Abstract
Serotype 19A strains have emerged as a cause of invasive pneumococcal disease after the introduction of the 7-valent pneumococcal conjugate vaccine (PCV7), and serotype 19A has now been included in the recent 13-valent vaccine (PCV13). Genetic analysis has revealed at least three different capsular serotype 19A subtypes, and nutritional environment-dependent variation of the 19A capsule structure has been reported. Pneumococcal vaccine effectiveness and serotyping accuracy might be impaired by structural differences in serotype 19A capsules. We therefore analyzed the distribution of 19A subtypes collected within a Swiss national surveillance program and determined capsule composition under different nutritional conditions with high-performance liquid chromatography (HPLC), gas chromatography-mass spectrometry (GC-MS), and nuclear magnetic resonance (NMR) spectroscopy. After the introduction of PCV7, a significant relative increase of subtype 19A-II and decrease of 19A-I occurred. Chemical analyses showed no difference in the composition as well as the linkage of 19A subtype capsular saccharides grown in defined and undefined growth media, which is consistent with a trisaccharide repeat unit composed of rhamnose, N-acetyl-mannosamine, and glucose. In summary, our study suggests that no structural variance dependent of the nutritional environment or the subtype exists. The serotype 19A subtype shift observed after the introduction of the PCV7 can therefore not be explained by selection of a capsule structure variant. However, capsule composition analysis of emerging 19A clones is recommended in cases where there is no other explanation for a selective advantage, such as antibiotic resistance or loss or acquisition of other virulence factors.
INTRODUCTION
The polysaccharide capsule is a major virulence factor of the human pathogen Streptococcus pneumoniae (pneumococcus), and more than 90 different capsular types are known today, which differ in the chemical structures of their capsular polysaccharides (1). These differences are reflected in the type-specific reaction with anticapsular antibodies, by which a serotype is determined and cross-reactive serotypes are pooled into serogroups.
Serogroup 19 contains, among others, serotypes 19F and 19A, which belong to the clinically most relevant serotypes (2). Based on both genetic background and chemical analyses, the serotype 19A and 19F oligosaccharide repeating unit structures have been determined to be trisaccharides of glucose (Glc), rhamnose (Rha), and N-acetyl-mannosamine (ManNAc), differing only in the glycosidic linkage between glucose and rhamnose:
(1, 3–5). Based on chemical analyses, two types of 19A oligosaccharide structures have been described (6, 7). In addition to the genetically proposed structure above, an alternative with a serotype 19F backbone and two side chains of β-d-GlcNAc-(1→3)-β-d-Gal-(1→P→2) and α-l-Fuc-(1→P→3) has been reported based on chemical analysis (5, 7). The polysaccharide structures appear to vary with different in vitro growth conditions (6). The influence of the nutritional environment on the pneumococcal polysaccharide capsule could have biological consequences, as this would potentially impair any intervention or test targeting the pneumococcal capsule. For example, the fungus Cryptococcus neoformans is known to be able to change the capsule structure in vitro and also during infection (8–11), and those changes have been shown to lead to altered antigenicity (8, 11).
Because polysaccharides for pneumococcal vaccine production are derived from in vitro cultures, a nutrient-dependent variation could lead to antigenic preparations that differ from the in vivo antigen.
Capsule variation could also impair diagnostic procedures such as classical serotyping, which is based on polysaccharide-specific antigen-antibody reaction.
Serotype 19A strains have emerged after the introduction of the 7-valent pneumococcal vaccine and were subsequently included in the 13-valent vaccine now recommended in most countries (12, 13). The emergence of serotype 19A after the introduction of the 7-valent vaccine was surprising, as a cross-protection was expected due to close chemical similarity to the serotype 19F capsule as observed for serotypes 6A and 6B, which also differ only by one glycosidic linkage (14, 15). Furthermore, 19F-19A cross-protection had been observed to a certain degree in an animal model (16). Recent work suggests a conformational difference in polysaccharide structure, which might explain the reduced cross-protection (17, 18).
At least three different 19A capsule subtypes are known based on the genetic arrangement of the capsule gene locus compared to a reference strain (19). In addition to various single nucleotide polymorphisms (SNPs) along the capsule operon, most characteristically, subtypes I and II have an inverted rmlD gene, which is the last gene in the rhamnose synthesis pathway (3). To our knowledge, no variations in the capsule structure have been described for different subtypes.
Given the recent discovery of 19A capsular subtypes and previous reports of structural variants as well as the introduction of serotype 19A in the 13-valent pneumococcal conjugate vaccine (PCV13), we aimed to determine the epidemiology and capsule composition of different 19A subtypes in different nutritional environments.
MATERIALS AND METHODS
Bacterial strains and serotype 19A subtype analysis.
Serotype 19A subtype strains were selected from a Swiss national pneumococcal surveillance program (20). In order to detect the different described 19A subtypes (19), we analyzed 158 pneumococcal serotype 19A isolates derived from the upper respiratory tract of infant and adult outpatients with signs of upper respiratory tract infection (20). Antimicrobial susceptibility for penicillin, sulfamethoxazole-trimethoprim (SXT), and erythromycin was determined as previously described (20). In brief, the MIC for penicillin nonsusceptibility was ≥0.06 mg/liter, while for erythromycin and SXT, the disk diffusion method was performed (with intermediate and resistant considered nonsusceptible). A two-step PCR protocol was used to determine the 19A subtype of each strain using the following conditions. The first PCR was done with two primer pairs: rmlb_1_f (GAT GGT GAG AAG AAC AAT AAG) and rmlb_2_f (GAC GGT GAG AAG AAC AAC AAG) and rmld_1_r (CTT CAT TAC GTT CAT CCA ATA) and rmld_2_r (CAG CTG AAG ACA CCA CTT GGT). The PCR conditions were initial heat activation for 6 min at 95°C, followed by 30 cycles of 30 s at 95°C, 20 s at 60°C, and 90 s at 72°C, with a final extension of 5 min at 72°C. The reaction mixture contained 2.5 μl of FastStart Taq reaction buffer without MgCl2, 2.7 μl of 25 mM MgCl2 stock solution, 4 μl of 1.25 mM deoxynucleoside triphosphates (0.2 mM final concentration), 0.2 μl (1 U) of FastStart Taq polymerase (all from Roche Molecular Biochemicals, Rotkreuz, Switzerland), and 0.25 μl of each primer (100 μM stock resulting in 1 μM final concentration, Microsynth AG, Balgach, Switzerland) in a total volume of 25 μl. PCR products were visualized on 1% agarose gels. A resulting PCR product of 560 bp indicated subtypes 19A-I and 19A-II, whereas a band of 425 bp indicated subtype 19A-III. For isolates within the first group (19A-I and 19A-II), a second PCR was performed to discriminate between the two groups using the following primers: wzg_2_f (AGT TGA TTC GTC CAT CCA CAC T), wzg_3_f (GGA ATT GAC ACA TAT GGT CCT), and wzh_r (GCC AAG AGA GCC TTG CTT TCC). The resulting PCR products were 654 bp and 833 bp for types 19A-I and 19A-II, respectively.
