Abstract
β-lactams are antibiotic molecules able to inhibit cell wall biosynthesis. Among other mechanisms, resistance in Gram-negative bacteria is mostly associated with production of β-lactamase enzymes able to bind and hydrolyze the β-lactam ring. Extended-spectrum β-lactamases extend this ability also to third- and fourth-generation cephalosporins, as well as to carbapenems and monobactams. Vibrio cholerae is the causative agent of epidemic cholera and a public health burden for developing countries like Bangladesh. Although appropriate oral or intravenous rehydration is the therapy of choice for cholera, severe infections and V. cholerae-associated septicemia are treated with antimicrobial drugs, including doxycycline, erythromycin, azithromycin, ciprofloxacin, and/or third-generation cephalosporins. In the years after the introduction of antibiotics in clinical practice, V. cholerae developed resistance to commonly used drugs worldwide mostly through gene acquisition via horizontal gene transfer. Reduced susceptibility of V. cholerae to third-generation cephalosporins has been occasionally documented. However, carbapenemase-producing V. cholerae has been reported at higher rates than resistance to extended-spectrum β-lactams, mainly associated with blaNDM-1 emergence and successful plasmid dissemination. Recent findings suggest limited β-lactam resistance is present in V. cholerae O1 isolates collected during ecological and epidemiological surveillance in Bangladesh. However, a trend to intermediate-susceptibility insurgence was observed. Horizontal gene transfer of β-lactam resistance from enteric pathogens to environmental microorganisms should not be underrated, given the ability of V. cholerae to acquire new genetic information.
Keywords: Vibrio cholerae, cholera, Bangladesh, extended-spectrum β-lactams, carbapenems, antibiotic resistance, aquatic environment, clinical environment
Introduction
β-lactams are assorted antibiotic molecules able to inhibit cell wall biosynthesis. Resistance to these drugs can be the result of altered permeability, antibiotic target site alteration, or antibiotic degradation (1). The latter represents the primary resistance mechanism in Gram-negative bacteria producing β-lactamase enzymes able to bind and hydrolyze the β-lactam ring (2). β-lactamases have extensively diversified in response to the clinical use of new generations of β-lactams, including the clinically significant extended-spectrum β-lactamases (ESBLs) CTX-M-, TEM-, and SHV-type enzymes (3). Carbapenemases are ESBLs that recognize almost all hydrolyzable β-lactams, including carbapenems (imipenem, ertapenem, meropenem, and doripenem), the last-line therapeutics to treat multidrug-resistant Gram-negative infections (4). Carbapenem resistance in Enterobacteriaceae constitutes an important and growing public health threat, especially since the appearance of the powerful enzyme NDM-1 (5), whose presence during the past years was reported worldwide (6).
The majority of diarrheal diseases in Bangladesh are endemic and waterborne since surface water can be heavily contaminated due to poor sanitation and hygiene (7) and access to safe drinking water is problematic (8). Together with enterotoxigenic Escherichia coli, Vibrio cholerae is one of the leading causes of enteric infections in the country. V. cholerae is a natural inhabitant of estuarine brackish waters and it can thrive in the human gut, causing mild to severe infections and cholera. Today, more than 200 serotypes of V. cholerae have been documented, with O1 and O139 being the only serotypes associated with epidemic cholera (9). Cholera can occur both as endemic disease with seasonal peaks and in epidemics associated with floods, droughts, and cyclones that occur in the country (10). Infections other than cholera are caused by non-epidemic V. cholerae serogroups, collectively referred to as V. cholerae non-O1/non-O139, with infections reported worldwide (11, 12).
According to World Health Organization guidelines, oral rehydration is the therapy of choice for V. cholerae infections, independent of serotype (13). It is recommended that severe infections and septicemia be treated with antimicrobial therapy, choosing an effective antibiotic according to local antibiotic susceptibility patterns. Doxycycline is recommended as the first-line treatment for V. cholerae O1 or O139 infections in adults, while erythromycin or azithromycin are recommended for children and pregnant women. Ciprofloxacin and/or third-generation cephalosporins (ceftazidime and ceftriaxone) are recommended for V. cholerae non-O1/non-O139 infections (14).
After the introduction of antibiotics into clinical practice, V. cholerae remained relatively susceptible till the end of the 1970s (15). Within a few years however, this scenario changed dramatically, with V. cholerae strains found to be resistant to commonly used drugs worldwide (16–18), a phenomenon likely attributable to indiscriminate use of antibiotics. Today, V. cholerae can be resistant virtually to all commonly used antibiotics, including ampicillin, quinolones, ciprofloxacin, tetracycline, cotrimoxazole, and macrolides (19). The reasons are multiple and rely on chromosomal mutations, enhanced efflux pumps, and acquisition of drug altering enzymes via horizontal gene transfer (20). The latter has proven to be the most powerful, as a result of a variety of mobile elements circulating amongst V. cholerae, such as conjugative plasmids (21, 22), integrative conjugative elements (23), and mobile genomic islands (24).
The full extent of antibiotic resistance in V. cholerae is not yet known because of limited data. Non-cholera Vibrio infections are not mandatorily notifiable in several countries and annual figures on cholera cases may be significantly under estimated, especially when labeled as “acute watery diarrhea,” in south-eastern and central Asia (10). The first clinical multi-resistant V. cholerae O1 in Bangladesh was isolated in 1979, displaying plasmid-mediated resistance to tetracycline, ampicillin, kanamycin, streptomycin, and trimethoprim–sulfamethoxazole (25). Several studies since and the current dissemination of carbapenemases and ESBLs make it mandatory to understand this phenomenon, especially because of the higher mortality, morbidity, and increased health treatment costs associated with resistance to β-lactams (26).
