Abstract
The majority of HIV infections in the United States can be traced back to a single introduction in late 1960s or early 1970s. However, it remains unclear whether subsequent introductions of HIV into the United States have given rise to onward transmission. Genetic transmission networks can aid in understanding HIV transmission. We constructed a genetic distance-based transmission network using HIV-1 pol sequences reported to the U.S. National HIV Surveillance System (n = 41,539) and all publicly available non-U.S. HIV-1 pol sequences (n = 86,215). Of the 13,145 U.S. persons clustered in the network, 457 (3.5%) were genetically linked to a potential transmission partner outside the United States. For internationally connected persons residing in but born outside the United States, 61% had a connection to their country of birth or to another country that shared a language with their country of birth. Bayesian molecular clock phylogenetic analyses indicate that introduced nonsubtype B infections have resulted in onward transmission within the United States.
Keywords: : HIV, transmission network, cluster, surveillance, molecular clock
Introduction
Genetic and phylogenetic approaches have succeeded in elucidating the spread of HIV-1 around the world.1,2 For example, phylogenetic approaches have demonstrated how HIV-1 subtype B migrated across Europe and East Asia.3,4 Moreover, these approaches have documented the introduction and continuous spread of non-B subtypes and circulating recombinant forms (CRFs), for example, in France and the United Kingdom.5,6 Importantly, international routes of HIV migration represent potential points of intervention to reduce the spread of the virus.
The HIV-1 epidemic in the United States likely began as the result of the introduction of a single subtype B founder variant from Haiti during the late 1960s or early 1970s.7 Currently, more than 1.2 million individuals are living with HIV infection in the United States,8 with subtype B accounting for ∼95% of infections.9–11 The remaining 5% of infections within the United States comprise a variety of non-B subtypes and CRFs, which are the result of multiple independent introductions of HIV-1 into the United States. However, evidence for onward transmission of introduced non-B HIV strains is sparse12 (e.g., a single heterosexual couple13).
Transmission networks inferred from genetic sequences allow the study of viral dynamics on a local or national level.14–19 Moreover, these networks provide the means to improve the effect of targeted treatment intervention.20 Recent work by Wertheim et al.2 has demonstrated how the global diversity of HIV can be efficiently queried for transmission links by comparing genetic distances between isolates. This approach can identify transmission clusters: groups of HIV-positive persons whose viral strains are highly genetically similar, implying a direct or indirect transmission link among these individuals.
Since 2001, the Centers for Disease Control and Prevention (CDC) has funded projects in selected jurisdictions to collect HIV sequences from persons newly diagnosed with HIV, which were subsequently reported to the National HIV Surveillance System. We used these surveillance data to characterize instances where a person diagnosed within the United States has a potential transmission partner genotyped in another country and identify clusters suggestive of onward transmission of HIV introduced into the United States.
Methods
Sequence data
We included HIV-1 pol (protease and reverse transcriptase) sequences reported during 2001 through 2012 to the U.S. National HIV Surveillance System through three projects: antiretroviral drug resistance testing (ARVDRT); the dried blood spot (DBS) project; and Variant, Atypical, and Resistant HIV Surveillance (VARHS).21 We included one sequence per person. When more than one sequence had been obtained from an individual, the longest sequence was used; 80% of these sequences were obtained within 1 year since diagnosis. Sequences <500 nucleotides in length were excluded. The resulting U.S. dataset comprised 41,549 sequences genotyped within the United States, representing ∼11% of all persons with a recent HIV diagnosis within the United States. A detailed description of the U.S. National HIV Surveillance System transmission network and related geographic sampling is available in Oster et al.22
All publicly available HIV-1 pol sequences (HXB2 coordinates 2253–3869), one sequence per individual, were downloaded from the Los Alamos National Laboratory (LANL) HIV Sequence Database (www.hiv.lanl.gov/). Again, sequences that were <500 nucleotides in length were excluded. Problematic sequences, defined by LANL as containing ≥5% ambiguous nucleotides, were excluded. Because the focus of this study was international linkage, sequences obtained within the United States were excluded from the LANL dataset. We also excluded LANL sequences without country of sampling information. The resulting non-US LANL dataset comprised 86,490 international sequences.
Constructing transmission networks
To construct a transmission network, we used a local implementation of HIV-TRACE (www.hivtrace.org), following the protocol outlined by Wertheim et al.2 First, sequences were aligned to an HIV-1 reference sequence (HXB2) in a pairwise fashion based on codon structure. Insertions relative to HXB2 were filtered from downstream analyses.
