Abstract
A glyco-array platform has been developed, in which glycans are attached to plasmonic nanoparticles through strain-promoted azide-alkyne cycloaddition. Glycan–protein binding events can then be detected in a label-free manner employing surface-enhanced Raman spectroscopy (SERS). As proof of concept, we have analyzed the binding of Gal1, Gal3, and influenza hemagglutinins (HAs) to various glycans and demonstrated that binding partners can be identified with high confidence. The attraction of SERS for optical sensing is that it can provide unique spectral signatures for glycan–protein complexes, confirm identity through statistical validation, and minimizes false positive results common to indirect methods. Furthermore, SERS is very sensitive and has multiplexing capabilities thereby allowing the simultaneous detection of multiple analytes.
Keywords: galectin, hemagglutinin, oligosaccharides, proteins, Raman spectroscopy
Almost all cell surface and secreted proteins are modified by covalently linked carbohydrates, and the glycan structures on these glycoproteins are essential mediators of processes, such as protein folding, cell signaling, fertilization, embryogenesis, neuronal development, hormone activity, and the proliferation of cells and their organization into specific tissues.[1] In addition, overwhelming data supports the relevance of glycosylation in pathogen recognition, inflammation, innate immune responses, the development of autoimmune diseases, and cancer.[2]
An important means to decipher the biological information encoded by the glycome is through specific interactions with endogenous glycan-binding proteins.[3] During the past decade, it has become clear that glycan-binding proteins can recognize with high selectivity relatively small oligosaccharide motifs that are often found at the termini of complex glycans.[4] For example, galectins recognize terminal LacNAc moieties; Siglecs bind 2,3- or 2,6-sialylated LacNAc and Selectins complex sialyl Lewisx (SLex) structures. A small number of recent studies indicate a more complex picture of protein–glycan recognition, in which the topology of a complex glycan modulates terminal glycan recognition.[5] For example, our studies have shown that the topology of N-glycans greatly impacts the binding of specific hemagglutinins (HAs) of influenza virus.[6] In another study, the binding of a mixture of 41 different oligosaccharides to various galectins was examined by affinity chromatography, and it was found that the degree of branching, length of repeating unit of LacNAc, and substitution patterns are important determinants of binding.[7] Thus, deciphering the glycome will require large number of precious complex glycans for binding studies.
Glycan arrays are attractive for analyzing minute amounts of large numbers of oligosaccharides for fast, systematic identification and characterization of carbohydrate–protein interactions.[4,8] Fluorescence-based methods are commonly employed to detect the binding of a protein to glycans on an array. However, this method is prone to false negative results, and the requirement of protein labeling can cause inactivation or interference with carbohydrate-binding events.[9] Moreover, fluorophore-labeled secondary reagents are not available for many proteins, especially newly discovered glycan-binding proteins (GBPs) and some secondary reagents can have glycan-binding properties of their own. For example, many polyclonal antibodies used to detect proteins contain carbohydrate-binding antibodies within the mixture.[10b] Therefore, one must ensure that proper controls are in place to correctly interpret results. Finally, most fluorophores are sensitive to light and prone to oxidative degradation. Therefore, signal intensity will decrease over time and can vary with experimental conditions. Therefore, it is attractive to develop label-free detection methods. Surface plasmon resonance (SPR) is a label-free detection method that eliminates some of the problems caused by fluorescence-based approaches. However, it is limited by the number of carbohydrate–lectin interactions that can simultaneously be evaluated.[10] Mass spectrometry is another attractive label-free detection method that has been employed to evaluate modifications of glycans by glycosyl transferases.[10b, 11] However, this method is less attractive for protein-binding studies.
Herein, we report that surface-enhanced Raman spectroscopy (SERS) of glycans linked to plasmonic gold nanoparticles offers a label-free approach to examine carbohydrate–protein binding events in an array format. The attraction of SERS for optical sensing is that it can provide unique spectral signatures for glycan–protein complexes, confirms identity through statistical validation, and minimizes false positive results common to indirect methods. Furthermore, SERS is very sensitive and has multiplexing capabilities, thereby allowing the simultaneous detection of multiple analytes. As proof of concept, we have analyzed the binding of galectin 1 and 3 (Gal1, Gal3) and influenza hemagglutinins (HAs) to various glycans and demonstrated that binding partners can be identified with high confidence.
