Abstract
Objectives
To determine the underlying biological basis for noninvasive prenatal testing (NIPT) results of multiple aneuploidies or autosomal monosomies.
Methods
Retrospective analysis of 113,415 tests to determine the study cohort, consisting of 138 (0.12%) cases reported as a single autosomal monosomy (n = 65), single trisomy with a sex chromosome aneuploidy (n = 36), or with multiple aneuploidies (n = 37). Clinical outcome information was reviewed and stratified into eight categories according to whether the karyotype or sonographic information agreed or disagreed with sequencing results.
Results
Of 67 cases with fetal or neonatal karyotypes available, 16 (24%) were partially or fully concordant with the NIPT result, 4 (6%) had aneuploidy on a reference chromosome, and 47 (70%) had normal fetal chromosomes, in which 5/47 had maternal malignancies reported. One case of maternal mosaic trisomy 8 was also detected. Of cases with no fetal karyotype information, ten had an abnormal clinical outcome, one was a normal live birth, and one reported maternal malignancy.
Conclusions
Noninvasive prenatal test results of autosomal monosomy or multiple aneuploidies are rare but have a diversity of underlying biologic causes. Some reflect the fetal karyotype; some reflect the presence of other maternal or fetal chromosome abnormalities, and a small number are linked to maternal disease. © 2016 Illumina. Prenatal Diagnosis published by John Wiley & Sons, Ltd.
Short abstract
What's already known about this topic?
Noninvasive prenatal testing (NIPT) has been validated for common autosomal trisomies (trisomy 21, 18, and 13), sex chromosome aneuploidies, and a selection of microdeletion syndromes.
NIPT findings that are discordant with the fetal karyotype can be because of biological reasons, such as confined placental mosaicism, maternal chromosome abnormalities, and other maternal conditions such as occult malignancy.
What does this study add?
Clinical and karyotype outcome information for cases that received an NIPT result indicating an autosomal monosomy or multiple aneuploidies. Some autosomal monosomy and multiple aneuploidy results reflect the true fetal karyotype, and some are explained by other factors, such as other fetal or maternal chromosomal abnormalities or maternal disease.
This information will help providers with post‐test counseling for these rare and unusual results.
Introduction
Cell‐free DNA (cfDNA)‐based noninvasive prenatal testing (NIPT) has been adopted into clinical care for pregnant women undergoing screening for the most common fetal aneuploidies: trisomies 13, 18, 21, and sex chromosome aneuploidies. More recently, test menus have expanded to include additional, elective test options, such as select microdeletion syndromes. Although most patients receive a negative (no aneuploidy detected) result or a positive (aneuploidy detected/aneuploidy suspected) result for one of the common aneuploidies, some patients receive unexpected findings, such as autosomal monosomy or multiple aneuploidies.
Most full fetal autosomal monosomies are not compatible with extrauterine life1; however, partial and mosaic cases of autosomal monosomy have been reported in liveborns.2 Although an estimated 0.16% of trisomy 21 cases involve a double aneuploidy with a sex chromosome (XXX, XXY, XYY, or monosomy X),3 conceptions with multiple aneuploidies are unlikely to result in a clinically recognized pregnancy or a viable birth.4, 5 A recent study of cytogenetic analysis following spontaneous miscarriage showed that 5% of samples were affected with multiple aneuploidies.5 Additionally, several reports have linked NIPT results of multiple aneuploidies and autosomal monosomy to occult maternal benign and malignant tumors.6, 7, 8, 9, 10 Thus, for clinicians and patients receiving these unexpected and uncommon results, there can be questions as to their clinical significance and confusion regarding interpretation and subsequent management.
Because of the rarity of autosomal monosomies and multiple aneuploidies in viable pregnancies and the recent studies linking NIPT results of autosomal monosomy and multiple aneuploidies to maternal disease, we performed a descriptive study to determine the underlying biological basis for these unusual results.