Next, the Tamura-Nei 93 (TN93)23 genetic distance was calculated for all sequence pairs (∼16.3 billion comparisons). TN93 is the most complex nucleotide substitution model that can be represented by a closed-form solution, allowing rapid pairwise genetic distance calculation. More complex nucleotide substitution models do not improve accuracy of short genetic distance estimates.24 Pairs whose genetic distance fell at or below a given TN93 distance threshold (1.5% in primary analyses) were connected (via an edge).
These pairs represent potential transmission partners: individuals whose sequences are far more similar than would be expected by chance. This genetic distance cutoff was selected because after a decade of longitudinal sampling, HIV pol sequences in mono-infected patients typically do not diverge more than 1% from baseline sequences.25 Therefore, a cutoff of 1.5% is slightly conservative compared with the expected 2% divergence between two transmission partners after a decade. We also explored the effects on clustering of more conservative and liberal genetic distance cutoffs (1.0% and 2.0%), and the effect of excluding 48 codons associated with drug resistance mutations.11 To prevent spurious linking of low-quality sequences (i.e., those with a high proportion of nucleotide ambiguities), genetic distances between ambiguous bases (e.g., R, Y, M) and known bases (e.g., A, C, T, G) were averaged over resolutions of the ambiguous base (i.e., Y is treated as 50% C and 50% T).
Finally, we assembled transmission clusters by connecting persons (nodes) who shared potential transmission partners. Therefore, every person in a transmission cluster is the potential transmission partner of at least one other person in that cluster.
Sequences that were unexpectedly similar (≤2.0% TN93 genetic distance) to the HXB2 reference sequence were excluded from the analysis as potential laboratory contaminants. In all, 10 U.S. sequences and 275 LANL sequences were excluded. Therefore, our final dataset included 127,754 sequences.
Among U.S. persons who clustered (either domestically or internationally), we compared the percentage that clustered internationally by demographic and geographic characteristics and transmission category. We also used multivariable regression in SAS, version 9.3 (SAS Institute, Cary, NC) to account for confounders; the model included age, race/ethnicity, transmission category, birthplace, population of area of residence, year of diagnosis, and U.S. census region. Transmission category was hierarchically assigned,8 and information was imputed for individuals missing these data.26
For foreign-born U.S. persons with a potential transmission partner genotyped in another country, we calculated the percentage linked to their country of birth. Among the remainder, we determined what percentage linked to a country that shared an official/dominant language with their country of birth, as listed in the Central Intelligence Agency's World Factbook (www.cia.gov/library/publications/the-world-factbook/).
Phylogenetics and molecular clock analyses
We identified non-B clusters that contained a minimum of four sequences, at least two of which were sampled within the United States and were potential transmission partners. Subtype classification was performed using COMET.27
Non-B clusters with multiple potential transmission partners within the United States were subjected to molecular clock analysis in a Bayesian Markov-chain Monte Carlo framework in BEAST v1.8.0.28,29 Each molecular clock analysis was run for 10,000,000 generations, sampling every 2,500 generations. The first 1,000,000 generations were discarded as burn-in. Analyses were performed using a simple nucleotide substitution model (HKY) and a strict molecular clock under a constant population size. More complicated models (e.g., GTR + Γ4, uncorrelated lognormal relaxed molecular clock, and exponential population growth) were explored, but these additional parameters were not supported by the data (e.g., the exponential growth rate parameter included zero). Convergence and mixing was assessed in Tracer v1.5, and all parameters achieved an effective sample size >200. Within these clusters, we identified strongly supported clades (i.e., posterior probability ≥0.95) of U.S. sequences, which provide evidence for onward transmission within the United States. For computational tractability, the largest non-B cluster with evidence on onward transmission within the United States (2,062 sequences) was broken into sub-clusters for molecular clock analysis.
Results
Of the 41,539 U.S. HIV-1 sequences included in our network analysis, 13,145 (32%) shared a link with another sequence either domestically or internationally and were part of the inferred transmission network (Table 1). Of the sequences from the U.S. National HIV Surveillance System that clustered within the network, 457 (3.5%) shared a link with a sequence sampled outside the United States in the LANL sequence database. Of these 457 U.S. persons, 360 (79%) linked to a subtype B sequence, and 97 (21%) linked to a nonsubtype B sequence. Forty-three U.S. persons linked to CRF01_AE, 28 to CRF02_AG, and 11 to a BG CRF.