Third-Generation Cephalosporin and Carbapenem Resistance in V. cholerae
The first reports of V. cholerae O1 showing strong reduced susceptibility to cefotaxime, ceftazidime, and/or aztreonam appeared during the first Argentinean cholera outbreak, which was caused by an ESBL-producing isolate in the 1990s (27). This phenotype was associated with two plasmid-mediated ESBLs of the CTX-M- and PER-2-type (28). After a long hiatus, new cases of reduced susceptibility to third-generation cephalosporins were described in V. cholerae O1, mostly in the Indian Subcontinent. Resistance to ceftriaxone was originally reported in pediatric cases from Puducherry, India in 2008–2010 (29, 30) and similar findings were described in Karnataka, South India, where cephalosporin-resistant strains were found to produce ESBLs (31). Due to lack of genetic analysis, the exact nature of the resistance mechanisms of the reduced susceptibilities is not clear. The most recent genetic characterization of multidrug-resistant, ESBL-producing V. cholerae was a plasmid-borne blaTEM-63 in V. cholerae O1 associated with a cholera outbreak in South Africa in 2008 (32), and an ISCR1-mediated blaPER-1 embedded in a class 1 integron on a conjugative IncA/C plasmid in a clinical V. cholerae non-O1/non-O139 isolate from human blood in China (33).
Clearly, extended-spectrum β-lactamases are uncommon in V. cholerae, as well as in other Vibrionaceae, with only two findings to date. Vibrio fluvialis isolated from cholera-like diarrheal patients in West Bengal, India in 2009 encoded a 150-kb plasmid harboring blaSHV and blaCTX-M-3, together with the quinolone resistance gene qnrA1, and ciprofloxacin-resistance gene aac(6)-Ib-cr (34). Vibrio parahaemolyticus of food origin from China was reported to carry either the AmpC β-lactamase blaCMY-2 on a 150-kb IncA/C-type conjugative plasmid, previously described in Enterobacteriaceae (35), or a 200-kb conjugative plasmid encoding blaPER-1, conferring resistance to both third- and fourth-generation cephalosporins (36, 37).
Carbapenemase-producing V. cholerae has been reported at a higher rate than ESBLs. The first description in Western countries was in southern France, where V. cholerae non-O1/non-O139 was isolated from a yellow-legged gull and found to encode both blaVIM-1 and blaVIM-4 on an IncA/C plasmid (38). Shortly after, the novel transferable carbapenemase blaVCC-1 was identified in a non-toxigenic strain of V. cholerae non-O1/non-O139 during antimicrobial resistance surveillance of food in Canada (39). VCC-1, the first class A carbapenemase to be found in a member of the Vibrionaceae, can hydrolyze penicillin, first-generation cephalosporins, aztreonam, and carbapenems but not second- and third-generation cephalosporins. Most notably, blaNDM-1 has been detected in environmental V. cholerae non-O1/non-O139 in southern Vietnam (40), in clinical V. cholerae O1 in India together with the AmpC β-lactamase blaDHA gene (41), in a polymicrobial infection (V. cholerae, Acinetobacter baumannii, Staphylococcus aureus, and Pseudomonas aeruginosa) in the UK (42), and from water seepage in New Delhi, India (43). The successful spread of blaNDM-1 can be attributed to its association with conjugative plasmids (44) and emphasizes the extent to which blaNDM-1 can disseminate among different species outside of the Enterobacteriaceae.
ESBL- and Carbapenemase-Mediated Resistance in Bangladesh
Antibiotic resistance is a serious threat in Bangladesh and has been for decades, most likely a result of unrestricted use of antimicrobial drugs to treat enteric infections, particularly those caused by V. cholerae, Salmonella, Shigella, and enterotoxigenic E. coli (45). In the recent years, ESBL- and carbapenemase-mediated resistance has been found to be ubiquitous and has been detected in a variety of bacterial hosts.
blaCTX-M-15 is the dominant ESBL variant circulating in Bangladesh. It has been reported mainly in E. coli found in wild birds and aquatic systems (46), in household pigeons (47), poultry (48), urban surface water (49), and in epidemic E. coli isolates from both patients and crows scavenging poorly managed hospital waste dumps (50). On occasion, other ESBL genes have been found to be associated with blaCTX-M-15, such as blaCTX-M-14 in wild birds (51), or blaCTX-M-27, blaSHV-2, and/or blaSHV-12 in E. coli and Enterobacter cloacae from environmental urban water (52). The clinical scenario is not very different, with blaCTX-M-15 prevailing (53), although bacterial species other than E. coli with the ESBL phenotype have been described (54–58). The first clinical Salmonella typhi positive for both blaTEM and blaCTX-M was recently reported to have been isolated from diarrheal patients in Dhaka (59). Overall, dissemination of ESBLs in Bangladesh seems to have reached all ecological niches, indicating that environmental contamination by antibiotic resistance is already quite high and probably widespread, from coastlines of the Bay of Bengal, to urban Dhaka, and to rural inland areas.
