of pharynx formation between 60 and 72 hpa is accompanied by the appearance of green autofluorescence that can be used as an in vivo assay to determine successful pharynx formation/regeneration. Taken together, this analysis laid down a basic framework to study oral regeneration after sub‐pharyngeal amputation in Nematostella juveniles by developing assays and defining specific landmarks of this process that can be used to analyze the effects of various perturbation experiments.32 While both adult or juvenile staging systems allow improved comparison of results obtained from different research groups investigating Nematostella regeneration, they are first drafts that need to be improved in the future with additional details (e.g., molecular markers). Importantly, the present information can also serve to define (or to compare with) different regeneration types, such as aboral regeneration that will provide additional information on the conservation/divergence of the overall mechanisms underlying the reformation of missing body parts in Nematostella at the molecular, cellular, and tissue level.
Figure 7.
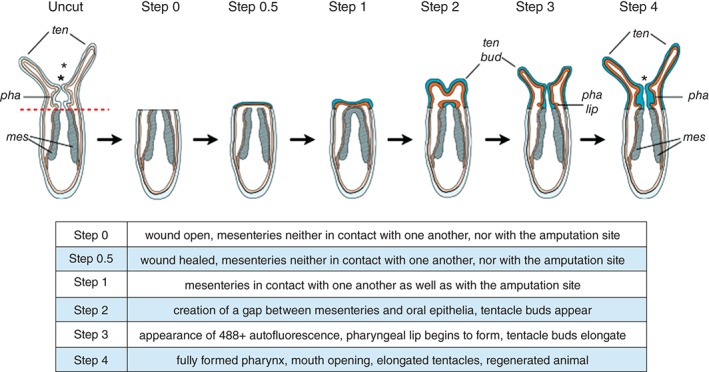
Diagram summarizing the morphological and cellular events underlying Nematostella oral regeneration. The table below the illustration provides definitions for the various regeneration steps. *, mouth opening; ten, tentacles; ten bud, tentacle bud; pha, pharynx; pha lip, pharyngeal lip; mes, mesenteries. (Reprinted with permission from Ref 32. Copyright 2015 International Journal of Molecular Sciences)
Patterning Mechanisms During Nematostella Regeneration
One of the main questions that can be addressed in Nematostella is how patterning during regeneration relates to patterning during development. To date only two papers have compared developmental and regenerative patterning in Nematostella. Briefly, Nematostella shows that major patterning molecules are similarly deployed during development and regeneration, but that subtle differences in regeneration are observed. In Nematostella, high Wnt activity is used to specify oral fates during development (discussed above). Hyperactivation of Wnt activity during regeneration is sufficient to induce complete ectopic oral regeneration in aboral territories.125 This observation argues that high Wnt activity also specifies oral fates during regeneration. A second study specifically assayed expression of a number of transcription factors with distinct patterns during development.33, 124 The authors of that work identified two classes of genes. One class displayed regenerative expression patterns that more or less resembled the spatial expression pattern each gene displayed during development. A second class, however, showed no expression or distinct expression patterns during regeneration that varied from their pattern during development. Although, studies of Nematostella regeneration are still preliminary, they do highlight that a common observation in animals is that regeneration varies albeit slightly from development. Given the fact that embryonic development is triggered by fertilization of the totipotent oocyte and regeneration is initiated by physical amputation of variable amounts of differentiated and stem cells, it is not surprising that regeneration does not simply recapitulate development. Regeneration requires identification of what is missing and what of the adult body remains, enacting a program to replace the missing cells/tissues, stopping regrowth at the appropriate time, and finally reintegration of new tissues and cell types into an already existing adult body. Identification and characterization of differences between development and regeneration will improve our understanding of what is required for successful regeneration and possibly aid in designing and deploying effective regenerative medical therapies.