Methods
This study consisted of a retrospective analysis of test reports on clinical samples submitted for the verifi® prenatal aneuploidy screening test at the College of American Pathologists‐accredited and Clinical Laboratory Improvement Act‐certified Illumina Laboratory (Redwood City, CA). This was a retrospective study performed on previously existing clinical data that were de‐identified and aggregated prior to analysis and thereby qualifies for exemption from investigational review board (IRB) review as per the regulatory requirements in Code of Federal Regulations Title 45: Public Welfare, part 46. For the maternal malignancy cases, an IRB review waiver was obtained from Tufts Medical Center, as previously described.6
The clinical cohort comprised 113,415 consecutive patient samples reported by the clinical laboratory between February, 2012 and August, 2014. cfDNA was analyzed using massively parallel whole‐genome sequencing to screen for fetal aneuploidies on chromosomes 13, 18, 21, X, and Y, as previously described.11, 12, 13, 14, 15, 16 The study cohort consisted of the subset of samples from women carrying singletons whose tests were reported as a single autosomal monosomy or multiple aneuploidies (two or more autosomal aneuploidies or an autosomal aneuploidy plus a sex chromosome aneuploidy) detected or suspected. This study cohort included six previously published cases originating from pregnancies with an occult maternal malignancy.6
Clinical outcome information was actively requested (by fax and phone) for all samples with abnormal results, according to our previously described standard laboratory practices and quality procedures.6, 11 Cases were categorized as (1) ‘fully concordant’, if NIPT results completely matched fetal, neonatal, or products of conception (POC) karyotype results [true positive (TP)]; (2) ‘partially concordant’, if NIPT reported two or more aneuploidies and at least one, but not all, were confirmed by fetal, neonatal, or POC karyotype; (3) ‘discordant’, if NIPT results did not match the fetal, neonatal, or POC karyotype or birth outcome [false positive (FP)]. If the placental karyotype matched the abnormal NIPT result (placental TP) but the fetal karyotype was euploid, indicating the presence of confined placental mosaicism (CPM), the case was classified as discordant (fetal FP); (4) ‘maternal malignancy’, if NIPT results were attributed to post‐test disclosure of a maternal malignancy; (5) ‘other’, if an aneuploidy (either fetal or maternal) was present on a chromosome other than the test chromosomes: 13, 18, 21, X, or Y; (6) ‘ultrasound findings, no karyotype’, if ultrasound findings supporting the NIPT result were reported, but karyotype confirmation was not available because of patient choice, a spontaneous miscarriage, or elective pregnancy termination; (7) ‘spontaneous pregnancy loss, no karyotype’, if a spontaneous miscarriage or fetal demise occurred without confirmatory karyotype analysis, and no abnormal ultrasound findings were reported (ultrasound evaluation may or may not have been performed); (8) ‘no information’, if outcome information was unavailable.
Results
Demographics
Within the sample of 113,415 reported cases, 138 (0.12%) fit the criteria for inclusion in this study. Of these, 65 were reported as a single autosomal monosomy, 36 with a single autosomal trisomy with sex chromosome aneuploidy, and 37 with multiple aneuploidies (Figure 1). The mean maternal age for these 138 cases was 35.4 years (range: 19–47 years), and the mean gestational age was 13 weeks (range: 10–34 weeks). Clinical follow‐up information was available for 79/138 cases (57%) (Table 1).
Figure 1.
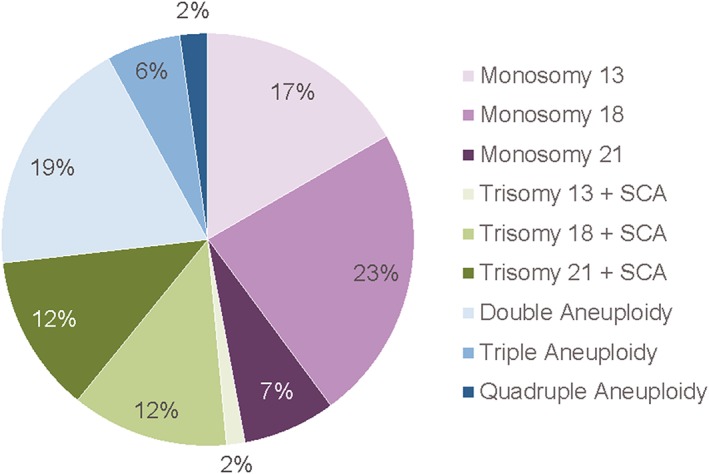
The relative percentages of the different types of aneuploidies represented in the study cohort (n = 138)
Table 1.