Emergence of blaNDM-1-mediated carbapenemase resistance was first described in Bangladesh in the Enterobacteriaceae in 2010 (60). Subsequent retrospective studies demonstrated the presence of blaNDM-1 in clinical Klebsiella pneumoniae isolated in 2008 (61). The same investigators also reported a 9% prevalence of fecal carriage of plasmid-encoded blaNDM-1 in diverse Enterobacteriaceae in the patient population (62). The problematic spread of carbapenemase in Bangladesh has been documented with the isolation of clinical A. baumannii, P. aeruginosa, and K. pneumoniae carrying genes encoding multiple enzymes, i.e., blaVIM-1, blaVIM-2, blaIMP-1, blaIMP-2, and/or blaNDM-1 (63). The same bacterial species have been detected in environmental water/sewage samples collected in Dhaka (64), documenting a high level of environmental distribution of blaNDM-1, a worrisome finding given the high levels of sewage-derived bacteria routinely isolated from drinking water in Bangladesh (65).
The role of plasmids in the successful spread of β-lactamase genes has been extensively described (66, 67) and their involvement in antibiotic resistance epidemiology in Bangladesh is no different. Although limited data are available, conjugative plasmids of various sizes (20–100 MDa) have been reported to carry ESBL genes (53, 61, 68), blaNDM-1, and/or other carbapenemases (60, 63), mostly in the metropolitan area of Dhaka.
Antimicrobial Resistance Surveillance in V. cholerae in Bangladesh
To date, ESBL or carbapenemase-producing V. cholerae in Bangladesh have not been reported. V. cholerae, E. coli, as well as other Enterobacteriaceae, can coexist in different ecological niches. Given the ability of conjugative plasmids to transfer naturally between enterobacterial populations in the intestinal gut (68), the aquatic environment is now an ideal setting for acquisition and dissemination of antibiotic resistance (69), and the horizontal transfer of ESBL/carbapenemase genes to V. cholerae cannot be excluded.
In this perspective, we investigated β-lactam resistance1 in V. cholerae O1 collected during ecological and epidemiological surveillance in Bangladesh, the sampling and isolation details of which are described elsewhere (70). A total of 460 V. cholerae O1 isolates were collected between 2009 and 2014 in the provinces of Mathbaria (MB; southwestern Bangladesh) and Chhatak (CH; northeastern Bangladesh) and analyzed. The set of strains included clinical (C) and environmental (E) isolates (MB: C = 178 and E = 120; CH: C = 141 and E = 21), either from fecal samples of cholera patients at local health-care facilities or from ponds used for drinking water and other domestic purposes.
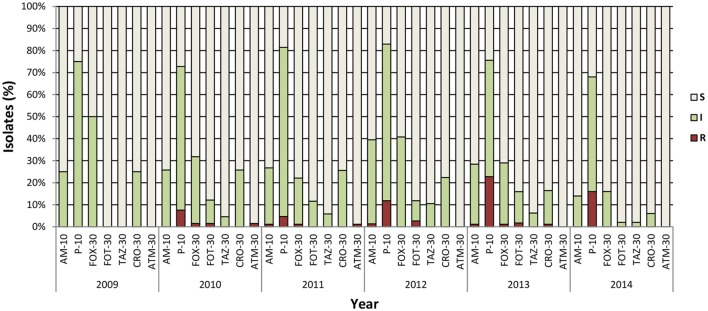
Resistance to penicillin (ampicillin and penicillin), monobactams (aztreonam), carbapenems (imipenem), second- (cefoxitin), third- (cefotaxime, ceftazidime, and ceftriaxone), and fourth- (cefepime) generation cephalosporins was tested by disk diffusion, and the results were interpreted according to CLSI clinical breakpoints, for V. cholerae (71) or Enterobacteriaceae (72). All 460 V. cholerae isolates were found to be susceptible to imipenem and to fourth-generation cephalosporin cefepime. Seventy-two isolates showed resistance to one or more third-generation cephalosporins (cefoxitin, cefotaxime, and ceftazidime) ampicillin, and aztreonam (Table 1). Of those isolates, 57 were resistant only to penicillin. Intrinsic resistance to penicillin has been observed in several V. cholerae strains, including V. cholerae N16961 (73), and it is likely to be mediated by penicillin insensitive transglycolase domains in penicillin binding proteins PBP1A and PBP1B. The majority of isolates were also resistant to carbenicillin (data not shown), very likely correlated with intrinsic resistance to penicillin, as described previously in V. parahaemolyticus (74). The second most common resistance was to third-generation cephalosporin, cefotaxime alone, or in combination with penicillin, ceftriaxone, or aztreonam. The latter was found only in Mathbaria. Overall, no remarkable difference was observed between the two geographical locations (Table 1) or during the 5 years of the study (Figure 1). As expected, the majority of resistant isolates were of clinical origin, compared to environmental sources (Table 1), and the most extensive resistance profile was observed in a clinical strain isolated in 2013 from Mathbaria (Am, Pen, Fox, and Cro). Interestingly, several clinical isolates showed an intermediate phenotype, indicating evolution toward a resistant phenotype in the clinical environment, where the selective pressure of antibiotic use/misuse is higher than in the aquatic environment, where antibiotics may be less prevalent or at lower concentration.
Table 1.
