Abstract

Reduced susceptibility to antimicrobials in Gram-negative bacteria may result from multiple resistance mechanisms, including increased efflux pump activity or reduced porin protein expression. Up-regulation of the efflux pump system is closely associated with multidrug resistance (MDR). To help investigate the role of efflux pumps on compound accumulation, a fluorescence-based assay was developed using fluorescent derivatives of trimethoprim (TMP), a broad-spectrum synthetic antibiotic that inhibits an intracellular target, dihydrofolate reductase (DHFR). Novel fluorescent TMP probes inhibited eDHFR activity with comparable potency to TMP, but did not kill or inhibit growth of wild type Escherichia coli. However, bactericidal activity was observed against an efflux pump deficient E. coli mutant strain (ΔtolC). A simple and quick fluorescence assay was developed to measure cellular accumulation of the TMP probe using either fluorescence spectroscopy or flow cytometry, with validation by LC-MS/MS. This fluorescence assay may provide a simple method to assess efflux pump activity with standard laboratory equipment.
Keywords: antibiotics, efflux pump, fluorescent probes, cellular accumulation, trimethoprim
The rise of multidrug-resistant (MDR) bacteria is causing significant healthcare issues.1 These MDR bacteria typically employ a combination of four main strategies to combat antibiotics: (i) prevention of intracellular antibiotic accumulation via efflux pumps and decreased outer membrane permeability; (ii) target modification; (iii) antibiotic inactivation; and (iv) acquisition of alternate metabolic pathways.2,3 Gram-negative bacteria are usually less susceptible to antibiotics compared to Gram-positive bacteria due to their dual membrane structure, combined with facile up-regulation of efflux pumps so that intracellular antibiotic concentrations do not reach cytotoxic concentrations.4,5 The lack of activity of many narrow-spectrum Gram-positive selective antibiotics against Gram-negative bacteria is often caused by efflux mechanisms.6 For example, it has been shown that Gram-negative bacteria become susceptible to linezolid, clarithromycin, and erythromycin7 when specific efflux pump genes have been deleted.8 In particular, the efflux effect has implications for the design of new antibiotic drugs,9 as potentially unique antibiotic structures or those acting on novel targets may be discarded on the basis of poor activity in minimum inhibitory concentration (MIC) assays resulting from high efflux. It may be possible to develop these as new antibiotics if co-administered with an efflux pump inhibitor or to optimize their structure to remove efflux pump susceptibility and increase their ability to diffuse through porin channels.10 Improving our understanding of how whole cell activity correlates with in vitro enzyme inhibition requires assessment of the intracellular accumulation of antibiotics and the role of efflux pumps.
Bacterial efflux pumps are membrane transporter proteins that facilitate the uptake and excretion of compounds11,12 and play important roles in cell-to-cell communication, bacterial pathogenicity, and biofilm formation.12,13 There are four main protein families for efflux pump systems: ATP-binding cassette superfamily (ABC) proteins, the major facilitator superfamily (MFS) transport proteins, small multidrug resistance (SMR) transporters, and multidrug and toxic compound extrusion (MATE) proteins, which are all present in both Gram-positive and Gram-negative bacteria. A fifth family, the resistance nodulation cell division (RND) pump, is found only in Gram-negative bacteria. Efflux pumps can contain multiple components, with an inner membrane transporter, a periplasmic adaptor protein, and an outer membrane channel that together are able to directly extrude various drugs from inside to outside the cell.4,8,13,14 Some efflux pumps are specific for particular substrates, whereas other efflux pumps can transport a wide range of substrates with various chemical structures. Up-regulation of efflux pumps contributes to the development of MDR bacteria.10,14
Fluorescent probes are versatile tools for biological study imaging, with different color combinations used to investigate specific target sites and to show biological events in living cells.15 For example, antibiotic fluorescent probes have been used to investigate cell division in Gram-positive bacteria.16−20 Detection of fluorescence can be used for both quantitative and qualitative analysis21 of the labeled cell,22 bacteria,23 or nanoparticles24 and also to detect bacterial infections in vivo.25
We have initiated a program to develop fluorescent probes based on all major classes of antibiotics. To prepare antibiotic-derived fluorescent probes, it is necessary to identify a suitable position on the antibiotic to link with the fluorophore so that the new derivative maintains antimicrobial activity. Published structure–activity relationship (SAR) studies can pinpoint sites that are able to tolerate structural changes. As antibiotics often possess multiple functional groups with various reactivities, we need to employ an orthogonal reaction that is compatible with various functional groups to systematically prepare fluorescent antibiotics of all major antibiotic classes. One such reaction is the Cu-catalyzed azide–alkyne cycloaddition (CuAAC) reaction that forms a stable and biocompatible triazole ring in the presence of unprotected functional groups (such as amines, hydroxyls, and thiols) or reactive moieties (such as lactams, epoxides, or α,β-unsaturated systems).26 The precursor azide and alkyne groups are known for their biocompatibility and selective reactivity as bio-orthogonal chemical reporters. For example, azides have been installed in the outer membrane (OM) of bacteria and also embedded within glycans. The resulting azido residues were labeled with various probes via cycloaddition with alkynes using either copper(I)-mediated or strain-promoted [3+2] cycloadditions.27,28 Meanwhile, terminal alkyne and cyclooctyne substituents have been widely used to link with dyes/markers or nanoparticles for therapeutic and diagnostic applications.29−31 In this study, the azide functionality was selected for antibiotic modification, given the more facile introduction of an azide group to highly diverse antibiotic cores compared to an alkyne. These azide-functionalized antibiotics also have potential for the development of diagnostics or sensing applications.32
We recently described an oxazolidinone-derived (linezolid) azide-functionalized antibiotic intermediate that was conjugated with fluorophores, with the resulting probes then used to image Gram-positive bacteria.33 This same strategy has now been employed to prepare TMP fluorescent derivatives. TMP is a synthetic antibiotic that acts on an intracellular bacterial target, dihydrofolate reductase (DHFR). In clinical use, it is normally administered in combination with sulfamethoxazole (SMZ), which inhibits another step in the folic acid pathway. The TMP–SMZ combination is used to treat a wide range of infections caused by Gram-negative bacteria (such as the Enterobacteriaceae family and Haemophilus influenza) and Gram-positive bacteria (such as Staphylococcus aureus and Streptococcus pneumoniae).34,35 TMP fluorescent probes have previously been applied to a general strategy for labeling targets in mammalian cell live cell imaging; the protein of interest was tagged with Escherichia coli DHFR (eDHFR) and then labeled with a cell-permeable TMP probe.36−43 Nevertheless, there have been no reports of applying TMP fluorescent probes to bacterial cells.
We also describe the development of a simple fluorescence-based assay using flow cytometry or fluorescence spectroscopy to assess efflux pump activity that affects the accumulation of fluorescent-labeled TMP in E. coli (Figure 1). The fluorescence-based assay was validated by liquid chromatography–mass spectrometry (LC-MS/MS) quantification that measured probe cellular concentrations. These methods that can measure the cellular concentration of the fluorescent probes could potentially be used to screen for efflux inhibitors, assess up-regulation of efflux in antibiotic-resistant bacteria, and assist in new antibiotic development.
Figure 1.
Use of fluorescently labeled antibiotics to assess efflux in Gram-negative bacteria without cell lysis: (A) in vitro enzyme inhibition (IC50), enzyme binding and inhibition by fluorescent antibiotic; (B) whole cell activity (MIC), fluorescent probe loses MIC activity due to efflux pumps. Fluorescence intensity can quantify the efflux effect.
Results and Discussion
Design and Synthesis of TMP-Linked Fluorophores
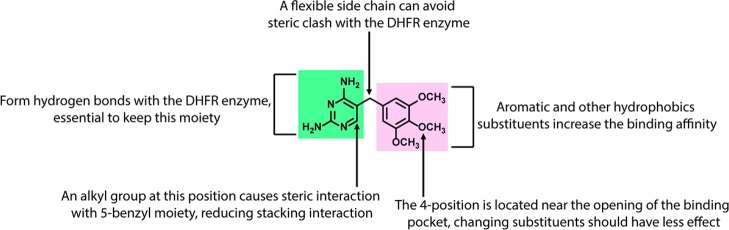
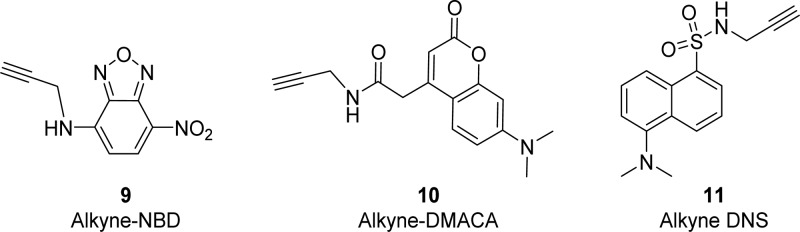
On the basis of published SAR of TMP derivatives and fluorescent TMP probes that have previously been prepared,44 we proposed that modification at position 4 of the TMP phenyl ring was possible without sacrificing significant activity (Figure 2).41,45,46 As antibiotics and fluorophores often possess reactive moieties and multiple functional groups with varied reactivity, an orthogonal reaction compatible with reactive moieties and many different functional groups was required. TMP has unprotected amines that can act as nucleophiles (reactive for alkylation and amide condensation reactions).47 Moreover, fluorophores such as coumarin-based fluorophores and rhodamine derivatives contain heterocyclic rings, which are sensitive to ring opening.48 In line with our overall goal of producing a “toolset” of azide-functionalized antibiotics that can be readily linked with different colored fluorophores, we selected the CuAAC reaction for antibiotic–probe conjugation. Low molecular weight fluorophores are more suitable for retaining antimicrobial activity as large highly hydrophobic fluorophores are likely to aggregate and have difficulty in penetrating through the bacterial membrane, as well as potentially disrupting DHFR target binding. Therefore, three comparatively small fluorophores, 7-(dimethylamino)-coumarin-4-acetic acid (DMACA), 7-nitrobenzofurazan (NBD), and dansyl (DNS, dimethylaminosulfonyl) (Figure 3), were each functionalized with an alkyne substituent for CuAAC reactions with the azide-substituted TMP derivatives.