Strains were further characterized by plyNCR-restriction fragment length polymorphism (RFLP) as previously described (21). We selected the following strains for further analysis of the capsule composition (Table 1): 109.44 and 501.14 (subtype 19A-I), 501.24 (subtype 19A-II), and 412.49 (subtype 19A-III). For all selected subtype test strains, the sequence type by multilocus sequence typing (MLST) was determined as previously described (22). In addition, we included the internationally spread Hungary-19A-6 strain (23), which was classified as 19A-III. Serotype 19F strains 505.32 and B201.73 were selected for comparison as the polysaccharide repeat unit contains the same backbone monosaccharides but different glycosidic linkages between the repeat units. Both strain are clinical isolates derived from Swiss national surveillance programs (20, 24). In addition a capsule knockout mutant of strain B201.73 was generated as previously described (25, 26) to assess the amount of background signal in capsule extracts. Commercially available capsule polysaccharide of serotype 19A from the American Type Culture Collection (ATCC, Molsheim Cedex, France) was used as a reference standard.
TABLE 1.
Streptococcus pneumoniae strains used in this studya
| Strain no. | Serotype | Subtypeb | plyNCR-RFLP typec | MLST |
|---|---|---|---|---|
| 109.44 | 19A | 19A-I | 16 | ST276 |
| 501.14 | 19A | 19A-I | 1 | ST416 |
| 501.24 | 19A | 19A-II | 1 | ST199 |
| 412.49 | 19A | 19A-III | 14 | ST1151 |
| Hungary 19A-6 (ATCC 700673) | 19A | 19A-III | 20 | ST268d |
| 505.32 | 19F | 19F | 4 | ST179 |
| B201.73 | 19F | 19F | 40 | ST43 |
MLST, multilocus sequencing typing; ST, sequence type; RFLP, restriction fragment length polymorphism.
Defined as previous described (19).
Done as previously described (21).
According to the MLST website (http://spneumoniae.mlst.net/sql/fulldetails.asp?id=689).
The chi-square and Fishers exact tests were used to calculate P values for epidemiological analyses. A P value of ≤0.05 was considered significant. In addition, we used a multivariate logistic regression model to ascertain the strength of the association between PCV7 era and 19A subtypes and adjusted for potential confounders like age (from 0 to 1 year [base], from 2 to 4 years, from 5 to 15 years, and >15 years), sex (male gender as base), penicillin resistance (susceptible as base), erythromycin resistance (susceptible as base), SXT resistance (susceptible as base), and geographical origin (east Switzerland as base). Adjusted odds ratios (aOR) with 95% confidence intervals (95% CI) were used (see Table S1 in the supplemental material). Trends over time in the prevalence of different 19A subtypes before the introduction of PCV7 were analyzed using linear regression.
Growth conditions, polysaccharide purification, hydrolysis, and HPLC.
Strains were handled and grown as described previously (25). The undefined growth medium pneumococcal inoculation medium (PIM), which has been reported to alter 19A capsule composition (6), and a chemically defined medium (CDM) by van den Rijn and Kessler (27) were used. CDM was supplemented with 5 mg/liter choline chloride (28), but made without monosaccharides to allow modification of the type and concentration of the sugar source for each experiment. Monosaccharides were added to the CDM, and the mixture was sterile filtered using a 0.22-μm-pore filter unit (TPP, Trasadingen, Switzerland). Two forms of CDM were created. One was supplemented with 55 mM glucose, and the other one mimicked the composition of the salivary mucin MG1: i.e., N-acetylneuraminic acid (NeuNAc)–fucose (Fuc)–galactose (Gal)–N-acetylglucosamine (GlcNAc)–N-acetylgalactosamine (GalNAc) at 1:5:4:3:1 (29). For one experiment, pooled human saliva from 10 healthy volunteers was collected using Salivette (Sarstedt, Nümbrecht, Germany). After centrifugation at 1,500 × g for 2 min, samples were pooled and sterile filtered with 0.22-μm-pore centrifugal filter units (Millipore, Billerica, MA) at 5,000 × g for 20 min as previously described (30).
Capsules of strains grown in different growth media were released by overnight incubation in 1% phenol, separated from the bacteria by centrifugation and filtration, and then purified by sodium acetate-ethanol precipitation, followed by protease and nuclease digestion of the remaining contaminants, and finally cutoff filtered as described previously (25). The extracted polysaccharides were then completely hydrolyzed by trifluoroacetic acid (TFA) (31). The monosaccharide composition of capsular polysaccharides of strains grown in CDM was determined by high-performance liquid chromatography (HPLC) analysis of fluorescently labeled monosaccharides as previously described (25, 32–35), whereas the PIM extracts were analyzed on a system consisting of an ASI-100 autosampler and P680 HPLC pump (Dionex, Sunnyvale, CA) with an injection volume of 20 μl per sample. Separation of the monosaccharide was done at a flow rate of 0.85 ml/min as follows: 6% solvent B isocratic for 35 min followed by a linear gradient from 6 to 12% solvent B over 20 min. Then, the column was washed with 100% solvent B for 10 min and 100% solvent A for 15 min followed by reequilibration of the system with 6% solvent B for 10 min. The total run time was 90 min, and data were collected for 55 min using an L-7480 fluorescence detector (Merck Hitachi, Darmstadt, Germany). A Luna 5-mm C18 column (Phenomenex, Torrance, CA) was used for separation, and the column temperature was maintained at 24°C using a TCC-100 column oven (Thermo Scientific Dionex, Reinach, Switzerland). Peaks were identified by comparing the retention time with those of monosaccharide standards analyzed in the same run (see Fig. S1A in the supplemental material). As N-acetylated amino sugars are deacetylated during hydrolysis with TFA (36), deacetylated amino sugars were used as standards for their acetylated counterpart (e.g., mannosamine for N-acetyl-mannosamine). The negative controls included in each experiment were the medium negative control for growth, extraction, hydrolysis, and HPLC. HPLC raw data were exported into GraphPad Prism (Version 5, GraphPad Software, Inc.) to create figures.
GC-MS.
Gas chromatography-mass spectrometry (GC-MS) analysis of alditol acetates of the polysaccharide hydrolysates from strains 109.44 grown in PIM and 501.24 grown in PIM, pooled saliva, and CDM glucose was performed as a control, as previously described (37, 38).
NMR spectroscopy.