Table 1.
Characteristics of Persons with HIV-1 Sequences and Percentage That Cluster Domestically and Internationally
| Total | Clustered domestically only | Clustered internationally | |||||
|---|---|---|---|---|---|---|---|
| Category | n | % | n | % of those clustering | n | % of those clustering | Adjusted prevalence ratioa |
| Total | 41,539 | 100 | 12,688 | 97 | 457 | 3 | — |
| Sex | |||||||
| Male | 32,400 | 78 | 10,987 | 96 | 438 | 4 | — |
| Female | 9,139 | 22 | 1,701 | 99 | 19 | 1 | — |
| Age at HIV diagnosis (year) | |||||||
| <13 | 245 | 1 | 29 | 94 | 2 | 6 | — |
| 13–19 | 2,282 | 5 | 1,154 | 99 | 16 | 1 | 1.8 (0.1–42.6) |
| 20–29 | 13,951 | 34 | 5,836 | 97 | 190 | 3 | 1.3 (0.7–2.1) |
| 30–39 | 11,507 | 28 | 3,090 | 95 | 152 | 5 | 1.3 (0.7–2.1) |
| 40–49 | 8,733 | 21 | 1,810 | 96 | 70 | 4 | 1.0 (0.6–1.8) |
| 50–59 | 3,749 | 9 | 645 | 97 | 18 | 3 | 0.9 (0.5–1.8) |
| ≥60 | 1,072 | 3 | 124 | 93 | 9 | 7 | 2.1 (0.9–4.8) |
| Race/ethnicity | |||||||
| Asian/NHOPI | 779 | 2 | 238 | 84 | 47 | 16 | 10.9 (6.8–17.6) |
| Black/African American | 19,645 | 47 | 5,460 | 99 | 37 | 1 | — |
| Hispanic/Latino | 9,307 | 22 | 2,810 | 95 | 136 | 5 | 3.7 (2.5–5.5) |
| White | 10,755 | 26 | 3,853 | 95 | 224 | 5 | 6.2 (4.3–8.9) |
| Other | 1,053 | 3 | 327 | 96 | 13 | 4 | 4.6 (2.4–8.6) |
| Transmission category | |||||||
| MSM | 25,142 | 61 | 9,596 | 96 | 400 | 4 | — |
| MSM and injection drug use | 1,621 | 4 | 487 | 98 | 12 | 2 | 0.5 (0.3–1.0) |
| Injection drug use (male) | 2,220 | 5 | 305 | 96 | 12 | 4 | 0.8 (0.4–1.6) |
| Injection drug use (female) | 1,692 | 4 | 269 | 99 | 4 | 1 | 0.5 (0.2–1.3) |
| Heterosexual contact (male) | 3,259 | 8 | 579 | 98 | 12 | 2 | 0.8 (0.5–1.5) |
| Heterosexual contact (female) | 7,306 | 18 | 1,418 | 99 | 14 | 1 | 0.5 (0.3–0.8) |
| Other | 300 | 1 | 35 | 92 | 3 | 8 | 1.7 (0.1–27.3) |
| Country of birth | |||||||
| United States | 26,524 | 64 | 8,592 | 97 | 234 | 3 | — |
| U.S. dependency | 574 | 1 | 98 | 92 | 8 | 8 | 1.6 (0.7–3.7) |
| Other (non-U.S.) | 6,170 | 15 | 1,381 | 92 | 120 | 8 | 2.0 (1.6–2.6) |
| Unknown | 8,271 | 20 | 2,617 | 96 | 95 | 4 | 1.3 (1.1–1.7) |
| Population of area of residence at HIV diagnosis | |||||||
| <50,000 | 2,357 | 6 | 680 | 99 | 8 | 1 | — |
| 50,000–499,999 | 4,019 | 10 | 1,200 | 98 | 24 | 2 | 1.4 (0.7–2.9) |
| ≥500,000 | 34,949 | 84 | 10,778 | 96 | 424 | 4 | 1.5 (0.7–3.4) |
| Year of diagnosis | |||||||
| 1999 or earlier | 2,745 | 7 | 108 | 93 | 8 | 7 | 4.4 (2.0–9.7) |
| 2000–2004 | 2,236 | 5 | 316 | 93 | 23 | 7 | 3.0 (1.9–4.7) |
| 2005–2009 | 22,768 | 55 | 7,138 | 96 | 261 | 4 | 1.4 (1.1–1.6) |
| 2010–2012 | 13,790 | 33 | 5,126 | 97 | 165 | 3 | — |
| Region of residence at HIV diagnosisb | |||||||
| Northeast | 12,509 | 30 | 3,130 | 95 | 156 | 5 | 3.4 (2.3–5.0) |
| Midwest | 8,689 | 21 | 3,941 | 98 | 65 | 2 | — |
| South | 12,344 | 30 | 3,034 | 94 | 200 | 6 | 1.6 (1.1–2.5) |
| West | 7,908 | 31 | 2,569 | 99 | 33 | 1 | 2.2 (1.5–3.3) |
Adjusted prevalence ratios were derived from a multivariable model that included all listed categories except sex, which is included as a stratification within transmission category. 95% confidence intervals are shown in parentheses.