Figure 2.
Known structure–activity relationships of trimethoprim.
Figure 3.
Alkyne-derivatized fluorophores.
The synthesis of the TMP analogues was based on a mild and efficient procedure reported by Ji et al. in 2013.49 In short, TMP derivatives 8 were synthesized from syringaldehyde 1 by alkylation with bromo–chloro alkane linkers in the presence of K2CO3 in DMF at 60 °C (Scheme 1). Treatment of 3 with sodium azide at 100 °C in the presence of sodium iodide gave azidobenzaldehyde 4. Aldol condensation and 1,3-prototropic isomerization between azidobenzaldehyde 4 and 3-morpholinopropionitrile in the presence of the catalytic amount of potassium hydroxide and methanol provided the intermediate 6, which was followed by in situ acid-mediated substitution of the 3-dimethylamino group with aniline with a convenient one-pot procedure that resulted in enamine 7. Cyclization of the enamine 7 with guanidine provided azide-TMPs 8a–c in high yield.
Scheme 1. Synthesis of Trimethoprim-Linked Azides 8a–c.
The alkyne derivatives 9 and 10 (Figure 3) of fluorophores NBD and DMACA were prepared according to literature procedures,33 whereas the DNS-alkyne 11 was synthesized via sulfonamide coupling of dansyl chloride and propargylamine in the presence of triethylamine.
TMP-azide derivatives 8a–c underwent CuAAC reaction with the alkyne-fluorophores 9–11 in the presence of sodium ascorbate and copper sulfate to produce probes 12–14a–c. At room temperature or even with heating at 50 °C, reactions often took days to produce the desired products despite the putative facile nature of the CuAAC reaction, so microwave heating at 100 °C was applied to accelerate the reaction rates (Scheme 2).
Scheme 2. Synthesis of Trimethoprim Probes Using Microwave-Assisted Click Chemistry.
Biological Activity
The TMP derivatives 8a–c were tested for their ability to inhibit eDHFR and their antimicrobial activities assessed against wild type and mutant E. coli strains (Table 1). The azide-TMPs retained moderate antimicrobial activity, with the shortest linker 8a showing the best activity, although still 8–16-fold less active than TMP itself. Unfortunately, all of the TMP fluorescent probes lost antimicrobial activity (>64 μg/mL), even though they were still able to inhibit eDHFR with comparable, or only 2–5-fold reduced, potency, relative to TMP. The ability to inhibit eDHFR tended to decrease with longer linker length for both azides and probes. These results suggested the loss of activity of TMP derivatives was due to an inability to access the DHFR target within the bacterial cell.
Table 1. MIC Activities of TMP Probes against E. coli and Inhibition of eDHFR Enzyme (n = 4)a.
| MIC (μg/mL) |
cytotoxicity
(CC50) (μM) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| compound | E. coli ATCC 25922 | E. coli clin isol | E. coliclin isol | E. coliMB4827 | E. coli DC2 mutant | E. colilpxC mutant | E. colitolC mutant | E. colilpxC/tolC mutant | S. aureus ATCC 25923 | HEK 293 | HepG2 | eDHFR inhibition IC50 (nM) | |
| TMP | 0.5 | 0.5 | 0.5 | 1 | 0.25 | 0.5 | 0.125 | 0.125 | 1 | >60 | >60 | 60 | |
| 8a | TMP-3C-N3 | 8 | 8 | 4 | 8 | 4 | 8 | 0.25 | 0.25 | 4 | >60 | >60 | 66 |
| 12a | TMP-3C-Tz-NBD | >64 | >64 | >64 | >64 | >64 | >64 | 2 | 1 | 64 | >60 | >60 | 254 |
| 13a | TMP-3C-Tz-DMACA | >64 | >64 | >64 | >64 | >64 | >64 | 8 | 8 | >64 | >60 | >60 | 83 |
| 14a | TMP-3C-Tz-DNS | >64 | >64 | >64 | >64 | >64 | >64 | 0.5 | 0.5 | >64 | 59 | >60 | 162 |
| 8b | TMP-4C-N3 | 32 | 8 | 16 | 16 | 8 | 8 | 0.125 | 0.25 | 4 | >60 | >60 | 97 |
| 12b | TMP-4C-Tz-NBD | >64 | >64 | >64 | >64 | >64 | >64 | 0.25 | 0.5 | 16 | >60 | >60 | 148 |
| 13b | TMP-4C-Tz-DMACA | >64 | >64 | >64 | >64 | >64 | >64 | 2 | 2 | >64 | >60 | >60 | 162 |
| 14b | TMP-4C-Tz-DNS | >64 | >64 | >64 | >64 | >64 | >64 | 0.5 | 1 | 64 | 59 | >60 | 226 |
| 8c | TMP-5C-N3 | 32 | 16 | 16 | 32 | 8 | 8 | 0.125 | 0.25 | 4 | >60 | >60 | 140 |
| 12c | TMP-5C-Tz-NBD | >64 | >64 | >64 | >64 | >64 | >64 | 0.5 | 0.5 | 8 | 46 | >60 | 208 |
| 13c | TMP-5C-Tz-DMACA | >64 | >64 | >64 | >64 | >64 | >64 | 1 | 1 | >64 | >60 | >60 | 668 |
| 14c | TMP-5C-Tz-DNS | >64 | >64 | >64 | >64 | >64 | >64 | 0.25 | 0.25 | 32 | 32 | 51 | 509 |
Clin isol, clinical isolate; wild type HEK 293, human embryonic kidney 293; HepG2, liver hepatocellular cells.
Interestingly, the antimicrobial activities of the TMP probe were restored against an E. coli ΔtolC mutant, lacking the TolC efflux pump component, with the probes 12–14a–c (0.25–8 μg/mL) showing potency almost as good as TMP (0.125 μg/mL) (Table 1). These MIC results indicated that the lack of whole cell activity was likely due to compound efflux. In addition, the TMP probes were tested against membrane-impaired mutant E. coli (LpxC and DC2), which are more permeable to many compounds. Somewhat unexpectedly, the antimicrobial activity of the TMP probes were not improved (TMP; MIC = 0.25–0.5, 12–14a–c; MIC > 64 μg/mL), implying that the ability to penetrate into the cells was not limiting their effectiveness. Although these two permeable membrane mutants provide general membrane disruption, they do not affect the porin channels, outer membrane proteins (OMPs) that largely control influx, so the ability of the TMP probes to penetrate by diffusion via porin channels has not been assessed. Porin penetration by the TMP probes would be expected to be affected by the increase in size caused by attachment of the fluorophore (e.g., 12b = 592 Da compared to TMP, 290 Da).
For the set of probes prepared, antimicrobial activity tended to improve for all fluorophore conjugates with a longer linker, whereas for a fixed linker the DMACA fluorophore was consistently the least active of the three fluorophores. DNS possessed the best activity for C3 (n = 1) and C5 (n = 3) linker lengths, but NBD the best for C4 (n = 2). Activity against mutant E. coli possessing both lpxC and tolC mutations was comparable to the single mutant E. colitolC, confirming that the TolC-dependent efflux pumps were most responsible for the poor MIC activity.
The TMP probes were not generally cytotoxic against mammalian kidney and liver cell lines at the highest concentration tested (CC50 > 60 μM). Therefore, these probes could also find utility for live cell fluorescence imaging studies in mammalian cell lines, for example, studying ATP-binding cassette superfamily (ABC) efflux pumps in cancer cells that are responsible for extruding chemotherapeutic agents, leading to the reduction of intracellular drug level and resistance.50
Use of the Fluorescent TMP Probe To Measure Efflux Pump Activity
Complete sequencing of bacterial genomes has revealed that E. coli contains at least 37 efflux pump transporters including 7 ABC, 19 MFS, 1 MATE, 5 SMR, and 7 RND.51 The source of energy utilized by MFS, SMR, and RND pumps is the proton motive force (PMF), whereas the ABC transporter obtains energy from ATP hydrolysis and the MATE pumps use a Na+ gradient as a driving force.52 However, the AcrAB-TolC system, which belongs to the tripartite RND family, is considered to be the predominant pump in E. coli for multidrug resistance.12 To further investigate the effects of the TolC-dependent efflux pump system and other efflux pumps influencing the susceptibility of E. coli to the TMP probes, we developed a fluorescence assay to measure cellular accumulation of the TMP probes in E. coli in the presence and absence of carbonyl cyanide m-chlorophenylhydrazone (CCCP). CCCP collapses the PMF energizing most E. coli efflux pumps,53 thus inactivating them, and has been used to study cellular accumulation in Gram-negative bacteria for a wide range of compounds.11,54,55
The most potent TMP probe, 12b (E. coli wild type MIC > 64 μg/mL, E. coli mutant deficient tolC MIC = 0.25 μg/mL, eDHFR IC50 = 148 nM), was selected to develop a fluorescent bacterial accumulation assay. Two strains of E. coli were tested: wild type (ATCC 25922) and mutant E. coli (ΔtolC). Various cell densities from wild type E. coli were evaluated to assess the number of bacteria required for significant fluorescence intensity: densities of OD600 = 2 were required to provide sufficient signal when 1 mL of bacteria was centrifuged, resuspended in 150 μL of PBS, and measured in a 96-well plate on a Tecan plate reader at 475 nm excitation/545 nm emission.