Nuclear magnetic resonance (NMR) spectroscopy data were collected on a Bruker Avance II spectrometer (500 MHz; 1H) equipped with a 1.7-mm triple-resonance (1H, 13C, 31P) microprobe head. The samples were extracted as described above and prepared as follows. The full amount of each capsule extract (∼4 to 5 mg) was dissolved in 50 μl of D2O, and 40 μl of the resulting mixtures was transferred into 1.7-mm NMR tubes. The water resonance was suppressed using a classical presaturation scheme. Heteronuclear single quantum coherence (HSQC) spectra were collected on a Bruker Avance III HD (600 MHz) spectrometer equipped with an inverse 5-mm TCI helium cryoprobe. All spectra were acquired at a regulated temperature of 298 K and calibrated to the residual water peak (4.766 ppm). For the interpretation of the received carbon and hydrogen shifts, results from previous studies were used as guidance for the capsule structure determination of the 19A capsule extracts (4, 39, 40).
RESULTS
Epidemiology of noninvasive 19A subtypes.
We first aimed at analyzing the distribution of the different pneumococcal serotype 19A subtypes within the Swiss Sentinel Network collection of upper respiratory tract isolates. PCV7 had been recommended in Switzerland since late 2005 to all children under the age of 24 months in a three-dose schedule given at 2, 4, and 12 months, and since August 2006, the vaccine has been fully reimbursed by the mandatory Swiss health insurance. Serotype 19A strains isolated from the upper respiratory tract between 1998 and 2011 were analyzed. In total, 158 19A isolates were screened for the three described subtypes using a two-step PCR protocol described above. Between 1998 and 2005, we identified 30 (57.7%), 5 (9.6%), and 17 (32.7%) isolates belonging to subgroups 19A-I, 19A-II, and 19A-III, respectively. After 2006 until 2011, a significant relative difference was noted overall (P = 0.02) but also individually for the subgroups 19A-I (P = 0.02) and 19A-II (P = 0.02) as 40 (37.7%), 28 (26.4%), and 38 (35.8%) isolates were detected with the 19A-I, 19A-II, and 19A-III subtypes, respectively (Fig. 1A). There was still a significant shift even after data from the PCV7 introduction year (2006) were excluded (P = 0.03). We found no evidence for a difference in distribution of 19A subtypes according to age (PCV7 pre- versus postintroduction eras over age groups of ≤1, 2 to 4, 5 to 15, and >15 years [Fisher's exact test; P = 0.3]). In addition, there was no indication of a time trend for the frequency of 19A subtypes before PCV7 was present (data not shown). As for molecular types derived by plyNCR-RFLP typing, plyNCR-RLPF types 1 (55.7%) and 16 (18.4%) were the most frequent within the 158 strains, but we did not detect a significant shift of molecular types between the PCV7 pre- and postintroduction eras (Fig. 1B). In addition, we revealed that antibiotic resistance within the 19A strains was generally high and that 19A II is significantly more susceptible to penicillin, erythromycin, and SXT than the other subtypes (Fig. 1C). However, calculating a multivariate logistic regression model, we confirmed that there was strong evidence that compared to 19A-I, the odds of observing subtype 19A-II after introduction of PCV7 was 6 times higher than before PCV7, and this association was independent of antimicrobial resistance, geographical region, age, or sex (P = 0.005) (see Table S1 in the supplemental material).
FIG 1.
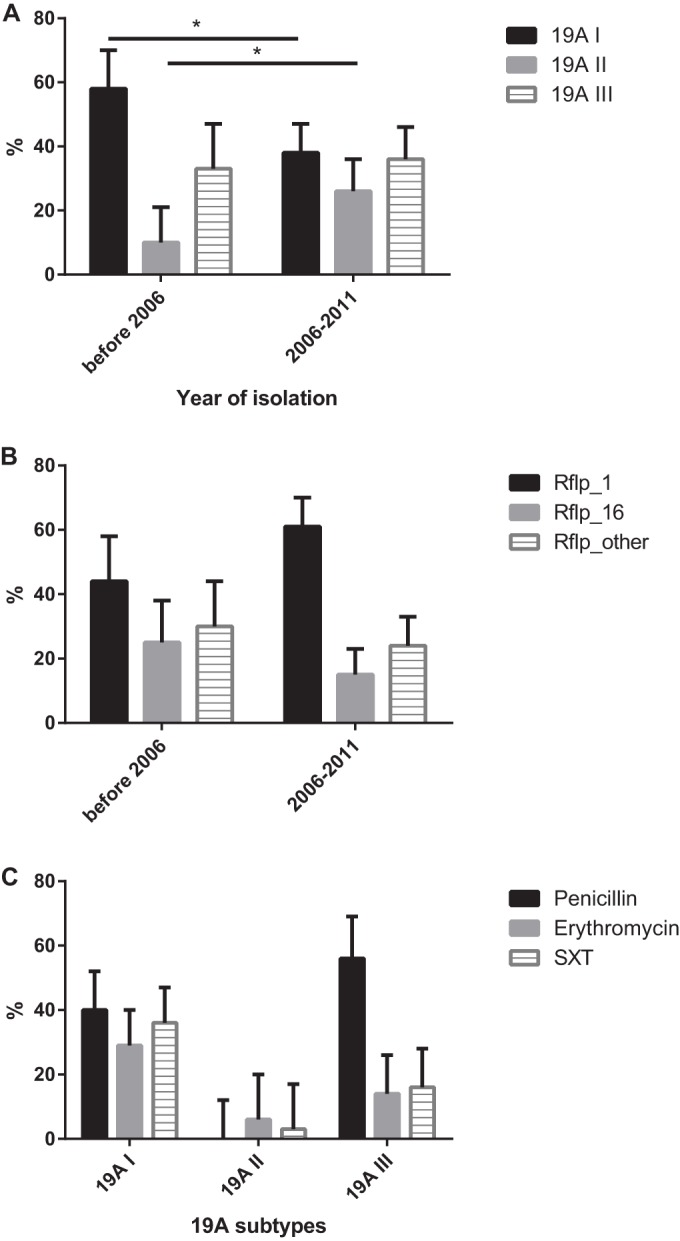
(A) Epidemiology of 19A subtypes (19A-I, 19A-II, and 19A-III) from 1998 to 2011. A total of 158 serotype 19A strains isolated within the Swiss Sentinel Network (outpatients with upper respiratory tract infections) were analyzed. The period from 1998 to 2005 was considered the preconjugate vaccine (PCV7) era. The percentage of tested isolates and subgroup assignment for both periods are shown. There was an overall significance derived by Fisher's exact test (P = 0.02). The chi-square test revealed significant changes in relative frequency between the two periods for subtypes 19A-I (P = 0.02) and 19A-II (P = 0.02). The 95% confidence intervals (CI) are indicated. *, P < 0.05. (B) Distribution of molecular types from 1998 to 2011 as determined by plyNCR-RFLP. Shown are the most frequent plyNCR-RFLP types (types 1 and 16). The remaining types were pooled within “others.” There was no overall significance as derived by Fisher's exact test for the two different eras. The 95% CI are indicated. (C) Antibiotic resistance of serotype 19A isolates against penicillin, erythromycin, and sulfamethoxazole-trimethoprim (SXT) from 1998 to 2011. The MIC for penicillin nonsusceptibility was ≥0.06 mg/liter, while for erythromycin and SXT, the disk diffusion method was performed (with intermediate and resistant considered nonsusceptible). 19A-II is significantly more susceptible to penicillin (Fisher's exact test, P < 0.001), erythromycin (P = 0.014), and SXT (P < 0.001) than the other subtypes. The 95% CI are indicated.