Clinical outcomes for 79 cases of autosomal monosomy and multiple aneuploidy calls with clinical outcome information
| Full concordance | Partial concordance | Discordant | Maternal malignancy | Other | Ultrasound findingsa | Spontaneous lossb | Total | |
|---|---|---|---|---|---|---|---|---|
| Single autosomal monosomy | ||||||||
| Monosomy 13 | 1 | 0 | 2 | 0 | 2 | 2 | 0 | 7 |
| Monosomy 18 | 0 | 0 | 13 | 1 | 1 | 0 | 1 | 16 |
| Monosomy 21 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 4 |
| Single autosomal trisomy + SCA | ||||||||
| Trisomy 13 + SCA | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 2 |
| Trisomy 18 + SCA | 1 | 0 | 7 | 0 | 0 | 2 | 0 | 10 |
| Trisomy 21 + SCA | 1 | 8 | 2 | 0 | 0 | 2 | 1 | 14 |
| Multiple aneuploidies | ||||||||
| Double aneuploidyc | 0 | 4 | 9 | 2 | 2 | 1 | 0 | 18 |
| Triple aneuploidyd | 0 | 0 | 4 | 1 | 0 | 0 | 0 | 5 |
| Quadruple aneuploidye | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 3 |
| Total | 3 | 13 | 42 | 6 | 5 | 8 | 2 | 79 |
SCA, sex chromosome aneuploidy.
Ultrasound findings were reported, but karotype information was not available because of patient choice, a spontaneous miscarriage, or elective pregnancy termination.
Patient experienced a spontaneous loss with no ultrasound findings reported and no karotype information.
Two autosomal aneuploidies (chromosomes 13 and 18, 13 and 21, or 18 and 21) or a single autosomal monosomy with a sex chromosome aneuploidy.
Three autosomal aneuploidies (chromosomes 13, 18, and 21) or two autosomal aneuploidies with a sex chromosome aneuploidy.
Three autosomal aneuploidies (chromosomes 13, 18, and 21) with a sex chromosome aneuploidy.
Fetal or neonatal karyotype information
Fifty‐seven patients had either chorionic villus sampling or amniocentesis, and one patient had both, for a total of 59 invasive diagnostic procedures. Ten CVS and 42 amniocenteses were performed; in seven patients, the type of procedure was not specified. Karyotypes were also available for four POCs and five neonates; thus, fetal or neonatal karyotypes were available for 67 total cases. Of the 67 cases with karyotype information, 20 (30%) had an aneuploid fetal karyotype: 3 (4%) were fully concordant and 13 (19%) were partially concordant with the NIPT results (Table 2), and 4 (6%) cases had an aneuploidy for a nontest chromosome (Table 3).
Table 2.
Clinical indications and outcomes for cases with full or partial concordance between the fetal karyotype and NIPT result
| NIPT resulta | NIPT indication | Karyotype method | Karyotype result | Concordance |
|---|---|---|---|---|
| Monosomy 13 | U/Sb | Postnatal blood | 46,XX: 9.3 Mb deletion on Chr 13q | Full |
| Trisomy 18, XXY | AMA, U/S | CVS | 48,XXY + 18 | Full |
| Trisomy 21, XXX | AMA | Amnio | 48,XXX + 21 | Full |
| Monosomy 21 | AMA | Amnio; maternal blood |
Fetal: 47,XX,del(21)(q11.2‐q21),+r(21)(p11.2‐q21) Maternal: 46,XX,del(21)(q11.2q21)[4]/46,XX,del(21)(q11.2q21)r(21)(p11.2‐q21)[26] |
Partial |
| Trisomy 21, monosomy X | U/S | Amnio | 47,XX + 21 | Partial |
| Trisomy 21, monosomy X | +MSS | Amnio | 47,XX + 21 | Partial |
| Trisomy 21, monosomy X | AMA | CVS | 47,XX + 21 | Partial |
| Trisomy 21, monosomy X | AMA | Amnio | 47,XX + 21 | Partial |
| Trisomy 21, monosomy X | AMA, U/Sc | CVS | 47,XX + 21 | Partial |
| Trisomy 21, monosomy X | +MSS | Amnio | 47,XX + 21 | Partial |
| Trisomy 21, monosomy X | AMA, +MSS, U/Sd | Amnio | 47,XX + 21 | Partial |
| Trisomy 21, monosomy X | AMA | Postnatal | 47 + 21, sex not reportede | Partial |
| Trisomy 13, trisomy 18 suspected | n.s. | POC | 47 + 13, sex not reported | Partial |
| Trisomy 18, trisomy 21 | +MSS, U/Sf | Postnatal blood |
Fetal: 47 + 21 Placental: mosaic 47 + 18 |
Partial |
| Trisomy 18, trisomy 21 suspected | U/Sg | POC | 47 + 18, sex not reported | Partial |
| Trisomy 18, trisomy 21 suspected | Previous History | POC | 47,XX + 18 | Partial |
AMA, advanced maternal age (>35 years at estimated date of conception); Amnio, amniocentesis; Chr, chromosome; CVS, chorionic villus sampling; n.s., not specified; +MSS, positive maternal serum screen; U/S, abnormal ultrasound findings.