To assess the effect of efflux pump inhibition, E. coli was pretreated with CCCP (100 μM) for 10 min before incubation with 12b. Under this condition, the fluorescence intensity of CCCP-treated wild type E. coli significantly increased compared to untreated E. coli (Figure 4A), confirming that accumulation of 12b increased when efflux pump systems were inhibited. The concentration dependence of compound accumulation was investigated by treating E. coli with concentrations of 12b ranging from 12.5 to 100 μM. Interestingly, a linear increase of cellular accumulation of 12b was observed, indicating a concentration-dependent cellular accumulation. Accumulation of 12b in the untreated E. coli also increased linearly even though the efflux pump systems were functional and capable of exporting the probe 12b to the outside of the bacterial cells. This linearity is consistent with a literature paper that showed a dose-dependent accumulation of sulfonyladenosine between 10 and 1000 μM in E. coli (ATCC 25922).54 Another study, examining accumulation of ciprofloxacin in wild type Pseudomonas aeruginosa, also showed an apparent linear accumulation, with a 4-fold cellular increase when the bacteria were treated with ciprofloxacin at 100 ng/mL compared to ciprofloxacin at 25 ng/mL.55
Figure 4.
Fluorescence spectroscopic measurement of cellular accumulation of 12b in E. coli. (A) Fluorescence intensity in E. coli (ATCC 25922) incubated with 12b at various concentrations with/without pretreatment with CCCP (100 μM at 37 °C for 10 min). Statistical significance was assessed using a linear regression of fluorescence intensity against concentration and experimental groups. The model was refitted without an interaction term, giving an estimated fluorescence intensity difference between the groups of 3622.250, with p value 2.52 × 108, ∗∗∗. (B) Fluorescence intensity in E. coli (ΔtolC) and E. coli (ATCC 25922) incubated with 12b at 50 μM with/without pretreatment with CCCP (100 μM at 37 °C for 10 min). Statistical significance (∗∗, p ≤ 0.01; ∗∗∗, p ≤ 0.001) is shown between the absence or presence of CCCP and between wild type and ΔtolCE. coli. Data reported are the mean ± SD for three experiments.
In Gram-negative bacteria, most OMPs are porins, water-filled open channels that allow passive diffusion of small hydrophilic molecules up to 600 Da across the OM. These porin channels are driven by a concentration gradient. Thus, the kinetics of uptake largely depend on the concentration gradient of the solute. With the size of 12b (592 Da) and its hydrophilicity, 12b may traverse the bacterial cell wall through porins, providing the observed linear concentration-dependent cellular accumulation.4,56,57 Although efflux pumps were functional in the untreated (no CCCP) wild type E. coli group, the overall accumulation depends on the relative rates of influx rate via porins versus efflux rate via efflux pumps. It is important to highlight that because porins discriminate solutes on the basis of the physicochemical properties of compounds, the size of potential fluorescent antibiotic conjugates for fluorescence assays must be considered; if they are too large to diffuse into bacterial cells via porins, they would be ineffective as probes of efflux pump activity.
A similar study was conducted with the TolC-deficient (ΔtolC) strain, but because 12b was much more active against this strain (MIC = 0.25 μg/mL = 0.37 μM), a maximum concentration of 50 μM 12b was employed (Figure 4B). Notably, CCCP-treated ΔtolCE. coli also showed higher fluorescence intensity compared to untreated ΔtolCE. coli. This result indicates that, in the ΔtolC strain, other TolC-independent efflux pumps that obtain energy from the PMF are exporting 12b.
As expected from the variation in antibiotic susceptibility, accumulation of 12b in wild type E. coli was significantly less than in ΔtolCE. coli when bacteria were incubated with the same concentration of 12b at 50 μM. This result showed the effect of TolC at reducing the cellular accumulation of 12b. The AcrAB-TolC efflux system, known as the predominant pump in multidrug-resistant E. coli, is composed of an inner membrane transporter, a periplasmic adaptor protein, and outer membrane channel (TolC), so this tripartite system becomes nonfunctional when any of these components is absent.58,59
Flow Cytometric Analysis
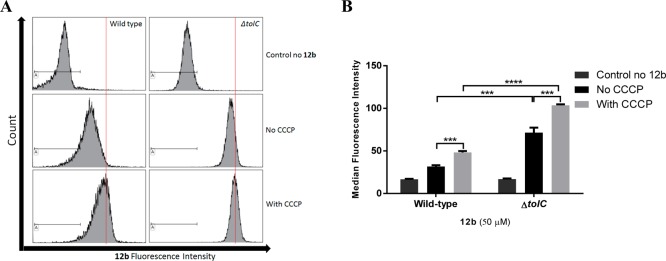
Flow cytometry can be used to assess bacterial characteristics related to size and intrinsic and extrinsic fluorescence in cells. Quantitative measurement of the staining intensity of probes in bacteria has been used for different purposes such as discrimination between Gram-positive and Gram-negative strains, bacteria viability, and antibiotic susceptibility testing.60−62 Recently, flow cytometry analysis was used to measure cellular fluorescence in B. subtilis to quantitatively evaluate labeling levels of amino acids on the cell surface.63 To investigate whether flow cytometric analysis could be used to measure cellular accumulation of fluorescent TMP, bacteria samples were prepared and treated with 12b (50 μM) in the same way as the fluorescence spectroscopy-based assay. Bacteria pellets were resuspended in 1 mL of PBS, with their fluorescence intensity measured using the flow cytometer. Variations in the fluorescence intensity led to shifts in the histogram peaks (Figure 5A), with measurement of the mean fluorescence intensity of the histograms providing a quantitative analysis.
Figure 5.
Flow cytometry measurement of cellular accumulation of 12b at 50 μM in E. coli (ATCC 25922) and E. coli (ΔtolC) with/without pretreatment with CCCP (100 μM at 37 °C for 10 min). (A) Histograms of 12b. Red line shows shift of fluorescence intensity. (B) Median fluorescence intensity of 12b. The data were collected from 10000 bacterial events. Statistical significance (∗∗∗, p ≤ 0.001; ∗∗∗∗, p ≤ 0.0001) is shown between the absence and presence of CCCP and between wild type and ΔtolCE. coli. Data reported are the mean ± SD for three experiments.
Flow cytometry showed that the fluorescence intensity of 12b in both CCCP-treated wild type and ΔtolCE. coli was greater than when the efflux pumps were not inhibited and that the ΔtolCE. coli intensity was greater than that of wild type E. coli. This result matched those provided by the fluorescence spectroscopy-based assay, although with more distinct differences between the groups (Figure 5B). Thus, either method can be used to assess cellular accumulation of fluorescent-labeled antibiotics.
LC-MS/MS-Based Bacterial Accumulation Assay
We wished to confirm that the fluorescence measurements correlated with the cellular concentration of the TMP probes. LC-MS/MS is an accurate quantification method that has previously been reported as a general method to measure compound accumulation within cells.54,55 Although it does not allow for visualization of compound distribution within a cell and is too time-consuming for rapid screening, the LC-MS/MS method does allow for direct measurement of unaltered parent compound, which is an advantage over the labeled probe methods requiring compound modifications. After probe treatment, the bacteria were first washed to remove any surface-bound compound and then lysed to release compound accumulated within the cell. Previous studies have applied sonication, mechanical cell lysis, and/or overnight incubation with glycine–HCl (pH ≤3) to lyse bacteria;54,55,64 however, these lysis procedures are time-consuming and not suitable for high-throughput analyses. Therefore, we used lysozyme, a muramidase that is able to lyse both Gram-positive and Gram-negative bacteria65,66 by cleaving bacterial peptidoglycan between the glycosidic β-1,4-linked N-acetylmuramic acid (MurNAc) and N-acetylglucosamine (GlucNAc).67 The lysozyme lysis buffer usually consists of Tris-HCl (pH 8), EDTA, and a detergent such as Triton X-100, but to improve our LC-MS analysis detection sensitivity, the detergent component was removed to avoid MS ion suppression and decreased HPLC resolution.68 Because the detergent component was required for disruption of the cell membrane, sonication and freeze–thaw steps were added to ensure that the bacterial cells were broken.