CONCLUSION
Nematostella was originally developed to better understand the origin and evolution of bilaterian characteristics and animals in general. Those studies yielded an understanding of the mechanisms that pattern the oral–aboral and directive axes of Nematostella, a preliminary GRN for formation of the bifunctional endoderm that likely gave rise to the endomesoderm of bilaterians, and the identification and understanding of conserved molecular programs that specify neurons in cnidarians and bilaterians. These three areas of research have been the primary focus of Nematostella research, and they provide an initial understanding of how the cnidarian body plan, tissues, and cell types are patterned during development. What we have learned is that while many of the most upstream components necessary to pattern similar structures and cell types in bilaterians are enacted in Nematostella, key differences are present. By further investigating these differences, we will not only gain a better understanding of the ancestral state from which bilaterian evolution occurred, but also gain critical insights into the mechanisms by which animals control patterning and cell specification during development to arise at the variety of forms present in extant taxa. In addition, with the emergence of Nematostella as a model system, it is now possible to focus on new questions such as understanding the relationship between mechanisms deployed during development and regeneration of identical structures. The future of Nematostella is a bright one. Ease of culture and embryo manipulation make Nematostella accessible to essentially any one interested in incorporating this exciting new model into their research program. New perspectives are likely to open novel avenues into which Nematostella can inform us about metazoan biology, evolution, and biomedical applications.
Conflict of interest: The authors have declared no conflicts of interest for this article.
References
FURTHER READING
- Rigo‐Watermeier T et al. Functional conservation of Nematostella Wnts in canonical and noncanonical Wnt‐signaling. Biol Open 2012, 1:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES
- 1. Dunn CW, Hejnol A, Matus MQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 2008, 452:745–749. [DOI] [PubMed] [Google Scholar]
- 2. Ryan JF, Pang K, Schnitzler CE, Nguyen A, Moreland RT, Simmons DK, Koch BJ, Francis WR, Havlak P, NISC Comparative Sequencing Program, et al. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 2013, 342:1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hejnol A, Obst M, Stamatakis A, Ott M, Rouse GW, Edgecombe GD, Martinez P, Baguna J, Bailly X, Jondelius U, et al. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc R Soc B Biol Sci 2009, 276:4261–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hand C, Uhlinger KR. The culture, sexual and asexual reproduction, and growth of the sea anemone Nematostella vectensis . Biol Bull 1992, 182:169–176. [DOI] [PubMed] [Google Scholar]
- 5. Houliston E, Momose T, Manuel M. Clytia hemisphaerica: a jellyfish cousin joins the laboratory. Trends Genet 2015, 26:159–167. [DOI] [PubMed] [Google Scholar]
- 6. Plickert G, Frank U, Müller WA. Hydractinia, a pioneering model for stem cell biology and reprogramming somatic cells to pluripotency. Int J Dev Biol 2012, 56:519–534. [DOI] [PubMed] [Google Scholar]
- 7. Fritzenwanker JH, Genikhovich G, Kraus Y, Technau U. Early development and axis specification in the sea anemone Nematostella vectensis . Dev Biol 2007, 310:264–279. [DOI] [PubMed] [Google Scholar]
- 8. Lee PN, Kumburegama S, Marlow HQ, Martindale MQ, Wikramanayake AH. Asymmetric developmental potential along the animal–vegetal axis in the anthozoan cnidarian, Nematostella vectensis, is mediated by Dishevelled. Dev Biol 2007, 310:169–186. [DOI] [PubMed] [Google Scholar]
- 9. Fritzenwanker JH, Technau U. Induction of gametogenesis in the basal cnidarian Nematostella vectensis (Anthozoa). Dev Genes Evol 2002, 212:99–103. [DOI] [PubMed] [Google Scholar]