Susceptibility vs. resistance (%) in V. cholerae O1 isolates from Bangladesh.
| Antibiotica | Mathbaria (n = 298) |
Chhatak (n = 162) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinical (n = 178) |
Environmental (n = 120) |
Clinical (n = 141) |
Environmental (n = 21) |
|||||||||
| S | I | R | S | I | R | S | I | R | S | I | R | |
| Ampicillin | 63 | 35 | 2 | 73 | 26 | 1 | 80 | 20 | 0 | 90 | 10 | 0 |
| Penicillin | 16 | 67 | 17 | 23 | 63 | 14 | 35 | 52 | 13 | 19 | 76 | 5 |
| Cefoxitin | 62 | 36 | 2 | 77 | 22 | 1 | 77 | 23 | 0 | 81 | 19 | 0 |
| Cefotaxime | 82 | 17 | 1 | 87 | 12 | 1 | 95 | 3 | 2 | 100 | 0 | 0 |
| Ceftazidime | 93 | 7 | 0 | 92 | 8 | 0 | 96 | 4 | 0 | 100 | 0 | 0 |
| Ceftriaxone | 75 | 24 | 1 | 84 | 16 | 0 | 84 | 16 | 0 | 95 | 5 | 0 |
| Aztreonam | 84 | 15 | 1 | 89 | 10 | 1 | 88 | 12 | 0 | 95 | 5 | 0 |
S, susceptible; I, intermediate; R, resistant.
aAll isolates were susceptible to cefepime and imipenem.
Figure 1.
Antibiotic sensitivity pattern of V. cholerae O1 during epidemiological surveillance (2009–2014). Number of isolates varied per year depending on results of sampling: 2009 (n = 4), 2010 (n = 66), 2011 (n = 86), 2012 (n = 76), 2013 (n = 176), and 2014 (n = 50). AM, ampicillin; P, penicillin; FOX, cefoxitin; FOT, cefotaxime; TAZ, ceftazidime; CRO, ceftriaxone; ATM, aztreonam. Concentrations are expressed as micrograms per disk. All isolates were susceptible to cefepime and imipenem. S, susceptible; I, intermediate; R, resistant.
Isolates showing reduced susceptibility were screened to detect the ESBL (blaCTX, blaTEM, and blaSHV), AmpC (blaMOX, blaCIT, blaDHA, blaACC, blaEBC, and blaFOX), and carbapenemase genes (blaIMP, blaSPM, blaVIM, blaBIC, blaNDM, blaKPC, blaAIM, blaSIM, blaDIM, and blaGIM), as previously reported (75–79). The results were negative for all isolates. The combination disk test results with clavulanic acid to detect ESBL for cefotaxime and ceftazidime-resistant isolates (72) and the phenotypic AmpC disk test with Tris–EDTA (80) were also negative, confirming the absence of an ESBL/AmpC phenotype. To date, the enzyme(s) responsible for the reduced susceptibility phenotype have not been identified. The presence of alternative resistance mechanisms or an intrinsic resistance cannot be ruled out, as observed earlier for V. cholerae non-O1/non-O139 in Germany (81) and for V. parahaemolyticus isolates from shellfish in Italy (82).
Concluding Remarks
Vibrio cholerae remains quite susceptible to β-lactams, despite the fact that other enteric pathogens, mostly Enterobacteriaceae, have developed this resistance in the same geographic regions (52, 59, 63). Yet, a trend showing an increase in intermediate-susceptible isolates was observed, especially in clinical settings, highlighting a developmental path to a resistance phenotype.
The contribution of conjugative plasmids, or other mobile elements, in the horizontal acquisition of genetic factors conferring β-lactam resistance must not be underestimated. It has been established that V. cholerae, independent of serotype, has a plastic genome and a long history of successful association with plasmids that have helped to shape the multi-resistant phenotype that now characterizes this pathogen (18, 22, 24). Plasmids encoding ESBLs have been shown to possess a wide bacterial host range (67) and the ICEs of the SXT/R391 family play a major role in antibiotic resistance acquisition by the Vibrionaceae (83). ICEPmiJpn1, encoding blaCMY-2 and conferring resistance to third-generation cephalosporins (77), can successfully be transferred among clinically relevant Enterobacteriaceae and readily disseminated to V. cholerae, one of its natural bacterial hosts. V. cholerae may very well act as an environmental reservoir for antibiotic resistance genes, contributing to genetic plasticity and dissemination. Finally, the presence of β-lactamase-producing bacteria in the aquatic environment renders environmental surveillance mandatory in order to monitor the role of the natural environment in the distribution of antibiotic resistance and to track potentially clinically relevant V. cholerae isolates.
Author Contributions
The work was conceived and performed by DC. All authors discussed, read, contributed to, and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Mr. Shah M. Rashed, Mrs. Sadaf Yahyai, and Miss. Mahasan Kalizadeh (present and past members of the Colwell/Huq lab at the University of Maryland College Park) for technical assistance, and all the members of Dr. MA’s group at the International Centre for Diarrheal Disease Research, Bangladesh (icddr,b) for their valuable and relentless clinical and environmental sampling throughout the years. icddr,b gratefully acknowledges the following donors: the governments of Australia, Bangladesh, Canada, Sweden, and UK for providing core/unrestricted support.
Footnotes
1Routine antibiotic resistance screening was also performed, results of which are beyond the scope of this article and will be reported elsewhere.