Following cell lysis, lysozyme (14.3 kDa) was removed using a filter membrane (10 kDa). The filtrate containing cell lysate (molecular mass < 10 kDa) and any cellular-localized probe were collected, lyophilized, and redissolved in the solution mixture containing 12a as an internal standard. Probe 12a was used as an internal standard as it has a structure similar to that of the analyte 12b and could be used to monitor the stability of 12b in LC-MS/MS samples. An LC-MS/MS method was developed for quantification of 12b, with the analogue 12a used as the MS internal standard.54,55
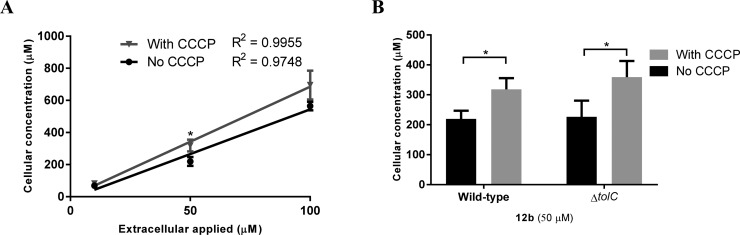
The bacteria were treated in the same way as for the fluorescence-based assay, then washed and lysed to recover 12b from the bacteria. Wild type E. coli (MIC > 64 μg/mL) was treated with 12b at 10, 50, and 100 μM, whereas the TolC-deficient (ΔtolC) strain (MIC = 0.25 μg/mL = 0.37 μM) was treated with 12b only at 50 μM. The results (Figure 6) again showed that both wild type and ΔtolCE. coli showed higher accumulation of 12b in CCCP-treated than in nontreated cells, with a linear concentration dependence (Figure 6A). However, the LC-MS/MS method was not able to distinguish between the accumulation of 12b in wild type E. coli versus ΔtolCE. coli when bacteria were incubated with a constant 50 μM (Figure 6B). The results from the LC-MS/MS assay were generally consistent with the results from fluorescence-based assay, apart from this lack of discrimination between the wild type and ΔtolC mutant strains.
Figure 6.
LC-MS measurement of accumulation of TMP derivative 12b in E. coli. (A) E. coli (ATCC 25922) was incubated with 12b at vaious concentrations with/without pretreatment with CCCP (100 μM at 37 °C for 10 min). (B) E. coli (ΔtolC) and E. coli (ATCC 25922) were incubated with 12b at 50 μM with/without pretreatment in the presence and absence of CCCP (100 μM at 37 °C for 10 min). A significant difference (∗, p ≤ 0.05) was shown between the absence and presence of CCCP. The cellular concentration was calculated from the lysate concentration of 2 × 109 bacteria cells. Data reported are the mean ± SD for three experiments.
The observation that cellular accumulations were higher than extracellular concentration (ranging from 4- to 8-fold) is consistent with previous results for accumulation assay using LC-MS/MS. Active import and Donnan potential have been suggested as mechanisms that may account for this accumulation.54,55
Fluorescence Microscopy
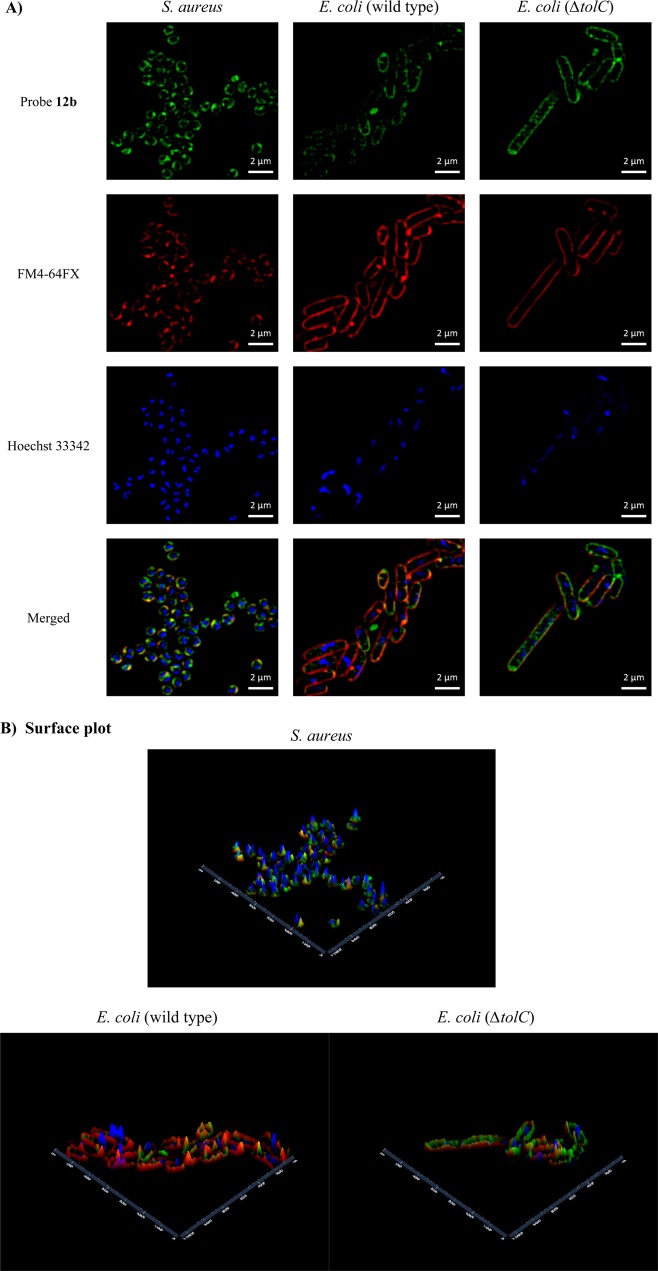
Super-resolution structured illumination microscopy (SR-SIM) has been used for imaging in bacteria to overcome the resolution barrier resulting from the small size of bacterial cells, which prevents good visualization by standard confocal microscopy.69,70 As TMP is active against both Gram-positive and Gram-negative bacteria, Staphylococcus aureus (ATCC 25923) and E. coli (ATCC 25922) were stained with TMP probe 12b. Both showed labeling at a concentration of 100 μM versus MIC values of >64 μg/mL (94 μM) and 16 μg/mL (24 μM), respectively (Figure 7). SR-SIM fluorescent imaging with membrane (FM4-64FX) and nucleic acid (Hoechst 33342) costaining showed that the membrane dye was quickly endocytosed in S. aureus, even though this Gram-positive bacteria was stained with FM4-64FX for only 1 min (Figure 7A). To compare fluorescence brightness, SR-SIM images were processed using the “enabling raw scale” option. A surface plot showed the relative fluorescence brightness of each fluorescent probe (Figure 7B).
Figure 7.
(A) SR-SIM fluorescence imaging of S. aureus (ATCC 25923) and E. coli (ATCC 25922 and ΔtolC): green, TMP-fluorophore 12b; red, FM4-64FX (bacterial membrane stain); blue, Hoechst 33342 (nucleic acid stain). (B) Surface plot: X, Y axes indicate distance (nm). The scale bar shown represents 2 μm.
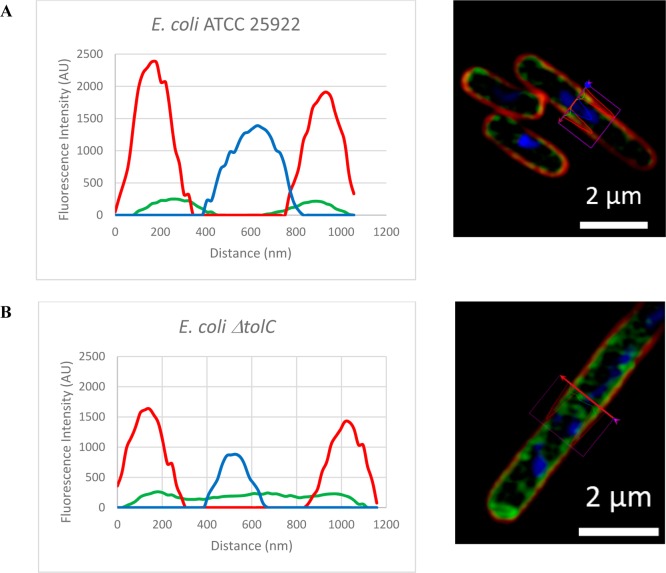
Cross sections of representative bacteria (Figure 8) clearly demonstrated that the 12b probe was largely localized inside the bacterial membrane when compared to the location of the red FM4-64FX dye, but generally remained associated with the cell wall/membrane structure, possibly due to either binding to DHFR or internal components of efflux pumps. Some internal cellular staining was also evident, which was notably stronger in the ΔtolC mutant, and was consistent with higher cellular accumulation. Cross sections of bacteria when images were processed without enabling raw scale are shown in Figure S2. Staining with the NBD-alkyne dye 9 alone did not result in significant bacterial labeling (Figure S1). The intracellular localization of DHFR has not been reported in the literature, and this study suggests that it may be associated with the internal cell wall/membrane structure, as opposed to freely distributed in the cytoplasm.
Figure 8.
Cross section of fluorescent imaging of (A) E. coli (ATCC 25922) (B) ΔtolCE. coli: green, TMP-fluorophore 12b; red, FM4-64FX (bacterial membrane stain); blue, Hoechst 33342 (nucleic acid stain). The images were processed using the “enabling raw scale” option.