- 10. Stefanik DJ, Friedman LE, Finnerty JR. Collecting, rearing, spawning and inducing regeneration of the starlet sea anemone, Nematostella vectensis . Nat Protoc 2013, 8:916–923. [DOI] [PubMed] [Google Scholar]
- 11. Fritz AE, Ikmi A, Seidel C, Paulson A, Gibson MC. Mechanisms of tentacle morphogenesis in the sea anemone Nematostella vectensis . Development 2013, 140:2212–2223. [DOI] [PubMed] [Google Scholar]
- 12. Renfer E, Amon‐Hassenzahl A, Steinmetz PRH, Technau U. A muscle‐specific transgenic reporter line of the sea anemone, Nematostella vectensis . Proc Natl Acad Sci USA 2010, 107:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Extavour CG, Pang K, Matus DQ, Martindale MQ. vasa and nanos expression patterns in a sea anemone and the evolution of bilaterian germ cell specification mechanisms. Evol Dev 2005, 7:201–215. [DOI] [PubMed] [Google Scholar]
- 14. Tucker RP, Shibata B, Blankenship TN. Ultrastructure of the mesoglea of the sea anemone Nematostella vectensis (Edwardsiidae). Invertebr Biol 2011, 130:11–24. [Google Scholar]
- 15. Leclère L, Rentzsch F. RGM regulates BMP‐mediated secondary axis formation in the sea anemone Nematostella vectensis . Cell Rep 2014, 9:1921–1930. doi:10.1016/j.celrep.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 16. Berking S. Generation of bilateral symmetry in Anthozoa: a model. J Theor Biol 2007, 246:477–490. [DOI] [PubMed] [Google Scholar]
- 17. Zenkert C, Takahashi T, Diesner M‐O, Ozbek S. Morphological and molecular analysis of the Nematostella vectensis cnidom. PLoS One 2011, 6:e22725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marlow HQ, Srivastava M, Matus DQ, Rokhsar D, Martindale MQ. Anatomy and development of the nervous system of Nematostella vectensis, an anthozoan cnidarian. Dev Neurobiol 2009, 69:235–254. [DOI] [PubMed] [Google Scholar]
- 19. Layden MJ, Boekhout M, Martindale MQ. Nematostella vectensis achaete‐scute homolog NvashA regulates embryonic ectodermal neurogenesis and represents an ancient component of the metazoan neural specification pathway. Development 2012, 139:1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sinigaglia C, Busengdal H, Leclère L, Technau U, Rentzsch F. The bilaterian head patterning gene six3/6 controls aboral domain development in a cnidarian. PLoS Biol 2013, 11:e1001488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nakanishi N, Renfer E, Technau U, Rentzsch F. Nervous systems of the sea anemone Nematostella vectensis are generated by ectoderm and endoderm and shaped by distinct mechanisms. Development 2011, 139:347–357. [DOI] [PubMed] [Google Scholar]
- 22. Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, Technau U, von Haeseler A, Hobmayer B, Martindale MQ, et al. Unexpected complexity of the Wnt gene family in a sea anemone. Nature 2005, 433:156–160. [DOI] [PubMed] [Google Scholar]
- 23. Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 2007, 317:86–94. [DOI] [PubMed] [Google Scholar]
- 24. Varmuza S, Sullivan JC, Finnerty JR. A surprising abundance of human disease genes in a simple ‘basal’ animal, the starlet sea anemone (Nematostella vectensis). Genome 2007, 50:689–692. [DOI] [PubMed] [Google Scholar]
- 25. Grimson ASrivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi‐interacting RNAs in animals. Nature 2008, 455:1193–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moran Y, Fredman D, Praher D, Li XZ, Wee LM, Rentzsch F, Zamore PD, Technau U, Seitz H. Cnidarian microRNAs frequently regulate targets by cleavage. Genome Res 2014, 24:651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schwaiger M, Schönauer A, Rendeiro AF, Pribitzer C, Schauer A, Gilles AF, Schinko JB, Renfer E, Fredman D, Technau U. Evolutionary conservation of the eumetazoan gene regulatory landscape. Genome Res 2014, 24:639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Helm RR, Siebert S, Tulin S, Smith J, Dunn CW. Characterization of differential transcript abundance through time during Nematostella vectensis development. BMC Genomics 2013, 14:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tulin S, Aguiar D, Istrail S, Smith J. A quantitative reference transcriptome for Nematostella vectensis early embryonic development: a pipeline for de novo assembly in emerging model systems. Evodevo 2013, 4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Genikhovich G, Technau U. Complex functions of Mef2 splice variants in the differentiation of endoderm and of a neuronal cell type in a sea anemone. Development 2011, 138:4911–4919. [DOI] [PubMed] [Google Scholar]