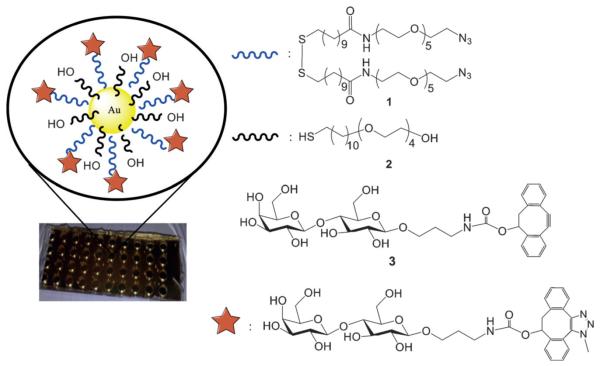
First, attention was focused on the development of a gold nanoparticle (AuNP) platform that allows controlled attachment of carbohydrate ligands and is suitable for SERS measurements. A post-functionalization approach was developed, in which AuNPs were covered with a monolayer of compound 1 to form nanoparticles having azides at the periphery (Figure 1). The latter functionality could be employed for the controlled attachment of glycans, such as 3, which contain a dibenzocyclooctynol (DIBO)[12] moiety for strain-promoted azide-alkyne cycloaddition (SPAAC). The use of compound 2, which terminates into a hydroxyl, made it possible to control the density of the glycans. The attraction of the two-step conjugation approach is that different glycans can be attached to the AuNPs at similar densities.
Figure 1.
Multiwell array substrate and formation of a monolayer of AuNPs modified with lactose through strain-promoted azide–alkyne cycloaddition for detection of galectins.
Compound 1 was readily synthesized by the coupling of 11-(10′-carboxy-decyldisulfanyl)undecanoic acid (4) with azido hexaethylene glycol amine (5) in the presence of 1-[bis(dime-thylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate (HATU) and N,N-diisopropylethylamine (DIPEA) in DMF. The latter compound was prepared by treatment of ethylene glycol with tosyl chloride in a mixture of dichloromethane and pyridine followed by reaction of the resulting product with sodium azide in DMF at 80 °C to give di-azido hexaethylene (71%). Selective reduction of one of the azide to an amine by triphenylphosphine in mixture of THF and water gave 5 in a yield of 47% (Scheme S1 in the Supporting Information).
A mixture of compounds 1 and 2 in DMF (1:5) was added to citrate-stabilized Au nanospheres having a diameter of 60 nm, and after a reaction time of 8–12 h, the particles were isolated by centrifugation. Monolayer formation was confirmed by SERS, which showed signals at 638 and 712 cm−1 corresponding to gauche and trans C—S vibrations, respectively (Figure S1 and Table S1 in the Supporting Information). The absence of bands for thiols and disulfides corroborated proper attachment of 1 and 2 To the Au nanoparticles. Reaction of the azide-modified nanoparticles with DIBO-lactose (3) for 8 h resulted in new SERS signals (679, 1004, 1029, 1567, and 1615 cm−1) that could be assigned to stretching vibrations of the aromatic ring and carbohydrate moiety (Figure 2). Signals corresponding to the alkyne moiety of DIBO (1900–2100 cm−1) were absent, supporting a successful cycloaddition. The formation of a monolayer by using the cycloaddition product of 1 with 3 (Scheme S2 in the Supporting Information) gave a similar SERS spectrum (Figure S2 in the Supporting Information), further supporting proper functionalization of the AuNP.
Figure 2.