Conclusion
We have synthesized azide-functionalized TMP analogues that strongly inhibited the TMP enzymatic target eDHFR in vitro and retained moderate antibacterial activity against E. coli strains. Reaction of these azide-containing TMPs with alkyne-fluorophores using CuAAC generated a TMP fluorescent probe library. These probes, despite generally good eDHFR inhibition, lost almost all ability to inhibit wild type E. coli growth. Activity was restored when tested against the TolC-deficient (ΔtolC) strain, indicating that in normal bacteria TMP probe accumulation at a sufficiently high concentration to inhibit the DHFR target was prevented due to removal by the TolC-dependent efflux pump system.
We then used the fluorescent TMP probe 12b to develop fluorescence-based assays to measure cellular accumulation of the probe. Notably, minimal sample manipulation was required for these fluorescence-based assays using either fluorescence spectrometry or flow cytometry. The cellular fluorescence intensity was directly measured without the need for cell lysis.
The development of alternative assays to show the roles of efflux pumps on intracellular accumulation provides important tools for antibiotic drug discovery and development. To date, fluorescent and radiometric detection methods have been used to determine intracellular drug concentration and efflux pump effects.71,72 Fluorescence was used to measure the accumulation of quinolone class compounds,73 but this method is only applicable to intrinsically fluorescent compounds. Radiometric detection needs radiolabeled analogues and a specific instrument for counting.55,71 An ethidium bromide-agar cartwheel method has also been used to assess the presence of MDR efflux pumps,74,75 but ethidium bromide is rigorously controlled in many countries.76,77 The fluorescent probes 1-N-phenylnaphthylamine (NPN) and 2-(4-dimethylaminostyryl)-1-ethylpyridinium iodide (DMP) (which are weakly fluorescent in an aqueous environment but fluoresce more intensely in a nonpolar environment such as the phospholipid layer of the outer membrane in Gram-negative bacteria) have been used to determine bacterial penetration of compounds and the effect of efflux pump inhibitors.11,78,79
Methods based on LC-MS analysis have been developed to quantify compound accumulation in bacterial cells.54,55,80,81 The LC-MS technique is a useful method for a wide range of structurally different compounds including unlabeled compounds, but is time-consuming as bacterial cells need to be lysed and the supernatant isolated to prepare samples for LC-MS. The development of a rapid fluorescence-based assay to assess inhibition of efflux pumps, as outlined in this study, is a potentially useful research tool in the search for new antibiotics and antibiotic adjuvants.
Methods
For chemical synthesis please see the Supporting Information.
Minimum Inhibitory Concentration Determination
Bacteria were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA), Merck Sharp & Dohme (Kenilworth, NJ, USA), the Coli Genetic Stock Center (CGSC, Yale University), and independent academic clinical isolate collections. Bacteria were cultured in Muller–Hinton broth (MHB) (Bacto Laboratories, catalog no. 211443) at 37 °C overnight. A sample of each culture was then diluted 50-fold in MHB and incubated at 37 °C for 1.5–3 h. The compounds were serially diluted 2-fold across the wells, with concentrations ranging from 0.06 to 128 μg/mL, plated in duplicate. The resultant mid log phase cultures were diluted to the final concentration of 5 × 105 CFU/mL, and then 50 μL was added to each well of the compound-containing 96-well plates (Corning; catalog no. 3641, NBS plates), giving a final compound concentration range of 0.03–64 μg/mL. All of the plates were covered and incubated at 37 °C for 18 h with the MIC defined as the lowest compound concentration at which no bacterial growth was visible (n = 4).
Cytotoxicity Assay (Resazurin Assay)
The compounds were provided at a stock concentration of 12 mM in 100% DMSO. A dilution series in DMEM + 10% FBS was carried out in a one in two step decrease to give the 2× concentrated range from 120 to 1.0 μM. Twenty microliters of each dilution was added to a 384-well black-wall clear-bottom tissue culture plate (Corning; catalog no. 3712) in n = 2. No cells served as negative control (“all cells are dead”), and in addition a serial dilution of tamoxifen (Sigma-Aldrich, catalog no. T5648), starting at 200 μM final concentration, was included as a dose response control. HEK293 and HepG2 cells were seeded into the plates containing the compounds, at 4000 and 5000 per well, respectively, in a volume of 20 μL in DMEM + 10% of FBS (Vtotal = 40 μL), giving the final compound concentration range from 60 to 0.5 μM. The cells together with the compounds were incubated for 20 h at 37 °C, 5% CO2. After the incubation, 5 μL of 100 μM resazurin (Sigma-Aldrich, catalog no. R7017) dissolved in PBS was added to each well (final concentration = 11 μM). The plates were then incubated for 3 h at 37 °C, 5% CO2. The fluorescence intensity was read using the TECAN Infinite M1000 PRO with excitation/emission 560/590 nm. The data were analyzed by GraphPad Prism 7.00 software. Results are presented as the average percentage of control ± SD for each set of duplicate wells using the following equation: cell survival % = (FIsample – FInegative/FIuntreated – FInegative) × 100.
DHFR Enzyme Inhibition assay
The inhibition of DHFR with TMP derivatives was investigated in a 96-well plate (flat-bottom wells, Nunclon Delta Cell Culture Treated Clear Polystyrene catalog no. 167008) using a dihydrofolate reductase assay kit (Sigma-Aldrich, product code CS0340). E. coli dihydrofolate reductase was prepared using an In Vitro protein synthesis kit (New England BioLabs; PURExpress In Vitro Protein Synthesis; NEB#E6800). The DHFR enzyme was diluted before using in the inhibition assay by adding 23 μL of DHFR in 1 mL of water. The assay was performed using the protocol supplied by Sigma-Aldrich; however, the procedure was adjusted for a reaction volume of 200 μL. Briefly, the compounds were provided at a stock concentration of 12 mM in 100% DMSO. The 1× buffer mixture for assay was composed of DHF and NADPH. The well containing 100 μM TMP served as negative control (“DHFR is completely inhibited”) and the well without TMP or the untreated well served as positive control (“DHFR is not inhibited”). A dilution series of compounds in water was carried out to give the 3× concentrated range from 120 to 0.018 μM. Ten microliters of each dilution was added to a 96-well plate containing 180 μL of buffer mixture. Then, 10 μL of diluted DHFR was added into each well, giving the final compound concentration range from 6 to 0.0009 μM. The samples were immediately read by the plate reader. The reaction progress was monitored by the decrease in absorbance at 340 nm, which was read every 60 s for 40 min using a POLARstar Omega plate reader, with enzymatic inhibition calculated using the equation DHRF activity % = (Abssample – Absnegative/Absuntreated – Absnegative) × 100. The data were analyzed by GraphPad Prism 7.00 software to determine IC50 values.
Preparation of Bacterial Cell for Fluorescence Measurement and LC-MS/MS
Bacteria were cultured in Luria broth (LB) (AMRESCO, catalog no. J106) at 37 °C overnight. A sample of each culture was then diluted 50-fold in LB and incubated at 37 °C for 1.5–2 h. The resultant mid log phase cultures were harvested at 4000 rpm for 25 min, washed once with PBS (4000 rpm, for 15 min), and resuspended in PBS to an OD600 of 2. Bacteria were treated with CCCP (Sigma-Aldrich, catalog no. C2759) (100 μM, 37 °C, 10 min) in PBS and then with the TMP probe at the desired concentration (37 °C, 30 min) and washed with 1 mL of cold water four times (13000 rpm, 4 °C, 7 min). For fluorescence intensity measurement using the plate reader, the flat-bottom black opti-plate 96 F (PerkinElmer, catalog no. 6005279) was chosen for this fluorescence assay as it shows less background noise and is suitable for measurements of fluorescence intensity.55 Bacteria cell pellets were transferred into a 96-well plate in a final volume of 150 μL in PBS. The fluorescence intensity was read using the TECAN Infinite M1000 PRO with excitation/emission at 475/545 nm. For fluorescence intensity measurement using the flow cytometer (Gallios flow cytometer from Beckman Coulter), bacteria cell pellets were resuspended in 1 mL of PBS. The sample was then read on the cytometer at a flow rate of approximately 60 μL/min, logarithmic amplification was used for the data acquisition, and the core diameter was calculated to be 6 μm. A total of 10000 events were collected, and the data were analyzed using Kaluza Analysis 1.3 software. Fluorescent intensity from FL1 (excitation, 488 nm; emission, 525/20 nm), was plotted against the number of events count. Median fluorescent intensity was estimated from the histogram peaks after the bacteria had been stained with the TMP probe.
Preparation of Bacterial Cell Extract for LC-MS/MS Analysis
Bacteria were lysed by adding 180 μL of enzymatic lysis buffer, 20 mM Tris-HCl, pH 8.0, and 2 mM sodium EDTA, followed by 70 μL of lysozyme (stock concentration = 72 mg/mL in H2O), then incubated at 37 °C for 30 min, freeze (−78 °C, 5 min)–thawed (34 °C, 15 min) for three cycles, sonicated for 20 min, and then heated to 65 °C for 30 min. The lysate was then centrifuged (14000 rpm, 8 min), and then the supernatants were filtered through a filter membrane 10 kDa (Merck, Amicon Ultra-0.5 centrifugal filter unit with Ultracel-10 membrane catalog no. UFC501096) (14000 rpm, 5 min), which was washed four times with 100 μL of water. The filtered supernatants were lyophilized and then redissolved in 150 μL of acetonitrile/methanol/10 mM 12a (internal control sample) in DMSO (1:1:0.0001) solution. The samples were kept at 4 °C for 1 h to precipitate impurities and centrifuged (14000 rpm, 8 min), and the supernatants were transferred to a vial for LC-MS/MS analysis.