Unless otherwise stated, all autosomal aneuploidies were reported as aneuploidy detected.
Dandy–Walker malformation, clinodactyly.
Increased nuchal translucency.
Absent nasal bone, duodenal atresia.
Co‐twin demise reported at 9 weeks.
Echogenic intracardiac focus, short long bones.
Choroid plexus cyst, unspecified congenital heart defect, agenesis of the corpus callosum, and diaphragmatic.
Table 3.
Clinical indications, karyotypes, and birth outcomes for cases with discordant clinical outcomes explained by maternal aneuploidy or other fetal aneuploidy
| NIPT resulta | NIPT indication | Karyotype source | Karyotype | Birth outcome |
|---|---|---|---|---|
| Monosomy 13 | AMA | Amnio | Fetal: mosaic 46/47 + 2b | Preterm delivery |
| Monosomy 13 | U/Sc | Amnio | Fetal: mosaic 46/47 + 6b | IUFD |
| Monosomy 18 | AMA | POC | Fetal: 47,XY + 14 | SAB |
| Monosomy 13, monosomy 18 | AMA | CVS | Fetal: mosaic 46,XY,t(2;10)(q21;q26)[14]/47,XY, t(2;10)(q21;q26),+7[6] | Unknown |
| Monosomy 13, monosomy 18 | AMA | Maternal blood | Maternal: mosaic 46,XX[26]/47,XX + 8[8] | Unknown |
AMA, advanced maternal age (>35 years at estimated date of conception); Amnio, amniocentesis; CVS, chorionic villus sampling; IUFD, intrauterine fetal demise; POC, products of conception testing; SAB, spontaneous abortion; U/S, abnormal ultrasound findings.
All autosomal monosomies results were reported as aneuploidy detected.
Sex chromosome results were not reported to the laboratory.
Unspecified congenital heart defect, pyelectasis, abnormal feet, echogenic bowel, clenched hands.
Within the group of 13 partially concordant cases, there were two with unusual findings that provide a biological explanation for the partial concordance. The first was reported as trisomies 18 and 21, in which the postnatal neonatal karyotype confirmed trisomy 21 and the placental analysis revealed mosaicism for trisomy 18. Circulating placental cfDNA with an extra copy of chromosome 18 was presumably the explanation for the test result of trisomy 18. In the second case, monosomy 21 was reported and the fetal karyotype was consistent with a partial loss and partial gain of chromosome 21 (47,XX,del(21)(q11.2‐q21),+r(21)(p11.2‐q21). In this case, the monosomy 21 call was explained by the partial deletion. The remaining cases with karyotype information had normal fetal karyotypes (n = 47). Unless otherwise specified, placental karyotypes were not routinely available.
A fetal origin for the NIPT result was present in 30% (20/67) of cases with karyotypes. The frequency of fetal concordance varied with the type of NIPT result (Figure 2). Fetal abnormalities (partial and full concordance) were highest in the cases reported as a single trisomy with a sex chromosome aneuploidy and lowest in the cases reported as a single autosomal monosomy (p < 0.05). Four cases of single autosomal monosomies could be explained by a fetal chromosome abnormality in a reference chromosome. Although the reported results differed from the fetal karyotypes, the positive screen results prompted the further diagnostic workup that detected the true abnormality.
Figure 2.