We then selected at least one strain of each subtype for further laboratory analysis to determine the capsular compositions of the different subtypes (Table 1). Selected subtype strains represented different genetic backgrounds, as indicated by MLST and plyNCR-RFLP analysis (Table 1).
Method validation.
To assess the degree of contamination of capsule extracts with cell wall and other components, we first compared the monosaccharide composition of hydrolyzed capsule extracts of a serotype 19F clinical isolate B201.73 with extracts of its isogenic capsule knockout mutant (B201.73Δcps) by HPLC with fluorescence detection (see Fig. S1B in the supplemental material). No rhamnose peaks were detected for the strain without a capsule, but small amounts of glucose and larger amounts of amino sugars, including mannosamine, could be detected, most likely deriving from cell wall polysaccharide (CWPS) and from the murein layer. Furthermore, hydrolysis with TFA is known to produce various amounts of hydrolysis-derived (di)saccharides (see Fig. S1B). This contamination was usually present in capsule extracts from tested strains and was in larger amounts than the commercially available purified pneumococcal polysaccharide (Fig. 2). Furthermore, after complete hydrolysis, a linkage analysis is not possible and only monosaccharide determination can be done (e.g., the same chromatogram for serotypes 19A and 19F). Therefore, we used the HPLC method only as a screening tool for additional neutral monosaccharides under different growth conditions (i.e., galactose and fucose).
FIG 2.
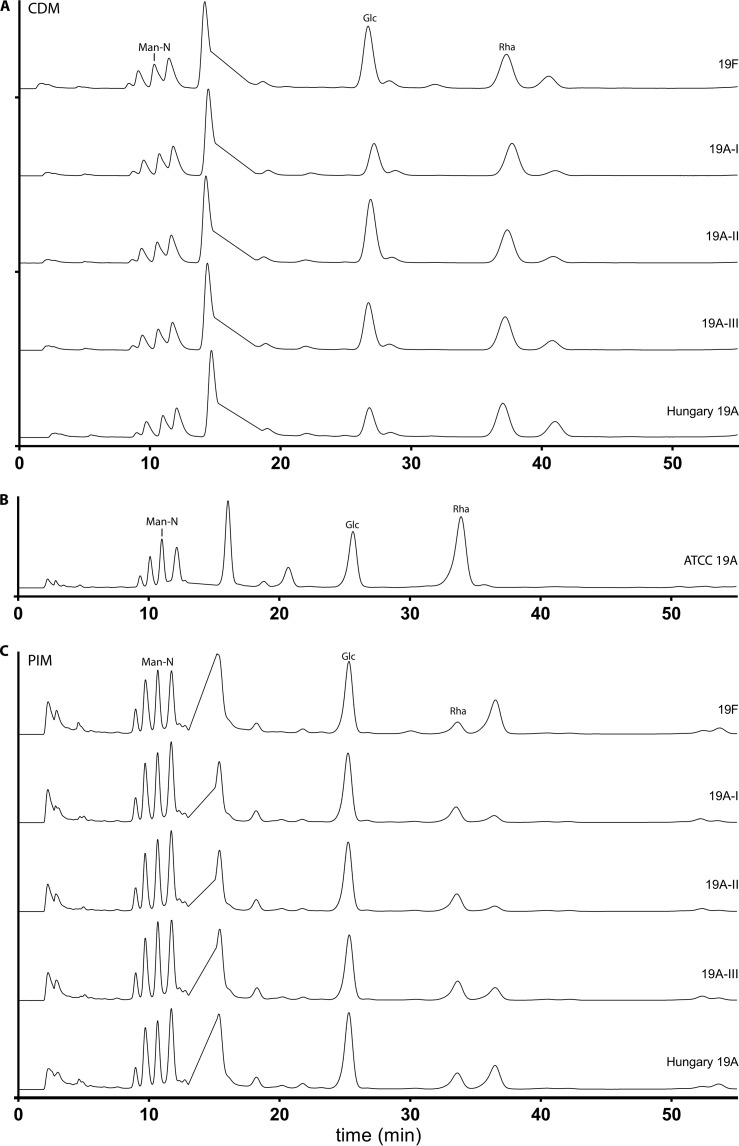
HPLC chromatograms of serotype 19A subtypes and 19F strain B201.73 grown in CDM and PIM. Results from HPLC composition analysis of hydrolyzed polysaccharide capsule of clinical isolates of serotype 19A subtypes grown in chemically defined medium with 55 mM glucose (A) compared to ATCC purified pneumococcal serotype 19A polysaccharide (B), and the pneumococcal inoculation medium PIM (C) are shown. The y axis shows fluorescence (fluorescence units), and chromatograms were stacked to facilitate comparison. Peaks of mannosamine (Man-N), rhamnose (Rha), and glucose (Glc) are labeled.
Capsule composition analysis of 19A subtypes by HPLC and GC-MS.
We then analyzed the capsule composition of a strain for each subgroup (109.44 and 501.14 (subtype 19A-I), 501.24 (subtype 19A-II), and Hungary-19A-6 and 412.49 (subtype 19A-III) grown in CDM supplemented with 55 mM glucose. We did not detect additional neutral monosaccharides, and comparison of the used isolates revealed no differences among the strains, indicating the same monosaccharide backbone built of glucose, rhamnose, and (N-acetyl)-mannosamine. As expected, HPLC chromatograms were not able to differentiate 19A from 19F as both serotypes contain the same monosaccharide compositions (Fig. 2A). We then analyzed the capsular extracts from the ATCC standard (Fig. 2B) and from 19A subtypes grown in the undefined medium PIM (Fig. 2C). Again, no additional neutral monosaccharides were detected, and thus reported additional monosaccharides (galactose and fucose would have been expected on the neutral side based on the literature [6]) were not seen in our preparations. As HPLC allows only the identification of peaks based on their retention time in comparison to standards, the presence of the neutral monosaccharides in capsule extracts was confirmed for 109.44 (19A-I) grown in CDM with 55 mM glucose (see Fig. S3A in the supplemental material) and PIM (see Fig. S3B) and 501.24 (19A-II) grown in PIM (see Fig. S3C) by GC-MS (which revealed glucose and rhamnose in all preparations). GC-MS also confirmed a lower degree of contamination with cell wall and other compounds in the ATCC standard compared to subtype capsule extracts (see Fig. S3D).