LC-MS/MS Conditions
Sample analysis was performed using an AB Sciex 4000 QTRAP system (USA) and a Shimadzu Nexara UPLC system (Japan) using a Waters Atlantis T3 column (2.1 × 50 mm, 5 μm) with a guard column. The column oven was set at 40 °C. Mobile phases were (A) water/formic acid (1:0.001, v/v) and (B) acetonitrile/formic acid (1:0.001, v/v). The gradient started at 2% B for 1 min, then 2–100% B for 8 min. The flow rate was set to 0.2 mL/min, and the autosampler was maintained at 12 °C. Injection volumes of 5 μL per sample were injected for uptake assays using 10 μM 12b, whereas 3 μL per sample was injected for uptake assays using 50 and 100 μM 12b. Source/gas parameters were as follows: curtain gas (CUR) at 35, ion spray voltage (IS) at 5500, temperature (TEM) at 500, ion source gas 1 (GS1) at 50, and ion source gas 2 (GS2) at 50. Multiple reaction monitoring (MRM) transitions were monitored in the positive mode. Collision energy (CE) and declustering potential (DP) were optimized to generate a good response signal. 12am/z was at 578.4 → 123.1 (DP = 90 and CE = 50), and 12bm/z was at 592.3 → 316.3 and 592.3 → 123.1 (DP = 45 and CE = 45).
Calibration curves were acquired by plotting the standard concentration against its peak area. Standard concentrations ranged from 0.01 to 6.67 μM for the uptake assay using 10 μM 12b, and standard concentrations from 0.07 to 22.22 μM for the uptake assay using 50 and 100 μM 12b were prepared in acetonitrile/methanol/10 mM 12a in DMSO (50:50:0.005, v/v) solution. For each assay, the standard calibration curve was obtained by plotting the standard concentration against the response peak area in the mass spectrometer. A linear regression of at least 0.99 was then established before using the standard curve in computations.
Cellular Concentration Calculation in E. coli
In the LC-MS/MS assay, each sample had 2 × 109 cells using 1 μm3 as the cellular volume of E. coli.55,82 The cellular concentration was calculated on the basis of the calculation in a previous study from Cai et al.55 Thus, the total volume of E. coli 2 × 109 cells is 2 μL [(2 × 109) × 1 μm3 = 2 × 109 μm3 = 2 μL (1 mL = 1 × 1012 μm3)]. Therefore, the cellular concentration was equal to the measured concentration using LC-MS/MS (in μM) multiplied by the extraction volume (150 μL) and divided by the total cellular volume per sample (2 μL). For example, for the measured 12b of 7.122 μM, the cellular concentration would be 534.15 μM [(7.122 μM × 150 μL)/2 μL = 534.15 μM].
Fluorescence Microscopy; Structured Illumination Microscopy (SIM)
SIM was performed using the Zeiss Elyra PS.1 SIM/STORM microscope. Images were analyzed with ZEN2012. To compare fluorescence intensity, the “raw scale” option was used to process images. VectaShield H1000 was used as a mounting medium. Coverslip glasses (Zeiss/Schott, 18 mm × 18 mm, no. 1.5H) were used to prepare samples. Hank’s balanced salt solution (HBSS) without phenol red, CaCl2, or MgSO4 (Sigma-Aldrich catalog no. H6648) was used for bacterial staining. Fluorescent dyes FM4-64FX and Hoechst 33342 (Life Technologies, Australia) were used for membrane staining and DNA staining, respectively.
S. aureus (ATCC 25923) and E. coli were cultured in LB at 37 °C overnight. A sample of each culture was then diluted 50-fold in LB and incubated at 37 °C for 1.5–2 h. One milliliter of the resultant mid log phase cultures was transferred to an Eppendorf tube and centrifuged. Bacteria were washed once with HBSS and then suspended in 20 μL of HBSS. Two microliters of this suspended bacteria solution was dropped onto a coverslip, spread, and dried. Probe 12b (200 μL, 100 μM) was then added to the bacteria, left for 30 min at room temperature, and then washed once with HBSS. An ice-cold solution (200 μL) of Hoechst 33342 (5 μg/mL in HBSS) was then dropped onto the bacteria, left for 10 min on ice, and then drained. This was followed by adding an ice-cold solution (200 μL) of FM4-64FX (5 μg/mL in HBSS) onto the bacteria, which was left for 5 min for E. coli and for 1 min for S. aureus on ice. The bacteria were then washed once with ice-cold HBSS. The bacteria were fixed with 4% paraformaldehyde for 20 min for E. coli and for 10 min for S. aureus on ice, followed by mounting on slides using VectaShield H1000 as a mounting medium.
Acknowledgments
M.A.C. is supported by a NHMRC Principal Research Fellowship (APP1059354), W.P. and S.K. are supported by a UQ International Scholarship (UQI) and IMB Postgraduate Award (IMBPA), and M.P. is supported by an Australian Postgraduate Award (APA) Ph.D. scholarship. R.P., M.A.T.B., M.S.B., and G.K. are supported in part by Wellcome Trust Strategic Grant WT141107. Imaging work was performed in the Queensland Brain Institute’s Advanced Microscopy Facility and generously supported by ARC LIEF LE130100078. We thank Luke Alexander Hammond for training and data analysis. The wild type and mutant tolC and lpxC and double-mutant E. coli strains MB4827, MB4902, MB5746, and MB5747 were generously supplied by Merck Sharp & Dohme (Kenilworth, NJ, USA).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.6b00080.
Methods, synthesis, compound characterization, and NMR spectrum; details of E. coli strains; fluorescent imaging of the control probe; fluorescent imaging of 12b without raw scale applied; excitation/emission spectra of TMP probes (PDF)
Author Contributions
M.A.T.B., M.S.B., M.A.C., and W.P. conceived the study. W.P., R.P., M.S.B., S.K., M.P., and G.K. performed the experiments and analyzed the data. W.P., M.S.B., and M.A.T.B. wrote the paper with input from all authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Xu Z.-Q.; Flavin M. T.; Flavin J. (2014) Combating multidrug-resistant Gram-negative bacterial infections. Expert Opin. Invest. Drugs 23, 163–182. 10.1517/13543784.2014.848853. [DOI] [PubMed] [Google Scholar]
- Cloete T. E. (2003) Resistance mechanisms of bacteria to antimicrobial compounds. Int. Biodeterior. Biodegrad. 51, 277–282. 10.1016/S0964-8305(03)00042-8. [DOI] [Google Scholar]
- Vranakis I.; Goniotakis I.; Psaroulaki A.; Sandalakis V.; Tselentis Y.; Gevaert K.; Tsiotis G. (2014) Proteome studies of bacterial antibiotic resistance mechanisms. J. Proteomics 97, 88–99. 10.1016/j.jprot.2013.10.027. [DOI] [PubMed] [Google Scholar]
- Blair J. M. A.; Webber M. A.; Baylay A. J.; Ogbolu D. O.; Piddock L. J. V. (2015) Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- Amaral L.; Martins A.; Spengler G.; Molnar J. (2014) Efflux pumps of Gram-negative bacteria: what they do, how they do it, with what and how to deal with them. Front. Pharmacol. 4, 1–11. 10.3389/fphar.2013.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. (2012) Efflux-mediated antimicrobial resistance. In Antibiotic Discovery and Development (Dougherty T. J., and Pucci M. J., Eds.), 1st ed., pp 349–395, Springer, New York. [Google Scholar]
- Chollet R.; Chevalier J.; Bryskier A.; Pagès J.-M. (2004) The AcrAB-Tolc pump is involved in macrolide resistance but not in telithromycin efflux in Enterobacter aerogenes and Escherichia coli. Antimicrob. Agents Chemother. 48, 3621–3624. 10.1128/AAC.48.9.3621-3624.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock L. J. V. (2006) Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19, 382–402. 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols W. W. (2012) Permeability of bacteria to antibacterial agents. In Antibiotic Discovery and Development (Dougherty T. J., and Pucci M. J., Eds.), 1st ed., pp 849–879, Springer: New York. [Google Scholar]
- Wang Y.; Venter H.; Ma S. (2016) Efflux pump inhibitors: a novel approach to combat efflux-mediated drug resistance in bacteria. Curr. Drug Targets 17, 702–719. 10.2174/1389450116666151001103948. [DOI] [PubMed] [Google Scholar]
- Lomovskaya O.; Warren M. S.; Lee A.; Galazzo J.; Fronko R.; Lee M.; Blais J.; Cho D.; Chamberland S.; Renau T.; Leger R.; Hecker S.; Watkins W.; Hoshino K.; Ishida H.; Lee V. J. (2001) Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45, 105–116. 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-Z.; Nikaido H. (2004) Efflux-mediated drug resistance in bacteria. Drugs 64, 159–204. 10.2165/00003495-200464020-00004. [DOI] [PubMed] [Google Scholar]
- Sun J.; Deng Z.; Yan A. (2014) Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochem. Biophys. Res. Commun. 453, 254–267. 10.1016/j.bbrc.2014.05.090. [DOI] [PubMed] [Google Scholar]