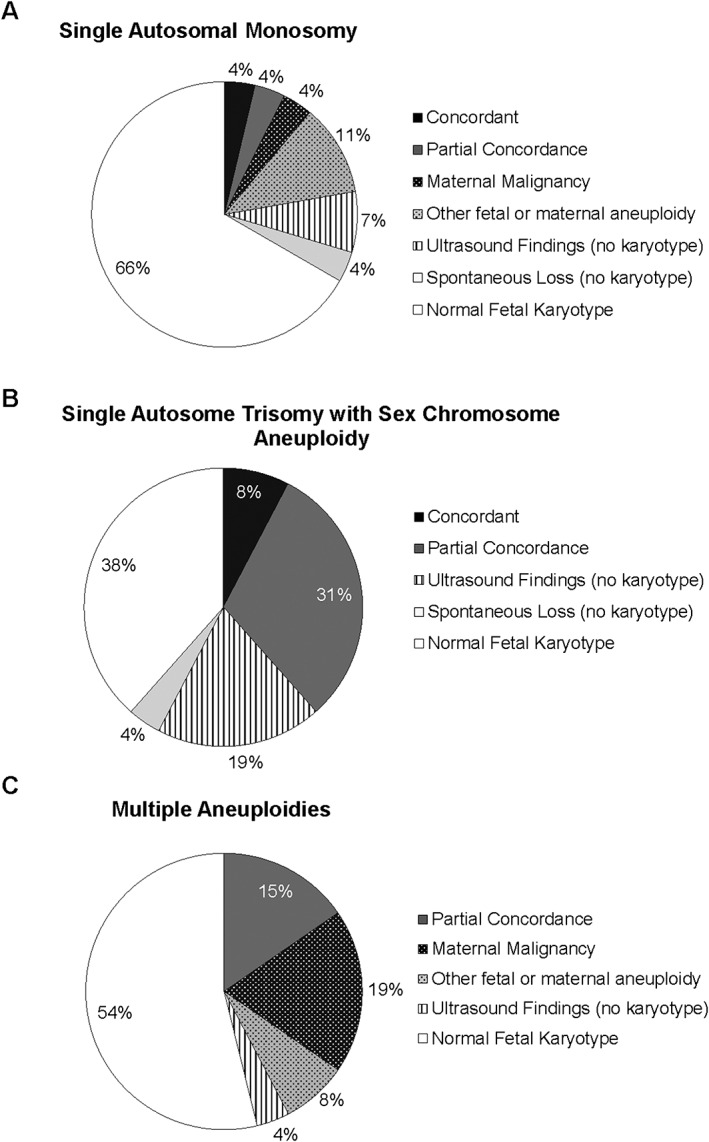
Clinical outcomes for NIPT cases reported as a single autosomal monosomy (A, n = 27), a single autosome trisomy with a sex chromosome aneuploidy (B, n = 26), and as multiple aneuploidies (C, n = 26)
Fetal and neonatal clinical information
There were eight cases with abnormal ultrasound findings and no karyotype information and two cases with a pregnancy loss but no karyotype information. The clinical outcomes for these cases are described in Table 4. This included one case with a known paternal Robertsonian translocation of chromosomes 13 and 21, supporting the NIPT result of monosomy 13. One additional case without karyotype information resulted in the birth of an apparently healthy newborn.
Table 4.
Clinical indications and history for cases with ultrasound findings or a spontaneous pregnancy loss without karyotype information
| NIPT resulta | NIPT indication | Ultrasound findings | Birth outcome |
|---|---|---|---|
| Monosomy 13 | U/S | SGA, oligohydramnios | TOP |
| Monosomy 13 | U/S | Cystic hygroma, encephalocele, known paternal translocation (45,XY, rob(13;21)(q22;q31.1) | TOP |
| Trisomy 13 suspected, monosomy X | U/S | Cystic hygroma, pleural effusion | TOP |
| Trisomy 18, monosomy X | +MSS | Increased nuchal translucency | IUFD |
| Trisomy 18, monosomy X | U/S | Cystic hygroma | SAB |
| Trisomy 21, monosomy X | AMA, +MSS, U/S | Increased nuchal translucency | TOP |
| Trisomy 21, XXX | U/S | SGA (with decreasing βHCG levels) | SAB |
| Trisomy 21 suspected, trisomy 13 suspected | AMA, +MSS, U/S | Possible VSD | IUFD |
| Monosomy 18 | n.s. | None reported | SAB |
| Trisomy 21, XXY | AMA | None reported | SAB |
AMA, advanced maternal age (>35 years at estimated date of conception); IUFD, intrauterine fetal demise; n.s., not specified; +MSS, positive maternal serum screen; SAB, spontaneous abortion; SGA, small for gestational age; TOP, termination of pregnancy; U/S, abnormal ultrasound findings; VSD, ventricular septal defect
Unless otherwise stated, all autosomal aneuploidies were reported as aneuploidy detected.