Representative SERS spectra and PLS-DA plots. (A) SERS spectra 1 + 2 (i), 1 + 2 + 3 (ii), and 1 + 2 + 3 + Gal1 (iii). Each spectrum is an average of ten individual spectra for each sample. The peaks marked with an arrow in spectrum (ii) could be assigned to the stretching of benzene rings and carbohydrates. (B) PLS-DA Y-CV predicted scores plots. Classes used to build the model were lactose (red), lactose + Gal1 (green), lactose + Gal3 (blue), and lactose + BSA (black). Galectin protein concentration used was 100 μg mL−1. Each plot predicts a sample as belonging to or not belonging to the specified classes. The defined classes were no protein and protein. The PLS-DA cross-validated results are shown in Tables S2 and S4 in the Supporting Information.
The nanoparticles were incubated with various concentrations of Gal1, Gal3, or BSA (5 to 100 μg mL−1) for 2 h at 37 °C followed by removal of the unbound protein by centrifugation and washing. The resulting nanoparticles were resuspended in water and placed into a gold-film multiwall substrate and allowed to dry overnight. Representative SERS spectra for exposure to Gal1are shown in Figure 2. Only minor spectral differences were observed in the normalized SERS spectra of the bound and unbound samples (for assignment of the peaks see Tables S2 and S3 in the Supporting Information). Therefore, to augment direct spectral analysis, partial least squares discrimination analysis (PLS-DA) was employed to differentiate the SERS spectra of the different molecular classes with statistical accuracy. PLS-DA is a supervised multivariate statistical method that employs prior knowledge of class membership to GIVE robust differentiation by minimizing class variations, while emphasizing latent variables between classes.[13] The model was generated by using the spectra of lactose, Gal1, Gal3, and BSA as the individual classes. The horizontal red line in each panel in Figure 2 is a calculation of a Bayesian threshold prediction value for each modeled class. Spectra with predicted values above the threshold level are determined to belong to a particular class, whereas spectra with the predicted values below are excluded. It is clear from all the panels that PLS-DA was able to classify each SERS spectrum in its class with full accuracy. Cross-validation by the venetian-blinds method gave root-mean square errors of 0.05–0.1 for all classes (Table S3 in the Supporting Information), further highlighting the robustness of the classification. Variables important in the projection (VIPs) was used to identify bands in the SERS spectra that can differentiate between protein and non-protein classes.[14] In particular, signals arising from aromatic amino acid residues exhibited differences (Table S2 in the Supporting Information), which is in agreement with the fact that galactose complexed to galectins makes stacking interactions with aromatic side chains. Furthermore, the SERS spectra of Gal1 and 3 exhibited minor differences that were corroborated by PLS-DA analysis as shown in Figure 3. Even at a concentration of 1 μg mL−1, SERS spectra of Gal1 and Gal3 were differentiated into two clearly separated clusters.
Figure 3.
Differentiation of Gal1 and Gal 3 by the comparison of their SERS spectra and PLS-DA analysis. (A) Overlap of normalized Savitzky–Golay SERS spectra of Gal1 (red) and Gal3 (blue) at 5 μg mL−1. (B–D) Cross-validated PLS-DA Y-prediction plots for Gal1 and Gal3 samples based on SERS spectra for each spectral class. The green asterisks and blue plus signs represent sample spectra of Gal1 and Gal3, respectively, at 100 μg mL−1 (B); 5 μg mL−1 (C); and 1 μg mL−1 (D). SERS signal assignments for Gal1 and Gal3 and the PLS-DA cross-validated results are shown in Tables S5 and S6 in the Supporting Information, respectively.
To demonstrate the sensitivity of the new methodology, anti-human Gal3-Alexa Fluor 488 or Cy5-streptavidin was added to the AuNP modified by the carbohydrates that had been incubated with Gal3 or BSA-biotin, respectively, and the fluorescent intensity was measured (Figure S4 in the Supporting Information). At a concentration of 5mm of Gal3, the fluorescent intensity was similar to that of the negative control (BSA), whereas SERS could detect binding at this concentration.
Next, the methodology was extended to a more convenient approach for glycan-array screening. For this purpose, arrays of glycans linked to AuNP were prepared by a layer-to-layer deposition and binding selectivities of human influenza HAs were determined. Influenza HAs recognize sialic acids as receptors, and it is well-documented that human and avian HAs preferentially bind to Neu5Acα(2-6)Gal and Neu5Acα(2-3)Gal linkages, respectively.[15] Glycan microarray binding studies are commonly employed to characterize new influenza strains,[16] and it was anticipated that a SERS-based detection methodology would aid such investigations.