To mimic saccharide nutrients present in the natural human environment of S. pneumoniae, 109.44 (19A-I) was grown in CDM with monosaccharides contained in human mucin (5.5 mM total concentration of the mucin building monosaccharides in ratios as determined for the salivary mucin MG1), and 501.24 (19A-II) was also grown in pooled human saliva collected from 10 healthy volunteers after sterile filtration. No additional neutral monosaccharides were identified to be present in the polysaccharide capsule extracted from the strains grown under these conditions compared to capsule extracted from strains grown in CDM supplemented with glucose or PIM, but the signal/noise ratio was much lower in saliva-grown capsule (see Fig. S2 and S3E in the supplemental material).
NMR analyses reveal no structural difference between subtypes.
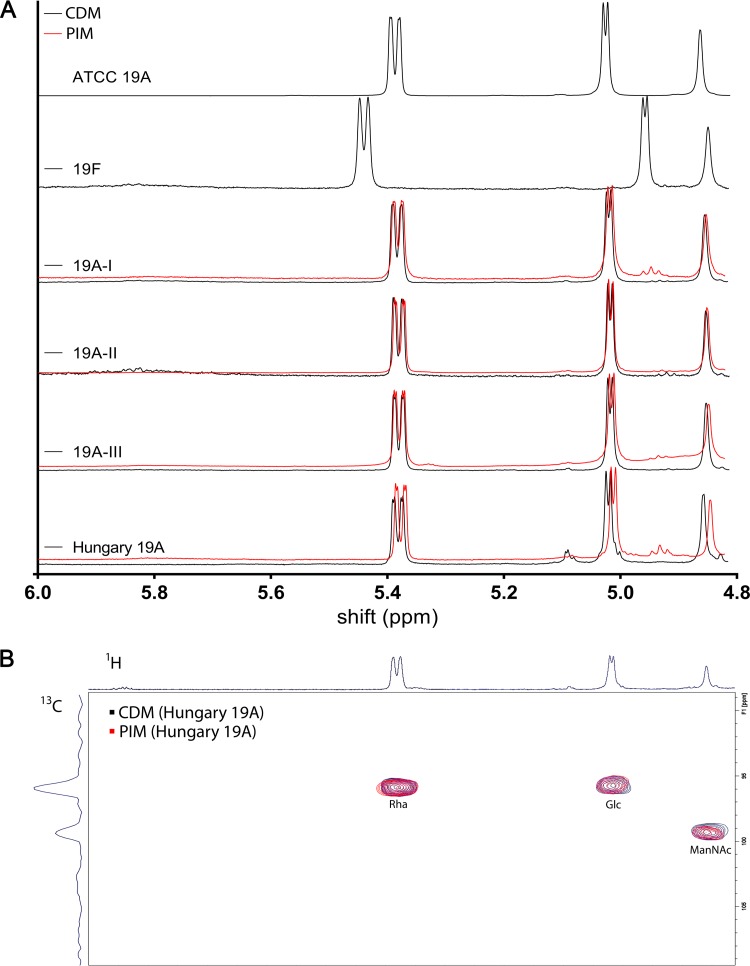
In order to confirm the proposed structures of the oligosaccharides, we compared the anomeric region (4.8 to 6.0 ppm) in the 1H-NMR of all strains grown in CDM supplemented with 55 mM glucose (Fig. 3A). We were able to clearly differentiate the capsule of serotype 19A strains from 19F capsule. Comparison of carbon and hydrogen shifts with previously reported NMR spectra confirmed the structures for serotype 19F of →2)-α-l-Rha-(1→P→4)-β-d-ManNAc-(1→4)-α-d-Glc-(1→ and for serotype 19A of →3)-α-l-Rha-(1→P→4)-β-d-ManNAc-(1→4)-α-d-Glc-(1→. In addition, no differences among the serotype 19A subtypes were detected. We then performed an identical 1H-NMR analysis of all strains grown in PIM, which has been reported to alter the composition of 19A capsule (6). The spectra were again consistent within 19A subtypes but different from serotype 19F capsular polysaccharide (Fig. 3A). Based on those results, it can be stated that the capsule composition is independent of tested growth medium as the patterns were identical to each other and to the ATCC reference 19A polysaccharide. Only a slight shift was observed for the capsule analysis of the Hungary 19A-6 strain. However, the overlay of the 2D 1H-13C-HSQC-NMR spectra of this isolate grown in PIM and CDM supplemented with glucose revealed identical patterns for the H atoms of the anomeric region, as illustrated for rhamnose, N-acetyl mannosamine, and glucose (Fig. 3B). The full 2D spectrum is shown in Fig. S4 in the supplemental material.
FIG 3.
(A) 1D NMR spectra of serotype 19A subtypes. Shown are 1H-NMR spectra of capsular polysaccharide purified from each subtype grown in CDM with 55 mM glucose (black) and PIM (red) compared to serotype 19F and ATCC 19A purified pneumococcal polysaccharide. (B) 2D NMR. Shown is a superimposition of 1H-13C-HSQC-NMR spectra of the anomeric region from PS from Hungary 19A-6 capsule grown in CDM and PIM.
DISCUSSION
In this study, we demonstrated that the distribution of pneumococcal serotype 19A subtypes changed after the implementation of PCV7 in Switzerland. However, all analyzed capsular extracts from serotype 19A subtypes grown in the defined medium CDM and the undefined medium PIM showed a capsule composition that was consistent with the one proposed based on genetic analysis. Furthermore, we did not find any evidence for additional side chains, altered repeat units, or linkage changes under different nutritional conditions.
During the time of widespread use of PCV7 in Switzerland, the serotype 19A subtype distribution changed substantially compared to the prevaccine period. Similar shifts with decreasing 19A-I and increasing 19A-II subtypes after the introduction of PCV7 have also been reported in The Netherlands (19). Furthermore, although classified as group III by PCR, based on its genome sequence, the Hungary 19A-6 strain contains an additional insertion element downstream of the last capsular polysaccharide gene (rmlD) and differs from the subgroup III strains as defined by Elberse et al. (19), which could suggest an additional serotype 19A subgroup.