- 31. Genikhovich G, Technau U. The starlet sea anemone Nematostella vectensis: an anthozoan model organism for studies in comparative genomics and functional evolutionary developmental biology. Cold Spring Harb Protoc 2009, 2009:pdb.emo129. [DOI] [PubMed] [Google Scholar]
- 32. Amiel AR, Johnston HT, Nedoncelle K, Warner JF, Ferreira S, Röttinger E. Characterization of morphological and cellular events underlying oral regeneration in the sea anemone, Nematostella vectensis . Int J Mol Sci 2015, 16:28449–28471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DuBuc TQ, Traylor‐Knowles N, Martindale MQ. Initiating a regenerative response, cellular and molecular features of wound healing in the cnidarian Nematostella vectensis . BMC Biol 2014, 214:134–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bossert PE, Dunn MP, Thomsen GH. A staging system for the regeneration of a polyp from the aboral physa of the anthozoan cnidarian Nematostella vectensis . Dev Dyn 2013, 242:1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ikmi A, McKinney SA, Delventhal KM, Gibson MC. TALEN and CRISPR/Cas9‐mediated genome editing in the early‐branching metazoan Nematostella vectensis . Nat Commun 2014, 5:5486. [DOI] [PubMed] [Google Scholar]
- 36. Magie CR, Daly M, Martindale MQ. Gastrulation in the cnidarian Nematostella vectensis occurs via invagination not ingression. Dev Biol 2007, 305:483–497. [DOI] [PubMed] [Google Scholar]
- 37. Rentzsch F, Fritzenwanker JH, Scholz CB, Technau U. FGF signalling controls formation of the apical sensory organ in the cnidarian Nematostella vectensis . Development 2008, 135:1761–1769. [DOI] [PubMed] [Google Scholar]
- 38. Layden MJ, Röttinger E, Wolenski FS, Gilmore TD, Martindale MQ. Microinjection of mRNA or morpholinos for reverse genetic analysis in the starlet sea anemone, Nematostella vectensis . Nat Protoc 2013, 8:924–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wikramanayake AH, Hong M, Lee PN, Pang K, Byrum CA, Bince JM, Xu R, Martindale MQ. An ancient role for nuclear β‐catenin in the evolution of axial polarity and germ layer segregation. Nature 2003, 426:446–450. [DOI] [PubMed] [Google Scholar]
- 40. Röttinger E, Dahlin P, Martindale MQ. A framework for the establishment of a cnidarian gene regulatory network for ‘endomesoderm’ specification: the inputs of β‐catenin/TCF signaling. PLoS Genet 2012, 8:e1003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sinigaglia C, Busengdal H, Lerner A, Oliveri P, Rentzsch F. Molecular characterization of the apical organ of the anthozoan Nematostella vectensis . Dev Biol 2015, 398:120–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wolenski FS, Layden MJ, Martindale MQ, Gilmore TD, Finnerty JR. Characterizing the spatiotemporal expression of RNAs and proteins in the starlet sea anemone, Nematostella vectensis . Nat Protoc 2013, 8:900–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Freeman G. The role of polarity in the development of the hydrozoan planula larva. Roux's Arch Dev Biol 1981, 190:168–184. [DOI] [PubMed] [Google Scholar]
- 44. Finnerty JR. Origins of bilateral symmetry: Hox and Dpp expression in a sea anemone. Science 2004, 304:1335–1337. [DOI] [PubMed] [Google Scholar]
- 45. Ryan JF, Mazza ME, Pang K, Matus DQ, Baxevanis AD, Martindale MQ, Finnerty JR. Pre‐bilaterian origins of the Hox cluster and the Hox code: evidence from the sea anemone, Nematostella vectensis . PLoS One 2007, 2:e153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kamm K, Schierwater B, Jakob W, Dellaporta SL, Miller DJ. Axial patterning and diversification in the cnidaria predate the Hox system. Curr Biol 2006, 16:920–926. [DOI] [PubMed] [Google Scholar]