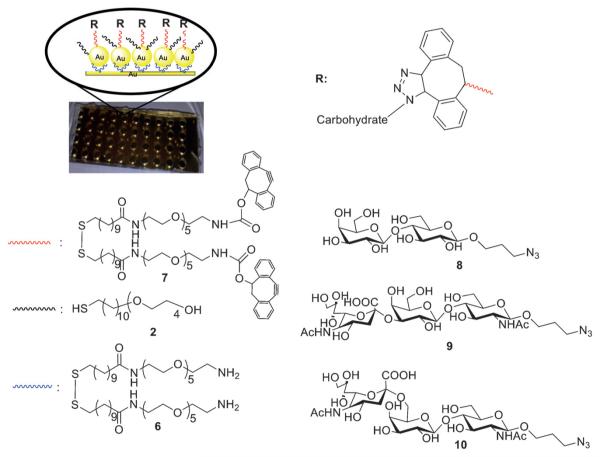
A SERS substrate for the analysis of HA binding to glycans was fabricated by first treating a glass microscope slide with the bifunctional reagent 6 for capturing AuNPs (Figure 4). A monolayer of AuNPs was self-assembled onto this treated glass slide by using an ionic surfactant-mediated Langmuir–Blodgett transfer methodology[17] (Figure S5a in the Supporting Information, SEM image of the AuNP monolayer). The reproducibility of the fabricated AuNP substrate was examined using 4-aminothiophenol (4-ATP). The SERS spectra of 4-aminothiophenol were averaged and the relative standard deviation (RSD) was calculated (see Figure S5b in the Supporting Information for a typical spectrum). A multiwell array was then patterned by using a polymer-molding technique.[18] Next, a mixture of (1-mercaptoundec-11-yl)tetra(ethylene glycol) and disulfide modified DIBO 7 was exposed to the substrate to form a nanoparticle surface that can be modified by SPAAC. Azido-modified glycans 8, 9, and 10 were added to functionalize the AuNPs nanolayer (Figure 4). Each stage of the array construction was monitored by SERS. A significant reduction of the peak at 1969 cm−1, which was assigned to the alkyne moiety, was observed after the addition of the azido-containing glycans (Figure S6 in the Supporting Information). Furthermore, spectral changes in the shape and intensity of peaks in the region 900–1200 cm−1 also supported successful immobilization of compound 7.
Figure 4.
Multiwell array substrate and the immobilization of the carbohydrates for influenza HA detection.
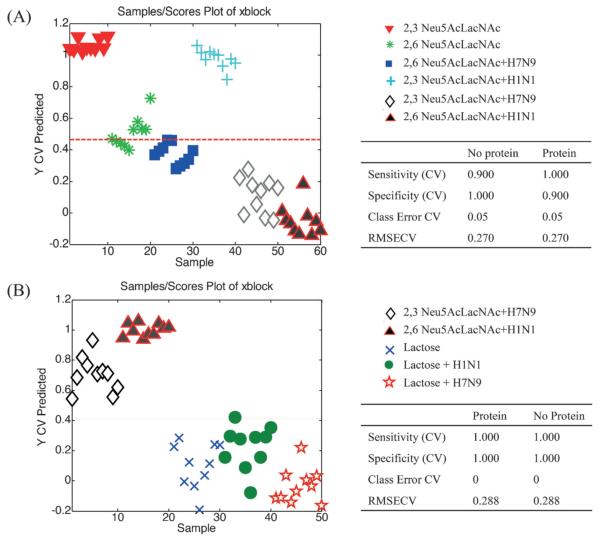
Next, the substrates modified with lactose (8), α(2,3)Neu5AcLacNAc (9) and α(2,6)Neu5AcLacNAc (10) were incubated with human H1N1 and avian H7N9 influenza HA at various concentrations (10 μL of 0.1 to 50 μg mL−1) for 8 h at 37 °C. Next, they were washed, dried, and analyzed by SERS. The spectra of specific substrates exhibited clear visual differences of bands at 1400–1600 and 1000–1200 cm−1 (Figure S7 in the Supporting Information), indicating binding. PLS-DA was employed to further analyze the data, and the results are shown in Figure 5. It was found that avian H7N9 HA binds strongly to the α-2,3-linked sialoside (9) and weakly to the α(2,6)-linked positional isomer (10). On the other hand, the human H1 HA only bound to α(2,6)-sialylated LacNAc derivative 10. These observations are in agreement with the known binding selectivities of HAs. In this respect, it has been reported that H7 avian viruses can transmit in mammalian model systems and exhibit an expanded receptor-binding capacity for the 2,6-sialoside form.[19]
Figure 5.