Funding
This research was supported by National Institute of Allergy and Infectious Disease (NIAID) grant no. 2RO1A1039129-11A2 from the National Institutes of Health (NIH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wilke MS, Lovering AL, Strynadka NC. Beta-lactam antibiotic resistance: a current structural perspective. Curr Opin Microbiol (2005) 8:525–33. 10.1016/j.mib.2005.08.016 [DOI] [PubMed] [Google Scholar]
- 2.Fisher JF, Meroueh SO, Mobashery S. Bacterial resistance to β-lactam antibiotics: compelling opportunism, compelling opportunity. Chem Rev (2005) 105:395–424. 10.1021/cr030102i [DOI] [PubMed] [Google Scholar]
- 3.Bush K, Fisher JF. Epidemiological expansion, structural studies, and clinical challenges of new β-lactamases from Gram-negative bacteria. Annu Rev Microbiol (2011) 65:455–78. 10.1146/annurev-micro-090110-102911 [DOI] [PubMed] [Google Scholar]
- 4.Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev (2007) 20:440–58. 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother (2009) 53:5046–54. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dortet L, Poirel L, Nordmann P. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res Int (2014) 249856:26. 10.1155/2014/249856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sirajul Islam M, Brooks A, Kabir MS, Jahid IK, Shafiqul Islam M, Goswami D, et al. Faecal contamination of drinking water sources of Dhaka city during the 2004 flood in Bangladesh and use of disinfectants for water treatment. J Appl Microbiol (2007) 103:80–7. 10.1111/j.1365-2672.2006.03234.x [DOI] [PubMed] [Google Scholar]
- 8.Grant SL, Tamason CC, Hoque BA, Jensen PK. Drinking cholera: salinity levels and palatability of drinking water in coastal Bangladesh. Trop Med Int Health (2015) 20:455–61. 10.1111/tmi.12455 [DOI] [PubMed] [Google Scholar]
- 9.Harris JB, Larocque RC, Qadri F, Ryan ET, Calderwood SB. Cholera. Lancet (2012) 379:2466–76. 10.1016/S0140-6736(12)60436-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO. Cholera in 2014. Wkly Epidemiol Rec (2015) 90:517–44. [PubMed] [Google Scholar]
- 11.Hao Y, Wang Y, Bi Z, Sun B, Jin Y, Bai Y, et al. A case of non-O1/non-O139 Vibrio cholerae septicemia and meningitis in a neonate. Int J Infect Dis (2015) 35:117–9. 10.1016/j.ijid.2015.05.004 [DOI] [PubMed] [Google Scholar]
- 12.Engel MF, Muijsken MA, Mooi-Kokenberg E, Kuijper EJ, Van Westerloo DJ. Vibrio cholerae non-O1 bacteraemia: description of three cases in the Netherlands and a literature review. Euro Surveill (2016) 21. 10.2807/1560-7917.ES.2016.21.15.30197 [DOI] [PubMed] [Google Scholar]
- 13.Global Task Force on Cholera Control. Cholera Outbreak: Assessing the Outbreak Response and Improving Preparedness. Geneva: World Health Organization; (2004). [Google Scholar]
- 14.Daniels NA, Shafaie A. A review of pathogenic Vibrio infections for clinicians. Infect Med (2000) 17:665–85. [Google Scholar]
- 15.O’Grady F, Lewis MJ, Pearson NJ. Global surveillance of antibiotic sensitivity of Vibrio cholerae. Bull World Health Organ (1976) 54:181–5. [PMC free article] [PubMed] [Google Scholar]
- 16.Hedges RW, Vialard JL, Pearson NJ, O’Grady F. R plasmids from Asian strains of Vibrio cholerae. Antimicrob Agents Chemother (1977) 11:585–8. 10.1128/AAC.11.4.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young HK, Amyes SG. Plasmid trimethoprim resistance in Vibrio cholerae: migration of the type I dihydrofolate reductase gene out of the Enterobacteriaceae. J Antimicrob Chemother (1986) 17:697–703. 10.1093/jac/17.6.697 [DOI] [PubMed] [Google Scholar]
- 18.Coppo A, Colombo M, Pazzani C, Bruni R, Mohamud KA, Omar KH, et al. Vibrio cholerae in the horn of Africa: epidemiology, plasmids, tetracycline resistance gene amplification, and comparison between O1 and non-O1 strains. Am J Trop Med Hyg (1995) 53:351–9. [DOI] [PubMed] [Google Scholar]
- 19.Kitaoka M, Miyata ST, Unterweger D, Pukatzki S. Antibiotic resistance mechanisms of Vibrio cholerae. J Med Microbiol (2011) 60:397–407. 10.1099/jmm.0.023051-0 [DOI] [PubMed] [Google Scholar]
- 20.Ghosh A, Ramamurthy T. Antimicrobials & cholera: are we stranded? Indian J Med Res (2011) 133:225–31. [PMC free article] [PubMed] [Google Scholar]
- 21.Ceccarelli D, Salvia AM, Sami J, Cappuccinelli P, Colombo MM. New cluster of plasmid-located class 1 integrons in Vibrio cholerae O1 and a dfrA15 cassette-containing integron in Vibrio parahaemolyticus isolated in Angola. Antimicrob Agents Chemother (2006) 50:2493–9. 10.1128/AAC.01310-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan JC, Ye R, Wang HQ, Xiang HQ, Zhang W, Yu XF, et al. Vibrio cholerae O139 multiple-drug resistance mediated by Yersinia pestis pIP1202-like conjugative plasmids. Antimicrob Agents Chemother (2008) 52:3829–36. 10.1128/AAC.00375-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spagnoletti M, Ceccarelli D, Rieux A, Fondi M, Taviani E, Fani R, et al. Acquisition and evolution of SXT-R391 integrative conjugative elements in the seventh-oandemic Vibrio cholerae lineage. MBio (2014) 5:e1356–1314. 10.1128/mBio.01356-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carraro N, Rivard N, Ceccarelli D, Colwell RR, Burrus V. IncA/C conjugative plasmids mobilize a new family of multidrug resistance islands in clinical Vibrio cholerae non-O1/non-O139 isolates from Haiti. MBio (2016) 7:509–16. 10.1128/mBio.00509-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glass RI, Huq I, Alim AR, Yunus M. Emergence of multiply antibiotic-resistant Vibrio cholerae in Bangladesh. J Infect Dis (1980) 142:939–42. 10.1093/infdis/142.6.939 [DOI] [PubMed] [Google Scholar]