- 47. Chiori R et al. Are Hox genes ancestrally involved in axial patterning? Evidence from the hydrozoan Clytia hemisphaerica (cnidaria). PLoS One 2009, 4:e4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Quiquand M et al. More constraint on ParaHox than Hox gene families in early metazoan evolution. Dev Biol 2009, 328:173–187. [DOI] [PubMed] [Google Scholar]
- 49. Yanze N, Spring J, Schmidli C, Schmid V. Conservation of Hox/ParaHox‐related genes in the early development of a cnidarian. Dev Biol 2001, 236:89–98. [DOI] [PubMed] [Google Scholar]
- 50. Kumburegama S, Wijesena N, Xu R, Wikramanayake AH. Strabismus‐mediated primary archenteron invagination is uncoupled from Wnt/β‐catenin‐dependent endoderm cell fate specification in Nematostella vectensis (Anthozoa, Cnidaria): implications for the evolution of gastrulation. Evodevo 2011, 2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee PN, Pang K, Matus DQ, Martindale MQ. A WNT of things to come: evolution of Wnt signaling and polarity in cnidarians. Semin Cell Dev Biol 2006, 17:157–167. [DOI] [PubMed] [Google Scholar]
- 52. Marlow H, Matus DQ, Martindale MQ. Ectopic activation of the canonical wnt signaling pathway affects ectodermal patterning along the primary axis during larval development in the anthozoan Nematostella vectensis . Dev Biol 2013, 380:324–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Momose T, Houliston E. Two oppositely localised frizzled RNAs as axis determinants in a cnidarian embryo. PLoS Biol 2007, 5:e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Momose T, Derelle R, Houliston E. A maternally localised Wnt ligand required for axial patterning in the cnidarian Clytia hemisphaerica. >Development 2008, 135:2105–2113. [DOI] [PubMed] [Google Scholar]
- 55. Plickert G, Jacoby V, Frank U, Müller WA, Mokady O. Wnt signaling in hydroid development: formation of the primary body axis in embryogenesis and its subsequent patterning. Dev Biol 2006, 298:368–378. [DOI] [PubMed] [Google Scholar]
- 56. Schmidt‐Rhaesa, A. The Evolution of Organs Systems. Oxford, UK: Oxford University Press; 2007. [Google Scholar]
- 57. Galliot B, Miller DJ. Origin of anterior patterning: how old is our head? Trends Genet 2000, 16:1–5. [DOI] [PubMed] [Google Scholar]
- 58. Aronowicz J, Lowe CJ. Hox gene expression in the hemichordate Saccoglossus kowalevskii and the evolution of deuterostome nervous systems. Integr Comp Biol 2006, 46:890–901. [DOI] [PubMed] [Google Scholar]
- 59. Darras S, Gerhart J, Terasaki M, Kirschner M, Lowe CJ. Catenin specifies the endomesoderm and defines the posterior organizer of the hemichordate Saccoglossus kowalevskii . Development 2011, 138:959–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wei Z, Yaguchi J, Yaguchi S, Angerer RC, Angerer LM. The sea urchin animal pole domain is a Six3‐dependent neurogenic patterning center. Development 2009, 136:1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tu Q, Brown CT, Davidson EH, Oliveri P. Sea urchin Forkhead gene family: phylogeny and embryonic expression. Dev Biol 2006, 300:49–62. [DOI] [PubMed] [Google Scholar]
- 62. Santagata S, Resh C, Hejnol A, Martindale MQ, Passamaneck YJ. Development of the larval anterior neurogenicdomains of Terebratalia transversa (Brachiopoda) provides insights into the diversification of larvalapical organs and the spiralian nervous system. Evodevo 2012, 3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Carlgren O. Studien ueber Nordische Actinien. Vetenskap‐Akademiens Handlinger 1893, 25:1–148. [Google Scholar]
- 64. Hyman, L. The Invertebrates: Protozoa through Ctenophora. New York: McGraw‐Hill; 1940, 538–565. [Google Scholar]
- 65. Matus DQ, Thomsen GH, Martindale MQ. Dorso/ventral genes are asymmetrically expressed and involved in germ‐layer demarcation during cnidarian gastrulation. Curr Biol 2006, 16:499–505. [DOI] [PubMed] [Google Scholar]