- Jamshidi S.; Sutton J. M.; Rahman K. M. (2016) An overview of bacterial efflux pumps and computational approaches to study efflux pump inhibitors. Future Med. Chem. 8, 195–210. 10.4155/fmc.15.173. [DOI] [PubMed] [Google Scholar]
- Bakshi S.; Siryaporn A.; Goulian M.; Weisshaar J. C. (2012) Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Mol. Microbiol. 85, 21–38. 10.1111/j.1365-2958.2012.08081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleurie A.; Lesterlin C.; Manuse S.; Zhao C.; Cluzel C.; Lavergne J.-P.; Franz-Wachtel M.; Macek B.; Combet C.; Kuru E.; VanNieuwenhze M. S.; Brun Y. V.; Sherratt D.; Grangeasse C. (2014) MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature 516, 259–262. 10.1038/nature13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss M. P.; Liew A. T. F.; Turnbull L.; Whitchurch C. B.; Monahan L. G.; Harry E. J. (2012) 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Biol. 10, e1001389. 10.1371/journal.pbio.1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel R. A.; Errington J. (2003) Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113, 767–776. 10.1016/S0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- Tiyanont K.; Doan T.; Lazarus M. B.; Fang X.; Rudner D. Z.; Walker S. (2006) Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc. Natl. Acad. Sci. U. S. A. 103, 11033–11038. 10.1073/pnas.0600829103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocaoglu O.; Calvo R. A.; Sham L.-T.; Cozy L. M.; Lanning B. R.; Francis S.; Winkler M. E.; Kearns D. B.; Carlson E. E. (2012) Selective penicillin-binding protein imaging probes reveal substructure in bacterial cell division. ACS Chem. Biol. 7, 1746–1753. 10.1021/cb300329r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei X.; Gu Y. (2009) Progress in modifications and applications of fluorescent dye probe. Prog. Nat. Sci. 19, 1–7. 10.1016/j.pnsc.2008.06.004. [DOI] [Google Scholar]
- Saffarian S.; Li Y.; Elson E. L.; Pike L. J. (2007) Oligomerization of the EGF receptor investigated by live cell fluorescence intensity distribution analysis. Biophys. J. 93, 1021–1031. 10.1529/biophysj.107.105494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C.; Jespersen L. (2003) A flow-cytometric Gram-staining technique for milk-associated bacteria. Appl. Environ. Microbiol. 69, 2857–2863. 10.1128/AEM.69.5.2857-2863.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurn K. T.; Paunesku T.; Wu A.; Brown E. M. B.; Lai B.; Vogt S.; Maser J.; Aslam M.; Dravid V.; Bergan R.; Woloschak G. E. (2009) Labeling TiO2 nanoparticles with dyes for optical fluorescence microscopy and determination of TiO2–DNA nanoconjugate stability. Small 5, 1318–1325. 10.1002/smll.200801458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosten M.; Schäfer T.; Gazendam J. A. C.; Ohlsen K.; Tsompanidou E.; de Goffau M. C.; Harmsen H. J. M.; Crane L. M. A.; Lim E.; Francis K. P.; Cheung L.; Olive M.; Ntziachristos V.; van Dijl J. M.; van Dam G. M. (2013) Real-time in vivo imaging of invasive- and biomaterial-associated bacterial infections using fluorescently labelled vancomycin. Nat. Commun. 4, 2584. 10.1038/ncomms3584. [DOI] [PubMed] [Google Scholar]
- Kolb H. C.; Sharpless K. B. (2003) The growing impact of click chemistry on drug discovery. Drug Discovery Today 8, 1128–1137. 10.1016/S1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- Prescher J. A.; Bertozzi C. R. (2005) Chemistry in living systems. Nat. Chem. Biol. 1, 13–21. 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- Tron G. C.; Pirali T.; Billington R. A.; Canonico P. L.; Sorba G.; Genazzani A. A. (2008) Click chemistry reactions in medicinal chemistry: applications of the 1,3-dipolar cycloaddition between azides and alkynes. Med. Res. Rev. 28, 278–308. 10.1002/med.20107. [DOI] [PubMed] [Google Scholar]
- Yoon H. I.; Yhee J. Y.; Na J. H.; Lee S.; Lee H.; Kang S.-W.; Chang H.; Ryu J. H.; Lee S.; Kwon I. C.; Cho Y. W.; Kim K. (2016) Bioorthogonal copper free click chemistry for labeling and tracking of chondrocytes in vivo. Bioconjugate Chem. 27, 927–936. 10.1021/acs.bioconjchem.6b00010. [DOI] [PubMed] [Google Scholar]
- Avvakumova S.; Colombo M.; Tortora P.; Prosperi D. (2014) Biotechnological approaches toward nanoparticle biofunctionalization. Trends Biotechnol. 32, 11–20. 10.1016/j.tibtech.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Yamagishi K.; Sawaki K.; Murata A.; Takeoka S. (2015) A Cu-free clickable fluorescent probe for intracellular targeting of small biomolecules. Chem. Commun. 51, 7879–7882. 10.1039/C4CC09947H. [DOI] [PubMed] [Google Scholar]
- Johansson L. B. G.; Simon R.; Bergström G.; Eriksson M.; Prokop S.; Mandenius C.-F.; Heppner F. L.; Åslund A. K. O.; Nilsson K. P. R. (2015) An azide functionalized oligothiophene ligand – a versatile tool for multimodal detection of disease associated protein aggregates. Biosens. Bioelectron. 63, 204–211. 10.1016/j.bios.2014.07.042. [DOI] [PubMed] [Google Scholar]
- Phetsang W.; Blaskovich M. A. T.; Butler M. S.; Huang J. X.; Zuegg J.; Mamidyala S. K.; Ramu S.; Kavanagh A. M.; Cooper M. A. (2014) An azido-oxazolidinone antibiotic for live bacterial cell imaging and generation of antibiotic variants. Bioorg. Med. Chem. 22, 4490–4498. 10.1016/j.bmc.2014.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliopoulos G. M.; Huovinen P. (2001) Resistance to trimethoprim-sulfamethoxazole. Clin. Infect. Dis. 32, 1608–1614. 10.1086/320532. [DOI] [PubMed] [Google Scholar]
- Cassir N.; Rolain J.-M.; Brouqui P. (2014) A new strategy to fight antimicrobial resistance: the revival of old antibiotics. Front. Microbiol. 5, 551. 10.3389/fmicb.2014.00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. Y.; Friedman; Larry J.; Gelles J.; Min W.; Hoskins A. A.; Cornish V. W. (2014) The covalent trimethoprim chemical tag facilitates single molecule imaging with organic fluorophores. Biophys. J. 106, 272–278. 10.1016/j.bpj.2013.11.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse H. E., and Miller L. W. (2012) Time-resolved luminescence resonance energy transfer imaging of protein–protein interactions in living cells. In Methods in Enzymology (Conn P. M., Ed.), 1st ed., pp 329–345, Academic Press, San Diego, CA, USA. [DOI] [PubMed] [Google Scholar]
- Chen Z.; Jing C.; Gallagher S. S.; Sheetz M. P.; Cornish V. W. (2012) Second-generation covalent TMP-tag for live cell imaging. J. Am. Chem. Soc. 134, 13692–13699. 10.1021/ja303374p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wombacher R.; Heidbreder M.; van de Linde S.; Sheetz M. P.; Heilemann M.; Cornish V. W.; Sauer M. (2010) Live-cell super-resolution imaging with trimethoprim conjugates. Nat. Methods 7, 717–719. 10.1038/nmeth.1489. [DOI] [PubMed] [Google Scholar]
- Gallagher S. S.; Jing C.; Peterka D. S.; Konate M.; Wombacher R.; Kaufman L. J.; Yuste R.; Cornish V. W. (2010) A trimethoprim-based chemical tag for live cell two-photon imaging. ChemBioChem 11, 782–784. 10.1002/cbic.200900731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher S. S.; Sable J. E.; Sheetz M. P.; Cornish V. W. (2009) An in vivo covalent TMP-tag based on proximity-induced reactivity. ACS Chem. Biol. 4, 547–556. 10.1021/cb900062k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. W.; Cai Y.; Sheetz M. P.; Cornish V. W. (2005) In vivo protein labeling with trimethoprim conjugates: a flexible chemical tag. Nat. Methods 2, 255–257. 10.1038/nmeth749. [DOI] [PubMed] [Google Scholar]
- Jing C.; Cornish V. W. (2013) Design, synthesis, and application of the trimethoprim-based chemical tag for live cell imaging. Curr. Protoc. Chem. Biol. 5, 131–155. 10.1002/9780470559277.ch130019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karoli T.; Mamidyala S. K.; Zuegg J.; Fry S. R.; Tee E. H. L.; Bradford T. A.; Madala P. K.; Huang J. X.; Ramu S.; Butler M. S.; Cooper M. A. (2012) Structure aided design of chimeric antibiotics. Bioorg. Med. Chem. Lett. 22, 2428–2433. 10.1016/j.bmcl.2012.02.019. [DOI] [PubMed] [Google Scholar]
- Yuthavong Y. (2002) Basis for antifolate action and resistance in malaria. Microbes Infect. 4, 175–182. 10.1016/S1286-4579(01)01525-8. [DOI] [PubMed] [Google Scholar]
- Sirichaiwat C.; Intaraudom C.; Kamchonwongpaisan S.; Vanichtanankul J.; Thebtaranonth Y.; Yuthavong Y. (2004) Target guided synthesis of 5-benzyl-2,4-diamonopyrimidines: their antimalarial activities and binding affinities to wild type and mutant dihydrofolate reductases from Plasmodium falciparum. J. Med. Chem. 47, 345–354. 10.1021/jm0303352. [DOI] [PubMed] [Google Scholar]