- 26.Pitout JD. Infections with extended-spectrum beta-lactamase-producing Enterobacteriaceae: changing epidemiology and drug treatment choices. Drugs (2010) 70:313–33. 10.2165/11533040-000000000-00000 [DOI] [PubMed] [Google Scholar]
- 27.Rossi A, Galas M, Binztein N, Rivas M, Caffer MI, Corso A, et al. Unusual multiresistant Vibrio cholerae O1 El Tor in Argentina. Lancet (1993) 342:1172–3. 10.1016/0140-6736(93)92155-M [DOI] [PubMed] [Google Scholar]
- 28.Petroni A, Corso A, Melano R, Cacace ML, Bru AM, Rossi A, et al. Plasmidic extended-spectrum beta-lactamases in Vibrio cholerae O1 El Tor isolates in Argentina. Antimicrob Agents Chemother (2002) 46:1462–8. 10.1128/AAC.46.5.1462-1468.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandal J, Dinoop KP, Parija SC. Increasing antimicrobial resistance of Vibrio cholerae O1 biotype E1 tor strains isolated in a tertiary-care centre in India. J Health Popul Nutr (2012) 30:12–6. 10.3329/jhpn.v30i1.11270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandal J, Preethi V, Vasanthraja R, Srinivasan S, Parija SC. Resistance to ceftriaxone in Vibrio cholerae. Indian J Med Res (2012) 136:674–5. [PMC free article] [PubMed] [Google Scholar]
- 31.Bhattacharya D, Dey S, Roy S, Parande MV, Telsang M, Seema MH, et al. Multidrug-resistant Vibrio cholerae O1 was responsible for a cholera outbreak in 2013 in Bagalkot, North Karnataka. Jpn J Infect Dis (2015) 68:347–50. 10.7883/yoken.JJID.2014.257 [DOI] [PubMed] [Google Scholar]
- 32.Ismail H, Smith AM, Sooka A, Keddy KH. Genetic characterization of multidrug-resistant, extended-spectrum-beta-lactamase-producing Vibrio cholerae O1 outbreak strains, Mpumalanga, South Africa, 2008. J Clin Microbiol (2011) 49:2976–9. 10.1128/JCM.00293-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J, Xie L, Zhang F, Ni Y, Sun J. Molecular characterization of ISCR1-mediated blaPER-1 in a non-O1, non-O139 Vibrio cholerae strain from China. Antimicrob Agents Chemother (2015) 59:4293–5. 10.1128/AAC.00166-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chowdhury G, Pazhani GP, Nair GB, Ghosh A, Ramamurthy T. Transferable plasmid-mediated quinolone resistance in association with extended-spectrum beta-lactamases and fluoroquinolone-acetylating aminoglycoside-6’-N-acetyltransferase in clinical isolates of Vibrio fluvialis. Int J Antimicrob Agents (2011) 38:169–73. 10.1016/j.ijantimicag.2011.04.013 [DOI] [PubMed] [Google Scholar]
- 35.Li R, Lin D, Chen K, Wong MH, Chen S. First detection of AmpC beta-lactamase bla(CMY-2) on a conjugative IncA/C plasmid in a Vibrio parahaemolyticus isolate of food origin. Antimicrob Agents Chemother (2015) 59:4106–11. 10.1128/AAC.05008-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong MH, Liu M, Wan HY, Chen S. Characterization of extended-spectrum-β-lactamase-producing Vibrio parahaemolyticus. Antimicrob Agents Chemother (2012) 56:4026–8. 10.1128/AAC.00385-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu M, Wong MH, Chen S. Molecular characterisation of a multidrug resistance conjugative plasmid from Vibrio parahaemolyticus. Int J Antimicrob Agents (2013) 42:575–9. 10.1016/j.ijantimicag.2013.08.014 [DOI] [PubMed] [Google Scholar]
- 38.Aberkane S, Compain F, Barraud O, Ouedraogo AS, Bouzinbi N, Vittecoq M, et al. Non-O1/non-O139 Vibrio cholerae avian isolate from France cocarrying the bla(VIM-1) and bla(VIM-4) genes. Antimicrob Agents Chemother (2015) 59:6594–6. 10.1128/AAC.00400-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mangat CS, Boyd D, Janecko N, Martz SL, Desruisseau A, Carpenter M, et al. Characterization of VCC-1, a novel Ambler class A carbapenemase from Vibrio cholerae isolated from imported retail shrimp sold in Canada. Antimicrob Agents Chemother (2016) 60:1819–25. 10.1128/AAC.00502-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diep TT, Nguyen NT, Nguyen TN, An HK, Nguyen TQ, Nguyen VH, et al. Isolation of New Delhi metallo-beta-lactamase 1-producing Vibrio cholerae non-O1, non-O139 strain carrying ctxA, st and hly genes in southern Vietnam. Microbiol Immunol (2015) 59:262–7. 10.1111/1348-0421.12248 [DOI] [PubMed] [Google Scholar]
- 41.Mandal J, Sangeetha V, Ganesan V, Parveen M, Preethi V, Harish BN, et al. Third-generation cephalosporin-resistant Vibrio cholerae, India. Emerg Infect Dis (2012) 18:1326–8. 10.3201/eid1808.111686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Darley E, Weeks J, Jones L, Daniels V, Wootton M, Macgowan A, et al. NDM-1 polymicrobial infections including Vibrio cholerae. Lancet (2012) 380:1358. 10.1016/S0140-6736(12)60911-8 [DOI] [PubMed] [Google Scholar]