Numbers do not sum to total due to persons residing outside the United States at time of diagnosis.
MSM, men who have sex with men; NHOPI, native Hawaiian or other Pacific islander.
Characteristics of persons with international potential transmission partners
In the multivariate analysis, persons in the U.S. National HIV Surveillance System born outside the United States were significantly more likely to have an international connection than those born inside the United States (Table 1). Of the 128 individuals born outside the United States who clustered internationally, 45 (35%) shared a link with a LANL sequence from their country of birth. Of the remaining individuals, 33 (40%) shared a link with a LANL sequence from a country that shared an official/dominant language with their country of birth. Within the United States, the percentage of persons with international links did not vary by population of the area of residence (Table 1).
Among persons in the U.S. National HIV Surveillance System who clustered with at least one other person, the percentage of persons with an international potential transmission partner varied significantly by race/ethnicity (Table 1). Approximately 5% of whites who clustered in the network had an international connection. In contrast, Asians, Native Hawaiians, or Other Pacific Islanders in the United States were more likely to have international connections (16%). Only 1% of clustered blacks/African Americans had an international connection. Hispanics/Latinos had a similar proportion of internationally clustered individuals as whites, around 5%.
Among individuals with different transmission categories, there were significant differences in the percentage that had a domestic transmission partner versus an international transmission partner (Table 1). However, the magnitude of these differences was smaller than those by race/ethnicity.
Improved ability to resolve domestic clusters with international sequences
In total, we inferred 9,814 transmission clusters in the network analysis. The majority, 6,083 clusters, contained only international sequences. Notably, international sequences improved our ability to connect persons in the U.S. National HIV Surveillance System. A total of 31 clusters containing persons genotyped within the United States were linked to other U.S. persons only via international sequences from the LANL database.
Onward transmission within the United States
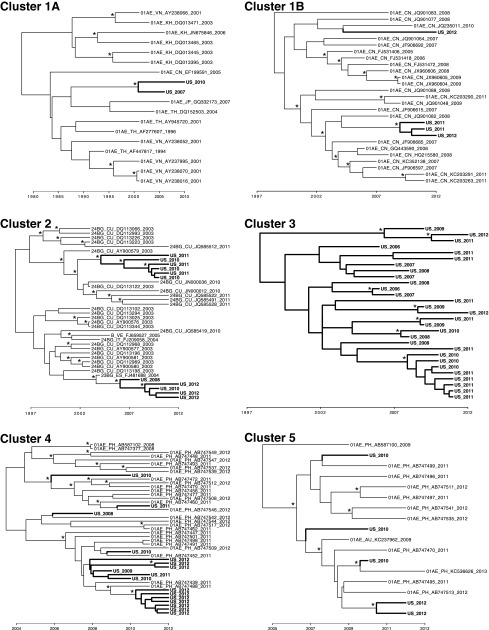
We found phylogenetic evidence for onward transmission of recently introduced non-B strains in the United States. Thirteen non-B clusters included at least four sequences, of which at least two were sampled within the United States and were potential transmission partners. Of these 13 clusters, five were amenable to molecular dating analyses (i.e., ≥10 sequences genotyped over multiple years; Figure 1 and Table 2). Our approach was biased toward detecting more recent introductions, since inclusion in clusters requires the genotyping of a potential transmission partner for every member of the cluster. Within these clusters, most of the U.S. clades (potentially representing onward transmission within the United States; bold in Fig. 1) had a time of most recent common ancestor (tMRCA) within the past decade (Table 2). However, cluster 3 suggests that transmission within the United States has been ongoing since the 1990s. In addition, three of these clusters (1, 2, and 4) have multiple distinct clades of U.S. sequences, pointing to at least two separate introductions of HIV into the United States from the same larger transmission cluster.