- 66. Matus DQ, Pang K, Marlow H, Dunn CW, Thomsen GH, Martindale MQ. Molecular evidence for deep evolutionary roots of bilaterality in animal development. Proc Natl Acad Sci USA 2006, 103:11195–11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hayward DC, Samuel G, Pontynen PC, Catmull J, Saint R, Miller DJ, Ball EE. Localized expression of a dpp/BMP2/4 ortholog in a coral embryo. Proc Natl Acad Sci USA 2002, 99:8106–8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rentzsch F, Anton R, Saina M, Hammerschmidt M, Holstein TW, Technau U. Asymmetric expression of the BMP antagonists chordin and gremlin in the sea anemone Nematostella vectensis: implications for the evolution of axial patterning. Dev Biol 2006, 296:375–387. [DOI] [PubMed] [Google Scholar]
- 69. Saina M, Genikhovich G, Renfer E, Technau U. BMPs and Chordin regulate patterning of the directive axis in a sea anemone. Proc Natl Acad Sci 2009, 106:18582–18597. doi:10.1073/pnas.0900151106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Genikhovich G et al. Axis patterning by BMPs: cnidarian network reveals evolutionary constraints. Cell Rep 2015, 10:1646–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cameron RA, Davidson EH. Cell type specification during sea urchin development. Trends Genet 1991, 7:212–218. [DOI] [PubMed] [Google Scholar]
- 72. Rodaway A, Patient R. Mesendoderm: an ancient germ layer? Cell 2001, 105:169–172. [DOI] [PubMed] [Google Scholar]
- 73. Kimelman D, Griffin KJ. Vertebrate mesendoderm induction and patterning. Curr Opin Genet Dev 2000, 10:350–356. [DOI] [PubMed] [Google Scholar]
- 74. Martindale MQ. Investigating the origins of triploblasty: ‘mesodermal' gene expression in a diploblastic animal, the sea anemone Nematostella vectensis (phylum, Cnidaria; class, Anthozoa). Development 2004, 131:2463–2474. [DOI] [PubMed] [Google Scholar]
- 75. Martindale MQ. The evolution of metazoan axial properties. Nat Rev Genet 2005, 6:917–927. [DOI] [PubMed] [Google Scholar]
- 76. Martindale, M. Q. & Finnerty, J. R. The Radiata and the evolutionary origins of the bilaterian body plan. Mol Phylogenet Evol 2002, 24: 358–365. [DOI] [PubMed] [Google Scholar]
- 77. Seipel K, Schmid V. Mesodermal anatomies in cnidarian polyps and medusae. Int J Dev Biol 2006, 50:589–599. [DOI] [PubMed] [Google Scholar]
- 78. Scholz CB, Technau U. The ancestral role of Brachyury: expression of NemBra1 in the basal cnidarian Nematostella vectensis (Anthozoa). Dev Genes Evol 2003, 212:563–570. [DOI] [PubMed] [Google Scholar]
- 79. Technau U. Brachyury, the blastopore and the evolution of the mesoderm. Bioessays 2001, 23:788–794. [DOI] [PubMed] [Google Scholar]
- 80. Burton PM. Insights from diploblasts; the evolution of mesoderm and muscle. J Exp Zool 2007, 310B:5–14. [DOI] [PubMed] [Google Scholar]
- 81. Byrum CA, Martindale MQ. Gastrulation. From Cells to Embryo. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2004, 33–50. [Google Scholar]
- 82. Jahnel, SM. , Walzl, M. & Technau, U. Development and epithelial organisation of muscle cells in the sea anemone Nematostella vectensis Front Zool 11, 44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mazza ME, Pang K, Martindale MQ, Finnerty JR. Genomic organization, gene structure, and developmental expression of three Clustered otx genes in the sea anemone Nematostella vectensis . J Exp Zool 2007, 308B:494–506. [DOI] [PubMed] [Google Scholar]
- 84. Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al. A provisional regulatory gene network for specification of endomesoderm in the sea urchin embryo. Dev Biol 2002, 246:162–190. [DOI] [PubMed] [Google Scholar]
- 85. Vonica A, Weng W, Gumbiner BM, Venuti JM. TCF is the nuclear effector of the β‐catenin signal that patterns the sea urchin animal–vegetal axis. Dev Biol 2000, 217:230–243. [DOI] [PubMed] [Google Scholar]
- 86. Imai K, Takada N, Satoh N, Satou Y. β‐Catenin mediates the specification of endoderm cells in ascidian embryos. Development 2000, 127:3009–3020. [DOI] [PubMed] [Google Scholar]