PLS-DA Y predicted plots and cross-validated results. Each plot predicts a sample as belonging to or not belonging to the specified classes. Classes used to build the model (A) were two forms of sialylated LacNAc and the addition of two HAs (50 μg mL−1). Classes used to build the model (B) were two HAs, lactose and lactose + HAs. The defined classes were protein and no-protein.
The results of PLS-DA analysis for the strong binding event are summarized in Table S7 in the Supporting Information for determining the sensitivity of the method. This method was able to discriminate spectra of the various classes as low as an HA concentration of 0.1 μg mL−1. Moreover, the spectra of the α(2,3)-sialylated LacNAc derivative (9) complexed with H7N9 and α(2,6)-sialylated LacNAc (10) with H1N1 separated in two different class (Figure S8 in the Supporting Information) at a concentration as low as 0.1 μg mL−1, demonstrating that these two sialylated LacNAc derivatives can be employed to differentiate the type of HA and their complex with sialosides (Figure S9 in the Supporting Information).
Previously, SERS has been used for the development of protein microarrays by directly absorbing proteins or antigens on silver nanoparticles. SERS spectra of such samples often exhibit modest reproducibility due to variations in denaturation and adsorption configuration. In addition, such analyses usually require purification of the target protein from biological fluids, in particular when it is of low abundance or exhibits weak adsorbing properties.[20] Some of these difficulties are being addressed by SERS measurements using a sandwich structure of an immunocomplex or protein-bridged bowl-shaped silver cavity (BSSC) array with silver nanoparticles (AgNPs).[21]
Herein, we have demonstrated that SERS of glycans linked to plasmonic nanoparticles offers a label-free approach to directly examine carbohydrate–protein binding events in an array format. The new plasmonic Au nanoparticle array described in this study addresses the difficulties previously noted when employing SERS-based protein microarrays by functionalizing the Au nanoparticle array with synthetic glycan-capture ligands. The attraction of the methodology is that it does not require an antibody as a capture reagent and directly identifies the protein of interest. The methodology represents the first example of detection of glycan–protein interactions by SERS. The attraction of SERS for optical sensing is that it can provide unique spectral signatures for glycan–protein complexes, confirm identity through statistical validation, and minimizes false positive results common to indirect methods. Furthermore, SERS detection is more sensitive than detection of fluorophore-labeled proteins directly or indirectly bound to glycan on the surface by using a fluorescence scanner. The method has multiplexing capabilities, thereby allowing the simultaneous detection of multiple analytes. It is the expectation that many different types of ligands can be attached to the nanoparticle array, allowing a wide range of proteins to be captured. Although quantification of the direct SERS signal has historically been difficult due to the known distance dependence of the SERS effect, it is possible to employ multivariate regression methods for calibration and quantification, as we have previously demonstrated in a different assay system.[22] Such methods are likely to be adaptable to the glycan–protein system studied herein as well.
Supplementary Material
Acknowledgements
This research was supported by the National Institute of General Medical Sciences from the US National Institutes of Health (R01M090269 and P01M107012 to G.-J.B, GM102546 to R.A.D.). Our research benefitted from instrumentation provided by an NIH grant (S10 RR027097). The sialyl transferases for the synthesis of compounds 9 and 10 were kindly provided by Prof. Dr. K.W. Moremen.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/chem.201602706.