Given the identical capsule structures for all tested 19A subtypes as revealed for the first time in our study using NMR, HPLC, and GC-MS, no alteration of vaccine effectiveness due to capsular differences between individual subtypes is suggested. Furthermore, the lack of anti-19F antibody cross-protection against serotype 19A can therefore not be explained by selection of 19A subtypes with altered capsule structures, which supports recent discoveries suggesting a general low protection due to conformational difference in polysaccharide structures of 19F and 19A (17, 18).
Various different factors could explain the observed subtype shift after the introduction of PCV7 in Switzerland. For example, changes in 19A subtypes could be due to changes in clonal distribution, although we did not detect a significant shift of molecular types as determined by plyNCR-RFLP. However, additional typing methods or whole-genome sequencing might be more appropriate to investigate this hypothesis. As for antibiotic resistance being a potential driver for the subtype shifts, we revealed that 19A-II is significantly more susceptible to antibiotics than the other subtypes indeed, but as 19A-II increases in the PCV7 era, increasing antibiotic resistance can therefore not explain the changes in the distribution, though antibiotic resistance within serotype 19A strains is generally high as previously shown (41). A subtype redistribution under increasing vaccine selection pressure after the introduction of PCV13 would be unexpected, although it has been speculated that there might be a difference in capsule thickness for different subtypes, which might lead to differences in opsonophagocytotic susceptibility (19).
Previously reported experiments showing additional side chains in 19A isolates grown in PIM could not be replicated (6). An explanation for this finding might be a strain-specific genetic alteration in previous studies. Indeed, it has been reported recently that serotype 11D has two different capsular polysaccharide repeating units in a ratio of 1:3 (25% and 75%, respectively) due to a bispecific transferase, WcrL (42). Although precursors of capsular galactose and N-acetylglucosamine might be available due to their synthesis in other pathways (43), the (environment-dependent) addition of 2 side chains comprised of three additional monosaccharides is expected to be reflected by at least additional glycosyltransferase enzymes within the capsule locus involved in their linkage, and a switch of the glycosidic linkage between rhamnose and glucose would suggest mutations resulting in bispecificity or two copies of the Wzy polymerase (3). To our knowledge, neither of these two possibilities has been detected in 19A strains (3). However, variation of the repeat unit structure would be suspected to have an influence on the antigenicity, thus resulting in suspect serotyping results (i.e., less reactivity with anti-19A antiserum), which were not observed in this study. However, given the increasing vaccine selection pressure after the introduction of PCV13, we recommend capsule structure determination of emerging clones and/or serotypes given recent discoveries of novel serotypes and capsule variants (42, 44–47). The importance of chemical capsule structure analysis is also highlighted by a recent analysis of serotype “6E” capsule, demonstrating that this potential new serotype determined at the genetic level produces capsular polysaccharide identical to 6B capsular polysaccharide (48).
A particular strength of this study is the use of multiple techniques for capsule structure determination, which enhances the reliability of the reported findings. We first used an HPLC-based approach to determine if additional neutral sugars were incorporated into the capsule and validated the methodology with GC-MS. As this technique requires complete hydrolysis, a linkage analysis was not possible in this first step. Furthermore, although this method has been demonstrated to have high normalized recoveries of neutral monosaccharides rhamnose and glucose of serotype 19F monosaccharides (32), TFA hydrolysis has also been reported to be less satisfactory for polysaccharides with amino sugar moieties (32), and it has been reported that disaccharides can be formed during hydrolysis based on different stabilities of intramolecular bonds (32), which also occurred in our experiments.
Results showed traces of contamination by cell wall components in our preparations: cell wall is a common coextracted compound in pneumococcal capsule preparations, probably because the capsule is covalently linked to the cell wall (32, 49, 50). To determine linkage analysis and further characterization of the capsule oligosaccharide repeat units, we performed additional 1D and 2D NMR analyses of polysaccharides from unhydrolyzed capsule extracts. The major limitation of this study is that the number of different 19A subtype strains analyzed was rather small. However, we chose at least one sample of each of the currently known subtypes, and it can therefore be expected that our study is representative. Although we did not find evidence for nutrition-dependent variations of capsule structures, this cannot be considered final proof for an absence of such variations. Furthermore, we did not have the same strains or capsule extracts for which a nutrient-dependent variation has been reported as a reference for the assays used (6).
In summary, the polysaccharide capsule composition for tested serotype 19A subtypes was consistently composed of the same trisaccharide repeat unit. Although we therefore do not expect a structural advantage for certain subtypes it remains to be determined how the 19A subtype distribution will be affected by PCV13. We did not detect any nutritional environment-dependent alterations of the capsule composition. However, given the genetic plasticity of S. pneumoniae and current vaccine selection pressure, we propose to test the capsule composition of emerging serotype 19A clones, especially in cases where there is no other explanation for a selective advantage such as antibiotic resistance or loss or acquisition of other virulence factors.
Supplementary Material
ACKNOWLEDGMENTS
We thank Adrian F. Brugger, Peter Bütikofer, Johannis P. Kamerling, Vincent Perreten, and Jean-Louis Reymond for advice and help with chemical questions and Suzanne Aebi for excellent technical assistance. We are grateful to Lindsey Bomar for critical comments and helpful discussions. We also thank Sandra Loss and Aitor Moreno from Bruker AG, Fällanden, Switzerland, for valuable support and providing access to their NMR facility and the Department of Anesthesiology, University Hospital Bern, Inselspital, for providing the HPLC instrument. We also acknowledge the physicians who participate in the Sentinella surveillance system and who provided noninvasive sample material.
The prospective surveillance study on colonizing pneumococci within Sentinella has been funded by the FOPH (Federal Office of Public Health).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00474-16.