Maternal information
Seven cases of discordance were likely of maternal origin. Of these, one was attributed to maternal mosaicism for trisomy 8 (Table 3); fetal karyotype is unknown. In the other six cases, maternal malignancy was subsequently detected: Five cases had normal fetal/neonatal karyotypes, and in one case, a full‐term infant was delivered with no phenotypic signs of aneuploidy (Supplementary Table 1). Five of these cases were described in detail in a recent publication; the leiomyosarcoma case was one of ten cases in that study that reported maternal malignancy back to the laboratory, but a detailed analysis was not performed for this patient.6 The frequency of maternal malignancy varied with the type of NIPT result (Figure 2), with the highest prevalence in cases with multiple aneuploidies and the lowest in cases reported as a single trisomy with a sex chromosome aneuploidy (p < 0.05).
Overall outcomes
Overall, 61/79 (77%) cases with known outcomes had only fetal cytogenetic information available (no maternal karyotype); 6/79 (8%) cases had both fetal cytogenetic and maternal information available (karyotype or disease); 2/79 (3%) cases had only maternal information (karyotype or disease) available (no fetal karyotype); 10/79 (13%) cases had either ultrasound findings or reported spontaneous loss without any karyotype information. Of these 79 cases with clinical or karyotype outcome information, 20/79 (25%) had a definite fetal abnormality as demonstrated by karyotype, 10/79 (13%) had a potential fetal abnormality as demonstrated by ultrasound abnormality or miscarriage, 7/79 (9%) had a maternal etiology for the abnormal NIPT results, and 42/79 (53%) were discordant with the fetal karyotype and remain unexplained.
Discussion
The increasing number of cfDNA sequencing tests being performed during pregnancy has led to the identification of rare and unusual results, including uncommon fetal aneuploidies and maternal malignancies.6, 8 In light of this and because fetal aneuploidies of this type have limited potential for extrauterine survival, we sought to determine the frequency for which there was a fetal versus maternal origin for these unusual NIPT results. In this study, we demonstrated that there were multiple etiologies for a single monosomy or multiple aneuploidies.
The strength of this study is that it provides a resource for providers' counseling patients with these unusual NIPT results. Despite the small cohort size, the outcomes do include several possible explanations for atypical NIPT results that providers may consider when addressing the potential underlying explanations for each patient's unique test result. The weakness is that the sample size is small and the outcomes were incomplete. However, obtaining outcomes for clinical cases remains a challenge for all clinical laboratories performing analysis of cfDNA. Our data demonstrate that diagnostic procedures that analyze the fetal karyotype, but not the placenta, likely result in an underestimation of biological causes for discordant NIPT results. Thus, as suggested by professional societies and others,17, 18 we support the development of national registries that link NIPT results with clinical follow‐up data.
For the 42 discordant cases with normal fetal karyotypes and no reported maternal disease, the NIPT result might reflect CPM,19 residual cfDNA from an unrecognized vanished twin,20 or a maternal copy number variant.21, 22 Grati et al. determined the potential contribution of CPM to the NIPT false‐positive rate, demonstrating that chromosomes 13 and X were more likely to be associated with CPM than chromosomes 18 and 21.23 There was one suspected case of CPM in our dataset, a partially concordant case that had postnatal placental analysis. However, postnatal placental analysis is not usually performed, so for patients with discordant fetal karyotypes, the contribution of placental aneuploidy is unknown. If concerns about CPM arise following confirmatory CVS, follow‐up amniocentesis could be considered. Here, 30/42 (71%) of discordant results without a reported maternal explanation were determined by amniocentesis, so undetected CPM might be a contributing factor. Possible alternative maternal explanations for the FP results include maternal copy number variations (CNVs)21, 22 and maternal 45,X/46,XX mosaicism, some of which may be secondary to maternal age‐related loss of the X chromosome.23, 24 Recent updates to the informatics algorithm used by the clinical laboratory enable algorithmic correction for maternal subchromosome CNVs, preventing these maternal events from causing false‐positive results.