References
- [1] a).Freeze HH. Curr. Mol. Med. 2007;7:389–396. doi: 10.2174/156652407780831548. [DOI] [PubMed] [Google Scholar]; b) Marth JD, Grewal PK. Nat. Rev. Immunol. 2008;8:874–887. doi: 10.1038/nri2417. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Clark GF. Hum. Reprod. 2013;28:566–577. doi: 10.1093/humrep/des447. [DOI] [PubMed] [Google Scholar]; d) Wolfert MA, Boons GJ. Nat. Chem. Biol. 2013;9:776–784. doi: 10.1038/nchembio.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Scott H, Panin VM. Glycobiology. 2014;24:407–417. doi: 10.1093/glycob/cwu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2] a).Dube DH, Bertozzi CR. Nat. Rev. Drug Discovery. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]; b) Ohtsubo K, Marth JD. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]; c) Dalziel M, Crispin M, Scanlan CN, Zitzmann N, Dwek RA. Science. 2014;343:1235681. doi: 10.1126/science.1235681. [DOI] [PubMed] [Google Scholar]; d) Hudak JE, Bertozzi CR. Chem. Biol. 2014;21:16–37. doi: 10.1016/j.chembiol.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Taylor ME, Drickamer K. Curr. Opin. Cell Biol. 2007;19:572–577. doi: 10.1016/j.ceb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- [4].Rillahan CD, Paulson JC. Annu. Rev. Biochem. 2011;80:797–823. doi: 10.1146/annurev-biochem-061809-152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5] a).Gabius HJ, Andre S, Jimenez-Barbero J, Romero A, Solis D. Trends Biochem. Sci. 2011;36:298–313. doi: 10.1016/j.tibs.2011.01.005. [DOI] [PubMed] [Google Scholar]; b) Lowary TL. Curr. Opin. Chem. Biol. 2013;17:990–996. doi: 10.1016/j.cbpa.2013.10.009. [DOI] [PubMed] [Google Scholar]
- [6].Wang Z, Chinoy ZS, Ambre SG, Peng W, McBride R, de Vries RP, Glushka J, Paulson JC, Boons GJ. Science. 2013;341:379–383. doi: 10.1126/science.1236231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hirabayashi J, Hashidate T, Arata Y, Nishi N, Nakamura T, Hirashima M, Urashima T, Oka T, Futai M, Muller WEG, Yagi F, Kasai K. Biochim. Biophys. Acta Gen. Subjects. 2002;1572:232–254. doi: 10.1016/s0304-4165(02)00311-2. [DOI] [PubMed] [Google Scholar]
- [8].Smith DF, Song X, Cummings RD. Methods Enzymol. 2010;480:417–444. doi: 10.1016/S0076-6879(10)80033-3. [DOI] [PubMed] [Google Scholar]
- [9] a).Park S, Lee MR, Shin I. Chem. Commun. 2008:4389–4399. doi: 10.1039/b806699j. [DOI] [PubMed] [Google Scholar]; b) Fei Y, Sun YS, Li Y, Lau K, Yu H, Chokhawala HA, Huang S, Landry JP, Chen X, Zhu X. Mol. Biosyst. 2011;7:3343–3352. doi: 10.1039/c1mb05332a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10] a).Unfricht DW, Colpitts SL, Fernandez SM, Lynes MA. Proteomics. 2005;5:4432–4442. doi: 10.1002/pmic.200401314. [DOI] [PubMed] [Google Scholar]; b) Park S, Gildersleeve JC, Blixt O, Shin I. Chem. Soc. Rev. 2013;42:4310–4326. doi: 10.1039/c2cs35401b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11] a).Su J, Mrksich M. Angew. Chem. Int. Ed. 2002;41:4715–4718. doi: 10.1002/anie.200290026. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2002;114:4909–4912. [Google Scholar]; b) Bryan MC, Fazio F, Lee HK, Huang CY, Chang A, Best MD, Calarese DA, Blixt O, Paulson JC, Burton D, Wilson IA, Wong CH. J. Am. Chem. Soc. 2004;126:8640–8641. doi: 10.1021/ja048433f. [DOI] [PubMed] [Google Scholar]; c) Beloqui A, Calvo J, Serna S, Yan S, Wilson IB, Martin-Lomas M, Reichardt NC. Angew. Chem. Int. Ed. 2013;52:7477–7481. doi: 10.1002/anie.201302455. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013;125:7625–7629. [Google Scholar]