REFERENCES
- 1.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hausdorff WP, Bryant J, Paradiso PR, Siber GR. 2000. Which pneumococcal serogroups cause the most invasive disease: implications for conjugate vaccine formulation and use, part I. Clin Infect Dis 30:100–121. doi: 10.1086/313608. [DOI] [PubMed] [Google Scholar]
- 3.Morona JK, Morona R, Paton JC. 1999. Comparative genetics of capsular polysaccharide biosynthesis in Streptococcus pneumoniae types belonging to serogroup 19. J Bacteriol 181:5355–5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katzenellenbogen E, Jennings HJ. 1983. Structural determination of the capsular polysaccharide of Streptococcus pneumoniae type 19A(57). Carbohydr Res 124:235–245. doi: 10.1016/0008-6215(83)88459-6. [DOI] [PubMed] [Google Scholar]
- 5.Kamerling JP. 2000. Pneumococcal polysaccharides: a chemical view, p 81–114. In Tomasz A. (ed), Streptococcus pneumoniae: molecular biology and mechanisms of disease. Mary Ann Liebert, Inc, Larchmont, NY. [Google Scholar]
- 6.Lee CJ, Fraser BA, Boykins RA, Li JP. 1987. Effect of culture conditions on the structure of Streptococcus pneumoniae type 19A(57) capsular polysaccharide. Infect Immun 55:1819–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee CJ, Fraser BA. 1980. The structures of the cross-reactive types 19 (19F) and 57 (19A) pneumococcal capsular polysaccharides. J Biol Chem 255:6847–6853. [PubMed] [Google Scholar]
- 8.Zaragoza O, Rodrigues ML, De Jesus M, Frases S, Dadachova E, Casadevall A. 2009. The capsule of the fungal pathogen Cryptococcus neoformans. Adv Appl Microbiol 68:133–216. doi: 10.1016/S0065-2164(09)01204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Hermoso D, Dromer F, Janbon G. 2004. Cryptococcus neoformans capsule structure evolution in vitro and during murine infection. Infect Immun 72:3359–3365. doi: 10.1128/IAI.72.6.3359-3365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherniak R, Morris LC, Belay T, Spitzer ED, Casadevall A. 1995. Variation in the structure of glucuronoxylomannan in isolates from patients with recurrent cryptococcal meningitis. Infect Immun 63:1899–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McFadden DC, Fries BC, Wang F, Casadevall A. 2007. Capsule structural heterogeneity and antigenic variation in Cryptococcus neoformans. Eukaryot Cell 6:1464–1473. doi: 10.1128/EC.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan TQ. 2012. Pediatric invasive pneumococcal disease in the United States in the era of pneumococcal conjugate vaccines. Clin Microbiol Rev 25:409–419. doi: 10.1128/CMR.00018-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gervaix A, Ansaldi F, Brito-Avo A, Azzari C, Knuf M, Martinon-Torres F, Tuerlinckx D, Tin Htar MT, Syrogiannopoulos GA. 2014. Pneumococcal vaccination in Europe: schedule adherence. Clin Ther 36:802–812.e1. doi: 10.1016/j.clinthera.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Whitney CG, Pilishvili T, Farley MM, Schaffner W, Craig AS, Lynfield R, Nyquist AC, Gershman KA, Vazquez M, Bennett NM, Reingold A, Thomas A, Glode MP, Zell ER, Jorgensen JH, Beall B, Schuchat A. 2006. Effectiveness of seven-valent pneumococcal conjugate vaccine against invasive pneumococcal disease: a matched case-control study. Lancet 368:1495–1502. doi: 10.1016/S0140-6736(06)69637-2. [DOI] [PubMed] [Google Scholar]
- 15.Vakevainen M, Eklund C, Eskola J, Kayhty H. 2001. Cross-reactivity of antibodies to type 6B and 6A polysaccharides of Streptococcus pneumoniae, evoked by pneumococcal conjugate vaccines, in infants. J Infect Dis 184:789–793. doi: 10.1086/322984. [DOI] [PubMed] [Google Scholar]
- 16.Jakobsen H, Sigurdsson VD, Sigurdardottir S, Schulz D, Jonsdottir I. 2003. Pneumococcal serotype 19F conjugate vaccine induces cross-protective immunity to serotype 19A in a murine pneumococcal pneumonia model. Infect Immun 71:2956–2959. doi: 10.1128/IAI.71.5.2956-2959.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuttel M, Gordon M, Ravenscroft N. 2014. Comparative simulation of pneumococcal serogroup 19 polysaccharide repeating units with two carbohydrate force fields. Carbohydr Res 390:20–27. doi: 10.1016/j.carres.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 18.Kuttel MM, Jackson GE, Mafata M, Ravenscroft N. 2015. Capsular polysaccharide conformations in pneumococcal serotypes 19F and 19A. Carbohydr Res 406:27–33. doi: 10.1016/j.carres.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 19.Elberse K, Witteveen S, van der Heide H, van de Pol I, Schot C, van der Ende A, Berbers G, Schouls L. 2011. Sequence diversity within the capsular genes of Streptococcus pneumoniae serogroup 6 and 19. PLoS One 6:e25018. doi: 10.1371/journal.pone.0025018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muhlemann K, Matter HC, Tauber MG, Bodmer T, Sentinel Working Group. 2003. Nationwide surveillance of nasopharyngeal Streptococcus pneumoniae isolates from children with respiratory infection, Switzerland, 1998–1999. J Infect Dis 187:589–596. doi: 10.1086/367994. [DOI] [PubMed] [Google Scholar]
- 21.Hathaway LJ, Brugger S, Martynova A, Aebi S, Muhlemann K. 2007. Use of the Agilent 2100 bioanalyzer for rapid and reproducible molecular typing of Streptococcus pneumoniae. J Clin Microbiol 45:803–809. doi: 10.1128/JCM.02169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou J, Enright MC, Spratt BG. 2000. Identification of the major Spanish clones of penicillin-resistant pneumococci via the Internet using multilocus sequence typing. J Clin Microbiol 38:977–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGee L, McDougal L, Zhou J, Spratt BG, Tenover FC, George R, Hakenbeck R, Hryniewicz W, Lefevre JC, Tomasz A, Klugman KP. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J Clin Microbiol 39:2565–2571. doi: 10.1128/JCM.39.7.2565-2571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meichtry J, Born R, Kuffer M, Zwahlen M, Albrich WC, Brugger SD, Muhlemann K, Hilty M. 2014. Serotype epidemiology of invasive pneumococcal disease in Swiss adults: a nationwide population-based study. Vaccine 32:5185–5191. doi: 10.1016/j.vaccine.2014.07.060. [DOI] [PubMed] [Google Scholar]