Interestingly, 23.3% (17/73) of the multiple aneuploidy results were reported as trisomy 21 with a sex chromosome aneuploidy (13 monosomy X, 3 XXX, 1 XXY). This combination has been previously described in surviving infants, with an estimated 0.16% of trisomy 21 cases also having a sex chromosome aneuploidy.3 Based on the single, confirmed case of trisomy 21 with a sex chromosome aneuploidy and the general prevalence of trisomy 21 detected/suspected results in this clinical cohort (data not shown), the overall prevalence of trisomy 21 with a sex chromosome aneuploidy is in line with published rates.3
The results reported here reinforce the importance of confirmatory diagnostic testing and review of the patient's clinical history following any abnormal or unusual NIPT result. For clinicians counseling patients on these unusual NIPT results, Figure 2 can be used as a guide to the potential rates of maternal disease and fetal concordance by result type. If diagnostic testing results indicate an unaffected fetus, then other potential biological explanations (CPM, vanishing twin, maternal chromosome abnormality, or maternal disease) can be considered. Furthermore, as suggested by Bianchi et al.,6 for cases with whole‐genome sequencing results available, an advanced bioinformatics review may be warranted if the fetal or neonatal karyotype is normal. This may demonstrate that the unusual NIPT result is because of genomic imbalance of a nontest chromosome.
Conclusion
In conclusion, although most abnormal NIPT test results indicate a single trisomy, it is important for patients and clinicians to be aware that in rare instances, other results, including autosomal monosomy and multiple aneuploidies, can be reported. Some of these results reflect the fetal karyotype, and a small number reflect maternal disease, but some may be explained by other factors, such as other maternal or fetal chromosome abnormalities or mosaicism. Importantly, NIPT is a screening test, so FPs can occur. By communicating the clinical outcomes of these cases, we aim to inform providers of the various underlying biological reasons for the results and therefore assist with post‐test counseling and management.
Supporting information
Supporting info item
Acknowledgements
The authors thank Meredith Halks‐Miller, Patricia Taneja, and Patricia Winters for their assistance with data generation and analysis.
What's Already Known About This Topic?
Noninvasive prenatal testing (NIPT) has been validated for common autosomal trisomies (trisomy 21, 18, and 13), sex chromosome aneuploidies, and a selection of microdeletion syndromes.
NIPT findings that are discordant with the fetal karyotype can be because of biological reasons, such as confined placental mosaicism, maternal chromosome abnormalities, and other maternal conditions such as occult malignancy.
What Does This Study Add?
Clinical and karyotype outcome information for cases that received an NIPT result indicating an autosomal monosomy or multiple aneuploidies. Some autosomal monosomy and multiple aneuploidy results reflect the true fetal karyotype, and some are explained by other factors, such as other fetal or maternal chromosomal abnormalities or maternal disease.
This information will help providers with post‐test counseling for these rare and unusual results.
Snyder, H. L. , Curnow, K. J. , Bhatt, S. , and Bianchi, D. W. (2016) Follow‐up of multiple aneuploidies and single monosomies detected by noninvasive prenatal testing: implications for management and counseling. Prenat Diagn, 36: 203–209. doi: 10.1002/pd.4778.
Funding sources: This study was based on samples submitted for clinical testing. Testing costs were paid by the patients in terms of copay and/or their insurance company to the Illumina Laboratory or one of their distributors.
Conflicts of interest: Ms. Snyder, Dr. Curnow, and Dr. Bhatt are employees of and hold equity in Illumina. Dr. Bianchi receives an honorarium for her role on the Clinical Advisory Board of the Reproductive and Genetic Health Unit at Illumina and sponsored research funding from Illumina that is administered through Tufts Medical Center.
References
- 1. Manolakos E, Peitsidis P, Eleftheriades M, et al. Prenatal detection of full monosomy 21 in a fetus with increased nuchal translucency: molecular cytogenetic analysis and review of the literature. J Obstet Gynaecol Res 2010;36(2):435–40. [DOI] [PubMed] [Google Scholar]