FIG. 1.
Maximum clade credibility trees for clusters suggesting onward transmission of nonsubtype B strains imported into the United States. Cluster 1 was subsampled into 1A and 1B for phylogenetic inference and display. U.S. sequences with evidence of onward transmission within the United States are indicated with bold branches. Posterior probability support for a given node ≥0.95 is designated with an asterisk. Time in indicated on the x-axis.
Table 2.
Information About Persons in U.S. Clades Who Are Part of Transmission Clusters with Evidence for Onward Transmission Within the United States
| Cluster | Cluster size | No. sequences in U.S. clade | Subtype/CRF | tMRCAa | Race/ethnicityb | Transmission categoryb | County of birth | U.S. region of residence at diagnosisb |
|---|---|---|---|---|---|---|---|---|
| 1Ac | 2,062 | 2 | CRF01_AE | 2001 (1996–2004) | Asian/NHOPI (2) | Female heterosexual (2) | U.S. (1), Unknown (1) | West (2) |
| 1Bc | 2,062 | 3 | CRF01_AE | 2009 (2007–2010) | Asian/NHOPI (2), White (1) | Female heterosexual (1), MSM (2) | China (2), Unknown (1) | Northeast (3) |
| 2 | 39 | 5 | CRF24_BG | 2006 (2004–2008) | Black (1), Hispanic (1), White (3) | MSM (5) | U.S. (3), Turkey (1), Unknown (1) | Northeast (5) |
| 2 | 39 | 6 | CRF24_BG | 2004 (2002–2006) | Hispanic (2), White (4) | MSM (6) | U.S. (4), Honduras (1), Reunion (1) | Northeast (1), Southeast (4), West (1) |
| 3 | 29 | 29 | CRF02_AG | 1998 (1986–2004) | Asian/NHOPI (1), Black (1), Hispanic (10), White (17) | Male heterosexual (2), MSM (26), MSM/IDU (1) | U.S. (19), Puerto Rico (1), Dominican Republic (2), Columbia (1), Unknown (6) | Midwest (2), Northeast (27) |
| 4 | 45 | 2 | CRF01_AE | 2011 (2010–2012) | White (1), Other (1) | MSM (2) | U.S. (1), Unknown (1) | Midwest (2) |
| 4 | 45 | 7 | CRF01_AE | 2010 (2009–2012) | Hispanic (4), White (3) | MSM (7) | U.S. (2), Mexico (3), Unknown (2) | West (7) |
| 5 | 17 | 2 | CRF01_AE | 2010 (2008–2011) | Asian/NHOPI (1), Hispanic (1) | MSM (2) | U.S. (2) | West (2) |
| 6 | 5 | 5 | A1 | — | Black (4), White (1) | Female heterosexual (5) | U.S. (2), Liberia (1), Jamaica (1), Kenya (1) | Midwest (5) |
| 7 | 5 | 5 | A1 | — | Hispanic (1), White (4) | MSM (5) | U.S. (1), Puerto Rico (1), Israel (1), Unknown (2) | Northeast (5) |
| 8 | 4 | 4 | C | — | Hispanic (3), White (1) | MSM (4) | Italy (1), Brazil (1), Unknown (2) | Northeast (3), West (1) |
| 9 | 4 | 4 | G | — | Black (4) | Female heterosexual (1), Male heterosexual (1), MSM (2) | U.S. (3), Unknown (1) | West (4) |
| 10 | 4 | 4 | CRF01_AE | — | Asian/NHOPI (3), Hispanic (1) | MSM (4) | U.S. (3), Unknown (1) | Northeast (1), West (3) |
| 11 | 7 | 7 | C | — | Black (5), Other (1), White (1) | Female heterosexual (3), Male heterosexual (1), Male IDU (1), MSM (2) | U.S. (3), Unknown (4) | Midwest (7) |
| 12 | 4 | 3 | CRF01_AE | — | Hispanic (2), Other (1) | Female heterosexual (1), Male heterosexual (1), MSM/IDU (1) | Indonesia (1), Columbia (1), Unknown (1) | Northeast (3) |
| 13 | 6 | 6 | CRF02_AG | — | Black (4), Other (1), White (1) | Female heterosexual (3), Female IDU (1), MSM (2) | U.S. (4), Unknown (4) | Midwest (6) |
Time of most recent common ancestor for clusters amenable to molecular clock analysis. Mean and 95% highest posterior density for U.S. potential transmission partners, which are shown in bold in Figure 1.