- Salvatore R. N.; Yoon C. H.; Jung K. W. (2001) Synthesis of secondary amines. Tetrahedron 57, 7785–7811. 10.1016/S0040-4020(01)00722-0. [DOI] [Google Scholar]
- Pak Y. L.; Swamy K. M. K.; Yoon J. (2015) Recent progress in fluorescent imaging probes. Sensors 15, 24374–24396. 10.3390/s150924374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y.-F.; Jiang J.-A.; Liu H.-W.; Liao D.-H.; Wei X.-Y. (2013) Practical preparation of trimethoprim: a classical antibacterial agent. Synth. Commun. 43, 1517–1522. 10.1080/00397911.2011.644380. [DOI] [Google Scholar]
- Fletcher J. I.; Haber M.; Henderson M. J.; Norris M. D. (2010) ABC transporters in cancer: more than just drug efflux pumps. Nat. Rev. Cancer 10, 147–156. 10.1038/nrc2789. [DOI] [PubMed] [Google Scholar]
- Nishino K.; Yamaguchi A. (2001) Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183, 5803–5812. 10.1128/JB.183.20.5803-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez B. (2005) Bacterial efflux systems and efflux pumps inhibitors. Biochimie 87, 1137–1147. 10.1016/j.biochi.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Ghoul M.; Pommepuy M.; Moillo-Batt A.; Cormier M. (1989) Effect of carbonyl cyanide m-chlorophenylhydrazone on Escherichia coli halotolerance. Appl. Environ. Microbiol. 55, 1040–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T. D.; Gerry C. J.; Tan D. S. (2014) General platform for systematic quantitative evaluation of small-molecule permeability in bacteria. ACS Chem. Biol. 9, 2535–2544. 10.1021/cb5003015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H.; Rose K.; Liang L.-H.; Dunham S.; Stover C. (2009) Development of a liquid chromatography/mass spectrometry-based drug accumulation assay in Pseudomonas aeruginosa. Anal. Biochem. 385, 321–325. 10.1016/j.ab.2008.10.041. [DOI] [PubMed] [Google Scholar]
- Tamber S.; Hancock R. E. W. (2003) On the mechanism of solute uptake in Pseudomonas. Front. Biosci., Landmark Ed. 8, 472–483. 10.2741/1075. [DOI] [PubMed] [Google Scholar]
- Galdiero S.; Falanga A.; Cantisani M.; Tarallo R.; Pepa M. E. D.; D’Oriano V.; Galdiero M. (2012) Microbe-host interactions: structure and role of Gram-negative bacterial porins. Curr. Protein Pept. Sci. 13, 843–854. 10.2174/138920312804871120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anes J.; McCusker M. P.; Fanning S.; Martins M. (2015) The ins and outs of RND efflux pumps in Escherichia coli. Front. Microbiol. 6, 587. 10.3389/fmicb.2015.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgurskaya H. I. (2009) Multicomponent drug efflux complexes: architecture and mechanism of assembly. Future Microbiol. 4, 919–932. 10.2217/fmb.09.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason D. J.; Shanmuganathan S.; Mortimer F. C.; Gant V. A. (1998) A fluorescent gram stain for flow cytometry and epifluorescence microscopy. Appl. Environ. Microbiol. 64, 2681–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth B. L.; Poot M.; Yue S. T.; Millard P. J. (1997) Bacterial viability and antibiotic susceptibility testing with SYTOX green nucleic acid stain. Appl. Environ. Microbiol. 63, 2421–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambriz-Aviña V.; Contreras-Garduño J. A.; Pedraza-Reyes M. (2014) Applications of flow cytometry to characterize bacterial physiological responses. BioMed Res. Int. 2014, 1–14. 10.1155/2014/461941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fura J. M.; Pidgeon S. E.; Birabaharan M.; Pires M. M. (2016) Dipeptide-based metabolic labeling of bacterial cells for endogenous antibody recruitment. ACS Infect. Dis. 2, 302–309. 10.1021/acsinfecdis.6b00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock L. J. V.; Johnson M. M. (2002) Accumulation of 10 fluoroquinolones by wild-type or efflux mutant Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46, 813–820. 10.1128/AAC.46.3.813-820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedov S. A.; Belogurova N. G.; Shipovskov S.; Levashov A. V.; Levashov P. A. (2011) Lysis of Escherichia coli cells by lysozyme: discrimination between adsorption and enzyme action. Colloids Surf., B 88, 131–133. 10.1016/j.colsurfb.2011.06.021. [DOI] [PubMed] [Google Scholar]
- Chan K. C.; Ho S.; Law J.; Yuen V. (2002) Microwave treatment as a substitute for EDTA in lysozyme-mediated bacterial cell lysis and its effects on bacterial protein release and β-galactosidase activity. J. Exp. Microbiol. Immunol. 2, 144–156. [Google Scholar]
- Wang G.; Lo L. F.; Forsberg L. S.; Maier R. J. (2013) Helicobacter pylori peptidoglycan modifications confer lysozyme resistance and contribute to survival in the host. mBio 3, e00409–e00412. 10.1128/mBio.00409-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung Y.-G.; Stanley E. R. (2010) Rapid detergent removal from peptide samples with ethyl acetate for mass spectrometry analysis. Curr. Protoc. Protein Sci. 59, 16.12.1–16.12.5. 10.1002/0471140864.ps1612s59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coltharp C.; Xiao J. (2012) Superresolution microscopy for microbiology. Cell. Microbiol. 14, 1808–1818. 10.1111/cmi.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull L.; Strauss M. P.; Liew A. T. F.; Monahan L. G.; Whitchurch C. B.; Harry E. J. (2014) Super-resolution imaging of the cytokinetic Z ring in live bacteria using fast 3D-structured illumination microscopy (f3D-SIM). J. Visualized Exp. 91, 51469. 10.3791/51469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Z.; Nikaido H.; Poole K. (1995) Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39, 1948–1953. 10.1128/AAC.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-Z.; Zhang L.; Nikaido H. (2004) Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48, 2415–2423. 10.1128/AAC.48.7.2415-2423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci V.; Piddock L. (2003) Accumulation of garenoxacin by Bacteroides fragilis compared with that of five fluoroquinolones. J. Antimicrob. Chemother. 52, 605–609. 10.1093/jac/dkg418. [DOI] [PubMed] [Google Scholar]
- Martins M.; McCusker M. P.; Viveiros M.; Couto I.; Fanning S.; Pagès J.-M.; Amaral L. (2013) A simple method for assessment of MDR bacteria for over-expressed efflux pumps. Open Microbiol. J. 7, 72–82. 10.2174/1874285801307010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins M.; Viveiros M.; Couto I.; Costa S. S.; Pacheco T.; Fanning S.; Pagès J.-M.; Amaral L. (2011) Identification of efflux pump-mediated multidrug-resistant bacteria by the ethidium bromide-agar cartwheel method. In Vivo 25, 171–178. [PubMed] [Google Scholar]
- Martins A.; Amaral L. (2012) Screening for efflux pump systems of bacteria by the new acridine orange agar method. In Vivo 26, 203–206. [PubMed] [Google Scholar]
- Paixão L.; Rodrigues L.; Couto I.; Martins M.; Fernandes P.; de Carvalho C. C. C. R.; Monteiro G. A.; Sansonetty F.; Amaral L.; Viveiros M. (2009) Fluorometric determination of ethidium bromide efflux kinetics in Escherichia coli. J. Biol. Eng. 3, 18–18. 10.1186/1754-1611-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helander I. M.; Mattila-Sandholm T. (2000) Fluorometric assessment of Gram-negative bacterial permeabilization. J. Appl. Microbiol. 88, 213–219. 10.1046/j.1365-2672.2000.00971.x. [DOI] [PubMed] [Google Scholar]
- Germ M.; Yoshihara E.; Yoneyama H.; Nakae T. (1999) Interplay between the efflux pump and the outer membrane permeability barrier in fluorescent dye accumulation in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 261, 452–455. 10.1006/bbrc.1999.1045. [DOI] [PubMed] [Google Scholar]
- Zhou Y.; Joubran C.; Miller-Vedam L.; Isabella V.; Nayar A.; Tentarelli S.; Miller A. (2015) Thinking outside the “bug”: a unique assay to measure intracellular drug penetration in Gram-negative bacteria. Anal. Chem. 87, 3579–3584. 10.1021/ac504880r. [DOI] [PubMed] [Google Scholar]
- Bhat J.; Narayan A.; Venkatraman J.; Chatterji M. (2013) LC–MS based assay to measure intracellular compound levels in Mycobacterium smegmatis: linking compound levels to cellular potency. J. Microbiol. Methods 94, 152–158. 10.1016/j.mimet.2013.05.010. [DOI] [PubMed] [Google Scholar]
- Smither R. (1975) Use of a coulter counter to detect discrete changes in cell numbers and volume during growth of Escherichia coli. J. Appl. Bacteriol. 39, 157–165. 10.1111/j.1365-2672.1975.tb00557.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.