- 87. Schohl A, Fagotto F. A role for maternal‐catenin in early mesoderm induction in Xenopus. EMBO J 2003, 22:3303–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Huang L, Li X, El-Hodiri M, Dayal S, Wikramanayake AH, Klein WH. Involvement of Tcf/Lef in establishing cell types along the animal‐vegetal axis of sea urchins. Dev Genes Evol 2000, 210:73–81. [DOI] [PubMed] [Google Scholar]
- 89. Matus DQ, Magie CR, Pang K, Martindale MQ, Thomsen GH. The Hedgehog gene family of the cnidarian, Nematostella vectensis, and implications for understanding metazoan Hedgehog pathway evolution. Dev Biol 2008, 313:501–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fritzenwanker JH, Saina M, Technau U. Analysis of forkhead and snail expression reveals epithelial–mesenchymal transitions during embryonic and larval development of Nematostella vectensis . Dev Biol 2004, 275:389–402. [DOI] [PubMed] [Google Scholar]
- 91. Ormestad M, Martindale MQ, Röttinger E. A comparative gene expression database for invertebrates. Evodevo 2011, 2:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Botman D, Kaandorp JA. Spatial gene expression quantification: a tool for analysis of in situ hybridizations in sea anemone Nematostella vectensis . BMC Res Notes 2012, 5:555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Botman D, Röttinger E, Martindale MQ, de Jong J, Kaandorp JA. A Computational approach towards a gene regulatory network for the developing Nematostella vectensis gut. PLoS One 2014, 9:e103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Loose M, Patient R. A genetic regulatory network for Xenopus mesendoderm formation. Dev Biol 2004, 271:467–478. [DOI] [PubMed] [Google Scholar]
- 95. Hinman VF, Nguyen A, Davidson EH. Caught in the evolutionary act: precise cis‐regulatory basis of difference in the organization of gene networks of sea stars and sea urchins. Dev Biol 2007, 312:584–595. [DOI] [PubMed] [Google Scholar]
- 96. Hinman VF, Nguyen AT, Davidson EH. Expression and function of a starfish Otx ortholog, AmOtx: a conserved role for Otx proteins in endoderm development that predates divergence of the eleutherozoa. Mech Dev 2003, 120:1165–1176. [DOI] [PubMed] [Google Scholar]
- 97. Oliveri P, Walton KD, Davidson EH, McClay DR. Repression of mesodermal fate by foxa, a key endoderm regulator of the sea urchin embryo. Development 2006, 133:4173–4181. [DOI] [PubMed] [Google Scholar]
- 98. Ghysen A. The origin and evolution of the nervous system. Int J Dev Biol 2003, 47:555–562. [PubMed] [Google Scholar]
- 99. Watanabe H, Fujisawa T, Holstein TW. Cnidarians and the evolutionary origin of the nervous system. Dev Growth Differ 2009, 51:167–183. [DOI] [PubMed] [Google Scholar]
- 100. Moran Y, Genikhovich G, Gordon D, Wienkoop S, Zenkert C, Özbeck S, Technau U, Gurevitz M. Neurotoxin localization to ectodermal gland cells uncovers an alternative mechanism of venom delivery in sea anemones. Proc R Soc B Biol Sci 2012, 279:1351–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kass‐Simon G, Scappaticci AA Jr. The behavioral and developmental physiology of nematocysts. Can J Zool 2002, 80:1772–1794. [Google Scholar]
- 102. Watanabe H, Kuhn A, Fushiki M, Agata K, Özbek S, Fujisawa T, Holstein TW. Sequential actions of β‐catenin and Bmp pattern the oral nerve net in Nematostella vectensis . Nat Commun 2014, 5:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Richards GS, Rentzsch F. Transgenic analysis of a SoxB gene reveals neural progenitor cells in the cnidarian Nematostella vectensis . Development 2014, 141:4681–4689. [DOI] [PubMed] [Google Scholar]
- 104. Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci 2002, 3:517–530. [DOI] [PubMed] [Google Scholar]
- 105. Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, Degnan BM, Vervoort M. Origin and diversification of the basic helix‐loop‐helix gene family in metazoans: insights from comparative genomics. BMC Evol Biol 2007, 7:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zhao, G , Skeath JB. The Sox‐domain containing gene Dichaete/fish‐hook acts in concert with vnd and ind to regulate cell fate in the Drosophila neuroectoderm. Development 129: 1165–1174 (2002). [DOI] [PubMed] [Google Scholar]