Number of persons in each category given in parentheses.
Cluster 1 was subsampled into clusters 1A and 1B for phylogenetic inference
CRF, circulating recombinant form; IDU, injection drug use; NHOPI, native Hawaiian or other Pacific islander; tMRCA, time of most recent common ancestor.
The source of the many transmission clusters introduced into the United States can be inferred from their phylogenies (Fig. 1). Cluster 1, which comprises 2,062 sequences, likely originated in Southeast and East Asia and has been previously characterized by Wertheim et al.2 as one of the largest international transmission clusters.2 The U.S. isolates (bold in Fig. 1) were found in Asians and whites residing in the Northeastern United States, two of whom were born in China (Table 2). Cluster 2 is primarily a Cuban cluster, with evidence for two distinct introduction events into the United States that resulted in onward transmission. Notably, none of the men who have sex with men (MSM) that comprise the U.S. portion of the cluster were born in Cuba. Clusters 4 and 5 appear to have originated in the Philippines, and the U.S. members of these clusters are MSM from a variety of racial/ethnic backgrounds, three of whom were born in Mexico.
The oldest introduced transmission cluster in the United States we inferred, cluster 3, does not include any international sequences that might shed light on its country of origin. The U.S. isolates were found in MSM residing in the Northeast United States and were linked to five CRF02_AG sequences from Great Britain15; however, these British sequences were not included in the dating analysis, because they were published without associated genotyping year. Notably, these British sequences were interspersed among the U.S. sequences, implying repeated transmission between these countries. No U.S. person in cluster 3 was born in a region with a high prevalence of CRF02_AG, such as West Africa (where CRF02_AG accounts for 50% of HIV-1 prevalence) or North Africa/Middle East (18% prevalence)9 (Table 2), and no potential transmission partner outside the United States and Great Britain was detected in the LANL database. Therefore, the geographic origin of this cluster remain unclear.
The nine smaller non-B clusters are also suggestive of onward transmission of recently introduced HIV-1 into the United States (clusters 6–13 in Table 2); however, these clusters were too small for molecular dating analyses to determine their tMRCA. These smaller clusters represent a wide array of persons of different race/ethnicities, different transmission risk factors, and HIV subtypes. Of these smaller clusters, only one (cluster 12) included a link to a sequence in the LANL database: an isolate from Singapore (GenBank Accession No. AY870205).
Sensitivity analysis
We explored the sensitivity of our findings on international clustering to the network inference parameters (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/aid). The total number of nodes and edges rises and falls with more liberal and conservative TN93 genetic distance cutoffs (2.0% and 1.0%, respectively) for potential transmission partners. However, the variables that are significantly associated with international linkage are relatively consistent among these cutoffs. Transmission category is not associated with international linkage using these other genetic distance cutoffs, although most transmission risk categories did not differ significantly from each other with respect to the extent of international linkage at the 1.5% TN93 cutoff. Excluding codons associated with drug resistance mutations11 did not have a notable effect on the transmission network or correlates of international linkage.
Discussion
Transmission networks can identify routes of international HIV migration. Of the 13,145 persons in the U.S. National HIV Surveillance System who were part of a transmission cluster, 457 linked to a person who was genotyped outside the United States. The majority of internationally linked U.S. persons who were born outside the United States had a potential transmission partner who was genotyped in either their country of birth or a country that shared an official/dominant language with their country of birth. The importance of shared language has also been demonstrated by work on the Swiss HIV Cohort Study, which found evidence of preferential within-group transmission among German and French speaking persons.16 Although we cannot exclude the possibility that the linked non-U.S. sequences from the LANL database were obtained from the same individuals in the U.S. National HIV Surveillance System, these linkages point to international travel of HIV to countries of birth and shared language.
We found instances of nonsubtype B HIV transmission clusters within the United States. Given that we do not have data on social/familial relationships for persons in the U.S. National HIV Surveillance System, we cannot exclude the possibility that some of the smaller clusters represent migration of multiple HIV-infected potential transmission partners into the United States (e.g., clusters 1A, 1B, and 5). Nevertheless, given the relative rarity of international linkage among persons in the National Surveillance System, the larger clusters (i.e., four or more potential transmission partners within the United States) can most parsimoniously be explained by onward transmission of HIV-1 recently introduced into the United States.