- 43.Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis (2011) 11:355–62. 10.1016/S1473-3099(11)70059-7 [DOI] [PubMed] [Google Scholar]
- 44.Nordmann P, Poirel L, Walsh TR, Livermore DM. The emerging NDM carbapenemases. Trends Microbiol (2011) 19:588–95. 10.1016/j.tim.2011.09.005 [DOI] [PubMed] [Google Scholar]
- 45.Sack RB, Rahman M, Yunus M, Khan EH. Antimicrobial resistance in organisms causing diarrheal disease. Clin Infect Dis (1997) 24:S102–5. 10.1093/clinids/24.Supplement_1.S102 [DOI] [PubMed] [Google Scholar]
- 46.Rashid M, Rakib MM, Hasan B. Antimicrobial-resistant and ESBL-producing Escherichia coli in different ecological niches in Bangladesh. Infect Ecol Epidemiol (2015) 5:26712. 10.3402/iee.v3405.26712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hasan B, Islam K, Ahsan M, Hossain Z, Rashid M, Talukder B, et al. Fecal carriage of multi-drug resistant and extended spectrum beta-lactamases producing E. coli in household pigeons, Bangladesh. Vet Microbiol (2014) 168:221–4. 10.1016/j.vetmic.2013.09.033 [DOI] [PubMed] [Google Scholar]
- 48.Hasan B, Sandegren L, Melhus Å, Drobni M, Hernandez J, Waldenström J, et al. Antimicrobial drug-resistant Escherichia coli in wild birds and free-range poultry, Bangladesh. Emerg Infect Dis (2012) 18:2055. 10.3201/eid1812.120513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kamruzzaman M, Shoma S, Bari SM, Ginn AN, Wiklendt AM, Partridge SR, et al. Genetic diversity and antibiotic resistance in Escherichia coli from environmental surface water in Dhaka City, Bangladesh. Diagn Microbiol Infect Dis (2013) 76:222–6. 10.1016/j.diagmicrobio.2013.02.016 [DOI] [PubMed] [Google Scholar]
- 50.Hasan B, Olsen B, Alam A, Akter L, Melhus Å. Dissemination of the multidrug-resistant extended-spectrum β-lactamase-producing Escherichia coli O25b-ST131 clone and the role of house crow (Corvus splendens) foraging on hospital waste in Bangladesh. Clin Microbiol Infect (2015) 21:.e1–4. 10.1016/j.cmi.2015.06.016 [DOI] [PubMed] [Google Scholar]
- 51.Hasan B, Melhus A, Sandegren L, Alam M, Olsen B. The gull (Chroicocephalus brunnicephalus) as an environmental bioindicator and reservoir for antibiotic resistance on the coastlines of the Bay of Bengal. Microb Drug Resist (2014) 20:466–71. 10.1089/mdr.2013.0233 [DOI] [PubMed] [Google Scholar]
- 52.Haque A, Yoshizumi A, Saga T, Ishii Y, Tateda K. ESBL-producing Enterobacteriaceae in environmental water in Dhaka, Bangladesh. J Infect Chemother (2014) 20:735–7. 10.1016/j.jiac.2014.07.003 [DOI] [PubMed] [Google Scholar]
- 53.Lina TT, Khajanchi BK, Azmi IJ, Islam MA, Mahmood B, Akter M, et al. Phenotypic and molecular characterization of extended-spectrum beta-lactamase-producing Escherichia coli in Bangladesh. PLoS One (2014) 9:e108735. 10.1371/journal.pone.0108735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rahman MM, Haq JA, Hossain MA, Sultana R, Islam F, Islam AHMS. Prevalence of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in an urban hospital in Dhaka, Bangladesh. Int J Antimicrob Agents (2004) 24:508–10. 10.1016/j.ijantimicag.2004.05.007 [DOI] [PubMed] [Google Scholar]
- 55.Mowla R, Imam KM, Asaduzzaman M, Nasrin N, Raihan SZ, Chowdhury AK. Emergence of multidrug resistant extended-spectrum beta-lactamase producing Escherichia coli associated with urinary tract infections in Bangladesh. J Basic Clin Pharm (2011) 3:225–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Begum S, Salam MA, Alam Kh F, Begum N, Hassan P, Haq JA. Detection of extended spectrum beta-lactamase in Pseudomonas spp. isolated from two tertiary care hospitals in Bangladesh. BMC Res Notes (2013) 6:7. 10.1186/1756-0500-6-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yasmin T, Hossain MA, Paul SK, Sarkar SR, Kabir MR, Rahman MM, et al. Detection of TEM, SHV and CTX-M in Mymensingh region in Bangladesh. Mymensingh Med J (2013) 22:465–72. [PubMed] [Google Scholar]
- 58.Masud MR, Afroz H, Fakruddin M. Prevalence of extended-spectrum beta-lactamase positive bacteria in radiologically positive urinary tract infection. Springerplus (2014) 3:216. 10.1186/2193-1801-3-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ahmed D, Ud-Din AIMS, Wahid SUH, Mazumder R, Nahar K, Hossain A. Emergence of blaTEM type extended-spectrum β-lactamase producing Salmonella spp. in the urban area of Bangladesh. ISRN Microbiol (2014) 2014:3. 10.1155/2014/715310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Islam MA, Talukdar PK, Hoque A, Huq M, Nabi A, Ahmed D, et al. Emergence of multidrug-resistant NDM-1-producing Gram-negative bacteria in Bangladesh. Eur J Clin Microbiol Infect Dis (2012) 31:2593–600. 10.1007/s10096-012-1601-2 [DOI] [PubMed] [Google Scholar]