- 25.Hathaway LJ, Brugger SD, Morand B, Bangert M, Rotzetter JU, Hauser C, Graber WA, Gore S, Kadioglu A, Muhlemann K. 2012. Capsule type of Streptococcus pneumoniae determines growth phenotype. PLoS Pathog 8:e1002574. doi: 10.1371/journal.ppat.1002574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trzcinski K, Thompson CM, Lipsitch M. 2003. Construction of otherwise isogenic serotype 6B, 7F, 14, and 19F capsular variants of Streptococcus pneumoniae strain TIGR4. Appl Environ Microbiol 69:7364–7370. doi: 10.1128/AEM.69.12.7364-7370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van de Rijn I, Kessler RE. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect Immun 27:444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desai BV, Reiter H, Morrison DA. 2003. Choline starvation induces the gene licD2 in Streptococcus pneumoniae. J Bacteriol 185:371–373. doi: 10.1128/JB.185.1.371-373.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomsson KA, Prakobphol A, Leffler H, Reddy MS, Levine MJ, Fisher SJ, Hansson GC. 2002. The salivary mucin MG1 (MUC5B) carries a repertoire of unique oligosaccharides that is large and diverse. Glycobiology 12:1–14. doi: 10.1093/glycob/12.1.1. [DOI] [PubMed] [Google Scholar]
- 30.Shelburne SA III, Sumby P, Sitkiewicz I, Granville C, DeLeo FR, Musser JM. 2005. Central role of a bacterial two-component gene regulatory system of previously unknown function in pathogen persistence in human saliva. Proc Natl Acad Sci U S A 102:16037–16042. doi: 10.1073/pnas.0505839102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albersheim P. 1968. A method for the analysis of sugars in plant cell wall polysaccharides by gas liquid chromatography. Carbohydr Res 5:340–345. [Google Scholar]
- 32.Talaga P, Vialle S, Moreau M. 2002. Development of a high-performance anion-exchange chromatography with pulsed-amperometric detection based quantification assay for pneumococcal polysaccharides and conjugates. Vaccine 20:2474–2484. doi: 10.1016/S0264-410X(02)00183-4. [DOI] [PubMed] [Google Scholar]
- 33.Anumula KR. 1994. Quantitative determination of monosaccharides in glycoproteins by high-performance liquid chromatography with highly sensitive fluorescence detection. Anal Biochem 220:275–283. doi: 10.1006/abio.1994.1338. [DOI] [PubMed] [Google Scholar]
- 34.Anumula KR, Dhume ST. 1998. High resolution and high sensitivity methods for oligosaccharide mapping and characterization by normal phase high performance liquid chromatography following derivatization with highly fluorescent anthranilic acid. Glycobiology 8:685–694. doi: 10.1093/glycob/8.7.685. [DOI] [PubMed] [Google Scholar]
- 35.Saddic GN, Dhume ST, Anumula KR. 2008. Carbohydrate composition analysis of glycoproteins by HPLC using highly fluorescent anthranilic acid (AA) tag. Methods Mol Biol 446:215–229. doi: 10.1007/978-1-60327-084-7_15. [DOI] [PubMed] [Google Scholar]
- 36.Kwon H, Kim J. 1993. Determination of monosaccharides in glycoproteins by reverse-phase high-performance liquid chromatography. Anal Biochem 215:243–252. doi: 10.1006/abio.1993.1582. [DOI] [PubMed] [Google Scholar]
- 37.Lehr T, Geyer H, Maass K, Doenhoff MJ, Geyer R. 2007. Structural characterization of N-glycans from the freshwater snail Biomphalaria glabrata cross-reacting with Schistosoma mansoni glycoconjugates. Glycobiology 17:82–103. [DOI] [PubMed] [Google Scholar]
- 38.Geyer R, Geyer H, Kuhnhardt S, Mink W, Stirm S. 1982. Capillary gas chromatography of methylhexitol acetates obtained upon methylation of N-glycosidically linked glycoprotein oligosaccharides. Anal Biochem 121:263–274. doi: 10.1016/0003-2697(82)90478-X. [DOI] [PubMed] [Google Scholar]
- 39.Abeygunawardana C, Williams TC, Sumner JS, Hennessey JP Jr. 2000. Development and validation of an NMR-based identity assay for bacterial polysaccharides. Anal Biochem 279:226–240. doi: 10.1006/abio.1999.4470. [DOI] [PubMed] [Google Scholar]
- 40.Jennings HJ, Rosell KG, Carlo DJ. 1980. Structural determination of the capsular polysaccharide of Streptococcus pneumoniae type-19 (F-19). Can J Chem 58:1069–1074. doi: 10.1139/v80-167. [DOI] [Google Scholar]
- 41.Hauser C, Kronenberg A, Allemann A, Muhlemann K, Hilty M. 26 May 2016. Serotype/serogroup-specific antibiotic non-susceptibility of invasive and non-invasive Streptococcus pneumoniae, Switzerland, 2004 to 2014. Euro Surveill doi: 10.2807/1560-7917.ES.2016.21.21.30239. [DOI] [PubMed] [Google Scholar]
- 42.Oliver MB, Jones C, Larson TR, Calix JJ, Zartler ER, Yother J, Nahm MH. 2013. Streptococcus pneumoniae serotype 11D has a bispecific glycosyltransferase and expresses two different capsular polysaccharide repeating units. J Biol Chem 288:21945–21954. doi: 10.1074/jbc.M113.488528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yother J. 2011. Capsules of Streptococcus pneumoniae and other bacteria: paradigms for polysaccharide biosynthesis and regulation. Annu Rev Microbiol 65:563–581. doi: 10.1146/annurev.micro.62.081307.162944. [DOI] [PubMed] [Google Scholar]
- 44.Park IH, Geno KA, Yu J, Oliver MB, Kim KH, Nahm MH. 2015. Genetic, biochemical, and serological characterization of a new pneumococcal serotype, 6H, and generation of a pneumococcal strain producing three different capsular repeat units. Clin Vaccine Immunol 22:313–318. doi: 10.1128/CVI.00647-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Camilli R, Spencer BL, Moschioni M, Pinto V, Berti F, Nahm MH, Pantosti A. 2014. Identification of Streptococcus pneumoniae serotype 11E, serovariant 11Av and mixed populations by high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and flow cytometric serotyping assay (FCSA). PLoS One 9:e100722. doi: 10.1371/journal.pone.0100722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oliver MB, van der Linden MP, Kuntzel SA, Saad JS, Nahm MH. 2013. Discovery of Streptococcus pneumoniae serotype 6 variants with glycosyltransferases synthesizing two differing repeating units. J Biol Chem 288:25976–25985. doi: 10.1074/jbc.M113.480152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calix JJ, Porambo RJ, Brady AM, Larson TR, Yother J, Abeygunwardana C, Nahm MH. 2012. Biochemical, genetic, and serological characterization of two capsule subtypes among Streptococcus pneumoniae serotype 20 strains: discovery of a new pneumococcal serotype. J Biol Chem 287:27885–27894. doi: 10.1074/jbc.M112.380451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burton RL, Geno KA, Saad JS, Nahm MH. 2016. Pneumococcus with the “6E” cps locus produces serotype 6B capsular polysaccharide. J Clin Microbiol 54:967–971. doi: 10.1128/JCM.03194-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sorensen UB, Henrichsen J, Chen HC, Szu SC. 1990. Covalent linkage between the capsular polysaccharide and the cell wall peptidoglycan of Streptococcus pneumoniae revealed by immunochemical methods. Microb Pathog 8:325–334. doi: 10.1016/0882-4010(90)90091-4. [DOI] [PubMed] [Google Scholar]
- 50.Talaga P, Bellamy L, Moreau M. 2001. Quantitative determination of C-polysaccharide in Streptococcus pneumoniae capsular polysaccharides by use of high-performance anion-exchange chromatography with pulsed amperometric detection. Vaccine 19:2987–2994. doi: 10.1016/S0264-410X(00)00535-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.