Studies of HIV transmission networks tend to focus on only a single country or geographic region.15,18,19 Here, we found that by including sequences representing the global diversity of HIV, we were able to infer indirect connections between persons within the United States through one or more international sequences. This finding demonstrates the added benefit of including the global diversity of HIV in transmission network analyses, even if the aim of an analysis is local in scope (as suggested by Wertheim et al.2). The relevance of this finding can be seen in the recent work by Takebe et al.,4 who looked at international migration of HIV in Japanese MSM.
Molecular surveillance has the potential to direct HIV intervention and therapies (e.g., treatment-as-prevention and pre-exposure prophylaxis).20 In a public health setting, demographics and transmission category may assist in identifying groups of highest priority for targeted intervention.18,30,31 Given the ability of HIV to spread across large geographic distances and establish onward transmission clusters, these surveillance efforts should be broad in scope.
The analysis presented here is likely affected by a number of limitations related to sequence acquisition both in the United States and abroad. First, the National HIV Surveillance System varies in completeness both by reporting jurisdiction and year. As the National HIV Surveillance System expands data collection to additional jurisdictions and completeness of reporting becomes more complete, this issue will diminish in importance. Moreover, although the National HIV Surveillance System reconciles duplicates within its database, it is possible that some sequences in the LANL database may be from the same person in the National HIV Surveillance System who was also genotyped outside the United States. It is also possible that multiple sequences in the LANL database may be from the same person, despite checks that are in place at LANL to ensure de-duplication.
The sequences deposited in the LANL database are not uniform with respect to geography or HIV burden, since these sequences are submitted as part of published research. Therefore, our approach is biased against finding potential transmission partners between persons in the United States and persons in under-sampled regions of the world, particularly in sub-Saharan Africa. This shortcoming may explain why we were unable to find the country of origin for cluster 3. Increased surveillance of viral genetic diversity in sub-Saharan Africa should help ameliorate this problem.32
Importantly, potential transmission partners identified in this study are not necessarily direct transmission partners. There are likely intermediate infections separating sequences that are connected in our network. Nonetheless, identifying closely related sequences is a good proxy for inferring the path of the virus across a social network.
Conclusion
Genetic transmission network analysis using HIV surveillance data is an important tool for understanding the dynamics of viral spread. Expanded surveillance efforts within the United States and abroad will increase our ability to trace the path of viral spread around the world.
Supplementary Material
Acknowledgments
We acknowledge the local and state health department staff instrumental in collecting HIV sequence data and other present and past members of CDC Incidence and Molecular Epidemiology Team, including Qian An, David Kim, Magan Pearson, Rebecca Ziebell, and Joseph “Buzz” Prejean. We also thank N. Lance Hepler and Sergei L. Kosakovsky Pond for their work on HIV-TRACE. JOW was funded in part by an NIH Career Development Award (K01AI110181).
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Tebit DM, Arts EJ: Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis 2011;11:45–56 [DOI] [PubMed] [Google Scholar]
- 2.Wertheim JO, Leigh Brown AJ, Hepler NL, et al. : The global transmission network of HIV-1. J Infect Dis 2014;209:304–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paraskevis D, Pybus O, Magiorkinis G, et al. : Tracing the HIV-1 subtype B mobility in Europe: A phylogeographic approach. Retrovirology 2009;6:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takebe Y, Naito Y, Raghwani J, et al. : Inter-continental dispersal of HIV-1 subtype B associated with transmission among men who have sex with men in Japan. J Virol 2014;88:9864–9876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aggarwal I, Smith M, Tatt ID, et al. : Evidence for onward transmission of HIV-1 non-B subtype strains in the United Kingdom. J Acquir Immune Defic Syndr 2006;41:201–209 [DOI] [PubMed] [Google Scholar]
- 6.Brand D, Cazein F, Lot F, et al. : Continuous spread of HIV-1 subtypes D and CRF01_AE in France from 2003 to 2009. J Clin Microbiol 2012;50:2484–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilbert MT, Rambaut A, Wlasiuk G, Spira TJ, Pitchenik AE, Worobey M: The emergence of HIV/AIDS in the Americas and beyond. Proc Natl Acad Sci U S A 2007;104:18566–18570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention: Monitoring selected national HIV prevention and care objectives by using HIV surveillance data—United States and 6 dependent areas—2013. HIV Surveillance Supplemental Report 2013;20 [Google Scholar]