- 61.Islam MA, Huq M, Nabi A, Talukdar PK, Ahmed D, Talukder KA, et al. Occurrence and characterization of multidrug-resistant New Delhi metallo-beta-lactamase-1-producing bacteria isolated between 2003 and 2010 in Bangladesh. J Med Microbiol (2013) 62:62–8. 10.1099/jmm.0.048066-0 [DOI] [PubMed] [Google Scholar]
- 62.Islam MA, Nabi A, Rahman M, Islam M, Ahmed D, Faruque AS, et al. Prevalence of faecal carriage of NDM-1-producing bacteria among patients with diarrhoea in Bangladesh. J Med Microbiol (2014) 63:620–2. 10.1099/jmm.0.064527-0 [DOI] [PubMed] [Google Scholar]
- 63.Farzana R, Shamsuzzaman S, Mamun KZ. Isolation and molecular characterization of New Delhi metallo-beta-lactamase-1 producing superbug in Bangladesh. J Infect Dev Ctries (2013) 7:161–8. 10.3855/jidc.2493 [DOI] [PubMed] [Google Scholar]
- 64.Toleman MA, Bugert JJ, Nizam SA. Extensively drug-resistant New Delhi metallo-beta-lactamase-encoding bacteria in the environment, Dhaka, Bangladesh, 2012. Emerg Infect Dis (2015) 21:1027–30. 10.3201/eid2106.141578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Talukdar PK, Rahman M, Rahman M, Nabi A, Islam Z, Hoque MM, et al. Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS One (2013) 8:e61090. 10.1371/journal.pone.0061090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother (2009) 53:2227–38. 10.1128/AAC.01707-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol (2013) 303:298–304. 10.1016/j.ijmm.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 68.Rashid H, Rahman M. Possible transfer of plasmid mediated third generation cephalosporin resistance between Escherichia coli and Shigella sonnei in the human gut. Infect Genet Evol (2015) 30:15–8. 10.1016/j.meegid.2014.11.023 [DOI] [PubMed] [Google Scholar]
- 69.Marti E, Variatza E, Balcazar JL. The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol (2014) 22:36–41. 10.1016/j.tim.2013.11.001 [DOI] [PubMed] [Google Scholar]
- 70.Alam M, Hasan NA, Sadique A, Bhuiyan NA, Ahmed KU, Nusrin S, et al. Seasonal cholera caused by Vibrio cholerae serogroups O1 and O139 in the coastal aquatic environment of Bangladesh. Appl Environ Microbiol (2006) 72:4096–104. 10.1128/AEM.00066-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.CLSI. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria; Approved Guideline – Second Edition M45-A2. Wayne, PA: Clinical and Laboratory Standards Institute; (2010). [Google Scholar]
- 72.CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twentieth Informational Supplement. CLSI Document M100-S20. Wayne, PA: Clinical and Laboratory Standards Institute; (2010). [Google Scholar]
- 73.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature (2000) 406:477–83. 10.1038/35020000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chiou J, Li R, Chen S. CARB-17 family of beta-lactamases mediates intrinsic resistance to penicillins in Vibrio parahaemolyticus. Antimicrob Agents Chemother (2015) 59:3593–5. 10.1128/AAC.00047-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perez-Perez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol (2002) 40:2153–62. 10.1128/JCM.40.6.2153-2162.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jemima SA, Verghese S. Multiplex PCR for bla(CTX-M) & bla(SHV) in the extended spectrum beta lactamase (ESBL) producing gram-negative isolates. Indian J Med Res (2008) 128:313–7. [PubMed] [Google Scholar]
- 77.Harada S, Ishii Y, Saga T, Tateda K, Yamaguchi K. Chromosomally encoded blaCMY-2 located on a novel SXT/R391-related integrating conjugative element in a Proteus mirabilis clinical isolate. Antimicrob Agents Chemother (2010) 54:3545–50. 10.1128/AAC.00111-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Poirel L, Dortet L, Bernabeu S, Nordmann P. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother (2011) 55:5403–7. 10.1128/AAC.00585-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis (2011) 70:119–23. 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 80.Black JA, Moland ES, Thomson KS. AmpC disk test for detection of plasmid-mediated AmpC beta-lactamases in Enterobacteriaceae lacking chromosomal AmpC beta-lactamases. J Clin Microbiol (2005) 43:3110–3. 10.1128/JCM.43.7.3110-3113.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bier N, Schwartz K, Guerra B, Strauch E. Survey on antimicrobial resistance patterns in Vibrio vulnificus and Vibrio cholerae non-O1/non-O139 in Germany reveals carbapenemase-producing Vibrio cholerae in coastal waters. Front Microbiol (2015) 6:1179. 10.3389/fmicb.2015.01179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ottaviani D, Leoni F, Talevi G, Masini L, Santarelli S, Rocchegiani E, et al. Extensive investigation of antimicrobial resistance in Vibrio parahaemolyticus from shellfish and clinical sources, Italy. Int J Antimicrob Agents (2013) 42:191–3. 10.1016/j.ijantimicag.2013.05.003 [DOI] [PubMed] [Google Scholar]
- 83.Wozniak RA, Fouts DE, Spagnoletti M, Colombo MM, Ceccarelli D, Garriss G, et al. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet (2009) 5:24. 10.1371/journal.pgen.1000786 [DOI] [PMC free article] [PubMed] [Google Scholar]