- 107. Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron 2003, 39:749–765. [DOI] [PubMed] [Google Scholar]
- 108. Sandberg M, Källström M, Muhr J. Sox21 promotes the progression of vertebrate neurogenesis. Nat Neurosci 2005, 8:995–1001. [DOI] [PubMed] [Google Scholar]
- 109. Jager M, Quéinnec E, Le Guyader H, Manuel M. Multiple Sox genes are expressed in stem cells or in differentiating neuro‐sensory cells in the hydrozoan Clytia hemisphaerica. Evodevo 2011, 2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Magie CR, Pang K, Martindale MQ. Genomic inventory and expression of Sox and Fox genes in the cnidarian Nematostella vectensis . Dev Genes Evol 2005, 215:618–630. [DOI] [PubMed] [Google Scholar]
- 111. Marlow H, Roettinger E, Boekhout M, Martindale MQ. Functional roles of Notch signaling in the cnidarian Nematostella vectensis . Dev Biol 2012, 362:295–308. doi:10.1016/j.ydbio.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Layden MJ, Martindale MQ. Non‐canonical Notch signaling represents an ancestral mechanism to regulate neural differentiation. Evodevo 2014, 5:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Matus DQ, Thomsen GH, Martindale MQ. FGF signaling in gastrulation and neural development in Nematostella vectensis, an anthozoan cnidarian. Dev Genes Evol 2007, 217:137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Marlow H et al. Larval body patterning and apical organs are conserved in animal evolution. BMC Biol 2014, 12:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Nielsen C. Larval and adult brains. Evol Dev 2005, 7:483–489. [DOI] [PubMed] [Google Scholar]
- 116. Martindale MQ, Hejnol A. A developmental perspective: changes in the position of the blastopore during bilaterian evolution. Dev Cell 2009, 17:162–174. [DOI] [PubMed] [Google Scholar]
- 117. Binari LA, Lewis GM, Kucenas S. Perineurial glia require notch signaling during motor nerve development but not regeneration. J Neurosci 2013, 33:4241–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Nacu E, Tanaka EM. Limb regeneration: a new development? Annu Rev Cell Dev Biol 2011, 27:409–440. [DOI] [PubMed] [Google Scholar]
- 119. Carlson MRJ, Komine Y, Bryant SV, Gardiner DM. Expression of Hoxb13 and Hoxc10 in developing and regenerating Axolotl limbs and tails. Dev Biol 2001, 229:396–406. [DOI] [PubMed] [Google Scholar]
- 120. Gemberling M, Bailey TJ, Hyde DR, Poss KD. The zebrafish as a model for complex tissue regeneration. Trends Genet 2013, 29:611–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Passamaneck YJ, Martindale MQ. Cell proliferation is necessary for the regeneration of oral structures in the anthozoan cnidarian Nematostella vectensis . BMC Dev Biol 2012, 12:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Darling JA et al. Rising starlet: the starlet sea anemone, Nematostella vectensis . Bioessays 2005, 27:211–221. [DOI] [PubMed] [Google Scholar]
- 123. Reitzel AM, Burton PM, Krone C, Finnerty JR. Comparison of developmental trajectories in the starlet sea anemone Nematostella vectensis: embryogenesis, regeneration, and two forms of asexual fission. Invertebr Biol 2007, 126:99–112. [Google Scholar]
- 124. Burton PM, Finnerty JR. Conserved and novel gene expression between regeneration and asexual fission in Nematostella vectensis . Dev Genes Evol 2009, 219:79–87. [DOI] [PubMed] [Google Scholar]
- 125. Trevino M, Stefanik DJ, Rodriguez R, Harmon S, Burton PM. Induction of canonical Wnt signaling by alsterpaullone is sufficient for oral tissue fate during regeneration and embryogenesis in Nematostella vectensis . Dev Dyn 2011, 240:2673–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Käsbauer T et al. The Notch signaling pathway in the cnidarian Hydra. Dev Biol 2007, 303:376–390. [DOI] [PubMed] [Google Scholar]