- 9.Hemelaar J, Gouws E, Ghys PD, Osmanov S, Isolation W-UNfH, Characterisation: Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 2011;25:679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pieniazek D, Ziebell RA, Bañez Ocfemia C, et al. : Phylogenetic surveillance of HIV-1 non-B subtypes among U.S.-born and foreign-born individuals living in the United States revealed similar viral genetic diversity but different race and male transmission patterns. Conference on Retroviruses and Other Opportunistic Infections (CROI) Seattle, WA; 2012 [Google Scholar]
- 11.Wheeler WH, Ziebell RA, Zabina H, et al. : Prevalence of transmitted drug resistance associated mutations and HIV-1 subtypes in new HIV-1 diagnoses U.S.-2006. AIDS 2010;24:1203–1212 [DOI] [PubMed] [Google Scholar]
- 12.Pyne MT, Hackett J, Jr., Holzmayer V, Hillyard DR: Large-scale analysis of the prevalence and geographic distribution of HIV-1 non-B variants in the United States. J Clin Microbiol 2013;51:2662–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Artenstein AW, Ohl CA, VanCott TC, Hegerich PA, Mascola JR: Transmission of HIV-1 subtype E in the United States. JAMA 1996;276:99–100 [PubMed] [Google Scholar]
- 14.Aldous JL, Pond SK, Poon A, et al. : Characterizing HIV transmission networks across the United States. Clin Infect Dis 2012;55:1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes GJ, Fearnhill E, Dunn D, et al. : Molecular phylodynamics of the heterosexual HIV epidemic in the United Kingdom. PLoS Pathog 2009;5:e1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kouyos RD, von Wyl V, Yerly S, et al. : Molecular epidemiology reveals long-term changes in HIV type 1 subtype B transmission in Switzerland. J Infect Dis 2010;201:1488–1497 [DOI] [PubMed] [Google Scholar]
- 17.Leigh Brown AJ, Lycett SJ, Weinert L, et al. : Transmission network parameters estimated from HIV sequences for a nationwide epidemic. J Infect Dis 2011;204:1463–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oster AM, Pieniazek D, Zhang X, et al. : Demographic but not geographic insularity in HIV transmission among young black MSM. AIDS 2011;25:2157–2165 [DOI] [PubMed] [Google Scholar]
- 19.Volz EM, Koopman JS, Ward MJ, Brown AL, Frost SD: Simple epidemiological dynamics explain phylogenetic clustering of HIV from patients with recent infection. PLoS Comput Biol 2012;8:e1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Little SJ, Kosakovsky Pond SL, Anderson CM, et al. : Using HIV networks to inform real time prevention interventions. PLoS One 2014;9:e98443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen SM, Gray KM, Ocfemia MC, Johnson AS, Hall HI: The status of the National HIV Surveillance System, United States, 2013. Public Health Rep 2014;129:335–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oster AM, Wertheim JO, Hernandez AL, Ocfemia MC, Saduvala N, Hall HI: Using molecular HIV surveillance data to understand transmission between subpopulations in the United States. J Acquir Immune Defic Syndr 2015;70:444–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura K, Nei M: Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 1993;10:512–526 [DOI] [PubMed] [Google Scholar]
- 24.Wertheim JO, Kosakovsky Pond SL: Purifying selection can obscure the ancient age of viral lineages. Mol Biol Evol 2011;28:3355–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hightower GK, May SJ, Perez-Santiago J, et al. : HIV-1 clade B pol evolution following primary infection. PLoS One 2013;8:e68188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison KM, Kajese T, Hall HI, Song R: Risk factor redistribution of the national HIV/AIDS surveillance data: An alternative approach. Public Health Rep 2008;123:618–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Struck D, Lawyer G, Ternes AM, Schmit JC, Bercoff DP: COMET: Adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res 2014;42:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drummond AJ, Suchard MA, Xie D, Rambaut A: Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 2012;29:1969–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drummond AJ, Rambaut A: BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 2007;7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whiteside YO, Song R, Wertheim JO, Oster AM: Molecular analysis allows inference into HIV transmission among young men who have sex with men in the United States. AIDS 2015;29:2517–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poon AF, Joy JB, Woods CK, et al. : The impact of clinical, demographic and risk factors on rates of HIV transmission: A population-based phylogenetic analysis in British Columbia, Canada. J Infect Dis 2015;211:926–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pillay D, Herbeck J, Cohen MS, et al. : PANGEA-HIV: phylogenetics for generalised epidemics in Africa. Lancet Infect Dis 2015;15:259–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.