- 2. Burgess T, Downie L, Pertile MD, et al. Monosomy 21 seen in live born is unlikely to represent true monosomy 21: a case report and review of the literature. Case Rep Genet 2014;2014965401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kovaleva NV, Mutton DE. Epidemiology of double aneuploidies involving chromosome 21 and the sex chromosomes. Am J Med Genet A 2005;134A(1):24–32. [DOI] [PubMed] [Google Scholar]
- 4. Reddy KS. Double trisomy in spontaneous abortions. Hum Genet 1997;101(3):339–45. [DOI] [PubMed] [Google Scholar]
- 5. Subramaniyam S, Pulijaal VR, Mathew S. Double and multiple chromosomal aneuploidies in spontaneous abortions: a single institutional experience. J Hum Reprod Sci 2014;7(4):262–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bianchi DW, Chudova D, Sehnert AJ, et al. Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA 2015;314(2):162–9. [DOI] [PubMed] [Google Scholar]
- 7. Osborne CM, Hardisty E, Devers P, et al. Discordant noninvasive prenatal testing results in a patient subsequently diagnosed with metastatic disease. Prenat Diagn 2013;33(6):609–11. [DOI] [PubMed] [Google Scholar]
- 8. McCullough RM, Almasri EA, Guan X, et al. Non‐invasive prenatal chromosomal aneuploidy testing‐‐clinical experience: 100,000 clinical samples. PLoS One 2014;9(10e109173). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Amant F, Verheecke M, Wlodarska I, et al. Presymptomatic identification of cancers in pregnant women during noninvasive prenatal testing. JAMA Oncol 2015;1(6):814–9. [DOI] [PubMed] [Google Scholar]
- 10. Dharajiya NG, Namba A, Horiuchi I, et al. Uterine leiomyoma confounding a noninvasive prenatal test result. Prenat Diagn 2015;35(10):990–3. [DOI] [PubMed] [Google Scholar]
- 11. Bianchi DW, Parsa S, Bhatt S, et al. Fetal sex chromosome testing by maternal plasma DNA sequencing: clinical laboratory experience and biology. Obstet Gynecol 2015;125(2):375–82. [DOI] [PubMed] [Google Scholar]
- 12. Futch T, Spinosa J, Bhatt S, et al. Initial clinical laboratory experience in noninvasive prenatal testing for fetal aneuploidy from maternal plasma DNA samples. Prenat Diagn 2013;33(6):569–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bianchi DW, Platt LD, Goldberg JD, et al. Genome‐wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol 2012;119(5):890–901. [DOI] [PubMed] [Google Scholar]
- 14. Rava RP, Srinivasan A, Sehnert AJ, et al. Circulating fetal cell‐free DNA fractions differ in autosomal aneuploidies and monosomy X. Clin Chem 2013;60(1):243–50. [DOI] [PubMed] [Google Scholar]
- 15. Sehnert AJ, Rhees B, Comstock D, et al. Optimal detection of fetal chromosomal abnormalities by massively parallel DNA sequencing of cell‐free fetal DNA from maternal blood. Clin Chem 2011;57(7):1042–9. [DOI] [PubMed] [Google Scholar]
- 16. Halks‐Miller M, Chudova D, Bianchi DW. Maternal malignancies detected with noninvasive prenatal testing‐reply. JAMA 2015;314(20):2192–3. [DOI] [PubMed] [Google Scholar]
- 17. Benn P, Borell A, Chiu R, et al. Position statement from the Aneuploidy Screening Committee on behalf of the Board of the International Society for Prenatal Diagnosis. Prenat Diagn 2013;33(7):622–9. [DOI] [PubMed] [Google Scholar]
- 18. Gregg AR, Gross SJ, Best RG, et al. ACMG statement on noninvasive prenatal screening for fetal aneuploidy. Genet Med 2013;15(5):395–8. [DOI] [PubMed] [Google Scholar]
- 19. Choi H, Lau TK, Jiang FM, et al. Fetal aneuploidy screening by maternal plasma DNA sequencing: 'false positive' due to confined placental mosaicism. Prenat Diagn 2013;33(2):198–200. [DOI] [PubMed] [Google Scholar]
- 20. Curnow KJ, Wilkins‐Haug L, Ryan A, et al. Detection of triploid, molar, and vanishing twin pregnancies by a single‐nucleotide polymorphism‐based noninvasive prenatal test. Am J Obstet Gynecol 2015;212(1):79 e1–9. [DOI] [PubMed] [Google Scholar]
- 21. Snyder MW, Simmons LE, Kitzman JO, et al. Copy‐number variation and false positive prenatal aneuploidy screening results. N Engl J Med 2015;372(17):1639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Flowers N, Kelley J, Sigurjonsson S, et al. Maternal mosaicism for a large segmental duplication of 18q as a secondary finding following non‐invasive prenatal testing and implications for test accuracy. Prenat Diagn 2015;35(10):986–9. [DOI] [PubMed] [Google Scholar]
- 23. Grati FR, Bajaj K, Malvestiti F, et al. The type of feto‐placental aneuploidy detected by cfDNA testing may influence the choice of confirmatory diagnostic procedure. Prenat Diagn 2015;35(10):994–8. [DOI] [PubMed] [Google Scholar]
- 24. Russell LM, Strike P, Browne CE, et al. X chromosome loss and ageing. Cytogenet Genome Res 2007;116(3):181–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting info item