- [12].Ning XH, Guo J, Wolfert MA, Boons GJ. Angew. Chem. Int. Ed. 2008;47:2253–2255. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2008;120:2285–2287. [Google Scholar]
- [13].Barker M, Rayens W. J. Chemometr. 2003;17:166–173. [Google Scholar]
- [14].Chong IG, Jun CH. Chemom. Intell. Lab. Syst. 2005;78:103–112. [Google Scholar]
- [15] a).Chandrasekaran A, Srinivasan A, Raman R, Viswanathan K, Raguram S, Tumpey TM, Sasisekharan V, Sasisekharan R. Nat. Biotechnol. 2008;26:107–113. doi: 10.1038/nbt1375. [DOI] [PubMed] [Google Scholar]; b) Imai M, Kawaoka Y. Curr. Opin. Virol. 2012;2:160–167. doi: 10.1016/j.coviro.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Paulson JC, de Vries RP. Virus Res. 2013;178:99–113. doi: 10.1016/j.virusres.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16] a).Tzarum N, de Vries RP, Zhu X, Yu W, McBride R, Paulson JC, Wilson IA. Cell Host Microbe. 2015;17:369–376. doi: 10.1016/j.chom.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang H, de Vries RP, Tzarum N, Zhu X, Yu W, McBride R, Paulson JC, Wilson IA. Cell Host Microbe. 2015;17:377–384. doi: 10.1016/j.chom.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Choi J, Martin SJ, Tripp RA, Tompkins SM, Dluhy RA. Analyst. 2015;140:7748–7760. doi: 10.1039/c5an00977d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18] a).Abell JL, Driskell JD, Dluhy RA, Tripp RA, Zhao YP. Biosens. Bio-electron. 2009;24:3663–3670. doi: 10.1016/j.bios.2009.05.039. [DOI] [PubMed] [Google Scholar]; b) Pienpinijtham P, Han XX, Ekgasit S, Ozaki Y. Phys. Chem. Chem. Phys. 2012;14:10132–10139. doi: 10.1039/c2cp41419h. [DOI] [PubMed] [Google Scholar]
- [19] a).Richard M, Schrauwen EJ, de Graaf M, Bestebroer TM, Spronken MI, van Boheemen S, de Meulder D, Lexmond P, Linster M, Herfst S, Smith DJ, van den Brand JM, Burke DF, Kuiken T, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Nature. 2013;501:560–563. doi: 10.1038/nature12476. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) van Riel D, Leijten LM, de Graaf M, Siegers JY, Short KR, Spronken MI, Schrauwen EJ, Fouchier RA, Osterhaus AD, Kuiken T. Am. J. Pathol. 2013;183:1137–1143. doi: 10.1016/j.ajpath.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Xu R, de Vries RP, Zhu X, Nycholat CM, McBride R, Yu W, Paulson JC, Wilson IA. Science. 2013;342:1230–1235. doi: 10.1126/science.1243761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lane LA, Qian X, Nie S. Chem. Rev. 2015;115:10489–10529. doi: 10.1021/acs.chemrev.5b00265. [DOI] [PubMed] [Google Scholar]
- [21].Gu X, Yan Y, Jiang G, Adkins J, Shi J, Jiang G, Tian S. Anal. Bioanal. Chem. 2014;406:1885–1894. doi: 10.1007/s00216-013-7587-5. [DOI] [PubMed] [Google Scholar]
- [22].Henderson KC, Sheppard ES, Rivera-Betancourt OE, Choi JY, Dluhy RA, Thurman KA, Winchell JM, Krause DC. Analyst. 2014;139:6426–6434. doi: 10.1039/c4an01141d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.